Abstract
Increasing evidence shows that protein phosphorylation on serine, threonine and tyrosine residues is a major regulatory post-translational modification in the bacteria. This review focuses on the implications of bacterial phosphoproteome in bacterial pathogenicity and highlights recent development of methods in phosphoproteomics and the connectivity of the phosphorylation networks. Recent technical developments in the high accuracy mass spectrometry have dramatically transformed proteomics and made it possible the characterization of a few exhaustive site-specific bacterial phosphoproteomes. The high abundance of tyrosine phosphorylations in a few bacterial phosphoproteomes suggests their roles in the pathogenicity, especially in the case of pathogen–host interactions; the high abundance of multi-phosphorylation sites in bacterial phosphoprotein is a compensation of the relatively small phosphorylation size and an indicator of the delicate regulation of protein functions.
Key words: protein phosphorylation, bacterium, pathogenicity, phosphoproteomics
Introduction
Post-translational modifications are essential for the rapid and reversible modification of the physiochemical properties of a protein, resulting in the changes of enzyme activity, oligomerization state, protein–protein interaction, subcellular localization or half-life (1). Protein phosphorylation is the most abundant and biologically the most important post-translational modification on the tyrosine, serine and threonine residues, and is catalyzed reversibly by specific protein kinases and phosphatases. Bacteria and some plants rely on histidine autophosphorylation of the sensory kinases and aspartate phosphorylation of the response regulators (thus two-component systems) (2). Protein phosphorylation is perhaps the best studied due to the close association between dys-regulated phosphorylation and human pathologies (3). Therefore, it is extremely important to determine the degree and the site of the in vivo protein phosphorylation. However, due to the very low stoichiometry, limited dynamic range, high complexity and quantitative difficulties of protein phosphorylations, highly selective enrichment procedures and sensitive mass spectrometry (MS) are required to decipher the phosphoproteome (4). Selective phosphopeptide enrichment has been accomplished in several ways by using anti-phosphotyrosine antibodies, immobilized metal affinity chromatography (IMAC), chemical modifications or strong cation exchange chromatography (5). The seamless combination of IMAC and nano-liquid chromatography enables reproducible separation and identification of phosphopeptides in a low-femtomole range 6, 7, and thus it is the most frequently used method in the study of cellular phosphorylation.
For some time, protein phosphorylation was considered to exist exclusively in eukaryotes until the first observation of protein phosphorylation occurring in Escherichia coli 8, 9. Indeed, protein phosphorylation is fundamental to the regulation of all kinds of physiological processes for the bacteria, especially for several key steps in the infection process, such as adhesion to the host, triggering and regulation of pathogenic functions as well as biochemical warfare, scrambling the host signaling cascades and impairing its defense mechanisms 10, 11. The global phosphoproteome has been established in a number of bacteria (Table 1), including Corynebacterium glutamicum (12), Campylobacter jejuni (13), Bacillus subtilis 14, 15, 16, E. coli (17), Latococcus lactis (18), Streptococcus pneumoniae (19), Klebsiella pneumoniae (20), Mycoplasma pneumoniae (21), Pseudomonas species (22), Mycobacterium tuberculosis (23), Streptomyces coelicolor (24) and Helicobacter pylori 13, 25. Phosphorylation in the bacteria was biased toward threonine compared with serine, while serine phosphorylation may account for 80%-90% of total phosphorylation sites in the eukaryotes (26). L. lactis, C. jejuni, and S. coelicolor contain more phosphorylation sites of threonine than serine (Table 1).
Table 1.
