Abstract
Transposable elements (TEs) are mobile genetic entities ubiquitously distributed in nearly all genomes. High frequency of codons ending in A/T in TEs has been previously observed in some species. In this study, the biases in nucleotide composition and codon usage of TE transposases and host nuclear genes were investigated in the AT-rich genome of Arabidopsis thaliana and the GC-rich genome of Oryza sativa. Codons ending in A/T are more frequently used by TEs compared with their host nuclear genes. A remarkable positive correlation between highly expressed nuclear genes and C/G-ending codons were detected in O. sativa (r=0.944 and 0.839, respectively, P<0.0001) but not in A. thaliana, indicating a close association between the GC content and gene expression level in monocot species. In both species, TE codon usage biases are similar to that of weakly expressed genes. The expression and activity of TEs may be strictly controlled in plant genomes. Mutation bias and selection pressure have simultaneously acted on the TE evolution in A. thaliana and O. sativa. The consistently observed biases of nucleotide composition and codon usage of TEs may also provide a useful clue to accurately detect TE sequences in different species.
Key words: transposable elements, transposase, codon usage, Arabidopsis thaliana, Oryza sativa
Introduction
Transposable elements (TEs) are mobile genetic elements that can move randomly from one position to another in the bacteria, animal and plant genome, with a great change in the copy number, type and distribution among different species. TEs have been reported to be present in most genomes with proportions ranging from a few percent in bacteria to more than 90% in some plant genomes 1., 2., 3., 4., 5., 6., 7., 8., 9.. The sequencings of large genomes have shown that TEs are a major constituent of these genomes, accounting for 15% of the genome of Drosophila melanogaster, 45% of the human genome, and more than 60% in Zea mays 4., 10.. However, studies indicated that small genomes, such as Caenorhabditis elegans, Arabidopsis thaliana and Saccharomyces cerevisiae, contain only 1.8%, 2% and 3.1% of TEs, respectively (4).
As a genome parasitic element, TEs are expected to have a different nucleotide composition than that of the host nuclear genes, considering the different origin and selection pressure during the evolution (11). In D. melanogaster, Shields and Sharp (12) observed an A/T preference in the third position of codons by comparing sequences of class I TEs to the host nuclear genes. A recent study also observed a high frequency of A/T-ending codons in TEs in five species (13). These observations indicated that this codon usage preference could be a general characteristic of TEs in certain species, regardless of nucleotide composition of their host genome. However, some studies suggested that TE codon usage bias is different in their families due to sequence characteristics, transmission pattern, insertion region and insertion history 11., 14., 15., 16., 17.. A similar codon usage pattern between P element and its host was observed in Drosophila willistoni and D. melanogaster (17), which suggested an accelerated evolution of P element in host genome. This type of TE is, therefore, subject to the selective pressure and/or mutation bias existed in the host genome after the insertion event.
In plants, TEs contribute to a large fraction of the DNA sequence amplification and rearrangement in addition to the more usual single nucleotide mutations (18). It is also known that TEs provide a substantial fraction of the regulatory elements and carry fragments of cellular genes 19., 20., which may intensively effect on the coding regions and the promoter regions of the host nuclear genes 21., 22.. Yet the coinfluence of evolutions of TEs and its host genome remains unclear. The proportion of TEs varied largely between A. thaliana and Oryza sativa, it therefore provides a chance to investigate the different patterns in nucleotide composition and codon usage in dicot and monocot plant species.
With the focus on the coding regions of TEs and their host genome, the conserved domains of TEs (including reverse transcriptase domains for class I TEs and transposase domains in the mariner superfamily) and coding regions of differentially expressed genes in A. thaliana and O. sativa were analyzed in this study. Base composition 23., 24., relative synonymous codon usage (RSCU) (25) and the effective number of codons (ENC) (26) were used to evaluate the compositional characteristics of studied sequences. These approaches coupled with the correspondence analysis (COA) on the synonymous codons 12., 17., 27. used by TEs and host nuclear genes allow us to investigate the difference of expression constraints and selective pressures acting on the TEs in A. thaliana and O. sativa.
