Abstract
Leptospirosis is an infectious bacterial disease caused by Leptospira species. In this study, we cloned and sequenced the gene encoding the immunodominant protein GroEL from L. interrogans serovar Autumnalis strain N2, which was isolated from the urine of a patient during an outbreak of leptospirosis in Chennai, India. This groEL gene encodes a protein of 60 kDa with a high degree of homology (99% similarity) to those of other leptospiral serovars. Recombinant GroEL was overexpressed in Escherichia coli. Immunoblot analysis indicated that the sera from confirmed leptospirosis patients showed strong reactivity with the recombinant GroEL while no reactivity was observed with the sera from seronegative control patient. In addition, the 3D structure of GroEL was constructed using chaperonin complex cpn60 from Thermus thermophilus as template and validated. The results indicated a Z-score of −8.35, which is in good agreement with the expected value for a protein. The superposition of the Cα traces of cpn60 structure and predicted structure of leptospiral GroEL indicates good agreement of secondary structure elements with an RMSD value of 1.5 Å. Further study is necessary to evaluate GroEL for serological diagnosis of leptospirosis and for its potential as a vaccine component.
Key words: GroEL, expression, homology modeling, leptospirosis, serodiagnosis
Introduction
Leptospirosis is a global zoonotic bacterial disease caused by Leptospira species, affecting both urban and rural areas with inestimable morbidity and mortality (1). The severe disease form, known as Weil’s syndrome, is an acute febrile illness associated with multiorgan system complications including jaundice, renal failure, meningitis and pulmonary haemorrhage, with a mortality rate exceeding 15% 2, 3. Antibiotic therapy, although effective in blood clearance, may not abolish leptospires from the kidney tubules. Case identification needs to be performed promptly so that rapid outbreak investigations and timely administration of antibiotic therapy can be implemented. However, failure of accurate diagnosis is frequent because of the broad spectrum of clinical presentations associated with leptospirosis. Ideally, early diagnosis requires an effective laboratory test that can be easily implemented in the field without dependence on reference laboratory settings. A diagnostic test applicable to the variety of epidemiological situations associated with human and veterinary leptospirosis requires an antigen that is highly conserved among diverse pathogenic leptospiral strains. One such candidate is the major outer membrane protein, LipL32, which is highly conserved across leptospiral species (4). A previous study of leptospiral proteins expressed during leptospiral infection of humans showed that the IgG response frequently targeted proteins of molecular weights 32, 41/42, 45, 58, 62, 76 and 82 kDa during the acute illness phase (5). During the convalescent phase, responses to the proteins were considerably increased. Antibody responses were greatest to p32 followed by p82, p41/42 and p62, respectively (5). Recent molecular characterization of leptospiral proteins, such as DnaK (6), GroEL 7, 8, the OmpL1 porin 9, 10, 11, LipL41 (12) and LipL32/MOMP (13), has resulted in antibody for definitive identification of major protein antigens.
Given this background, we undertook cloning, expression and characterization of the GroEL protein of Leptospira interrogans serovar Autumnalis N2 from the urine of an infected human (14) as a means of obtaining the protein in quantity and with acceptable purity. The results show that recombinant GroEL is a useful candidate for evaluation in serodiagnosis and immunoprotection.
Results
Cloning and sequencing of groEL
A fragment of 1.65 kb was generated by PCR amplification of genomic DNA. Following digestion of the PCR product with Nde1 and BamH1, the fragment was cloned between Nde1 and BamH1 site of pET15b to generate the plasmid pET15b-GroEL. Successful cloning of the gene was confirmed by restriction endonuclease analysis (REA), colony PCR and sequencing (Figure S1). Sequencing and BLAST analysis of the open reading frame fragments revealed a high degree of homology (99%) to the groEL gene of L. interrogans serovar Copenhageni (GenBank Accession No. L14682).
