Abstract
Small RNAs (sRNAs) are non-coding transcripts exerting their functions in the cells directly. Identification of sRNAs is a difficult task due to the lack of clear sequence and structural biases. Most sRNAs are identified within genus specific intergenic regions in related genomes. However, several of these regions remain un-annotated due to lack of sequence homology and/or potent statistical identification tools. A computational engine has been built to search within the intergenic regions to identify and roughly annotate new putative sRNA regions in Enterobacteriaceae genomes. It utilizes experimentally known sRNA data and their flanking genes/KEGG Orthology (KO) numbers as templates to identify similar sRNA regions in related query genomes. The search engine not only has the capability to locate putative intergenic regions for specific sRNAs, but also has the potency to locate conserved, shuffled or deleted gene clusters in query genomes. Because it uses the KO terms for locating functionally important regions such as sRNAs, any further KO number assignment to additional genes will increase the sensitivity. The PsRNA server is used for the identification of putative sRNA regions through the information retrieved from the sRNA of interest. The computing engine is available online at http://bioserver1.physics.iisc.ernet.in/psrna/ and http://bicmku.in:8081/psrna/.
Key words: small RNA, KEGG Orthology, flanking genes, intergenic regions
Introduction
The un-translated non-coding RNAs (ncRNAs) have recently been discovered in all life forms to play vital roles in different physiological processes such as transcriptional regulation, chromosome replication, RNA processing and modification, messenger RNA stability, protein degradation and translocation (1). Thus by coordinating important processes, ncRNAs control essential functions in eukaryotes such as developmental gene regulation and disease progression (2). Prokaryotic small RNAs (sRNAs) are counterparts of the eukaryotic ncRNAs, which are 50 to 400 nucleotides in length (3). They predominantly act by base pairing with their specific target mRNAs, thereby affecting the stability and/or translation of the message (4). They also modify the activity of RNA-binding regulatory proteins through binding and sequestering them (5).
Different computational and experimental approaches have been carried out for the identification of sRNAs (3). However, computational identification of sRNAs is not accurate when compared to the prediction of coding genes (6). Recently, accumulated sRNA data in various public databases like Rfam (7), NONCODE (8), KEGG (9) and GenBank (10) open a possibility of new context based prediction methods. An analysis of the existing sRNA data from Enterobacteriaceae family indicates that 21 different sRNA groups do not show sequence homology within closely related genomes (11). However, such homologous and non-homologous sRNA regions of a specific sRNA group can be identified using their specific conserved flanking genes 12, 13. In this study, an automated computing server is constructed and successfully employed to identify regions of both homologous and non-homologous sRNAs in completely sequenced enterobacterial genomes. We have used the KEGG Orthology (KO) numbers of the sRNA specific conserved flanking gene pairs for the automated identification of putative sRNA regions in the query genomes. Data mining using bio-ontology terms has recently been used for identifying disease related genes in eukaryotes (14), and for adding functional annotations of coding genes 13, 15. A comparison among prokaryotic controlled vocabularies from COG, KEGG and TIGR databases shows that KO datasets (9) have higher quality of annotations than other available ontology datasets (16). The KO system classifies both orthologous genes and orthologous relationships of paralogous gene groups (17), and the current version of KO dataset (dated on 9/30/2009) has been used to identify orthologous relationships between known sRNA specific conserved flanking genes and query genomes. Using KO terms, the prediction server has been designed to locate potentially important putative intergenic regions for prokaryotic sRNAs. In order to remove false positives, simultaneous occurrence of the orthologous sRNA specific flanking gene pairs that follow gene synteny and genomic backbone retention rule in query genomes (11) is reported. The co-existence of the flanking genes assigned with KO numbers is being re-analyzed by their gene locus numbers, and pairs found beyond the limit of five genes are excluded to minimize the false positives. The proposed putative sRNA identification strategy is a context based methodology that looks for the occurrence of sRNA specific flanking gene pairs alone, but not for any other promoter or terminator signals, thereby this tool is restricted in predicting the starts and ends of the sRNA regions residing within the intergenic regions. In some of the sRNAs, the transcriptional signals were not traceable either due to the lack of potent statistical biases or weak transcriptional signals, and such sRNA regions need to be verified using biochemical approaches. We are not attempting to find out the starts and ends of the “novel” sRNAs like QRNA (18) and RNAz (19) tools, but this is a novel approach towards the identification of putative intergenic regions/locations of the sRNA of interest.