Results
AT content of TEs and host nuclear genes
In this study, the global AT content of coding sequences, intron and intergenic regions of A. thaliana and O. sativa are highly coincident with two previous studies 13., 28. (Table 1). The TEs of both species show a higher AT content compared to the host nuclear genes at all of the three codon positions. The first position AT content of TEs in both species is 5.5%–10% lower than that of the second and third position (Table 1). A great difference of AT composition between TEs and host nuclear genes was observed in O. sativa (P=0.03) but not in A. thaliana, which is mainly caused by a trend in G/C-ending codons in the nuclear genes of O. sativa. The global AT content of TEs is 6.1%–9.7% and 5.7%–12% lower than that of the non-coding DNA in A. thaliana and O. sativa, respectively. This observation suggests that varied evolution constraints may be adopted by different functional regions in host genomes.
Table 1.
Comparison of the AT content between A. thaliana and O. sativa across different genomic regions and genetic components
| Region | Total number of codons used | %AT at the first position | %AT at the second position | %AT at the third position | Over all %AT |
|---|---|---|---|---|---|
| Coding sequence of host genes | |||||
| A. thaliana | 394,465 | 48.8 | 59.0 | 56.6 | 54.8 |
| O. sativa | 45,173,142 | 42.3 | 56.2 | 35.5 | 44.7 |
| Coding sequence of transposases | |||||
| A. thaliana | 153,997 | 51.2 | 61.3 | 61.3 | 57.9 |
| O. sativa | 230,627 | 45.1 | 57.3 | 50.6 | 51.0 |
| Non-coding sequences | |||||
| Intron regions | |||||
| A. thaliana | / | / | / | / | 67.6 |
| O. sativa | / | / | / | / | 63.0 |
| Intergenic regions | |||||
| A. thaliana | / | / | / | / | 64.0 |
| O. sativa | / | / | / | / | 56.7 |
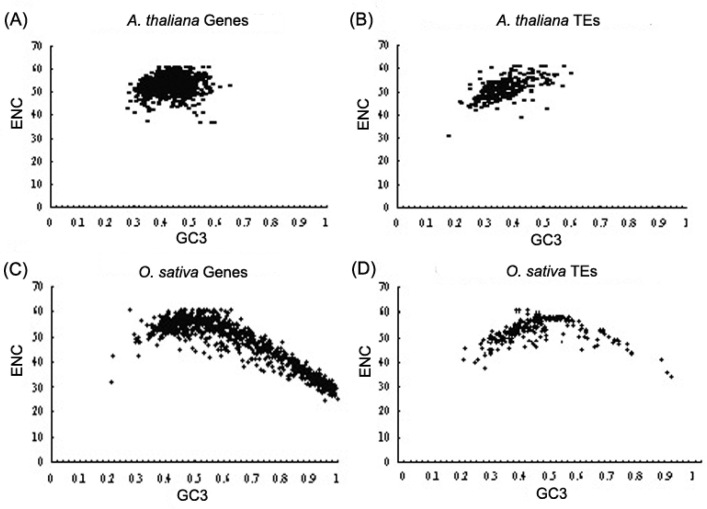
In A. thaliana, we observed that ENC values of both TEs and host nuclear genes are narrowly distributed in a range of 40 to 61. A- and T-ending codons are frequently used by A. thaliana coding sequences, and are positively correlated with ENC and GC3 (r=0.177 and 0.585 in the host nuclear genes and TEs, respectively, P<0.0001) (Figure 1, Figure 1). On the contrary, ENC and GC3 are remarkably negatively correlated in the host nuclear genes of O. sativa (r=−0.906, P<0.0001) (Figure 1C) due to the high G/C preference in the third codon position. This codon usage feature was also observed in other monocotyledon plants (29). However, the similar trend was not observed in the coding sequences of TEs in O. sativa (r=0.34, P<0.59) (Figure 1D). Although the GC3 of both host nuclear genes and TEs is varied widely in O. sativa, its TEs still prefer to use A- and T-ending codons as observed in A. thaliana.
Figure 1.
Distribution and correlation of GC3 and ENC in host nuclear genes and TEs. A. A. thaliana nuclear genes; B. A. thaliana TEs; C. O. sativa nuclear genes; D. O. sativa TEs.
Determination and comparison of optimal codons in TEs and host nuclear genes
The frequency of each synonymous codon was estimated from highly and weakly expressed host nuclear genes and TEs by using RSCU (Table 2). The identified preferred codons in A. thaliana TEs are highly in agreement with Lerat’s study (14 out of 18 amino acids) (13). It was observed that TEs prefer to use the same codons as those weakly expressed genes in both A. thaliana and O. sativa. This pattern is even more prominent in O. sativa. In rice genome, those highly expressed genes prefer to use codons ending in C/G (14 out of 18 amino acids), whereas no significant difference of preferred codons was observed between the differentially expressed host nuclear genes and TEs in A. thaliana. Moreover, some degenerated codons are almost equally used by both TEs and low-expressed genes in A. thaliana (e.g., lysine and leucine).