The bacterial phosphoproteomes identified so far
| Bacterium* | No. of phosphopeptides | No. of phospho sites | % of serine | % of threonine | % of tyrosine |
|---|---|---|---|---|---|
| Escherichia coli(17) | 105 | 81 | 67.9 | 23.5 | 8.6 |
| Bacillus subtilis(16) | 103 | 78 | 69.2 | 20.5 | 10.3 |
| Latococcus lactis(18) | 102 | 79 | 46.5 | 50.6 | 2.7 |
| Pseudomonas putida(22) | 56 | 53 | 52.8 | 39.6 | 7.5 |
| Pseudomonas aeruginosa(22) | 57 | 55 | 52.7 | 32.7 | 14.5 |
| Campylobacter jejuni(13) | 58 | 35 | 30.3 | 72.7 | 9.1 |
| Streptococcus pneumoniae(19) | 102 | 163 | 47.2 | 43.8 | 9.0 |
| Streptomyces coelicolor(24) | 44 | 44 | 34.1 | 52.3 | 13.6 |
| Klebsiella pneumoniae(20) | 117 | 93 | 31.2 | 15.1 | 25.8 |
| Mycoplasma pneumoniae(21) | 15 | 15 | 53.3 | 46.7 | 0 |
| Helicobacter pylori(25) | 80 | 124 | 42.8 | 38.7 | 18.5 |
Bacterial name followed by the reference in brackets.
The most abundant subset of phosphorylated proteins is the enzymes involved in the central carbon/protein/nucleotide metabolism, and some other phosphorylated housekeeping proteins are helicases, chaperones, ribosomal proteins and amino acyl tRNA-synthetases (27). One quantitative phosphoproteomic analysis on the model bacterium B. subtilis has been performed with stable isotope labeling by amino acids in cell culture (SILAC) (28). The striking distinction between the known metazoan and bacterial phosphoproteomes are the extent: 30%-50% of proteins are phosphorylated in humans 29, 30, whereas it is at least one order of magnitude lower in bacteria (27). The only notable exception is M. tuberculosis, whose yield was significantly high: 516 phosphorylation sites in 301 phosphoproteins, accounting for >7% of M. tuberculosis proteins (23). Differential roles were proposed for the protein phosphorylations in eukaryotes and prokaryotes: protein phosphorylation in the eukaryotes is extensively used for transduction of signals inter- and intra-cellularly, whereas the function may be less central in the prokaryotes (16). Bacterial phosphorylation sites can be conferred by two major search algorithms NetPhosBac (31) and Diphos (32), which were summarized in a recent review paper (27). Here we will focus on an overview of the recent advances in the field of bacterial phosphoproteome, highlighting recent methods in phosphoproteomics, connectivity of the phosphorylation networks, as well as the correlation between pathological potentials and the known bacterial phosphoproteomes.
Methods in Phosphoproteomic Analysis
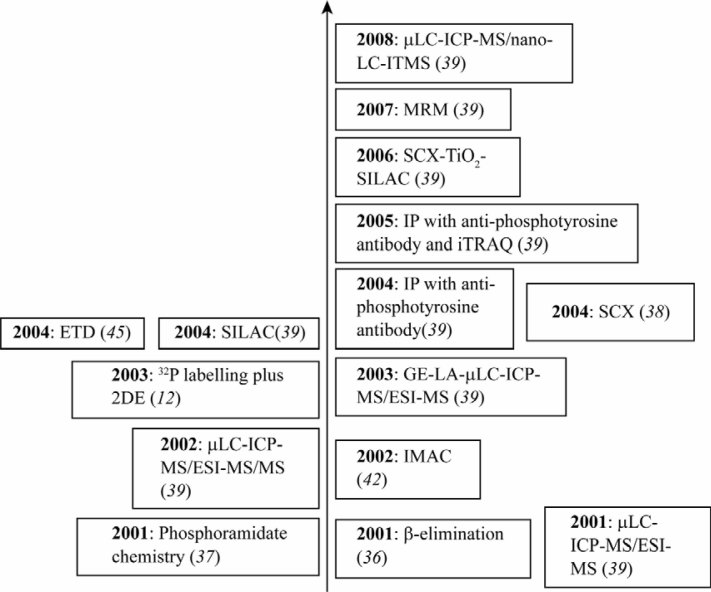
The major challenges in phosphoproteomics are the generally very low stoichiometry and high complexity of phosphorylation, limited dynamic range of detection methods available, and quantitative difficulties (4). As for the complexity, the phosphosite database (www.phosphosite.org) lists over 99,000 non-redundant phosphorylation sites up to August 2011, and this number is expanding continually. The conservation of the phosphorylation sites in the bacterial phosphoproteome is extremely low, even in the case of proteins phosphorylated in most bacterial species, such as the enzymes of the central carbon metabolism (27), which suggests the diverse kinase specificities to adapt to different environmental niches. The dynamic range of the phosphoproteome is quite large (~109), from hundreds of millions to a few copies per cell. Therefore, it is mission impossible to detect all the phosphopeptides upon proteolytic digestion of the whole cell lysates or tissue samples. Selective enrichment of the phosphoproteins/peptides is required and has been accomplished in several ways: anti-phosphotyrosine antibodies (33), IMAC 34, 35, chemical modifications 36, 37 and strong cation exchange chromatography (SCX) (38). Figure 1 summarizes several of the most significant work over the past decade 5, 39. For a global view of serine, threonine and tyrosine phosphorylations, IMAC may be the best choice (6), with peptide recovery