Abstract
Here, we evaluate the contribution of two major biological processes—DNA replication and transcription—to mutation rate variation in human genomes. Based on analysis of the public human tissue transcriptomics data, high-resolution replicating map of Hela cells and dbSNP data, we present significant correlations between expression breadth, replication time in local regions and SNP density. SNP density of tissue-specific (TS) genes is significantly higher than that of housekeeping (HK) genes. TS genes tend to locate in late-replicating genomic regions and genes in such regions have a higher SNP density compared to those in early-replication regions. In addition, SNP density is found to be positively correlated with expression level among HK genes. We conclude that the process of DNA replication generates stronger mutational pressure than transcription-associated biological processes do, resulting in an increase of mutation rate in TS genes while having weaker effects on HK genes. In contrast, transcription-associated processes are mainly responsible for the accumulation of mutations in highly-expressed HK genes.
Key words: replication, transcription, mutational pressure, genetic variation
Introduction
DNA replication and transcription are the dominant mechanisms responsible for mutation rate variation across human genomes 1, 2, 3, 4, 5. In the process of DNA replication, the accumulation of single-strand DNA leads to an increased mutation rate in late-replicating regions 6, 7. In the process of transcription, DNA (it becomes single-stranded when serving as template) damage and transcription-coupled repair (TCR) are also believed to accelerate mutation in frequently-transcribed genes 8, 9, 10, 11, 12, 13, 14. While the entire genome is replicated all the time, it is not transcribed in the same way in each cell or at least each cell type. Therefore, the relationship between the two sources is complex and dynamic. Here, to differentiate the contribution of these two processes to mutation rate variation in human genomes, we correlated mutation rate, as reflected in SNP density, to tissue specificity, replication progress, and expression level of all human genes. We first determined the SNP density of human genes (human SNPs from NCBI dbSNP, build 130) (15) measured as the number of SNP per base pair (Table S1), and expression breadth and levels (Table S2) based on RNA-Seq data from 10 human tissues (16). We then determined replication progress or timing of these human genes (Table S3) based on the high-resolution replication map of human genomes in HeLa cells (6).
Results
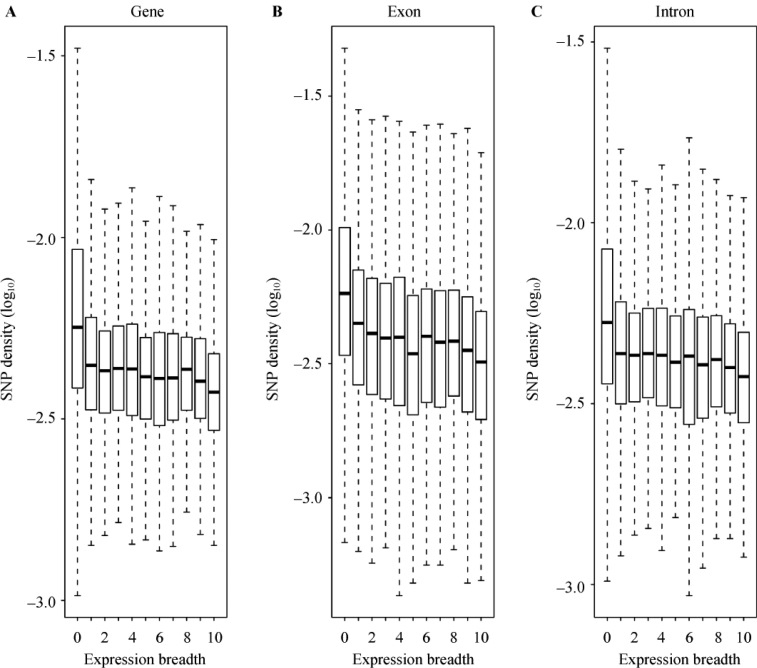
To explore the relationship between gene mutation rate and tissue specificity, we examined the expression breadth of 17,288 human genes across 10 tissues. Genes expressed in all 10 tissues examined are considered as housekeeping (HK) genes, while genes expressed in only one tissue examined are considered as tissue-specific (TS) genes. We observed that there is a significant correlation between SNP density and expression breadth among human genes (Figure 1A). The SNP density of TS genes is significantly higher than that of HK genes (Wilcoxon test, P<0.001) (Table 1). Moreover, the increase of SNP density can be observed not only in coding sequences of genes (Figure 1B), but also in intron regions (Figure 1C). Since most intronic sequences are thought to be non-functional, this pattern reflects both spontaneous and expression-related mutation processes rather than a result of selection. These observations suggested that mutation rate is remarkably increased in TS genes but remains relatively low in HK genes. Therefore, mutation rate closely associates with tissue specificity.
