Abstract
MicroRNAs (miRNAs) are a newly identified class of small non-protein-coding post-transcriptional regulatory RNA in both plants and animals. The use of computational homology based search for expressed sequence tags (ESTs) with the Ambros empirical formula and other structural feature criteria filter is a suitable combination towards the discovery and isolation of conserved miRNAs from tea and other plant species whose genomes are not yet sequenced. In the present study, we blasted the database of tea (Camellia sinensis) ESTs to search for potential miRNAs, using previously known plant miRNAs. For the first time, four candidate miRNAs from four families were identified in tea. Using the newly identified miRNA sequences, a total of 30 potential target genes were identified for 11 miRNA families; 6 of these predicted target genes encode transcription factors (20%), 16 target genes appear to play roles in diverse physiological processes (53%) and 8 target genes have hypothetical or unknown functions (27%). These findings considerably broaden the scope of understanding the functions of miRNA in tea.
Key words: Camellia sinensis, EST, miRNA, tea
Introduction
Gene expression is regulated at several levels and a recently discovered post-transcriptional mechanism involves small RNA (sRNA) molecules (1). In the class of plant sRNAs, microRNAs (miRNAs) represent a newly identified class of non-protein-coding small (~20 nt) RNAs, which negatively regulate the gene expression at the post-transcriptional level by repressing gene translation or degrading targeted mRNAs (2). miRNAs play an important role in many plant biological processes, including leaf development (3), stem development (4), root development (5), signal transduction (6), developmental timing (7) and responses to different environmental stresses 8, 9. Interest in miRNA identification has attracted the attention of many scientists to understand the evolution of miRNAs and miRNA targeted gene regulation.
There are two major approaches for identifying miRNAs: genetic approach by direct cloning and computational approach by comparative genomics. Although the direct cloning approach can be used to identify new miRNAs, it has several disadvantages such as (1) difficulty in finding miRNAs that are expressed at lower levels, (2) difficulty in cloning due to their physical properties including sequence composition or post-transcriptional modifications such as editing or methylation, (3) RNA degradation during sample separation, and (4) tissue or stage specific expression. On the other hand, the computational or bioinformatic prediction is an effective alternative for large-scale discovery of miRNAs from different plants and animals. Nowadays, publicly available databases play a central role in in silico biology. Homology based search of these databases using ~21 conserved plant miRNA families can help to identify orthologs and paralogs of miRNAs in plants (10).
Tea [Camellia sinensis (L.) O. Kuntze], an important commercial beverage crop of the world, is an evergreen woody perennial grown in different agro-climatic zones. It also has great value as a source of secondary metabolic products, such as tea polyphenols, catechins, caffeine, theanine, and saponin, which have medicinal properties (11).
Recently, molecular biology of tea plants has been one of the most active and kinetic research fields of tea science. The recent progress in functional genomics research based on large-scale expressed sequence tag (EST) generation, analysis and cloning of genes in tea plant has provided a critical significance on elucidating the molecular mechanism of growth, development, differentiation, metabolism, quality, yield, and stress resistance, as well as genetic manipulation via biotechnological approaches in the foreseeable future (11).
Hundreds of miRNAs have been identified in recent years but there has been no report on miRNAs in tea. Therefore, we introduce the computational approach for identifying miRNAs in the tea plant. In the present study, we used all known plant miRNAs (so far publicly available) from Arabidopsis, rice and other plants to search the conserved C. sinensis miRNA homologues in publicly available EST databases. A total of four potential miRNAs were detected with predicted stem-loop precursor structure. Using the potential miRNA sequences, we further blasted C. sinensis mRNA database and found 30 potential miRNA target genes for 11 miRNA families.