Table 2.
Average relative frequency (RSCU) of 59 degenerated codons for highly and weakly expressed host nuclear genes and TEs in A. thaliana and O. sativa
| No. | Amino acid | Codon |
A. thaliana |
O. sativa |
||||
|---|---|---|---|---|---|---|---|---|
| Genes (high) | Genes (weak) | TEs | Genes (high) | Genes (weak) | TEs | |||
| 1 | K | AAA | 0.30 | 0.48 | 0.47* | 0.02 | 0.47 | 0.40 |
| 2 | K | AAG | 0.70 | 0.52 | 0.53 | 0.98 | 0.53 | 0.60 |
| 3 | N | AAU | 0.27 | 0.63 | 0.60* | 0.03 | 0.66 | 0.56 |
| 4 | N | AAC | 0.73 | 0.37 | 0.40 | 0.97 | 0.34 | 0.44 |
| 5 | I | AUA | 0.10 | 0.30 | 0.26 | 0.02 | 0.28 | 0.23 |
| 6 | I | AUU | 0.32 | 0.46 | 0.45* | 0.02 | 0.48 | 0.43 |
| 7 | I | AUC | 0.57 | 0.24 | 0.29 | 0.96 | 0.24 | 0.33 |
| 8 | T | ACA | 0.18 | 0.34 | 0.36* | 0.02 | 0.38 | 0.30 |
| 9 | T | ACU | 0.32 | 0.39 | 0.36 | 0.02 | 0.36 | 0.30 |
| 10 | T | ACC | 0.38 | 0.16 | 0.16 | 0.48 | 0.19 | 0.25 |
| 11 | T | ACG | 0.12 | 0.11 | 0.12 | 0.49 | 0.07 | 0.15 |
| 12 | R | AGA | 0.05 | 0.13 | 0.14* | 0.01 | 0.12 | 0.13 |
| 13 | R | AGA | 0.10 | 0.06 | 0.07 | 0.49 | 0.08 | 0.16 |
| 14 | R | CGU | 0.09 | 0.09 | 0.07 | 0.31 | 0.08 | 0.13 |
| 15 | R | CGC | 0.20 | 0.24 | 0.21 | 0.17 | 0.24 | 0.21 |
| 16 | R | CGG | 0.33 | 0.11 | 0.14 | 0.01 | 0.15 | 0.17 |
| 17 | R | CGG | 0.23 | 0.36 | 0.38 | 0.01 | 0.33 | 0.20 |
| 18 | S | AGA | 0.24 | 0.08 | 0.10 | 0.36 | 0.12 | 0.15 |
| 19 | S | AGU | 0.15 | 0.24 | 0.24 | 0.01 | 0.24 | 0.19 |
| 20 | S | UCU | 0.11 | 0.08 | 0.08* | 0.33 | 0.06 | 0.12 |
| 21 | S | UCC | 0.07 | 0.20 | 0.20 | 0.00 | 0.19 | 0.16 |
| 22 | S | UCC | 0.26 | 0.29 | 0.28 | 0.01 | 0.27 | 0.22 |
| 23 | S | UCG | 0.16 | 0.11 | 0.11 | 0.29 | 0.12 | 0.15 |
| 24 | Y | UAU | 0.27 | 0.63 | 0.61* | 0.01 | 0.64 | 0.51 |
| 25 | Y | UAC | 0.73 | 0.37 | 0.39 | 0.99 | 0.36 | 0.49 |
| 26 | L | CUA | 0.07 | 0.11 | 0.12 | 0.00 | 0.12 | 0.13 |
| 27 | L | CUA | 0.33 | 0.11 | 0.13 | 0.60 | 0.11 | 0.16 |
| 28 | L | CUU | 0.07 | 0.14 | 0.11 | 0.36 | 0.14 | 0.13 |
| 29 | L | CUC | 0.19 | 0.23 | 0.24* | 0.03 | 0.21 | 0.26 |
| 30 | L | UUG | 0.28 | 0.25 | 0.22 | 0.01 | 0.27 | 0.22 |
| 31 | L | UUG | 0.05 | 0.16 | 0.17 | 0.00 | 0.15 | 0.10 |
| 32 | F | UUU | 0.69 | 0.38 | 0.40* | 0.99 | 0.40 | 0.46 |
| 33 | F | UUC | 0.31 | 0.62 | 0.60 | 0.01 | 0.60 | 0.54 |
| 34 | C | UGU | 0.41 | 0.66 | 0.68* | 0.00 | 0.55 | 0.51 |
| 35 | C | UGC | 0.59 | 0.34 | 0.32 | 1.00 | 0.45 | 0.49 |
| 36 | Q | CAA | 0.44 | 0.55 | 0.65* | 0.03 | 0.60 | 0.48 |
| 37 | Q | CAG | 0.56 | 0.45 | 0.35 | 0.97 | 0.40 | 0.52 |