Figure 1.
Correlation between SNP density and expression breadth. SNP density is shown as a box-plot for genes in each expression breadth group. The boxes depict data between the 25th and 75th percentiles with central horizontal lines representing the median values; extreme values are indicated by dots outside the boxes. SNP density is negatively correlated with expression breadth, which can be equally observed in entire gene (A, Spearman ρ=−0.26, P<0.001), exons (B, Spearman ρ=−0.25, P<0.001) and introns (C, Spearman ρ=−0.18, P<0.001). SNP density is measured as the number of SNP per base pair. Expression breadth is defined as the number of unique tissues where a gene is expressed, which ranged from 0 (no expression detected in any tissue) to 10 (detected in all 10 tissues including muscle, heart, adipose, colon, breast, liver, kidney, lymphnode, brain and testis).
Table 1.
Comparison of SNP density among different classes of human genes
| HKG | TSG | P | LRG | ERG | P | LHKG | HHKG | P | |
|---|---|---|---|---|---|---|---|---|---|
| Gene | 0.0042 | 0.0062 | 1.79×10−61 | 0.0057 | 0.0045 | 2.20×10−16 | 0.0037 | 0.0073 | 0.0001 |
| Exon | 0.0041 | 0.0066 | 4.74×10−71 | 0.0061 | 0.0045 | 8.76×10−15 | 0.0039 | 0.0102 | 0.0012 |
| Intron | 0.0042 | 0.0059 | 6.65×10−30 | 0.0047 | 0.0046 | 0.0146 | 0.0036 | 0.0062 | 1.58×10−9 |
Note: HKG, HK genes; TSG, TS genes; LRG, late-replicating genes; ERG, early-replicating genes; LHKG, lowly-expressed HK genes; HHKG, Highly-expressed HK genes; P values were calculated by Wilcoxon test.
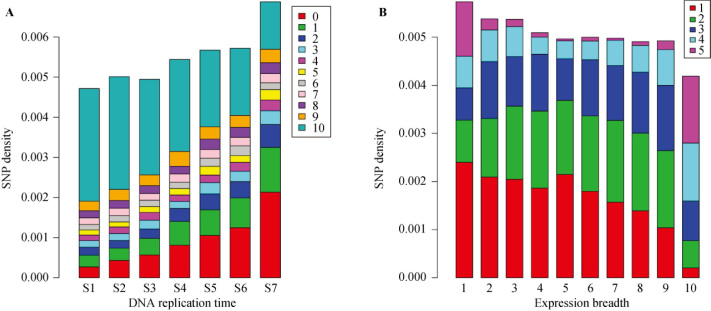
To examine whether this mutation rate variation is associated with the process of DNA replication, we first divided the whole genome into 7 sequential temporal zones (S1-S7) based on the global timing map of DNA replication in the human genome (6). We then surveyed the distribution of genes in each time zone and correlated it to the expression breadth of the genes. We found that HK genes tended to concentrate in early-replicating genomic regions, whereas TS genes were enriched in the late-replicating regions (Figure 2A). Therefore, we hypothesized that the increase of mutation rate in the late-replicating genomic regions should be responsible for the mutation rate variation between HK and TS genes. As expected, we found a significant trend correlating SNP density with the progression of DNA replication (Figure 2A). The mutation rate of the late-replicating genes is significantly higher than that of early-replicating genes (Wilcoxon test, P<0.001) (Table 1). This result suggests that replication-associated processes predominantly contribute to this mutation rate variation across human genomes.