Results
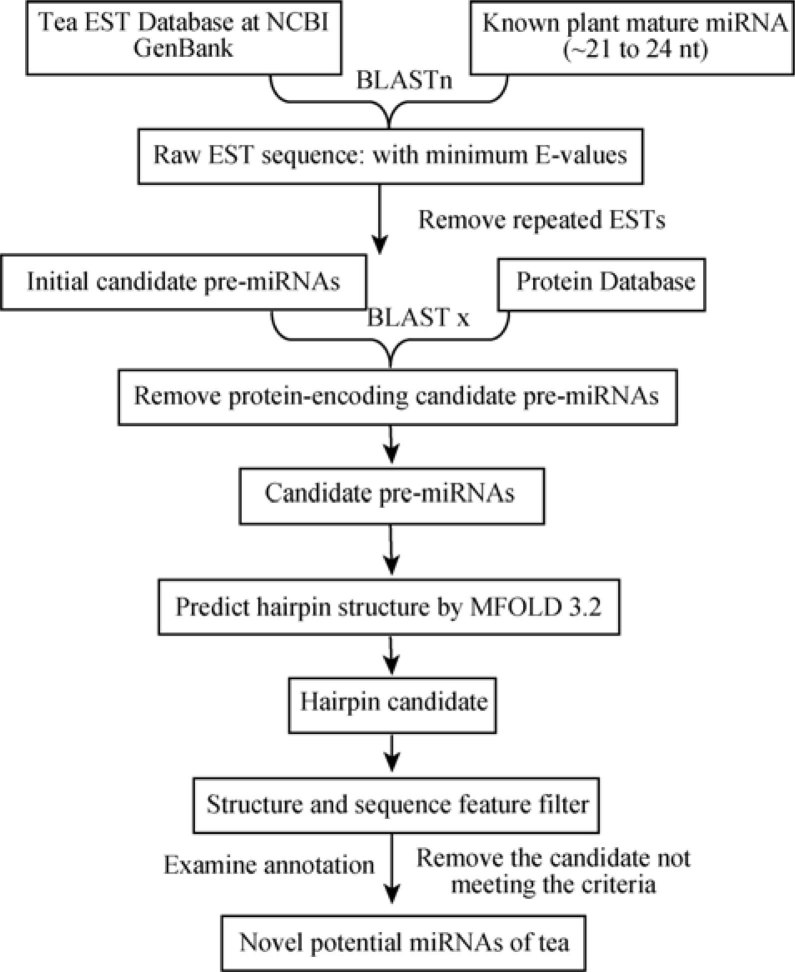
To predict new miRNAs in tea by computational methods, we used defined sequence and structural properties of known miRNAs to screen candidate miRNAs in the EST database of tea. Figure 1 shows the search and filtering procedure for identifying potential miRNAs in tea. Since most of the known mature miRNAs are conserved within the plant species, it is possible to perform a computational search for new miRNAs (10).
Figure 1.
Schematic representation of the tea miRNA search procedure used to identify homologues of known plant miRNAs.
A total of 299 EST sequences of tea were identified by BLAST search using all known plant mature miRNA sequences. These sequences were further analyzed for the presence of miRNAs using mirEval miRNA prediction software, and 159 ESTs were filtered. Out of the 159 ESTs, 43 met the preliminary screening criteria for mature miRNA sequences against all known plant mature miRNA sequences. The mature miRNAs predicted from precursor miRNAs are 21-24 nt long and with 0-4 mismatches to known miRNAs. Then, the initially predicted mature miRNAs containing the full EST sequence were further subjected to BLASTx against protein databases to exclude coding sequences. A total of 23 non-protein-coding homologs were predicted as potential miRNAs and their secondary structures were predicted using MFold 3.1 program. After this filtering based on secondary structure, four folded miRNA precursors were predicted from tea EST sequences, which confirmed to the criteria mentioned in Materials and Methods (Table 1 and Figure 2).
Table 1.