up to 90% as determined by 32P or 33P-radioactivity measurements (40). Phosphopeptides are retained on nitriloacetic acid (NTA) or iminodiacetic acid (IDA)-linked hard metal ions, such as Fe3+, Ga3+, Al3+ (39) and Zr4+ (41), through their negatively charged phosphate group. Non-specific binding of peptides containing Glu and Asp could be minimized either by carefully controlling experimental conditions or through methyl-esterification of carboxylic acids by HCl-saturated dry methanol (42). Recently, titanium dioxide (TiO2) chromatography has emerged as the most common method for the enrichment of global phosphoproteins/peptides (43). This technique requires shorter preparation time and has increased capacity compared to IMAC resins. Advances in the high accuracy MS allow for the identification of thousands of phosphorylation sites in a single experiment (44). Both IMAC and TiO2 columns can be coupled seamlessly with MS for reproducible separation, detection and identification of phosphopeptides in a low-femtomole range 7, 45, 46, 47. Upon the identification of the phosphoproteome, bioinformatics analysis through Gene Ontology (GO) annotation (48) or the bacterial localization prediction tool pSORTb (49) is often applied to get the relevant information of cellular function, localization and protein–protein interaction network (50).
Figure 1.
Timeline in phosphoproteomics with chosen milestones based on the implementation of a new method during the last decade. Abbreviations used are: μLC, micro-liquid chromatography; ICP, inductively coupled plasma; ITMS, ion trap mass spectrometer; MRM, multiple reaction monitoring; SCX, strong cation exchange chromatography; SILAC, stable isotope labeling by amino acids in cell culture; IP, immunoprecipitation; iTRAQ, isobaric tag for relative and absolute quantitation; ETD, electron-transfer dissociation; 2DE, two-dimensional gel electrophoresis; GE, gel electrophoresis; LA, laser ablation; ESI, electrospray ionization; IMAC, immobilized metal affinity chromatography.
Tyrosine Phosphorylation
As a Gram-negative, spiral-shaped bacterium that colonizes in the gastric mucosa of humans (51), H. pylori has been frequently associated with atrophic gastritis, peptic ulcer disease, functional dyspepsia and gastric carcinomas (52). Cytotoxin-associated antigen A (CagA) is a major pathogenicity protein in H. pylori and plays key roles in inducing gastric inflammation, ulcer and carcinogenesis. CagA protein is translocated from the bacterium, undergoes tyrosine phosphorylation at the Glu-Pro-Ile-Tyr-Ala (EPIYA) motif in the host cells and induces a cellular hummingbird phenotype of transformation (53). In addition, non-phosphorylated CagA interacts with host proteins, such as epithelial tight junction-scaffolding protein zonulin (ZO-1), cell adhesion protein E-cadherin, hepatocyte growth factor receptor c-Met, cadherin-associated protein β-catenin, adaptor protein GRB-2 and the kinase PAR1, leading to a loss of cell polarity and inducing pro-inflammatory and mitogenic responses (54). Besides the knowledge about CagA phosphorylation in the host cells, there is no information available about the in vivo phosphorylation state of H. pylori intracellular proteins. Bioinformatics indicated that the genome of H. pylori strain 26695 contains at least one protein kinase (HP0432) and one PPM-family protein phosphatase (HP0431) (55). Up to now, three independent studies intended to discover the intracellular phosphorylation in H. pylori: (1) eight proteins phosphorylated at serine residues through SDS-PAGE and autoradiography (56); (2) 57 proteins identified through Fe3+-IMAC enrichment, 2D gel electrophoresis and MALDI-TOF MS analysis, albeit without further phosphorylation site mapping (13); and (3) 82 phosphopeptides from 67 proteins with 79 class I (with localization probability higher than 0.75) phosphorylation sites: 33 (42.8%) on serine, 31 (38.7%) on threonine and 15 (18.5%) on tyrosine (25).