Amino acid sequence analysis
The groEL gene encodes a protein of 546 amino acid residues with a predicted molecular mass of 58.5 kDa. This protein is highly homologous to the GroEL proteins of other leptospires, including L. interrogans serovars Copenhageni (100%) and Lai (99%), L. borgpetersenii serovar Hardjo-bovis (97%), L. bifelxa serovar Patoc (87%) and other spirochaetes (69%) with identities of 68% to 100%. GroEL contains C-terminal GGM and AAVEEGIVPGGG motifs, forming a signature pattern with a well-conserved C-terminal region of twelve residues 15, 16. In addition, there is also a GroEL-like chaperone apical domain, an ATPase domain and an intermediate domain (Figure S2).
Overexpression and purification of recombinant GroEL
Expression of His-tagged GroEL was induced by IPTG and the His6-GroEL recombinant protein was purified by metal affinity resin. A higher molecular mass protein co-purified with GroEL was removed by size exclusion filtration. This protein may belong to the caseinolytic peptidases (Clp) involved in degradation of misfolded proteins following heat shock (17). The apparent molecular mass of the recombinant protein as determined by SDS-PAGE was consistent with the molecular mass predicted from its amino acid composition (Figure S3).
Immunoblot analysis
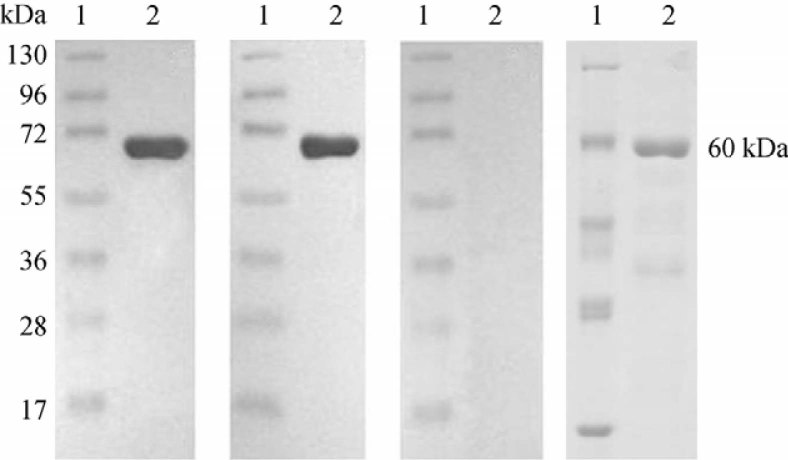
Immunoblot analysis performed with polyclonal goat antibodies against recombinant GroEL revealed a band of 60 kDa (Figure 1A). Immunoblot developed with pooled sera from leptospirosis patients showed reactivity with the recombinant GroEL (Figure 1B). However, no reactivity was observed on immunoblots probed with sera from controls that were negative in the microscopic agglutination test (MAT) (Figure 1C), indicating the specificity of the reaction. In addition, the size of immuno-reactive protein agreed well with that revealed by protein staining with Coomassie Brilliant Blue (Figure 1D).
Figure 1.
Immunoblot analysis of the purified recombinant GroEL. Purified GroEL protein was separated by electrophoresis and subjected to immnunoblotting after transferred to membrane. Membranes were incubated with GroEL specific hyperimmune serum (A), leptospirosis patients’ sera (B), or serum from a seronegative control patient (C), followed by incubation with secondary antibodies for visualization. Another set of gel was stained with Coomassie Brilliant Blue after separation and served as a positive control (D). Lane 1: protein ladder; Lane 2: purified GroEL protein.