| 38 | H | CAU | 0.37 | 0.70 | 0.65* | 0.05 | 0.72 | 0.52 |
| 39 | H | CAC | 0.63 | 0.30 | 0.35 | 0.95 | 0.28 | 0.48 |
| 40 | P | CCA | 0.34 | 0.35 | 0.39* | 0.03 | 0.41 | 0.30 |
| 41 | P | CCU | 0.29 | 0.44 | 0.35 | 0.02 | 0.40 | 0.32 |
| 42 | P | CCC | 0.17 | 0.09 | 0.11 | 0.27 | 0.11 | 0.18 |
| 43 | P | CCG | 0.21 | 0.12 | 0.14 | 0.68 | 0.08 | 0.20 |
| 44 | E | GAA | 0.32 | 0.53 | 0.58* | 0.03 | 0.57 | 0.42 |
| 45 | E | GAG | 0.68 | 0.47 | 0.42 | 0.97 | 0.43 | 0.58 |
| 46 | D | GAU | 0.46 | 0.74 | 0.70* | 0.03 | 0.74 | 0.59 |
| 47 | D | GAC | 0.54 | 0.26 | 0.30 | 0.97 | 0.26 | 0.41 |
| 48 | V | GUA | 0.05 | 0.17 | 0.18 | 0.00 | 0.19 | 0.15 |
| 49 | V | GUU | 0.32 | 0.46 | 0.39* | 0.01 | 0.43 | 0.32 |
| 50 | V | GUC | 0.39 | 0.12 | 0.17 | 0.47 | 0.15 | 0.22 |
| 51 | V | GUG | 0.24 | 0.26 | 0.26 | 0.51 | 0.23 | 0.31 |
| 52 | A | GCA | 0.17 | 0.35 | 0.34 | 0.01 | 0.36 | 0.27 |
| 53 | A | GCU | 0.41 | 0.44 | 0.42* | 0.01 | 0.40 | 0.32 |
| 54 | A | GCC | 0.28 | 0.11 | 0.13 | 0.48 | 0.15 | 0.24 |
| 55 | A | GCG | 0.14 | 0.10 | 0.12 | 0.49 | 0.09 | 0.18 |
| 56 | G | GGA | 0.34 | 0.34 | 0.38* | 0.03 | 0.30 | 0.27 |
| 57 | G | GGU | 0.39 | 0.36 | 0.33 | 0.02 | 0.33 | 0.29 |
| 58 | G | GGC | 0.17 | 0.12 | 0.13 | 0.68 | 0.20 | 0.24 |
| 59 | G | GGG | 0.09 | 0.18 | 0.16 | 0.27 | 0.17 | 0.20 |
Note: A codon with the highest RSCU value for each amino acid is indicated in boldface.
Codons reported by Lerat et al. (13).
Major factors of variations in synonymous codon usages in TEs and host nuclear genes
Synonymous codon-based COA analysis is commonly used to detect explanatory axes of major codon usage variations from a group of given sequences. In this study, this method was expected to further identify the major factors that affect codon usage frequencies and synonymous codon preferences observed from TEs and host nuclear genes. As shown in Table 3, the first explanatory axis accounts for 9.89% and 35.17% of total variations of synonymous codons in A. thaliana nuclear genes and TEs, respectively, and 50.78% and 30.14% of total variations of synonymous codons in rice nuclear genes and TEs, respectively. The first explanatory axis is closely and positively correlated with C3 or GC3 in all cases except TEs of A. thaliana. Different influence factors were detected in the second explanatory axis. G3 becomes a major variation factor in all of the host nuclear genes in this axis, whereas the codon usage biases of both O. sativa and A. thaliana TEs are mainly affected by T-ending codons.
Table 3.