Tyrosine phosphorylation has generally been regarded as an exclusively eukaryotic phenomenon and plays roles in multicellularity, and metazoans have a higher proportion of tyrosine phosphorylation than unicellular eukaryotes (57). This is based on the virtual absence of tyrosine phosphorylation in unicellular eukaryotes, such as yeasts. Most bacterial phosphorylation sites are on serine (70%) and threonine (20%), while tyrosine phosphorylation sites account for less than 10%. A noteworthy feature of H. pylori phosphoproteome is the significantly high overall abundance (~18.5%) of tyrosine phosphorylation. There are three bacterial phosphoproteomes with comparable tyrosine phosphorylation percentage: S. coelicolor, 13.6% (24); P. aeruginosa, 14.5% (22); and K. pneumoniae, 25.8% (20); while most other bacterial model organisms with known phosphoproteomes have less than 10% phosphorylation sites on tyrosine (Table 1). The level of tyrosine phosphorylation seems to be positively correlated with the pathogenicity. P. aeruginosa phosphoproteome has a much higher tyrosine phosphorylation level (14.5%) compared with the non-pathogenic P. putida species (7.5%) (22). In fact, various findings have supported the contribution of tyrosine phosphorylation to the bacterial pathogenicity, such as pedestal formation (Tir of enteropathogenic E. coli and Citrobacter) (58), cell elongation, scattering and inflammation (CagA of Helicobacter) 53, 59, capsular polysaccharide biosynthesis (20), cell invasion (Tarp of Chlamydia) (60), pro-inflammatory responses and cell proliferation (BepD-F of Bartonella) (61).
The association has been established between protein tyrosine phosphorylation and the control of surface polysaccharide production or transport (62), as well as between protein serine/threonine phosphorylation and cell envelope biosynthesis (63). Surface polysaccharides are believed to be involved in the early steps of the infection process and are considered potent virulent factors (64). Some proteins from various species are homologous to a family of enzymes involved in exopolysaccharide synthesis grouped together as BY-kinases (Bacterial tYrosine kinases) and show similar auto-phosphorylation activities 65, 66. BY-kinases, albeit only a few copies per bacterial genome, are widespread in bacteria (66) and are usually encoded by genes in the large operons involved in biosynthesis and export of sugar polymers (67). For instance, the tyrosine kinase Wzc of E. coli is essential for the synthesis of the exopolysaccharide colonic acid and the assembly of group 1 capsular polysaccharide (62). It is essential for the pathogenicity of S. pneumonia to produce capsular polysaccharide (CPS). The CPS biosynthesis proteins CpsB, CpsC and CpsD regulate CPS production via tyrosine phosphorylation of CpsD, a homologue of Wzc. CpsC is required for CpsD tyrosine phosphorylation (68). The mutations in some key elements of CpsC produced wild-type levels of CPS, but were unable to cause bacteremia in mice upon intranasal challenge (69). It seems that the relatively high abundance of tyrosine phosphorylation in the phosphoproteomes of several pathogenic bacteria reflects in some way the pathogenicity of the bacteria, such as in the direct pathogen–host interactions.