Figure 2.
Replication timing, SNP density, expression breadth and expression levels of human genes. A. Average SNP density is significantly correlated with the progression of DNA replication (Spearman ρ=−0.26, P<0.001). The late-replicating genes show higher SNP density than the early-replicating genes. The progression of DNA replication in S phase was divided equally into 7 sequential time zones (S1 to S7). Furthermore, genes are classified into 11 groups according to their expression breadth as indicated in Figure 1. The fractions of each category are plotted over replication timing, showing that narrowly-expressed genes are concentrated in late-replicating regions. B. The widely-expressed genes show higher expression levels. Genes are classified into 5 groups according to expression level, which was defined as the average RPKM value of genes expressed in each tissue, and the fractions of each category are plotted over expression breadth.
HK genes are early-replicating and thus considered to bear weaker pressure from replication-associated mutations. In fact, among HK genes as well as genes with broader expression spectrum, we were not able to conclude that there is correlation between SNP density and the progression of DNA replication (Figure S1A-G). On the contrary, there is a weak correlation in TS genes (Figure S1J-K). We next assessed the contribution of transcription-associated processes, such as transcription-coupled DNA damage or TCR to this mutation rate variation. Correlating the expression level to the expression breadth of genes, we found that HK genes had higher expression levels than TS genes (Figure 2B), which is consistent with a previous observation (17). However, HK genes had lower SNP density than TS genes, which is contradictory with the viewpoint that mutation rate should be accelerated among highly-expressed genes under the process of transcription-coupled DNA damage or TCR 9, 12. Therefore, we are now able to draw two conclusions. First, considering the weaker effect from transcription-associated processes, we believe that transcription should make a minor contribution to this mutation rate variation in a global sense, i.e., when evaluated in the context of the whole genome and all the genes. Second, we believe that replication-associated processes generate stronger mutational pressure than transcription-associated processes, which results in the observed augmentation in mutation rate in the lowly-expressed TS genes.
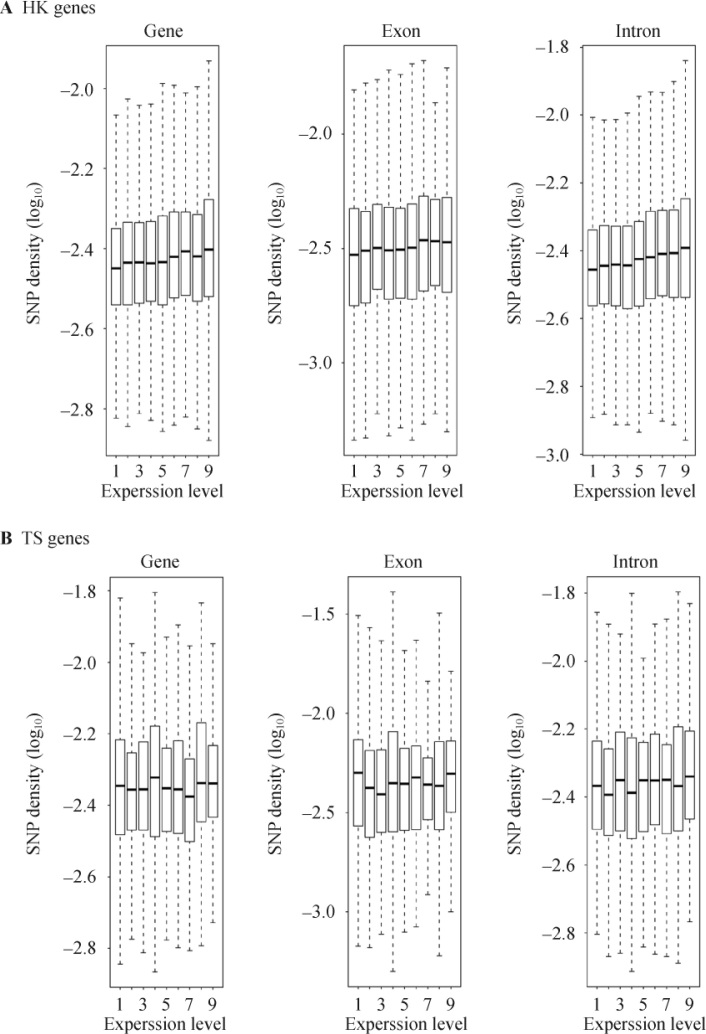
Although transcription-associated processes are suggested to have a weaker effect on the global variation of mutation rate, we noticed, however, when examining HK genes, that transcription-associated mutation pressure mainly affected the highly-expressed genes. We noted that SNP density was actually positively correlated with expression level among HK genes (Figure 3A). The average SNP density in the highly-expressed HK genes is significantly higher than that in the lowly-expressed HK genes (Wilcoxon test, P<0.01) (Table 1). However, when we further examined this effect among TS genes, we failed to find any correlation between SNP density and expression level among TS genes or genes that were expressed in limited tissues (Figure 3B and Figure S2). These results suggest that SNP density of TS genes is not associated with transcription, but replication. In addition, since TS genes are lowly-expressed in general, the SNP gradient around transcriptional start sites (TSS) of genes, known as a consequence of transcription-coupled TCR or DNA damage, exhibited a weaker effect in TS genes than in HK genes (Figure S3). Therefore, the above results suggest that transcription-associated processes are mainly responsible for the accumulation of more mutations in HK genes, especially for highly-expressed genes.