Predicted miRNAs of tea
| miRNA family | EST ID | miRBase best hit ID | Mature sequencea | ML | MN | Ab | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| miR164*** | CV013669 | osa-miR164e | UGGAGAAGCAGGGCACGUGAG | 21 | 2 | 3’ | |||||
| miR169*** | GE650220 | osa-miR169n | AAGCCAAGAAUGACUUGCCa | 21 | 3 | 5’ | |||||
| miR1846* | DN976181 | osa-miR1846-5p | AUUGAGGAGGCCGGGaCuUCaU | 22 | 3 | 5’ | |||||
| miR1863* | GH623864 | osa-miR1863 | AGCUCUGAUACCAUaUUAGAaUAa | 24 | 3 | 5’ | |||||
| miRNA family | Precursor miRNAc |
PL (nt) | A (%) | C (%) | G (%) | U (%) | A+U (%) | G+C (%) | MFE | ||
| Start | End | ||||||||||
| miR164*** | 309 | 376 | 68 | 28 | 20 | 28 | 23.5 | 51.5 | 48.53 | 20 | |
| miR169*** | 442 | 492 | 51 | 22 | 18 | 25 | 35.3 | 56.9 | 43.14 | 7.8 | |
| miR1846* | 11 | 228 | 218 | 28 | 12 | 20 | 39.5 | 67.9 | 32.11 | 40.5 | |
| miR1863* | 377 | 448 | 72 | 33 | 8.3 | 25 | 35.3 | 66.7 | 33.33 | 19.2 | |
unique miRNA family registered in only one plant species in miRBase
highly conserved miRNA family registered in more than two plant species in miRBase.
Mature sequence of predicted miRNA. Lowercase represents the mismatch with the known miRNA sequence hit.
Location of the mature miRNA in the arm of the precursor sequence.
Location of the precursor sequence in the EST. ML, length of the mature miRNA sequence; MN, number of mismatches; PL, length of the precursor; A, C, G, U (%), adenine, guanine, cytosine, uracil nucleotide composition in the precursor miRNA; MFE, minimum free energy.
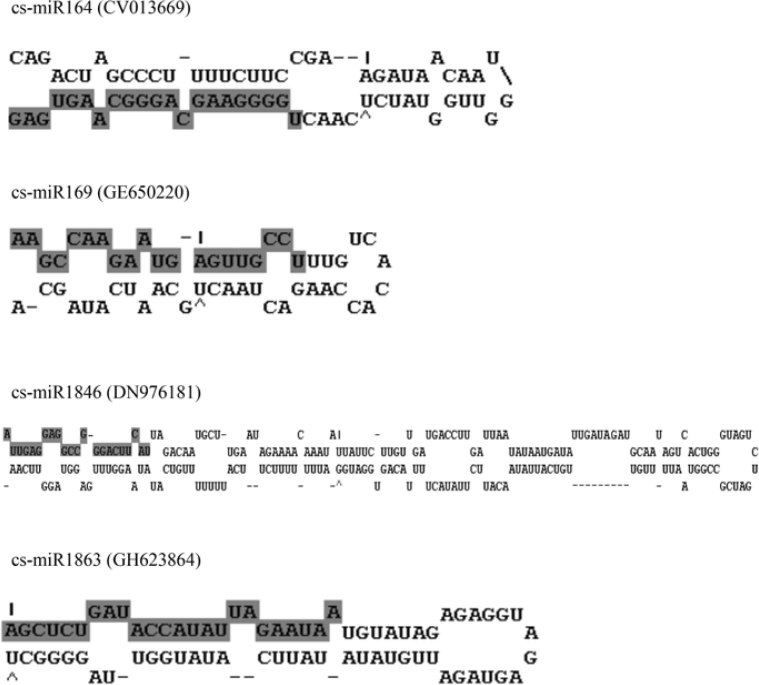
Figure 2.
Folded hairpin structures for miRNA precursors from tea. The region of the mature miRNA sequence is shown in bold and highlighted in gray. The EST sequence accession numbers are presented in parenthesis.
Table 1 summarizes the miRNA family, mature miRNA sequence, precursor miRNA length and their position, minimum free energy (MFE), and number of nucleotide difference. The newly discovered tea precursor miRNAs have MFE values ranging from -7.8 to -40.5 kcal/mol according to MFOLD. The lengths of the identified precusor miRNAs range from 51 to 218 nt and the mature sequences range from 21 to 24 nt. Of the four miRNAs identified in tea, two belong to highly conserved miRNA families of miR164 and miR169, and the other two belong to unique miRNA families of miR1846 and miR1863.