Multiple Phosphorylation Sites
In eukaryotic cells, multiple phosphorylated proteins are common and serve as molecular switches for signal fine-tuning. For example, five phosphorylation sites are present in the eukaryotic initiation factor (eIF) 2B catalyzed by four different protein kinases (70): two conserved C-terminus serine sites are phosphorylated by casein kinase 2 and required for the in vivo interaction of eIF2Bɛ with eIF2 and in vitro eIF2B activity; the third site (Ser539) is required for the recruition of the glycogen synthase kinase 3 (GSK3) to the fourth site (Ser535); the fifth site is phosphorylated by casein kinase 1 and lies out of the catalytic domain of eIF2Bɛ. One distinctive feature of H. pylori phosphoproteome is the high occurrence of multiple phosphorylation sites: 35 out of the identified 84 phosphopeptides contain at least two phosphorylation sites. One example is the peptide SAKANDASEITA LLNTIAYETISTLSK from the enzyme of alanine racemase, which has six phosphorylation sites. This phenomenon has been noted for proteins accumulated when the bacteria were under stress or with overloaded proteolytic systems (71), and was also reported in the characterization of the phosphoproteome of the Gram-positive pathogenic bacterium S. pneumoniae (19) as well as the non-pathogenic L. lactis (18). These observations indicated that one protein may be phosphorylated on multiple sites to fulfill differential roles or to function together to achieve delicate micro-regulations of the pathogenicity mechanisms, such as adhesion to the host, stimulation and regulation of pathogenic functions and impairing the host defense mechanisms (10). Multiple phosphorylation sites may have similar biological implications, at least for the species of L. lactis (18) and H. pylori: both L. lactis and H. pylori have smaller genome sizes, simpler transcriptional machinery and fewer (two and three, respectively) sigma factors compared with other model bacteria such as E. coli, suggesting that more regulations are needed through delicate post-translational modifications.
Protein–Protein Interaction Network of Phosphorylated Proteins
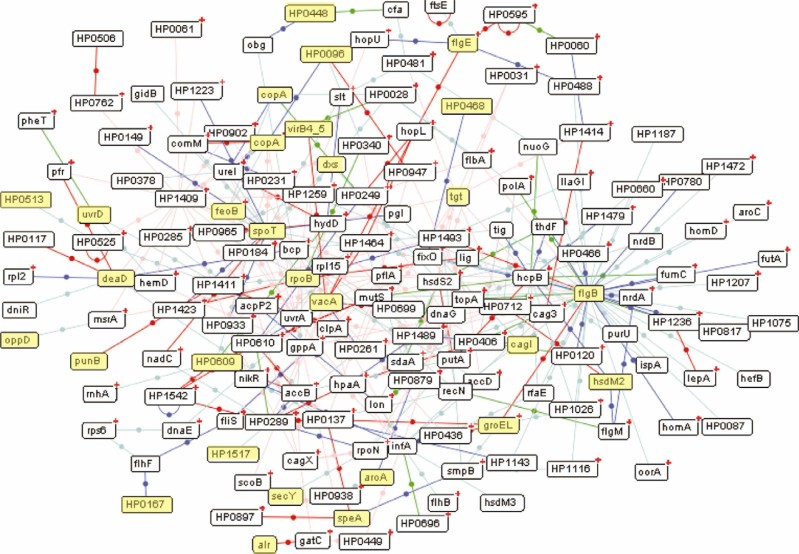
Protein–protein interaction map with the involvement of phosphoproteins is important to understand the regulatory mechanisms of post-translational modifications in the bacteria. As shown in Figure 2, the phosphoprotein interaction map of H. pylori consisted of a large network covering 163 proteins, among which there are 28 identified phosphoproteins (25). The major H. pylori pathogenicity factor VacA is centered on this interaction map. The possible partners for VacA in the A category include an outer membrane porin protein HopL, a hypothetical protein HP0699, a predicted ABC transporter HP1464 and a predicted ATPase/DNA transfer protein VirB4_5. VacA is secreted from H. pylori through the syringe-like VirB/VirD4-like type IV secretion apparatus. It is noteworthy that VirB4_5 is also identified to be phosphorylated, implicating the involvement of phosphorylation in the regulation of VacA secretion through the bacterial membrane. The two partner proteins GroEL and FlgB were determined to be phosphorylated and were predicted to interact directly with each other (25). The in vivo interaction between GroEL and FlgB was previously observed in B. subtilis (72), albeit without any assignment of the in vivo function for such interaction. H. pylori produces flagella to fulfill its requirement for the bacterial colonization in the human gastric mucosa. As a member in the HSP60 family, H. pylori GroEL has been shown to increase the risk of gastric carcinoma (73). Previous studies demonstrated that the phosphorylation and dephosphorylation of GroEL regulated its binding and dissociation from unfolded proteins (74), possibly through the switch between the oligomeric states mediated by phosphorylation (75). Further experimental investigations are needed to reveal the contribution of the GroEL and FlgB partners to the pathology of the bacterium with regards to how the phosphorylation of these two proteins regulates the bacterial colonization. Phosphoprotein interaction network through bioinformatics or experimental determinations provides some hints for the connectivity of the phosphorylation networks and the correlation between pathological potentials and the known bacterial phosphoproteomes.