Model construction and refinement
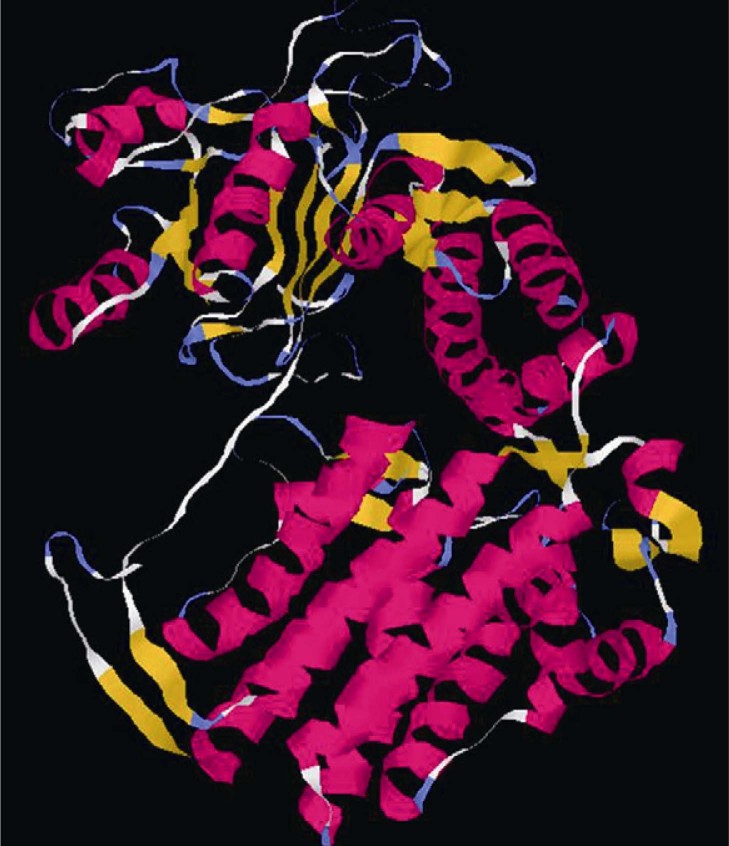
The template for modeling leptospiral GroEL was determined using PSI-BLAST against Protein Data Bank (PDB). The crystal structure of chaperonin complex cpn60 from Thermus thermophilus (PDB: 1WE3) provided a sequence identity of 68% (Figure S4). Therefore, it was used as the template to generate the initial model of GroEL using Modeler software. The initial model was refined using two-stage energy minimization protocol in Gromacs (Figure 2).
Figure 2.
Final predicted 3D structure of L. interrogans GroEL. The initial model of the putative 3D structure of GroEL from L. interrogans Autumnalis N2 was build using Modeller and refined by energy minimizing. The α-helix is indicated in pink and the β-sheet is depicted in yellow. The turns and coils are indicated in blue and white, respectively.
Validation of the model
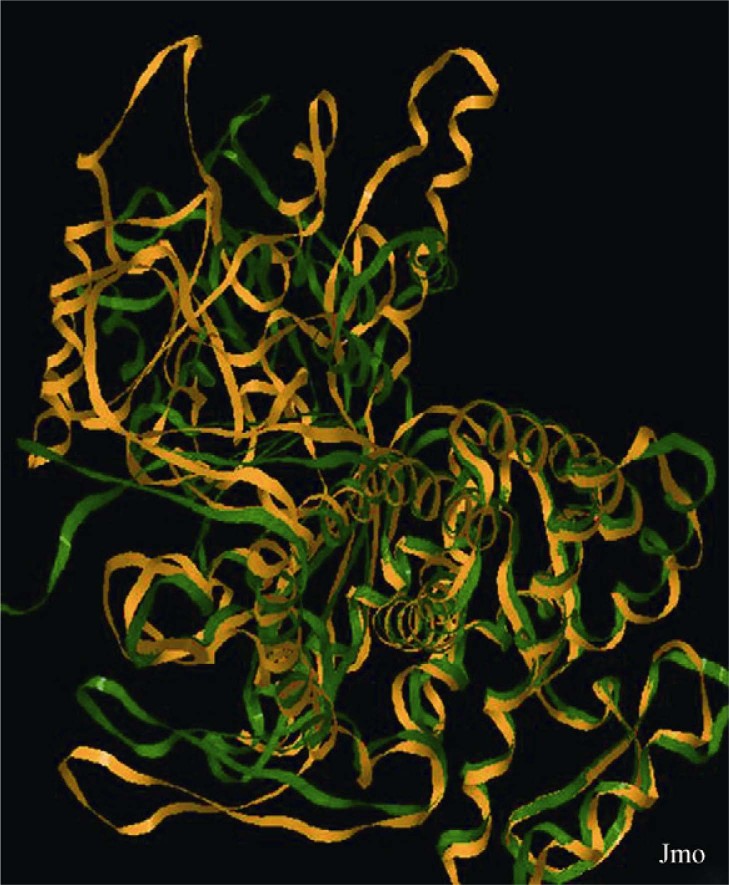
The stereochemical quality of the predicted model was evaluated by PROCHECK program and correlated favorably with the template. About 77.05% of the residues fall in the most favorable region of the Ramachandran plot while 20.4% are in the additional regions. The accuracy of the model was tested using 3D profile program PROSA, which calculates the conformational energy of each residue of the protein from a compilation of potentials of mean force for all amino acid pairs. The energy profiles of GroEL are negative, corresponding to a correctly folded structure. Although higher than that obtained for the X-ray structures of GroEL of T. thermophilus (around −10.37), the combined pair interaction-surface energy Z-score was determined as −8.35 for leptospiral GroEL, which is in good agreement with the expected value for a protein (18). The superposition of the Cα traces of the template structure and the predicted structure clearly shows that there is better agreement between the secondary structure elements than between the variable loops and the C-terminal regions (RMSD value = 1.5 Å) (Figure 3). Sequence alignment was performed for amino acid residues involved in binding of polypeptide, Mg2+ and ATP/ADP in cpn60 from T. thermophilus with that of GroEL from L. interrogans Autumnalis N2 (Table S1). It was shown that only the residues in ATPase and Mg2+ binding sites were highly conserved while residues important for polypeptide binding were not well conserved between the two sequences.
Figure 3.
Superposition of 3D structure of L. interrogans GroEL and T. thermophilus cpn60. The generated 3D structure of GroEL of L. interrogans Autumnalis N2 (green) was superposed with the crystal structure of cpn60 of T. thermophilus (PDB: 1WE3) (yellow).
Discussion