Application
Implementation and utilities
The web interface for the identification of putative sRNA locations against query genomes is created and implemented using PERL and CGI scripts. The design of the input page and the validation is done using HTML and JavaScripts, respectively. The computing engine is developed and optimized for Fedora core (Version 9.0), and is driven by 3.0 GHz dual core processor equipped with 2 GB DDR RAM. It is compatible with Windows 95/98/2000/XP/NT and Linux operating systems through Netscape and Mozilla web browsers. Users need to choose the reference genome from the list of available microbial genomes. The server allows any one of the experimentally proved sRNA coordinates or flanking genes as training data to predict the putative sRNA regions in the query genomes.
Availability
The PsRNA computing engine is freely accessible at the following locations: http://bioserver1.physics.iisc.ernet.in/psrna/ and http://bicmku.in:8081/psrna/.
Algorithmic description and evolvability of the server
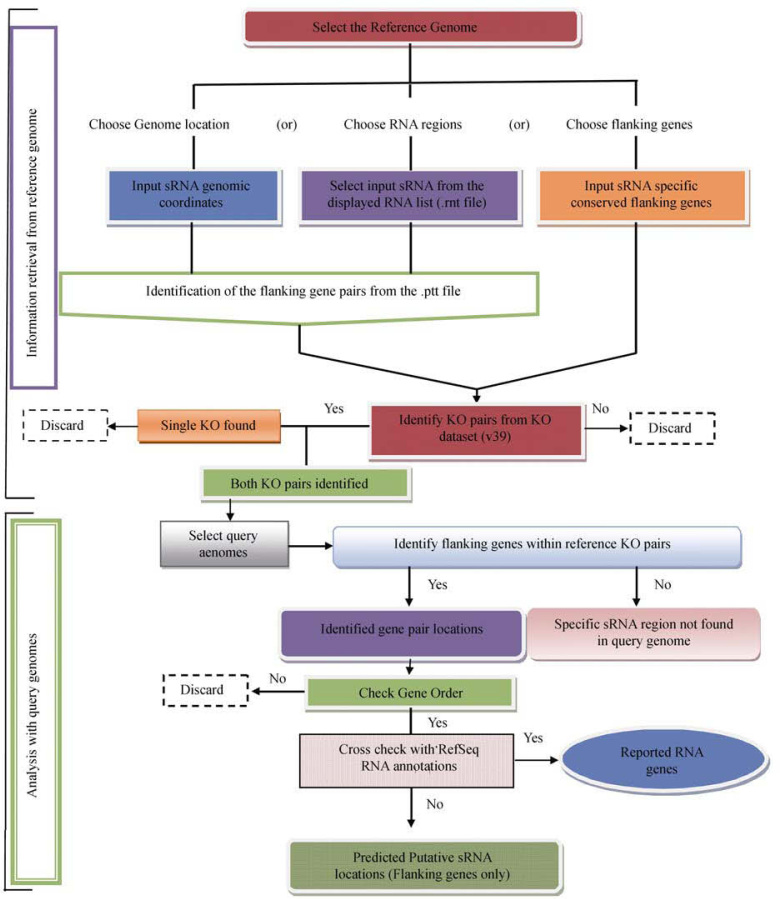
The web server accepts sRNA information from the selected reference genome through any one of the displayed forms: (1) the reference sRNA genomic coordinates, (2) the gene IDs of conserved gene pairs that flank the known sRNA, or (3) selection of RNA genomic coordinate from the .rnt files of the reference genome obtained from GenBank (10). For the first option, users have to enter the genomic coordinates obtained from other databases, such as Rfam (www.sanger.ac.uk/Software/Rfam) (20), NONCODE (8), KEGG (9) and/or collected sRNA data from literature, in the ‘from’ and ‘to’ boxes within a range of 0 to 500 bases. For the second option, users need to specify the conserved gene pair IDs that flank the known sRNA of the reference genome. Finally, the users can load the reference genome of interest and simply select the known sRNA genomic coordinates that are displayed in the scroll down menu. The sRNA genomic coordinates (.rnt files) are obtained from GenBank (10).