Figure 2.
Protein–protein interaction network of the identified phosphoproteins in H. pylori constructed with PIMRider software (http://pim.hybrigenics.com) (76). Proteins in yellow represent the identified phosphoproteins. Interactions with different levels of reliability are assigned with different colors in the following sequence: red>blue>green>cyan>pink. A small red cross “+” is drawn on the top right of the protein whenever a protein contains partners that are not currently displayed within the map. The figure is reprinted with permission from Ref. 25.
Conclusion
Phosphorylation is biologically the most important post-translational modification. It is essential to determine the degree and the site of the in vivo protein phosphorylation in order to understand the underlying mechanisms between dys-regulation of phosphorylation and human pathologies. Highly selective enrichment procedures and sensitive MS are required to decipher the phosphoproteome due to the very low stoichiometry, limited dynamic range, high complexity and quantitative difficulties of protein phosphorylations. Recent technical developments made it possible to uncover some whole-cell bacterial phosphoproteomes, which revealed the correlation between bacterial pathological potentials and phosphorylations: the high abundance of tyrosine phosphorylations in a few bacterial phosphoproteomes indicates their roles in the pathogenicity, especially in the case of pathogen-host interactions; the high abundance of multi-phosphorylation sites in bacterial phosphoprotein is a compensation of the relatively small phosphorylation size and an indicator of the delicate regulation of protein functions. With further development in the MS-based techniques, more information will be obtained about bacterial phosphoproteomes and the correlation between bacterial phosphorylations and potential pathogenicity. This will help us to develop protein phosphorylation-targeted prodrugs in the control of bacterial infections.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 20801061), Guangdong Natural Science Foundation (Grant No. 8451027501001233), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, Ministry of Education, and the Fundamental Research Funds for the Central Universities (Grant No. 10lgpy19).
References
- 1.Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem. Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 2.Ninfa A.J. Use of two-component signal transduction systems in the construction of synthetic genetic networks. Curr. Opin. Microbiol. 2010;13:240–245. doi: 10.1016/j.mib.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson L.N. The regulation of protein phosphorylation. Biochem. Soc. Trans. 2009;37:627–641. doi: 10.1042/BST0370627. [DOI] [PubMed] [Google Scholar]
- 4.Mann M. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 5.Ge R., Sun H. Metallomics: an integrated biometal science. Sci. China Ser. B-Chem. 2009;52:2055–2070. [Google Scholar]
- 6.Paradela A., Albar J.P. Advances in the analysis of protein phosphorylation. J. Proteome Res. 2008;7:1809–1818. doi: 10.1021/pr7006544. [DOI] [PubMed] [Google Scholar]
- 7.Li X. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 8.Manai M., Cozzone A.J. Analysis of the protein-kinase activity of Escherichia coli cells. Biochem. Biophys. Res. Commun. 1979;91:819–826. doi: 10.1016/0006-291x(79)91953-3. [DOI] [PubMed] [Google Scholar]
- 9.Garnak M., Reeves H.C. Phosphorylation of Isocitrate dehydrogenase of Escherichia coli. Science. 1979;203:1111–1112. doi: 10.1126/science.34215. [DOI] [PubMed] [Google Scholar]
- 10.Jers C. Phosphoproteomics in bacteria: towards a systemic understanding of bacterial phosphorylation networks. Expert Rev. Proteomics. 2008;5:619–627. doi: 10.1586/14789450.5.4.619. [DOI] [PubMed] [Google Scholar]
- 11.Soufi B. Insights from site-specific phosphoproteomics in bacteria. Biochim. Biophys. Acta. 2008;1784:186–192. doi: 10.1016/j.bbapap.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Bendt A.K. Towards a phosphoproteome map of Corynebacterium glutamicum. Proteomics. 2003;3:1637–1646. doi: 10.1002/pmic.200300494. [DOI] [PubMed] [Google Scholar]
- 13.Voisin S. The cytoplasmic phosphoproteome of the Gram-negative bacterium Campylobacter jejuni: evidence for modification by unidentified protein kinases. Proteomics. 2007;7:4338–4348. doi: 10.1002/pmic.200700483. [DOI] [PubMed] [Google Scholar]