Major factors of variations in synonymous codon usages in TEs and host nuclear genes
| Subject | Source of variation | Axis 1 |
Axis 2 |
||
|---|---|---|---|---|---|
| Total variability | Correlation coefficient (r-value) | Total variability | Correlation coefficient (r-value) | ||
| A. thaliana nuclear genes | A3 | 9.89 | −0.64 | 7.42 | 0.11 |
| C3 | 0.85 | −0.20 | |||
| G3 | – | 0.37 | |||
| T3 | −0.50 | −0.26 | |||
| GC3 | 0.81 | 0.11 | |||
| GC | 0.71 | – | |||
| ENC | – | 0.34 | |||
| A. thaliana TEs | A3 | 35.17 | 0.31 | 7.47 | −0.53 |
| C3 | −0.93 | – | |||
| G3 | −0.15 | – | |||
| T3 | 0.76 | 0.51 | |||
| GC3 | −0.83 | – | |||
| GC | −0.80 | – | |||
| ENC | −0.67 | – | |||
| O. sativa nuclear genes | A3 | 50.78 | −0.96 | 4.64 | 0.13 |
| C3 | 0.94 | −0.23 | |||
| G3 | 0.84 | 0.27 | |||
| T3 | −0.98 | – | |||
| GC3 | 1.00 | – | |||
| GC | 0.96 | – | |||
| ENC | −0.91 | – | |||
| O. sativa TEs | A3 | 30.14 | −0.79 | 9.47 | −0.47 |
| C3 | 0.94 | – | |||
| G3 | 0.76 | – | |||
| T3 | −0.92 | 0.32 | |||
| GC3 | 0.99 | – | |||
| GC | 0.95 | – | |||
| ENC | 0.21 | −0.29 | |||
Note: Only significant correlation coefficients are listed (P<0.0001).
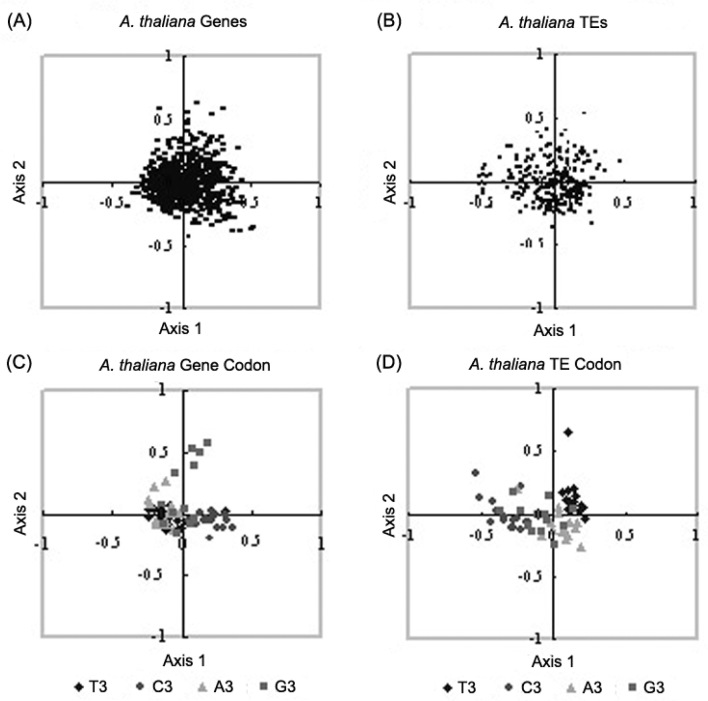
In A. thaliana, TEs and host nuclear genes mainly clustered at the center of the first and second explanatory axes, suggesting a weak codon usage bias of these coding sequences (Figure 2, Figure 2). The similar pattern can also be observed in the COA plot of 59 synonymous codons in A. thaliana TEs and coding sequences (Figure 2, Figure 2). It is noticed that G-ending codons (r=0.370, P<0.0001) are a major variation contributor of host genes, whereas T-ending codons (r=0.528, P<0.0001) account for the codon usage bias observed from TE sequences.
Figure 2.
Correspondence analysis plots of the major explanatory axes of A. thaliana nuclear genes and TEs. A. A. thaliana nuclear genes; B. A. thaliana TEs; C. 59 synonymous codons of A. thaliana nuclear genes; D. 59 synonymous codons of A. thaliana TEs.