In this study, we report the cloning, expression and characterization of GroEL of L. interrogans serovar Autumnalis N2. In earlier studies, a protein of 62 kDa characterized as a molecular chaperone was recognized by many acute and convalescent phase sera from patients with confirmed leptospirosis 5, 19. Moreover, acute phase sera recognized GroEL more frequently than other proteins. This might be explained by the elevated expression of leptospiral GroEL during the elevated temperature encountered within the infected host 7, 8, 20. Previous studies also suggested that enhanced immunoreactivity may be explained by a pre-existing, possibly cross reactive memory response (20). Significant seroreactivity has been reported among control sera to GroEL, which may reflect a ubiquitous expression of these proteins in eubacteria (21). In addition, many different infections are associated with an immune response to heat shock proteins (22). It was also reported that the dominant antigenic determinant in leptospiral GroEL consists of 20 amino acid residues highly conserved among the GroEL family in prokaryotes (7). These findings suggest that cross-reactivity of GroEL proteins could limit the feasibility of using leptospiral GroEL as a marker for leptospiral seroreactivity. Thus, GroEL are often dismissed a priori for serodiagnostic utility because of their potential for low sensitivity. Conversely, a protein microarray of Burkholderia pseudomallei has shown that GroEL is a significantly differentially reactive antigen for melioidosis (23). The performance of ELISA with purified recombinant leptospiral GroEL produced a sensitivity and specificity of 90.65% and 94.9%, respectively (24). Sensitive and rapid detection utilizing recombinant leptospiral GroEL based immunoreactions will be a logical next step in evaluating the usefulness of this protein for laboratory confirmation in the field.
In addition to its use in serodiagnosis, leptospiral GroEL may also have immunoprotective potential. Due to its ability to elicit a memory T-cell response, GroEL could be a potent stimulator of immune response even in the absence of CD4+ T cells (25). The use of this protein as immunoreactant can further be supported from the active site analysis of the protein. A high level of identity between cpn60 of T. thermophilus and GroEL of L. interrogans Autumnalis N2 was observed in regions corresponding to the ATP-binding site and Mg2+-binding site. In contrast, the relatively low identity shared in the polypeptide-binding region suggests that the polypeptide-binding sites of leptospiral GroEL have diverged from the template sequence, which may explain the immunodominant potency of the leptospiral GroEL. The result of the present study was also supported by earlier studies showing that cytosolic chaperonins have a highly conserved ATPase domain but diverged polypeptide-binding domains (26).