Information retrieval from the reference genome
The genomic coordinate input will be used by the server to search the corresponding up-stream and down-stream flanking genes and their gene ID codes from the protein coding table (.ptt file) obtained from GenBank (10). These gene ID codes of the reference genome are used to search and obtain orthologous KO number assignments from KEGG datasets (9). The server will use the above identified KO numbers to search and display the presence or absence of orthologous genes in the selected query genomes. The intergenic region that is flanked by the orthologous flanking gene pairs in the query genomes having similar KO term assignments will be identified as putative sRNA locations. The users can simultaneously select one or more query genomes for every search.
Analysis with the selected query genomes
The first step in the proposed methodology (represented as a flow chart in Figure 1) is the conversion of the selected query genomes from the list into universal three-letter KEGG genome codes (9). Then, the PsRNA server attempts to identify the simultaneous occurrence of a pair of sRNA specific orthologous conserved flanking genes for every query genome that matches with the orthologous KO number pair obtained from the reference genome. The reference gene identification codes are used here to obtain their orthologous KO number pairs in the query genomes using current KO dataset from KEGG ftp site (ftp://ftp.genome.jp/pub/kegg/) (9). If the orthologous KO number pair is found within the query genome, the particular intergenic region is selected by the server as putative sRNA region. In order to remove false positives, the server selects and displays orthologous gene pairs that are separated within five genes (maximum), maintaining their gene orders. The server further attempts to compare these selected putative sRNA locations with already reported RNA annotations of the query genome and marks the unknown putative sRNA locations. Significant subsets of the genes are yet to be assigned with KO number. Further KO assignments to the existing gene annotations will increase the sensitivity of the designed algorithm/server to identify any sort of the flanking gene pairs in the available query genomes in future.
Figure 1.
Flow chart of the methodology used in the identification of putative sRNA locations within the intergenic regions of the query genomes.
Applications and performances
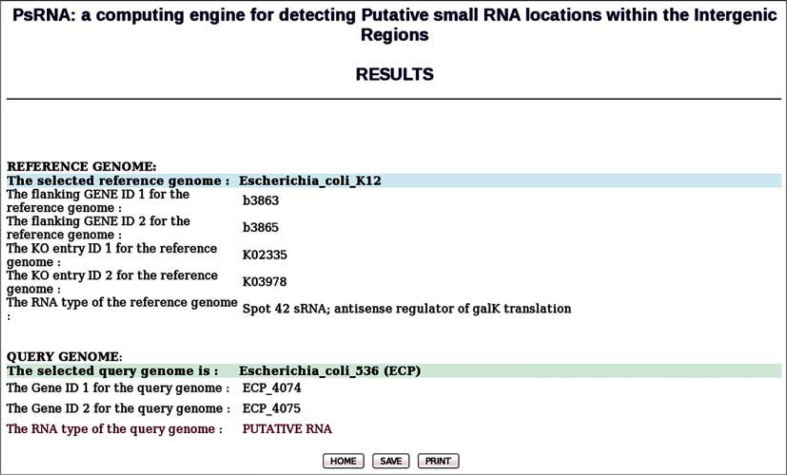