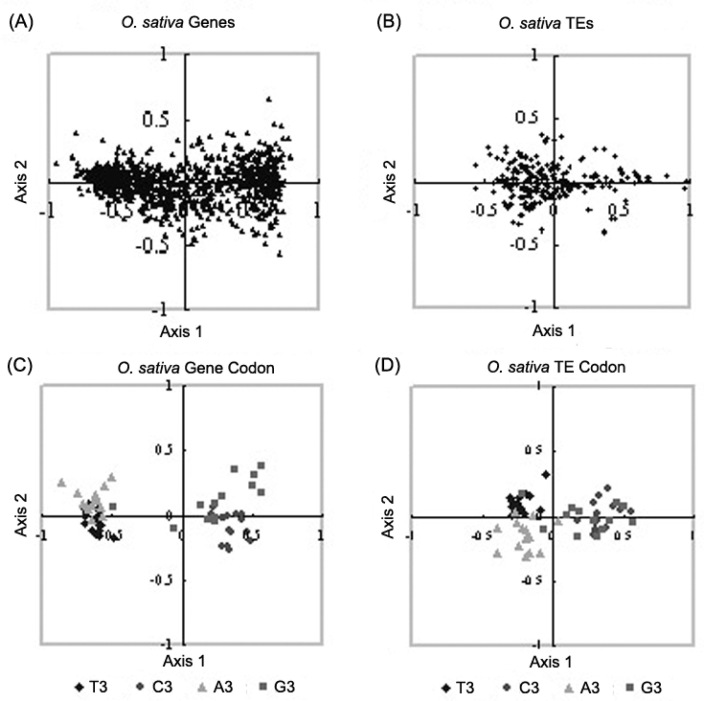
In O. sativa, both TEs and host nuclear genes are widely distributed along the first explanatory axis (Figure 3, Figure 3). A further COA analysis of synonymous codons in rice is surprised to find that only G-ending codons are weakly but not significantly associated with the host nuclear genes (r=0.268). Nevertheless, the rice TEs show a clear trend of using T-ending synonymous codons (r=0.320, P<0.0001), which is coincided with A. thaliana TEs (Figure 3, Figure 3).
Figure 3.
Correspondence analysis plots of the major explanatory axes of O. sativa nuclear genes and TEs. A. O. sativa nuclear genes; B. O. sativa TEs; C. 59 synonymous codons of O. sativa nuclear genes; D. 59 synonymous codons of O. sativa TEs.
Taken together, the host nuclear genes of both A. thaliana and O. sativa show a varied codon usage bias regarding to G/C-ending codons, whereas TEs prefer to use T-ending codons in both species.
Discussion
In this study, the biases in nucleotide composition and codon usage of TE transposases and host nuclear genes were investigated in the AT-rich genome of A. thaliana and the GC-rich genome of O. sativa. We observed by comparing sequences of TEs and host nuclear genes that TEs have a higher A/T content compared with their host nuclear genes. More precisely, in TEs the T-ending codons are more frequently used in both O. sativa and A. thaliana, whereas for host nuclear genes, only A. thaliana shows the similar trend. Lerat et al. (13) previously reported the similar observation that TEs of H. sapiens, D. melanogaster, S. cerevisiae, C. elegans and A. thaliana preferred the A/T-ending codons. In addtion, we noticed that codon usage in TEs is less biased than in nuclear genes in rice (mean ENC=57.0 versus 41.3). Moreover, the AT content at third codon position in TEs (50.6% and 61.3% in O. sativa and A. thaliana, respectively) is much closer to the intergenic AT content (56.7% and 64.0% in O. sativa and A. thaliana, respectively), suggesting a lower effectiveness of selection on synonymous sites of TEs than on host nuclear genes. It is argued that the high AT content at third codon position of TEs may be possibly caused by natural selection on the silent codon locus (30), AT-biased gene conversion, or GC to AT mutational bias (31). The non-independent duplication event of retrotransposons may also contribute to the changing of the AT content in their coding regions (32). In our RSCU analysis, TEs and host nuclear genes derived from O. sativa and A. thaliana adopt the same synonymous codons in 15 and 16 amino acids, respectively. A certain mutation pressure is therefore implied. However, in TEs the association between evolution and selection pressures on AT-rich sequences and the high AT content features observed from studied species remains to be validated.