In conclusion, groEL from a local isolate of L. interrogans serovar Autumnalis strain N2 has been cloned, expressed and purified. The recombinant protein has been evaluated for serological diagnosis of leptospirosis (24). Further studies are warranted for the potential use of GroEL as a vaccine candidate.
Materials and Methods
Bacterial strains and media
L. interrogans serovar Autumnalis strain N2 was isolated from a patient during an outbreak of leptospirosis in Chennai, India (14) and maintained at 30°C in Ellinghausen-Mc Cullough-Johnson-Harris (EMJH) medium (Difco Laboratories, USA) with 1% BSA and subcultured every 7 days. E. coli strains were grown on Luria-Bertani agar supplemented with 100 µg/mL ampicillin (Sigma). E. coli DH5a (Invitrogen) was used for transformation experiments and His-tagged recombinant proteins were expressed in E. coli BL-21 (Invitrogen).
Patients’ sera
Sera were obtained from patients who fulfilled the leptospirosis case definition including laboratory based confirmation (IgM ELISA / isolation of leptopsires / four-fold rise in MAT titer).
Goat antisera
A yearling goat was immunized by intramuscular injection of 200 µg of recombinant protein dissolved in 1 mL of phosphate-buffered saline and 60 µL of Quil A (10 mg/mL). Two additional injections were performed at days 35 and 70 using same amount of antigen and adjuvant. The antibody titer of the hyperimmune serum was determined by ELISA.
Isolation of groEL
Genomic DNA was prepared from L. interrogans Autumnalis N2 as described earlier (27) and used as template to amplify the groEL gene with the primers groelF and groelR (forward primer: 5’-GAGTTAACA TATGGCGAAAGATA-3’; reverse primer: 5’-GCGGA TCCCGGATGAATTACATCATTC-3’) containing a NdeI and BamHI restriction enzyme site, respectively, designed using Primer 2 (Scientific and Educational software). PCR reaction was carried out in a thermal cycler (Eppendorff, Germany) with denaturation at 94°C for 2 min before 30 cycles of 94°C for 1 min, 57°C for 30 s, 72°C for 1 min 30 s and 72°C for 7 min for final extension.
Plasmid construction
Standard recombinant DNA techniques were used to construct all plasmids. PCR products were purified (QIAquick PCR purification kit, Qiagen), then digested with NdeI and BamHI and ligated in frame with His6 tag sequence at the N-terminus of pET15b expression vector with T4 DNA ligase (MBI fermentas). pET15b-groEL was introduced into E. coli DH5α by CaCl2 transformation and the insert was confirmed by colony PCR, REA and DNA sequencing (Macrogen, South Korea).
Expression of recombinant GroEL
pET15b-groEL was transformed into E. coli BL21 (DE3) and the expression of His6-GroEL was induced by IPTG (isopropylthio-β-D-galactoside). Briefly, 100 mL of super optimal culture broth containing 100 µg/mL ampicillin was inoculated with 2 mL of overnight culture and incubated at 37°C at 150 rpm until the OD600 reaches 0.5. IPTG was then added at a final concentration of 1 mM and the induction continued at 37°C for 2.5-3 h.
Purification of recombinant GroEL
Cells were pelleted by centrifugation and resuspended in 2.5 mL ice-cold TEN buffer [50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 100 mM NaCl]. The resultant cell suspension was sonicated for 15 min and then centrifuged at 5000× g for 10 min. The His6-GroEL recombinant protein was purified by TALON superflow metal affinity resin (Clontech, USA) as per supplier’s instruction. After removal of unbound proteins from the column, the His-tagged recombinant protein was eluted with 0.02-0.1 M gradient of imidazole (pH 8.0), and was further purified by size-exclusion chromatography in an Äkta Purifier system (Amersham Biosciences). The eluted fractions were directly loaded on a Superose 6 10/300 GL column and run in isocratic mode at 0.5 mL/min. The column was equilibrated with 50 mM sodium phosphate buffer containing 150 mM NaCl at pH 7.0. The fractions were collected and then dialysed against 0.1 M phosphate-buffered saline (pH 7.2) containing 10% (v/v) glycerol, 0.3% (v/v) Triton X-100, and 0.025% (w/v) sodium azide. Protein concentration was determined using bicinchoninic-acid protein assay (Pierce).