The percentage composition of the four nucleotides (A, C, G and U) in tea pre-miRNAs is presented in Table 1. Uracil is dominant in the identified pre-miRNA sequences and ranges from 23.5% to 39.5% of total nucleotide composition followed by adenine, guanine and cytosine.
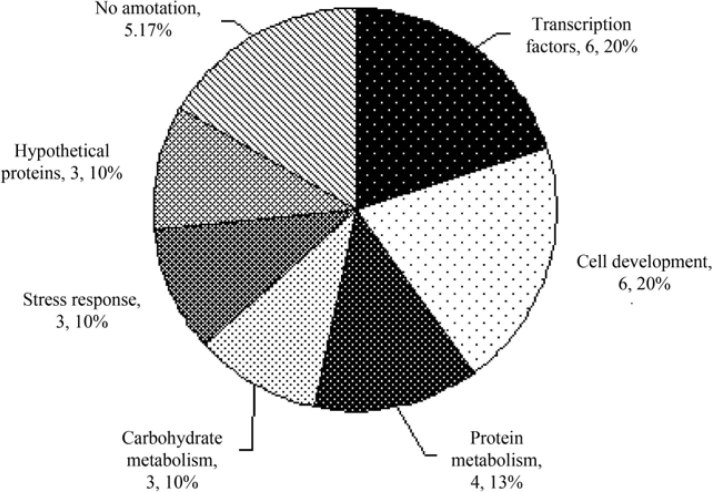
To identify potential miRNA targets for known plant miRNAs, we used miRU software, tea mRNA/ cDNA from NCBI and nucleotide database from TIGR along with the parameters as mentioned in Materials and Methods. In this study, we identified a total of 30 targets for 11 miRNA families (Table S1). These targets belong to several gene families with different biological functions (Figure 3), including the control of cell development (six genes; 20%), transcription factors (six genes; 20%), metabolic pathways (three and four genes for carbohydrate and protein metabolisms, respectively; 10% and 13%), response to stress (three genes; 10%) and unknown or hypothetical function (eight genes; 27%) (Figure 3). The frequency of pri-miRNAs in the tea EST collection was found to be approximately 0.23% (23 out of 10,000 ESTs). The target genes of miRNAs and the list of these predicted targets are presented in Table S1.
Figure 3.
Functional categorization of miRNA target genes in tea. The functional categorization of target genes was presented along with the number of genes in each group and their percentage.
Discussion
Most mature miRNAs are evolutionarily conserved from species to species within the plant kingdom. This piece of information enables us to computationally predict new miRNA homologs or orthologs in different plant species (10). Therefore, we used all previously known plant mature miRNAs from miR registry to search for homologs of miRNAs and their target genes in tea in the publicly available EST database of tea.
By computational predictions, we found four pre-miRNAs belonging to four families (miR 164, 169, 1846 and 1863) for the first time in tea. Application of the criteria for filtering offers the following advantages: (1) the number of RNAs analysed is reduced; (2) the likelihood of inclusion of non-miRNAs in subsequent analyses is minimized; and (3) the total number of predicted false miRNAs is significantly reduced. All four predicted miRNAs were considered to be valid candidates by satisfying criteria B, C and D of the empirical formula for biogenesis and expression of the miRNAs as suggested by Ambros et al. (12). According to Ambros et al. (12), criterion D alone is enough for homologous sequences to validate the new miRNAs in different species.
In the present study, the length of predicted miRNA precursors varies from 51 to 218 nt. The different sizes of the identified miRNAs within the different families suggest that they may perform unique functions in the regulation of miRNA biogenesis or gene expression (10).