The highest numbers of the homologous and non-homologous sRNA locations are identified in Enterobacteriaceae genomes 11, 21. Among them, significant number of sRNAs are identified and studied in E. coli K12-MG1655 20, 21. The PsRNA server is successfully used in this study to demonstrate the identification of putative intergenic sRNA locations in the recently sequenced 20 enterobacterial genomes (Table 1) using E. coli K12 (NC_000913) as the reference genome. Although more than 82 sRNAs are experimentally reported in E. coli K12 genome 21, 22, only 45 of them are documented in GenBank (NC_000913.rnt). This dataset (NC_000913.rnt) is used as sRNA reference input for the computational identification of similar putative sRNA regions in the query enterobacterial genomes listed in Table 1. To search the putative sRNA region, E. coli K12 is used as reference genome from the list of genomes in the drop down menu, choosing the option “select the particular sRNA displayed from the list”. It has a list of available RNAs from the .rnt file. For example, one of the known sRNAs, spf (gene ID: b3864), whose genomic coordinates lie between 4047922 and 4048030, is annotated as “Spot 42 sRNA; antisense regulator of galK translation”. This spf (23) sRNA is sandwiched between conserved flanking gene pair b3863 (DNA polymerase I) and b3865 (GTP-binding protein) of E. coli K12. The orthologous KO terms for b3863 and b3865 obtained from KO dataset are KO2335 (DNA polymerase I) and KO3978 (GTP-binding protein), respectively. Based on the above KO pair, a search is performed to obtain similar orthologous KO gene pairs in the selected 20 query genomes.
Table 1.
Reference and query genomes used
| No. | Organism | Genome code | GenBank ID | Gene ID code*1 |
|---|---|---|---|---|
| 1 | Escherichia coli K-12 MG1655*2 | eco | NC_000913 | Bxxxx |
| 2 | Escherichia coli 536 (UPEC) | ecp | NC_008253 | ECP_xxxx |
| 3 | Escherichia coli APEC O1 | ecv | NC_008563 | APECO1_xxxx |
| 4 | Escherichia coli UTI89 (UPEC) | eci | NC_007946 | UTI89_Cxxxx |
| 5 | Salmonella enterica serovar Choleraesuis str. SC-B67 | sec | NC_006905 | SCxxxx |
| 6 | Shigella boydii Sb227 | sbo | NC_007613 | SBO_xxxx |
| 7 | Shigella dysenteriae Sd197 | sdy | NC_007606 | SDY_xxxx |
| 8 | Shigella sonnei Ss046 | ssn | NC_007384 | SSON_xxxx |
| 9 | Shigella flexneri 5 str. 8401 | sfv | NC_008258 | SFV_xxxx |
| 10 | Sodalis glossinidius str. morsitans | sgl | NC_007712 | SGxxxx |
| 11 | Yersinia pestis Antiqua | ypa | NC_008150 | YPA_xxxx |
| 12 | Yersinia pestis Nepal516 | ypn | NC_008149 | YPN_xxxx |
| 13 | Yersinia enterocolitica subsp. enterocolitica 8081 | yen | NC_008800 | YExxxx |
| 14 | Yersinia pestis PestoidesF | ypp | NC_009381 | YPDSF_xxxx |
| 15 | Yersinia pseudotuberculosis IP31758 | ypi | NC_009708 | YPSIP31758_xxxx |
| 16 | Candidatus Blochmannia pennsylvanicus str. BPEN | bpn | NC_007292 | BPEN_xxxx |
| 17 | Candidatus Blochmannia floridanus | bfl | NC_005061 | BFLxxx |
| 18 | Buchnera aphidicola Cc | bcc | NC_008513 | BCC_xxx |
| 19 | Klebsiella pneumonia subsp. pneumonia MGH78578 | kpn | NC_009648 | KPN_xxxxx |
| 20 | Enterobacter sp. 638 | ent | NC_009436 | ENT638_xxxx |
| 21 | Escherichia coli K-12 substr. W3110 | ecj | NC_000091 | JWxxxx |
Gene ID codes are as per KEGG database (9).