Figure 3.
Relationships between SNP density and expression level. A. SNP density is shown as a box-plot for genes in each expression breadth group. The boxes depict data between the 25th and 75th percentiles with central horizontal lines representing the median values; extreme values are indicated by dots outside the boxes. SNP density is positively correlated with expression level in HK genes, which can be equally observed in entire gene (Spearman ρ=−0.26, P<0.001), exons (Spearman ρ=−0.25, P<0.001) and introns (Spearman ρ=−0.18, P<0.001). B. Correlation between SNP density and expression level is absent among TS genes, all P>0.05. Genes are classified into 9 groups according to expression level, which was defined as the average RPKM value of genes expressed in each tissue.
Discussion
In conclusion, we find that DNA replication and transcription exert distinct impacts on mutation rate variations among human genes. First, mutational pressure from DNA replication-associated processes is stronger than that from transcription-associated processes since the former leave sequence signatures over the entire genome whilst the latter affect only transcriptionally-active genes that are highly-expressed and in particular, sequences around TSS. Second, the mutation pressure from replication-associated processes has distinct influences on human genes, such as significantly increased mutation rates in TS genes but a weaker effect on HK genes. Third, mutation pressure from transcription-associated processes contributes more to the mutation rate of HK genes but exhibits weaker effect on TS genes. Our results further elucidate the inter-related relationships concerning how DNA replication and transcription machineries commonly act on mutation rate variation across the human genome and in the context of genes and their expression/regulation. Our results are consistent with recent reports, such as the increased SNP density near late-replicating genes (18). In addition, we also took into account the effect of natural selection pressure on this mutation rate variation. Genes involved in certain tissue-specific functions, such as immunity and reproduction, are highly variable or fast-evolving, and they may behave differently in this type of analysis (19). However, we believe that this effect should be weaker, since all the correlations of SNP density to replication progress and expression levels were observed in intronic regions that are supposed to be free from selection. Finally, it is noted that if mutations can be propagated, they must arise in the germline (1). We can’t rule out the possibility that some of our results might be different from data in germline cells and tissues, since our analysis was mainly based on data of replication timing and gene expression from somatic cells and tissues. Nevertheless, our results should facilitate further validation/comparison of mutation rate variations in the germline.