MFE is an important characteristic that determines the secondary structure of nucleic acids (DNA and RNA). The lower the MFE, the higher the thermodynamically stable secondary structure of the corresponding sequence. We observed that the MFEs of the precursor miRNAs ranged from -7.8 to -40.5 kcal/mol, which are almost equal to the values of other plant precursor miRNAs and much lower than those of tRNA and ribosomal RNA (13). All the mature sequences of tea miRNAs are in the stem portion of the hairpin structures, as shown in Figure 2. The predicted miRNA hairpin structures show that there are at least 12-21 nt engaged in Watson-Crick or G/U pairings between the miRNA/miRNA* in the stem region and do not contain large internal loops or bulges. These findings are in accordance with those described by Zhang et al. (10).
The frequency of pri-miRNAs in the tea EST collection was found to be approximately 0.23%, which is higher than the rate of 0.01% (1 in 10,000 ESTs) reported (10). This could likely overestimate the frequency of pri-miRNAs, because the precursors identified during the preliminary screening steps include sequences that are targets, mRNAs for coding sequence that match the miRNA in the wrong orientation, and duplicate sequences derived from the same genes.
The miRNA families found in miRBase are classified as highly conserved or unique depending on the number of species in which they have been identified. We considered miRNA families registered in single species as unique, and those registered in two species and three or more species as moderately and highly conserved, respectively (14). The predicted tea miRNAs, cs-miR164 and cs-miR169, are conserved as in Arabidopsis thaliana, Oryza sativa (rice), Zea mays (maize), Triticum aseativum (wheat) and Saccharum officinarum (sugarcane) miRNAs, while cs-miR1846 and cs-miR1863 are uniquely conserved with rice miRNAs (10).
The predicted miRNAs were further classified as non-coding ESTs through the BLASTx analysis. The EST based identification of miRNA shows a relationship between the miRNAs and their tissue, organ or developmental stage to which the ESTs belong. Based on the expression in predicted miRNAs, the highly conserved cs-miR164 and unique cs-miR1846 and cs-miR1863 are expressed in the leaves of tea. The highly conserved cs-miR169 is expressed in the roots. Thus, the findings relating to the conservation of miRNA families, their non-coding nature, and the expression pattern of EST based identification strongly validated our identified sequences as candidate miRNAs in tea.
To understand the biological function of miRNAs in plant development, it is necessary to identify their targets. No high-throughput experimental techniques for target site identification have been reported yet. Two strategies have been employed towards this end: (1) genetic approach, which is based on the abnormal expression of target mRNAs in the miRNA loss-of-function mutants, and (2) computational approaches, which have been successful in plants. MIRcheck (8), findMiRNA (15) and miRU (16) are the available software for identifying miRNA targets in plants. The predicted targets can be subsequently verified by adopting PCR based strategies.
Plant miRNAs generally show a near-perfect complementarity with their targets on mRNAs, which immensely facilitates computational searches 6, 17. Taking advantage of this unique property, we identified antisense hits of known miRNAs on tea mRNA/cDNA database.
In miRNA target prediction, the screening criteria were set according to the description in Materials and Methods. Finally, we found 30 target EST/mRNAs for the 11 miRNA families (Table S1). There were 22 target ESTs encoding functional proteins and another 8 target ESTs coding hypothetical or unknown proteins (Figure 3). The presence of targets in the EST database of tea provides additional evidence for the existence of other miRNA families in tea.
As reported in Arabidopsis, MIR164 showed positive correlation with its target CUC2, suggesting that cs-miR164 may determine the young leaf primordia serration by translational repression (18). The regulation mechanism of conserved cs-miR164 was conserved throughout monocot and dicot plants.
The targets of cs-miR397 and cs-miR408 were conserved between Arabidopsis and rice, and these were involved in copper homeostasis regulation (Table S1) by guiding the cleavage of mRNAs of plastacyanins, copper/zinc superoxide dismutases and laccases, respectively (19), and/or the ROS levels in rice embryo (20).
Additionally, cs-miR472 and cs-miR782 both target 50S, 60S ribosomal proteins L16 and L44, which in turn constitute the structural component of ribosomes and regulate the translation process (21).