Reference genome used in this study (gray shade).
Figure 2 shows the results of orthologous KO gene pair ECP_4074 (DNA polymerase I) and ECP_4075 (Probable GTP-binding protein EngB) obtained for E. coli 536 strain (ecp). The intergenic region between these two genes is reported as putative spf sRNA location for the query E. coli 536 (ecp) genome (24). A similar search using 31 known sRNAs of the E. coli K12 (eco) reference genome resulted in identification of 294 putative sRNA locations in 20 query genomes. The search had to be restricted to 31 sRNAs due to the absence of KO terms from the KO dataset for the remaining sRNA specific flanking gene pairs. The search results can be further improved with more KO assignments to the KO datasets in future.
Figure 2.
A snap shot of the results page from PsRNA server for spf sRNA with eco reference and ecp as the query genome. The putative sRNA region is identified between flanking genes ECP_4074 and ECP_4075. The KO pair obtained based on the reference genome eco is also displayed.
Comparison with the predictions available in Rfam database
We took 31 sRNA specific conserved flanking genes having KO numbers from 82 sRNAs of E. coli K12-MG1655 (NC_000913) as test datasets. The predictions made by PsRNA server in query genomes (Table 1) using the above datasets were compared with the predictions made by QRNA, RNAz and INFERNAL approaches available in Rfam database (7). However, only 23 out of the 31 sRNA families (having KO pairs) were reported in Rfam database and used to make a comparison with PsRNA predictions. The remaining 8 sRNA families, including ryeF, sraA, tp2, tpke11, C0664, rybD, ryjB and sokC, were not documented in Rfam database, but we also analyzed those sRNA regions in the query genomes (Table 1) using PsRNA server.
Above comparison resulted in the identification of most of the sRNA locations (their flanking gene pairs) reported in Rfam database. Interestingly, 18 unique sRNA regions predicted by PsRNA server in 9 query genomes were only located by this method but not reported by Rfam approach or any other tools (Tables S1 and S2). Analysis of the 8 new sRNA groups using the PsRNA server resulted in the identification of 77 new sRNA regions in 20 query genomes collectively (Tables S1 and S2). Such comparison with the predictions made by Rfam approach confirms the reliability of PsRNA server in predicting the functionally important sRNA regions in bacterial genomes using KO terms. Above computational approaches simply look for possible conserved secondary structures and predict some of the mRNA regions (CDS) as sRNA regions false positively (11). But the proposed approach predicts the putative intergenic sRNA regions alone.
Results and Discussion
Twenty recently completed enterobacterial genomes (Table 1) are selected for the analysis using PsRNA server with E. coli K12 MG1655 (22) as the reference genome and its known sRNA information as reference dataset. The 31 sRNAs of E. coli K12 (NC_000913.rnt) having flanking gene pairs with KO numbers are used to identify putative sRNA locations in the selected 20 enterobacterial genomes using the PsRNA computing engine. The selected 31 sRNAs with their conserved flanking genes and KO terms obtained from PsRNA server (shaded gray) for the reference genome eco are listed in Table S1. The table also lists the newly identified 124 orthologous gene pairs that sandwich the putative intergenic sRNA regions from five query genomes (ecp, ecv, eci, sec and sbo) obtained from the PsRNA server. The current study uses reference flanking genes or KO terms as footprints. Table S2 lists the results of 170 putative sRNA locations in 15 more enterobacterial query genomes. Most of the sRNA regions (their flanking gene pairs/genomic regions) available in Rfam were also retained by PsRNA server, except the sRNAs having shuffled/rearranged/deleted flanking gene pairs.