A remarkable positive correlation between highly expressed nuclear genes and C/G-ending codons were detected in O. sativa (r=0.944 and 0.839, respectively, P<0.0001) but not in A. thaliana. This observation suggests a close association between the GC content and gene expression level in monocot species. In both species TE codon usage biases are similar to that of weakly expressed genes. A study of active autonomous TEs in the genomes of Drosophila identified the low median numbers of potentially active TE copies per family in the species of D. melanogaster, D. simulans and D. yakuba (5.5, 1.0 and 2.5, respectively) (33). It is suggested that host can adjust active TEs through methylation, chromatin-mediated silencing and homology-dependent gene silencing or co-suppression (34). In order to resist to its potential harmful effects of the genome, the expression and activity of TEs may be strictly controlled in both O. sativa and A. thaliana genomes (35). On the other hand, TEs may adapt a specific selection pressure due to this non-activation defense, retaining it in the host genome. As a transferred DNA, TE evolution may be simultaneously affected by mutation bias and selection pressure of its host.
In summary, the study of codon usage bias of TEs in monocot and dicot plant species enriched our knowledge at the point of regulation and organism adaptability across the different genomic regions and genetic components in several studied species 14., 36.. The consistently observed biases of nucleotide composition and codon usage of TEs may also provide a useful clue to accurately detect TE sequences.
Materials and Methods
Datasets
The completely annotated sequences of host nuclear genes (28,585 in A. thaliana and 56,056 in O. sativa) were downloaded from The Arabidopsis Information Resource (http://www.arabidopsis.org/) and the Rice Genome Annotation Database (http://rice.plantbiology.msu.edu/), respectively. Only those well-annotated genes were used in this analysis. Genes annotated with “unknown, putative and hypothetical” were eliminated from the original datasets. In addition, highly redundant genes, such as histone, rRNAs, tRNAs and transposases as well as genes derived from the mitochondria and chloroplast were also eliminated. We further removed genes with products shorter than 100 amino acids in order to avoid the sequence length influence in codon usage (22). Finally, 903 and 1,000 genes were selected from A. thaliana and O. sativa, respectively. Using the same selection criteria, 268 and 256 transposases were collected from A. thaliana and O. sativa, respectively. Non-coding sequences, represented by intron and intergenic regions of two hosts, were used to compare the compositional difference between coding and non-coding sequences.
Computation of base composition
In this study, the computation of base composition was classified into two types: (1) the whole gene GC content (GCall); (2) base frequency at the third codon position, including G+C content at the third codon position (GC3) and the frequency of A-, T-, C-, G-ending codons (A3, T3, C3, G3). This analysis was carried out by using CodonW 1.4 (http://www.molbiol.ox.ac.uk/cu). A Perl script was developed in computing the whole gene AT content (ATall), the frequency of A and T in all of three codon positions (AT1, AT2 and AT3), and the relative frequency of synonymous codon usage of host nuclear genes and transposases 23., 24..
Relative synonymous codon usage
RSCU is a statistical estimation approach of the relative frequency of each synonymous codon (25). RSCU reflects the number of times that a particular codon is observed relative to the number of times that the codon would be observed in the absence of any codon usage bias. In the absence of any codon usage bias, the RSCU value is 1.00. A codon that is used less frequently than expected will have a value of less than 1.00 and vice versa for a codon that is used more frequently than expected. RSCU uses only 59 degenerated codons of the 64 existing, while three stop codons (TAG, TGG and TGA) and two initiation codons (ATG and TGG) are not taken into account.
Effective number of codons
ENC is commonly used to measure the preferred codon usage from a give coding sequence. The value of ENC ranges from 20 to 61 and a small value indicates a high degree of bias in synonymous codon usage (26). It is known that such kind of bias is correlated significantly with the level of gene expression due to the translational selection in both single and multiple cellular organisms 37., 38., 39.. The value of ENC, therefore, can be used to identify those high-or low-expressed genes. In this study, the ENC values of host nuclear genes were calculated from CodonW. Genes ranked in the top and bottom 5% in the ENC calculation were considered as highly and weakly expressed host nuclear genes according to the suggestion of CodonW. The most preferred synonymous codons that occurred more frequently in both highly and weakly expressed genes were determined from these two groups of genes 37., 40..
Correspondence analysis
COA is one of the most popular multivariate methods for studying codon usage variation 12., 17., 27.. It calculates the position of the sequences in a multidimensional space according to codon usage frequency, identifies the major trends in the variation of the synonymous codon usage from a group of genes, and distributes these genes along continuous axes in accordance with these trends. In this study, we also calculated the position of the codons in a similar fashion. The linear association between identified major axes and nucleotide compositional properties, including A3, G3, C3, T3, GC3, GC and ENC, were further analyzed. This analysis was carried out by SPSS 11.0 (www.spss.com).