DNA methylation at specific loci is induced by heterochromatic histone modifications and siRNAs. RNA-dependent DNA methylation (RdDM) depends on the components of siRNA biogenesis and methylation complex. In this study, the predicted cs-miR852 target gene encodes histone H2B, which has also been found to regulate RNA-directed gene silencing by de-ubiquitination (22).
The identified target gene of unique cs-miR1134 encodes β-1,3 glucanase, which is a plasmadesmata targeted protein (23). In plants, cell-to-cell communication through plasmodesmata (Pds) is vital and establishes a symplastic continuum in the plant cell. Callose deposition at Pds is stimulated by physical and physiological stresses, which is correlated with β-1,3 glucanase expression. The plants with reduced accumulation of the glycolytic enzyme β-1,3 glucanase had increased callose accumulation and a reduction in the experimental molecular size exclusion limit (24). Hence, the control of callose synthesis and turnover by miRNA mediated β-1,3 glucanase activity is proposed to provide a mechanism for regulating Pd flux. We also found perfect or near-perfect complementary sites for cs-miR1134 in protein processing associated proteins like 10 kDa chaperonin and serine carboxypeptidase cluster protein and peptidase S10 (Table S1).
Among the pool of mRNA targets, six genes are transcriptional factors, whereas others are associated with metabolism and response to environmental stress. Transcriptional factors are important components in the transcriptional process and play an important role in a variety of biological functions including plant development, hormone signaling and metabolism. There are 16 potential targets of cs-miR414 identified in tea (Table S1). Among those, three were predicted to be transcriptional factors, and two are hypothetical or unknown proteins. Plant high-mobility-group (HMG) chromosomal proteins are the most abundant, ubiquitous non-histone proteins found in the nuclei of higher eukaryotes and conserved target genes for miR414 family (25). Due to their high binding affinity to DNA, it is suggested that post-transcriptional regulation of the HMG protein family by cs-miR414 may be involved in genetic recombination and transcription in nuclei.
In this study, we found that cs-miR828 and cs-miR414 appear to target the transcription factor genes such as DNA binding domain containing protein, zinc finger protein-B box, CONSTANS-like 5 related cluster protein and NAC alpha subunit-like protein as in Arabidopsis and rice, which control diverse functions ranging from control of floral morphology and flowering time (cs-miR414), translational control and protein targeting (cs-miR414) to plant developmental processes (cs-miR828) (Table S1). CONSTANS-like (COL) proteins are plant-specific nuclear regulators of gene expression but do not contain a known DNA-binding motif and can use the CBF element to interact DNA and regulate transcription. CONSTANS (CO) plays a central role in the photoperiod response pathway by mediating between the circadian clock and the floral integrators (26). Functional studies have shown that CONSTANS (AtCO) protein mediates photoperiodic induction of flowering in Arabidopsis (27). cs-miR414 also targets alpha subunit of nascent polypeptide associated complex (4). These results are in accordance with studies observed in wheat (14).
Here we found that tea cs-miR414 is perfectly complementary to the mRNAs encoding α-tubulin protein and calmodulin-binding heat shock protein, suggesting that the miRNA may regulate genes involved in structural integrity of the cell during stress response. In addition, cs-miR414 was found to target the gene coding for plastidic aldolase and fructose biphosphate aldolase (At4g38970; Table S1), which undergoes glutathionylation/deglutathionylation in response to illumination and in turn facilitates the calvin cycle during oxidative inhibition and stress condition 28, 29.
The non-conserved miRNA targets identified in this study may be involved in processes that are species specific or tissue specific (Table S1). Eight of the newly identified potential targets (Table S1) are annotated as hypothetical proteins, having unknown function and/or no BLASTx hit. Thus there remain many avenues of investigation that will enhance the understanding of the role of miRNAs in the regulation of gene expression. These findings considerably broaden the scope of understanding the function of miRNA in tea.