SDS-PAGE and immunoblotting
SDS-PAGE (10% gel) was performed as described in literature (28). The proteins were separated by electrophoresis and then electrophoretically transferred to Protean Nitrocellulose membrane (Schleicher and Schuell, USA) and blocked with 4% non-fat dry milk in TBST (20 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.5). Membranes were incubated with GroEL specific hyperimmune serum or patients’ sera followed by incubation with protein G conjugated to horseradish peroxidase (Zymed, USA). Bands were visualized using 4-chloro-α-naphthol (Sigma).
Targets and templates
Nucleotide sequences were translated using translation tool on the ExPASY proteomics server (www.expasy.org/tool/dna.html). Amino acid sequences were subjected to BLAST search against PDB (NCBI) to select template structures for homology modeling. After identification of structurally conserved regions common to target and templates, sequence alignment was performed using Clustal W.
Homology modeling
Modeller is an implementation of automated approach for comparative modeling by satisfaction of special restrains 29, 30, 31, 32. Modeller 9v8 (release April 2010) was used in this study to build an initial model of the putative 3D structure of GroEL from L. interrogans Autumnalis N2.
Refinement of the model
The refinement process was accomplished in several stages. Firstly, 100 iterations of steepest descent followed by conjugated gradient calculations were adopted. All energy calculations were computed in vacuo with Gromas 96 43B1 parameters set without reaction field. Energy computations were done with Gromas 96 implementation of Swiss-PDB viewer including certain parameters such as bond length and angle deviation (33).
Evaluation of the model
A good model corresponds to a low value of the objective function. The profile 3D program (PROSA) tests the validity of hypothetical protein structure by measuring the compatibility of the structure with its own amino acid sequence. To verify the protein model, the coordinates of the model were submitted to PROCHECK (34). The stereochemcial quality of the protein structure was examined by Ramachandran plot 2.0. The number of residues in the allowed and disallowed regions determines the quality of the model. The RMSD (root mean square deviation) value of the model with respect to Cα atoms of the template was measured using a combinatorial extension method (35).
Authors’ contributions
KN designed the study, carried out the cloning and drafted the manuscript. SS carried out the sequence and bioinformatics analysis. SV raised the goat hyperimmune serum. SCA designed the primers used in the study. JFT revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
KN is thankful to the Department of Biotechnology (DBT), New Delhi, India for funding and providing DBT Overseas Associateship Training in Niche areas of Biotechnology. The authors are also thankful to the Vice-Chancellor, Bharathidasan University and the President, University of Kentucky for the research facilities provided to carry out this work.
Supplementary Material
Figures S1-S4; Table S1
References
- 1.Vinetz J.M. Leptospirosis. Curr. Opin. Infect. Dis. 2001;14:527–538. doi: 10.1097/00001432-200110000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Faine S. second edition. MediSci; Melbourne, Australia: 1999. Leptospira and Leptospirosis. [Google Scholar]
- 3.Marotto P.C. Acute lung injury in leptospirosis: clinical and laboratory features, outcome, and factors associated with mortality. Clin. Infect. Dis. 1999;29:1561–1563. doi: 10.1086/313501. [DOI] [PubMed] [Google Scholar]
- 4.Haake D.A. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 2000;68:2276–2285. doi: 10.1128/iai.68.4.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natarajaseenivasan K. Leptospiral proteins expressed during acute and convalescent phases of human leptospirosis. Indian J. Med. Res. 2004;120:151–159. [PubMed] [Google Scholar]
- 6.Ballard S.A. Molecular analysis of the dnaK locus of Leptospira interrogans serovar Copenhageni. Gene. 1998;216:21–29. doi: 10.1016/s0378-1119(98)00329-1. [DOI] [PubMed] [Google Scholar]
- 7.Park S.H. Expression and immunologic characterization of recombinant heat shock protein 58 of Leptospira species: a major target antigen of the humoral immune response. DNA Cell Biol. 1999;18:903–910. doi: 10.1089/104454999314764. [DOI] [PubMed] [Google Scholar]