Authors’ contributions
JJ collected the datasets, conducted data analyses and prepared the manuscript. QX supervised the project and co-wrote the manuscript. Both authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 30571146) and the Rice Project (04–06) of Zhejiang Province of China.
References
- 1.Flavell R.B. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem. Genet. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- 2.Morescalchi A., Olmo E. Single-copy DNA and vertebrate phylogeny. Cytogenet. Cell Genet. 1982;34:93–101. doi: 10.1159/000131797. [DOI] [PubMed] [Google Scholar]
- 3.SanMiguel P. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 4.Lander E.S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Rizzon C. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 2002;12:400–407. doi: 10.1101/gr.210802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waterston R., Sulston J. The genome of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1995;92:10836–10840. doi: 10.1073/pnas.92.24.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.M. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 9.Mahillon J., Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feschotte C. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 11.Smit A.F. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 12.Shields D.C., Sharp P.M. Evidence that mutation patterns vary among Drosophila transposable elements. J. Mol. Biol. 1989;207:843–846. doi: 10.1016/0022-2836(89)90252-0. [DOI] [PubMed] [Google Scholar]
- 13.Lerat E. Codon usage by transposable elements and their host genes in five species. J. Mol. Evol. 2002;54:625–637. doi: 10.1007/s00239-001-0059-0. [DOI] [PubMed] [Google Scholar]
- 14.Silva J.C., Kidwell M.G. Horizontal transfer and selection in the evolution of P elements. Mol. Biol. Evol. 2000;17:1542–1557. doi: 10.1093/oxfordjournals.molbev.a026253. [DOI] [PubMed] [Google Scholar]
- 15.Karlin S., Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 16.Lerat E. Sequence divergence within transposable element families in the Drosophila melanogaster genome. Genome Res. 2003;13:1889–1896. doi: 10.1101/gr.827603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell J.R., Gleason J.M. Codon usage and the origin of P elements. Mol. Biol. Evol. 1996;13:278–279. doi: 10.1093/oxfordjournals.molbev.a025564. [DOI] [PubMed] [Google Scholar]
- 18.Morgante M. Plant genome organisation and diversity: the year of the junk! Curr. Opin. Biotechnol. 2006;17:168–173. doi: 10.1016/j.copbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Hancock J.F. Contributions of domesticated plant studies to our understanding of plant evolution. Ann. Bot. 2005;96:953–963. doi: 10.1093/aob/mci259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang N. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 21.Kazazian H.H., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 22.Long M. Evolution of novel genes. Curr. Opin. Genet. Dev. 2001;11:673–680. doi: 10.1016/s0959-437x(00)00252-5. [DOI] [PubMed] [Google Scholar]
- 23.Sueoka N. Translation-coupled violation of Parity Rule 2 in human genes is not the cause of heterogeneity of the DNA G+C content of third codon position. Gene. 1999;238:53–58. doi: 10.1016/s0378-1119(99)00320-0. [DOI] [PubMed] [Google Scholar]
- 24.Wu C.I., Maeda N. Inequality in mutation rates of the two strands of DNA. Nature. 1987;327:169–170. doi: 10.1038/327169a0. [DOI] [PubMed] [Google Scholar]
- 25.Sharp P.M. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright F. The “effective number of codons” used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 27.Grantham R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 29.Kawabe A., Miyashita N.T. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet. Syst. 2003;78:343–352. doi: 10.1266/ggs.78.343. [DOI] [PubMed] [Google Scholar]
- 30.Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. USA. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordaux R., Batzer M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartolome C. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10:R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matzke M.A. Host defenses to parasitic sequences and the evolution of epigenetic control mechanisms. Genetics. 1999;107:271–287. [PubMed] [Google Scholar]
- 35.Jensen S. Cosuppression of I transposon activity in Drosophlia by I-containing sense and antisense transgenes. Genetics. 1999;153:1767–1774. doi: 10.1093/genetics/153.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrieu O. Detection of transposable elements by their compositional bias. BMC Bioinformatics. 2004;5:94. doi: 10.1186/1471-2105-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 38.Stenico M. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 1994;22:2437–2446. doi: 10.1093/nar/22.13.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moriyama E.N., Powell J.R. Codon usage bias and tRNA abundance in Drosophila. J. Mol. Evol. 1997;45:514–523. doi: 10.1007/pl00006256. [DOI] [PubMed] [Google Scholar]
- 40.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]