Conclusion
The present study is based on the computational approach for new miRNA identification from plant species whose genome is not yet sequenced. We have identified four mature miRNAs along with their target genes in tea. This is a first step towards the identification of miRNAs in tea and further experimental and in silico studies leading to understand the function and processing of miRNAs are in progress in our laboratory. Thus, the identification of miRNAs and their target genes can serve as an initial point for the characterization of their roles in gene regulation in this important economic crop.
Materials and Methods
Reference set of miRNAs
To search potential miRNAs in tea, a total of previously known 1,024 miRNAs and their precursor sequences from Arabidopsis thaliana, Oryza sativa, Glycine max, Sorghum bicolor, Zea mays, Saccharum officinarum, and Vitis vinifera were obtained from miRNA Registry Database (Release 9.0, October 2006; http://miRNA.sanger.ac.uk) (30). These miRNAs were defined as a reference set of miRNA sequences. To avoid the redundant or overlapping miRNAs, the repeated sequences of miRNAs within the above species were removed and the remaining sequences were used as query sequences for BLAST search.
Tea ESTs, cDNAs and mRNAs
Tea mRNA, cDNA and EST sequences were obtained from the GenBank nucleotide databases at NCBI (http://www.ncbi.nlm.nih.gov; March 2009) and the tea nucleotide databases were from The Institute for Genome Research (http://www.tigr.org). A total of 10,000 tea ESTs were deposited in the EST database and all of these ESTs were screened against the known plant miRNAs.
Prediction of potential miRNAs and their precursors using EST-based comparative genomics
Figure 1 summarizes the major steps for identifying potential miRNA sequences in tea. The mature sequences of all known plant miRNAs were subjected to BLASTn search in the tea EST databases using BLASTn 2.2.9 (31). Adjusted BLASTn parameter settings were as follows: expect values were set at 1,000; low complexity was chosen as the sequence filter; the number of descriptions and alignments was raised to 1,000. The default word-match size between the query and database sequences was seven. All BLASTn results were saved. If the matched sequence was shorter than the queried miRNA sequence, the aligned and non-aligned parts were manually inspected and compared to determine the number of matching nucleotides. RNA sequences were considered miRNA canditates only if they fit the following criteria: (1) at least 18 nt length were adopted between the predicted mature miRNAs and (2) allowed to have 0-3 nt mismatches in sequence with all previously known plant mature miRNAs. The ESTs that closely matched the previously known plant mature miRNAs were included in the set of miRNA candidates and used for additional characterization based on the following criteria: (1) the entire EST sequence was selected to predict the secondary structures and to screen for miRNA precursor sequences; (2) the selected ESTs were further compared with each other to eliminate redundancies; and (3) these sequences were subjected to evaluation for miRNA prediction properties using mirEval software (32). These precursor sequences were used for BLASTx analysis for removing the protein-coding sequences and retained only the non-protein sequences.
Prediction of secondary structure
Precursor sequences of these potential miRNA homologs were used for hairpin structure predictions using the Zuker folding algorithm with MFOLD 3.1 (33), which is publicly available at http://www.bioinfo.rpi.edu/applications/mfold/old/rna/. The following parameters were used in predicting the secondary structures: (1) linear RNA sequence; (2) folding temperatures fixed at 37°C; ionic conditions of 1M NaCl and with no divalent ions; (3) percent suboptimility number of 5; (4) maximum interior/bulge loop size of 30; (5) the grid lines in energy dot plot turned on. All other parameters were set with default values.
In brief, the following criteria were applied in designating the RNA sequence as an miRNA homolog as described by Zhang et al. (34): (1) pre-miRNA sequence can fold into an appropriate stem-loop hairpin secondary structure; (2) it contains the ~22 nt mature miRNA sequence within one arm of the hairpin; (3) predicted secondary structures had higher negative minimal free energies and minimal free energy index (MFEI) than other different types of RNAs; (4) an MFEI of greater than 0.85; (5) 30%-70% A+U content; (6) predicted mature miRNAs had no more than six mismatches with the opposite miRNA* sequence in the other arm; (7) maximum size of 3 nt for a bulge in the miRNA sequence; and (8) no loop or break in miRNA sequences was allowed. These criteria significantly reduced false positives and required that the predicted miRNAs fit the criteria proposed by Ambros and co-workers (12).