The major difference between previous manual studies 11, 25 and automated PsRNA server is that it looks for sRNA specific flanking gene pairs having KO number pairs alone, which may miss some of the sRNA regions without generic locus IDs (Example: JW5407/JW2541 for “sroF” sRNA in “ecj” genome) or lack of KO assignments. The homologs and partial homologs could be identified by simple BLAST searches, but the “unique” non-homologous sRNA regions were only predicted by this server. The sRNA regions predicted by PsRNA server and reported in this study were a subset of our earlier manually curated data in the listed 20 query genomes (25). The automated PsRNA server works based on the existence of KO numbers alone, which restricts the coverage of this server in query genomes when compared to our previous studies 11, 25, but it saves lots of time and opens a new way of predicting functionally important regions.
Conclusion
The proposed PsRNA server can be used to fish out regions of interest based on the KO information collected from positive training data. Current KO dataset has ~75% of KO assignments. Any further KO assignments to this dataset will increase the sensitivity of this computing engine. Interestingly, some specific sRNAs are associated with a single conserved gene instead of a pair of conserved flanking genes, and such regions were missed by PsRNA server. The enterobacterial sRNAs have been shown as possible hot spots of genetic pool integrations recently (12). These spots show gene rearrangements and are reported as possible “alien” gene integration sites. Obviously, the rearrangement or break in the gene synteny could affect the prediction of such sRNA regions by PsRNA server due to lack of coexistence of KO pairs within the limit. The proposed computing engine, PsRNA, is an effective tool for locating all such functionally important regions in prokaryotic genomes.
Authors’ contributions
JS conceived and coordinated the construction of the server. GS participated in the programming and assisted in testing the server. ZAR and KS improved the algorithm and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
The authors thank the use of the Bioinformatics Centre, Supercomputer Education and Research Centre, Indian Institute of Science and the Interactive Graphics Facility, Indian Institute of Science, funded by the Department of Biotechnology (DBT), Government of India. JS thanks the DBT for a research fellowship. ZAR thanks the DBT for the financial support.
Supplementary Material
Tables S1 and S2
References
- 1.Storz G. An expanding universe of noncoding RNA. Science. 2002;296:1260–1263. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- 2.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Wassarman K.M. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel J., Wagner E.G.H. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 2007;10:262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke P., Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Eddy S.R. Computational genomics of noncoding RNA genes. Cell. 2002;139:137–140. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson D.A. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sridhar J., Rafi Z.A. Small RNA identification in Enterobacteriaceae using synteny and genomic backbone retention. OMICS. 2007;11:74–99. doi: 10.1089/omi.2006.0006. [DOI] [PubMed] [Google Scholar]
- 12.Sridhar J., Rafi Z.A. Identification of novel genomic islands associated with small RNAs. In Silico Biol. 2007;7:601–611. [PubMed] [Google Scholar]
- 13.Sridhar J., Rafi Z.A. Functional annotations in bacterial genomes based on small RNA signatures. Bioinformation. 2008;2:284–295. doi: 10.6026/97320630002284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiffin N. Integration of text- and data-mining using ontologies successfully selects disease gene candidates. Nucleic Acids Res. 2005;33:1544–1552. doi: 10.1093/nar/gki296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao X. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 16.Konstantinidis K.T., Tiedje J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh M. Identification of ortholog groups in KEGG/SSDB by considering domain structures. Genome Informatics. 2002;13:342–343. [Google Scholar]
- 18.Rivas E., Eddy S.R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Washietl S. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershberg R. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blattner F.R. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 23.Joyce C.M., Grindley N.D. Identification of two genes immediately downstream from the polA gene of Escherichia coli. J. Bacteriol. 1982;152:1211–1219. doi: 10.1128/jb.152.3.1211-1219.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brzuszkiewicz E. How to become a uropathogen: comparative genomic anlaysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. USA. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sridhar J. Small RNA identification in Enterobacteriaceae using synteny and genomic backbone retention II. OMICS. 2009;13:261–284. doi: 10.1089/omi.2008.0067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2