- 14.Eymann C. Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics. 2007;7:3509–3526. doi: 10.1002/pmic.200700232. [DOI] [PubMed] [Google Scholar]
- 15.Levine A. Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics. 2006;6:2157–2173. doi: 10.1002/pmic.200500352. [DOI] [PubMed] [Google Scholar]
- 16.Macek B. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics. 2007;6:697–707. doi: 10.1074/mcp.M600464-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Macek B. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Soufi B. The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics. 2008;8:3486–3493. doi: 10.1002/pmic.200800069. [DOI] [PubMed] [Google Scholar]
- 19.Sun X. Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J. Proteome Res. 2010;9:275–282. doi: 10.1021/pr900612v. [DOI] [PubMed] [Google Scholar]
- 20.Lin M.H. Phosphoproteomics of Klebsiella pneumoniae NTUH-K2044 reveals a tight link between tyrosine phosphorylation and virulence. Mol. Cell. Proteomics. 2009;8:2613–2623. doi: 10.1074/mcp.M900276-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidl S.R. The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae: analysis of the complete known Ser/Thr kinome suggests the existence of novel kinases. Mol. Cell. Proteomics. 2010;9:1228–1242. doi: 10.1074/mcp.M900267-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravichandran A. Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics. 2009;9:2764–2775. doi: 10.1002/pmic.200800655. [DOI] [PubMed] [Google Scholar]
- 23.Prisic S. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. USA. 2010;107:7521–7526. doi: 10.1073/pnas.0913482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker J.L. Analysis of the phosphoproteome of the multicellular bacterium Streptomyces coelicolor A3(2) by protein/peptide fractionation, phosphopeptide enrichment and high-accuracy mass spectrometry. Proteomics. 2010;10:2486–2497. doi: 10.1002/pmic.201000090. [DOI] [PubMed] [Google Scholar]
- 25.Ge R. Phosphoproteome analysis of the pathogenic bacterium Helicobacter pylori reveals over-representation of tyrosine phosphorylation and multiply phosphorylated proteins. Proteomics. 2011;11:1449–1461. doi: 10.1002/pmic.201000649. [DOI] [PubMed] [Google Scholar]
- 26.Ubersax J.A., Ferrell J.E., Jr. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 27.Kobir A. Protein phosphorylation in bacterial signal transduction. Biochim. Biophys. Acta. 2011;1810:989–994. doi: 10.1016/j.bbagen.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Soufi B. Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis. J. Proteome Res. 2010;9:3638–3646. doi: 10.1021/pr100150w. [DOI] [PubMed] [Google Scholar]
- 29.Manning G. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 30.Cohen P. The origins of protein phosphorylation. Nat. Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 31.Miller M.L. NetPhosBac—a predictor for Ser/Thr phosphorylation sites in bacterial proteins. Proteomics. 2009;9:116–125. doi: 10.1002/pmic.200800285. [DOI] [PubMed] [Google Scholar]
- 32.Iakoucheva L.M. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey A. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet–derived growth factor receptors. Proc. Natl. Acad. Sci. USA. 2000;97:179–184. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersson L., Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 35.Sun X. Application of immobilized metal affinity chromatography in proteomics. Expert Rev. Proteomics. 2005;2:649–657. doi: 10.1586/14789450.2.5.649. [DOI] [PubMed] [Google Scholar]
- 36.Oda Y. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 37.Zhou H. A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 38.Beausoleil S.A. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nita-Lazar A. Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics. 2008;8:4433–4443. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubrovska A., Souchelnytskyi S. Efficient enrichment of intact phosphorylated proteins by modified immobilized metal-affinity chromatography. Proteomics. 2005;5:4678–4683. doi: 10.1002/pmic.200500002. [DOI] [PubMed] [Google Scholar]
- 41.Feng S. Immobilized zirconium ion affinity chromatography for specific enrichment of phosphopeptides in phosphoproteome analysis. Mol. Cell. Proteomics. 2007;6:1656–1665. doi: 10.1074/mcp.T600071-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Ficarro S.B. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 43.Larsen M.R. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Collins M.O. Analysis of protein phosphorylation on a proteome-scale. Proteomics. 2007;7:2751–2768. doi: 10.1002/pmic.200700145. [DOI] [PubMed] [Google Scholar]