Prediction of potential miRNA targets
To predict the potential miRNA targets, we used miRU software publicly available at http://bioinfo3.noble.org/miRNA/miRU.htm (16). The following parameters were adjusted: 3 as score for each 20 nt, 6 for G:U Wobble pairs, 0 for indel and 3 for other mismatches.
Authors’ contributions
GRP and AKAM conceived the project. GRP collected the data and conducted the computational analysis. AKAM supervised the work. GRP and AKAM interpreted the data and prepared the manuscript. Both authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We thank Dr. P. Mohankumar, Director and Dr. N. Muraleedharan, Adviser of UPASI Tea Research Foundation for their encouragement and support during the course of study.
Supplementary Material
Table S1
References
- 1.Herr A.J. Pathways through the small RNA world of plants. FEBS Lett. 2005;579:5879–5888. doi: 10.1016/j.febslet.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Palatnik J.F. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 4.Mallory A.C. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 5.Guo H.S. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhoades M.W. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 7.Pasquinelli A.E., Ruvkun G. Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 8.Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Chiou T.J. The role of microRNAs in sensing nutrient stress. Plant Cell Environ. 2007;30:323–332. doi: 10.1111/j.1365-3040.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 11.Ma C.L., Chen L. Research progress on isolation and cloning of functional genes in tea plants. Front. Agric. China. 2007;1:449–455. [Google Scholar]
- 12.Ambros V. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnet E. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 14.Dryanova A. Data mining for miRNAs and their targets in the Triticeae. Genome. 2008;51:433–443. doi: 10.1139/G08-025. [DOI] [PubMed] [Google Scholar]
- 15.Adai A. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendes N.D. Current tools for the identification of miRNA genes and their targets. Nucleic Acid Res. 2009;37:2419–2433. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikovics K. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdel-Ghany S.E., Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008;283:15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue L.J. Characterisation and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 2008;37:916–930. doi: 10.1093/nar/gkn998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manuell A. Regulation of chloroplast translation: interactions of RNA elements, RNA-binding proteins and the plastid ribosome. Biochem. Soc. Trans. 2004;32:601–605. doi: 10.1042/BST0320601. [DOI] [PubMed] [Google Scholar]
- 22.Sridhar V.V. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 23.Levy A. A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J. 2007;49:669–682. doi: 10.1111/j.1365-313X.2006.02986.x. [DOI] [PubMed] [Google Scholar]
- 24.Simpson C. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell. 2009;21:581–594. doi: 10.1105/tpc.108.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W.S. Interaction of wheat high-mobility group proteins with four-way-junction DNA and characterization of the structure and expression of HMGA gene. Arch. Biochem. Biophys. 2003;409:357–366. doi: 10.1016/s0003-9861(02)00630-6. [DOI] [PubMed] [Google Scholar]
- 26.Searle I., Coupland G. Induction of flowering by seasonal changes in photoperiod. EMBO J. 2004;23:1217–1222. doi: 10.1038/sj.emboj.7600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben-Naim O. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- 28.Yamada S. Differential expression of plastidic aldolase genes in Nicotiana plants under salt stress. Plant Sci. 2000;154:61–69. doi: 10.1016/s0168-9452(00)00188-6. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto M., Ogawa K. New insight into the calvin cycle regulation glutathionylation of fructose bisphosphate aldolase in response to illumination. In: John F.A., editor. Photosynthesis. Energy from the Sun. Springer; Netherlands: 2008. pp. 872–874. [Google Scholar]
- 30.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul S.F. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie W. Mireval: a web tool for simple microRNA prediction in genome sequences. Bioinformatics. 2008;24:1394–1396. doi: 10.1093/bioinformatics/btn137. [DOI] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B.H. Identification and characterisation of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1