- 8.Ballard S.A. Molecular analysis of the hsp (groE) operon of Leptospira interrogans serovar Copenhageni. Mol. Microbiol. 1993;8:739–751. doi: 10.1111/j.1365-2958.1993.tb01617.x. [DOI] [PubMed] [Google Scholar]
- 9.Haake D.A. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J. Bacteriol. 1993;175:4225–4234. doi: 10.1128/jb.175.13.4225-4234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang E.S. The rare outer membrane protein, OmpL1, of pathogenic Leptospira species is a heat-modifiable porin. Infect. Immun. 1995;63:3174–3181. doi: 10.1128/iai.63.8.3174-3181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vedhagiri K. Evolutionary implication of outer membrane lipoprotein-encoding genes ompL1, lipL32 and lipL41 of pathogenic Leptospira species. Genomics Proteomics Bioinformatics. 2009;7:96–106. doi: 10.1016/S1672-0229(08)60038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang E.S. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 1996;64:2322–2330. doi: 10.1128/iai.64.6.2322-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haake D.A. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 2000;68:2276–2285. doi: 10.1128/iai.68.4.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natarajaseenivasan K., Ratnam S. Recent Leptospira isolates from Tamilnadu. Indian J. Anim. Sci. 2000;70:551–555. [Google Scholar]
- 15.Ellis R.J., van der Vies S.M. Molecular chaperones. Annu. Rev. Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- 16.Zielsta-Ryalls J. The universally conserved GroE (Hsp60) chaperonins. Annu. Rev. Microbiol. 1991;45:301–325. doi: 10.1146/annurev.mi.45.100191.001505. [DOI] [PubMed] [Google Scholar]
- 17.Farrar D.M. Heat shock protein and inflammatory acne vulgaris: molecular cloning, overexpression and purification of a propionibacterium acnes GroEL and DnaK homologue. FEMS. Microbiol. Lett. 2000;191:183–186. doi: 10.1111/j.1574-6968.2000.tb09337.x. [DOI] [PubMed] [Google Scholar]
- 18.Ensgraber M., Loos M. A 66-kilodalton heat shock protein of Salmonella typhimurium is responsible for binding of the bacterium to intestinal mucus. Infect. Immun. 1992;60:3072–3078. doi: 10.1128/iai.60.8.3072-3078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerreiro H. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 2001;69:4958–4968. doi: 10.1128/IAI.69.8.4958-4968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamm L.V. Heat shock response of spirochetes. Infect. Immun. 1991;59:1572–1575. doi: 10.1128/iai.59.4.1572-1575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta R.S. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol. Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 22.Zugel U., Kaufmann S.H. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 1999;12:19–39. doi: 10.1128/cmr.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felgner P.L. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc. Natl. Acad. Sci. USA. 2009;106:13499–13504. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natarajaseenivasan K. Surface-associated Hsp60 chaperonin of Leptospira interrogans serovar Autumnalis N2 strain as an immunoreactive protein. Eur. J. Clin. Microbiol. Infect. Dis. 2011;30:1383–1389. doi: 10.1007/s10096-011-1232-z. [DOI] [PubMed] [Google Scholar]
- 25.Huang Q. In vivo cytotoxic T lymphocyte elicitation by mycobacterial heat shock protein 70 fusion proteins maps to a discrete domain and is CD4+ T-cell independent. J. Exp. Med. 2000;191:403–408. doi: 10.1084/jem.191.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S. Cystosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem. Sci. 1994;19:543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 27.Boom R. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Sali A., Overington J.P. Derivation of rules for comparative protein modeling from a database of protein structure alignments. Protein Sci. 1994;3:1582–1596. doi: 10.1002/pro.5560030923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sali A. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 32.Sali A. Modelling mutations and homologous proteins. Curr. Opin. Biotechnol. 1995;6:437–451. doi: 10.1016/0958-1669(95)80074-3. [DOI] [PubMed] [Google Scholar]
- 33.W.F., van Gunsteren, et al. 1996. Biomolecular Simulation: The GROMOS 96 Manual and User Guide. Zurich, Switzerland.
- 34.Laskowski R.A. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 35.Shindyalov I.N., Bourne P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein. Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S4; Table S1