- 45.Syka J.E. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinkse M.W. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 47.Ficarro S.B. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 48.Conesa A. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 49.Gardy J.L. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 2003;31:3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaab C. Analysis of phosphoproteomics data. Methods Mol. Biol. 2011;696:41–57. doi: 10.1007/978-1-60761-987-1_3. [DOI] [PubMed] [Google Scholar]
- 51.Marshall B.J., Warren J.R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;323:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 52.Blaser M.J. Ecology of Helicobacter pylori in the human stomach. J. Clin. Invest. 1997;100:759–762. doi: 10.1172/JCI119588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer. 2004;4:688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 54.Wen S., Moss S.F. Helicobacter pylori virulence factors in gastric carcinogenesis. Cancer Lett. 2009;282:1–8. doi: 10.1016/j.canlet.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi L. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 1998;22:229–253. doi: 10.1111/j.1574-6976.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 56.Grangeasse C. Protein kinase activity in Helicobacter pylori. FEMS Microbiol. Lett. 1999;176:327–332. doi: 10.1111/j.1574-6968.1999.tb13679.x. [DOI] [PubMed] [Google Scholar]
- 57.Ullrich A., Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 58.Kenny B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 59.Segal E.D. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clifton D.R. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. USA. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schulein R. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. USA. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cozzone A.J. Bacterial tyrosine kinases: novel targets for antibacterial therapy? Trends Microbiol. 2009;17:536–543. doi: 10.1016/j.tim.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 63.Molle, V. and Kremer, L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75: 1064–1077. [DOI] [PubMed]
- 64.Mazmanian S.K., Kasper D.L. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 65.Grangeasse C. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem. Sci. 2007;32:86–94. doi: 10.1016/j.tibs.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 66.Jadeau F. Identification of the idiosyncratic bacterial protein tyrosine kinase (BY-kinase) family signature. Bioinformatics. 2008;24:2427–2430. doi: 10.1093/bioinformatics/btn462. [DOI] [PubMed] [Google Scholar]
- 67.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 68.Morona J.K. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 2000;35:1431–1442. doi: 10.1046/j.1365-2958.2000.01808.x. [DOI] [PubMed] [Google Scholar]
- 69.Morona J.K. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc. Natl. Acad. Sci. USA. 2006;103:8505–8510. doi: 10.1073/pnas.0602148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X. Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon-subunit and their functions in vivo. EMBO J. 2001;20:4349–4359. doi: 10.1093/emboj/20.16.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosen R. Highly phosphorylated bacterial proteins. Proteomics. 2004;4:3068–3077. doi: 10.1002/pmic.200400890. [DOI] [PubMed] [Google Scholar]
- 72.Endo A., Kurusu Y. Identification of in vivo substrates of the chaperonin GroEL from Bacillus subtilis. Biosci. Biotechnol. Biochem. 2007;71:1073–1077. doi: 10.1271/bbb.60640. [DOI] [PubMed] [Google Scholar]
- 73.Suerbaum S. Helicobacter pylori hspA-hspB heat-shock gene cluster: nucleotide sequence, expression, putative function and immunogenicity. Mol. Microbiol. 1994;14:959–974. doi: 10.1111/j.1365-2958.1994.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 74.Sherman M., Goldberg A.L. Heat shock-induced phosphorylation of GroEL alters its binding and dissociation from unfolded proteins. J. Biol. Chem. 1994;269:31479–31483. [PubMed] [Google Scholar]
- 75.Kumar C.M. Facilitated oligomerization of mycobacterial GroEL: evidence for phosphorylation-mediated oligomerization. J. Bacteriol. 2009;191:6525–6538. doi: 10.1128/JB.00652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rain J.C. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]