Materials and Methods
All analyses were done on the March 2006 assembly of the human genome (versions NCBI 36, hg18). The gene annotation is from NCBI RefSeq database (20). All SNPs measured in the HapMap3 project were annotated using the UCSC genome browser (21) on the dbSNP build 130 to retrieve their physical locations. SNP density was calculated as the number of SNPs per base pair. We also calculated SNP densities for exons, introns and the entire gene.
A high-resolution replication timing profile of the human genome in HeLa cells was obtained from the published data (6). In this analysis, we divided equally the progression of DNA replication in S phase into 7 sequential time zones.
RNA-seq data was collected from 10 human tissues including muscle, heart, adipose, colon, breast, liver, kidney, lymphnode, brain and testis as described previously (16). We re-mapped the reads onto hg18 using MAQ (22). Uniquely mapped sequence reads were annotated according to Refseq-defined genes. Reads per kilobase-of-exon-model-per million-mapped-reads, or RPKM, was calculated to quantitate mRNA expression (23). Since the 5′ portion of mRNAs is frequently truncated in the process of RNA-seq library construction, the RPKM value of the last exon is often preferred as a measure of gene expression levels. However, when there are no reads mapped at the last exon, expression levels are defined by looking at RPKM values from the entire gene. A threshold RPKM value of 0.3 was used to filter out background noise (24). Expression breadth is defined as the number of unique tissues where a gene is expressed, which ranged from 1 (TS) to 10 (HK) with decreasing tissue specificity. Expression level was defined as the average RPKM value of genes expressed in each tissue.
Wilcoxon tests were performed by wilcox.test function in R software (version 2.5.1), where a normal approximation was used owing to the existence of ties. Spearman ρ (rank correlation coefficient) between the density of SNP and the expression breadth and level were calculated by cor function. The P values associated were calculated by cor.test function.
Authors’ contributions
JY and SH designed the experiments. PC, QL, FD, LZ, AL and ZZ collected the public data and performed the analysis. PC, QL and JY wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have no competing interests to declare.
Acknowledgements
The work was supported by grants from the National Basic Research Program (973 Program; 2006CB910401 and 2006CB910403) awarded to JY and SH.
Contributor Information
Songnian Hu, Email: husn@big.ac.cn.
Jun Yu, Email: junyu@big.ac.cn.
Supplementary Material
Figures S1-S3; Tables S1-S3
References
- 1.Arnheim N., Calabrese P. Understanding what determines the frequency and pattern of human germline mutations. Nat. Rev. Genet. 2009;10:478–488. doi: 10.1038/nrg2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer C.F. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- 3.Ellegren H. Mutation rate variation in the mammalian genome. Curr. Opin. Genet. Dev. 2003;13:562–568. doi: 10.1016/j.gde.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Hellmann I. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith N.G. Deterministic mutation rate variation in the human genome. Genome Res. 2002;12:1350–1356. doi: 10.1101/gr.220502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C.L. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–457. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stamatoyannopoulos J.A. Human mutation rate associated with DNA replication timing. Nat. Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comeron J.M. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics. 2004;167:1293–1304. doi: 10.1534/genetics.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green P. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 10.Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 11.Klapacz J., Bhagwat A.S. Transcription promotes guanine to thymine mutations in the non-transcribed strand of an Escherichia coli gene. DNA Repair (Amst.) 2005;4:806–813. doi: 10.1016/j.dnarep.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am. J. Hum. Genet. 2003;73:688–692. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polak P., Arndt P.F. Transcription induces strand-specific mutations at the 5′ end of human genes. Genome Res. 2008;18:1216–1223. doi: 10.1101/gr.076570.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimori S. GC-compositional strand bias around transcription start sites in plants and fungi. BMC Genomics. 2005;6:26. doi: 10.1186/1471-2164-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherry S.T. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang E.T. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe Y. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: disease-related genes in timing-switch regions. Hum. Mol. Genet. 2002;11:13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- 19.Wang D.P. Gamma-MYN: a new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biol. Direct. 2009;4:20. doi: 10.1186/1745-6150-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruitt K.D. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent W.J. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortazavi A. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 24.Ramskold D. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S3; Tables S1-S3