Abstract
Since the human genome is mostly transcribed, genetic variations must exhibit sequence signatures reflecting the relationship between transcription processes and chromosomal structures as we have observed in unicellular organisms. In this study, a set of 646 ubiquitous expression-invariable genes (EIGs) which are present in germline cells were defined and examined based on RNA-sequencing data from multiple high-throughput transcriptomic data. We demonstrated a relationship between gene expression level and transcript-centric mutations in the human genome based on single nucleotide polymorphism (SNP) data. A significant positive correlation was shown between gene expression and mutation, where highly-expressed genes accumulate more mutations than lowly-expressed genes. Furthermore, we found four major types of transcript-centric mutations: C→T, A→G, C→G, and G→T in human genomes and identified a negative gradient of the sequence variations aligning from the 5′ end to the 3′ end of the transcription units (TUs). The periodical occurrence of these genetic variations across TUs is associated with nucleosome phasing. We propose that transcript-centric mutations are one of the major driving forces for gene and genome evolution along with creation of new genes, gene/genome duplication, and horizontal gene transfer.
Key words: RNA-seq, genetic variations, sequence signatures
Introduction
DNA damage results in sequence variations through processes of damage-repair and genome replication 1, 2, 3, 4, 5. Such variations when occurring in germline cells and early developmental processes are expected to be passed on to the next generation. It is well-established that both germline and somatic mutations may lead to diseases, such as inheritable disorders and cancers 6, 7. Therefore, it is critical to understand how genetic variations arise and what these disease-causing mutations are. Owing to the development of the next-generation sequencers, discovery of sequence variations has entered a new phase. It is now time to understand mechanisms and rules governing the generation and the inheritance of genetic variations. Here, we systemically investigated the transcript-centric genetic variations in human genomes by analyzing data from both high-throughput RNA-sequencing and single nucleotide polymorphisms (SNPs, dbSNP build 128) in human populations. Analysis of the relationship between the intensity of gene transcription and the occurrence of genetic variations supported our previous finding that a mutation gradient exists in a transcript-centric manner. Furthermore, we noticed a periodicity along the gradient, which was associated with nucleosome occupancy. Our study provides novel insights into the very basic set of laws of genetic mutations in the human genome.
Results
Defining invariable housekeeping genes
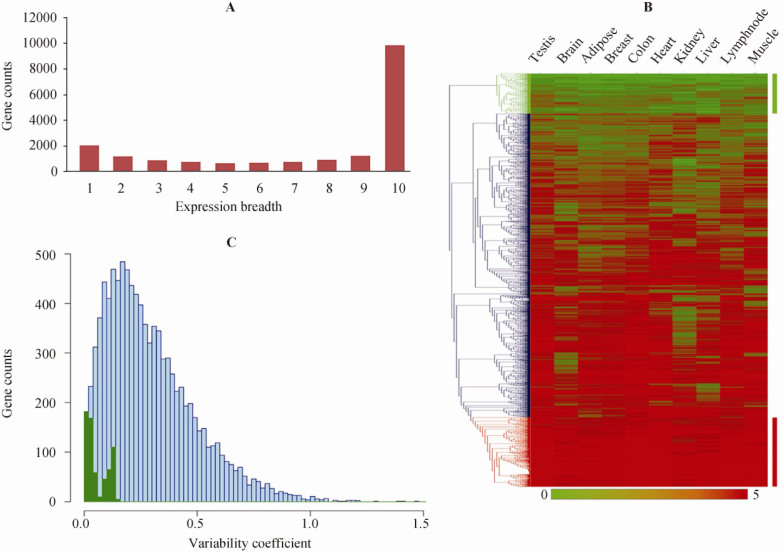
There are several important points that we must address when assessing correlations between gene expression levels and genetic variations. First, gene expression is precisely compartmentalized at the cellular level and can be both tissue-specific and condition-associated. Second, gene expression levels are highly regulated and vary between different tissues and under different conditions. Third, genetic variations are inherited through germline cells and relevant genes must be expressed in such tissues or cells. We started our analysis with publicly-available RNA-Seq data from 10 human tissues, including testis, brain, adipose, breast, colon, heart, kidney, liver, lymph-node and muscle 8, 9, 10. We estimated gene expression levels by calculating read densities of the last exons using Refseq-annotated genes (11), using a background threshold RPKM (Reads Per Kilobase of exon model per Million mapped reads) value of 0.3 (8). The number of genes expressed among the 10 tissues varied from 11,891 to 16,708 (Table S1). Among these genes, expression of 9,732 genes was found in all tissues. These genes therefore are defined as ubiquitous or housekeeping (HK) (Figure 1A and Table S2). Although the expression levels of these HK genes differed significantly among tissues, we managed to identify a set of 646 expression-invariable genes (EIGs) (Table S3), using hierarchical clustering analysis (Figure 1B). In addition, after estimating the variation coefficients (CVs) for gene expression levels in all 10 tissues, we noticed that the CVs in this dataset deviated very narrowly (Figure 1C). Gene ontology analysis indicated that most of these EIGs are involved in important HK functions, such as forming cell skeletons, processing RNA, translating proteins, or participating in metabolic pathways (Table S4). This definition of EIGs is consistent with previous studies based on data from microarray and EST experiments 12, 13. An excellent example of an EIG is the gene encoding GAPDH, which is known to be constantly expressed in all tissues and is often used for sample normalization in quantitative PCR assays.
Figure 1.
Genes with invariable expression in 10 human tissues. A. Expression breadth analysis of genes expressed in 10 human tissues. Gene expression was examined in testis, brain, adipose, breast, colon, heart, kidney, liver, lymph-node and muscle. There are 9,732 genes categorized and shared by all 10 tissues. B. Hierarchical clustering analysis of 9,732 genes from 10 human tissues. Relative gene expression levels were indicated by RPKM (see details in Methods) to define expression intensity and 646 expression-invariable genes (EIGs) in all tissues were chosen. The EIGs were partitioned into lowly- (green) and highly-expressed (red) genes. C. Distribution of variation coefficients for genes expressed in 10 human tissues. 646 EIGs selected using clustering analysis in panel B are highlighted in green.
Correlating expression level with genetic variation
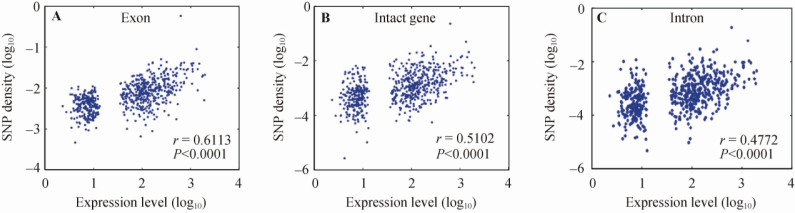
To investigate the effects of transcription-associated biological processes on genetic variations, we re-examined the relationship between gene expression level and genetic variation in human genomes using our defined EIGs. Since only germline mutations are passed on to the next generation, we believe that gene expression in germline cells should have a direct influence on genetic variations. We thus used EIGs that are expected to have a consistent expression pattern in germline cells to perform this correlation analysis. We took advantage of the public dbSNP data (Build 128)(14) to calculate SNP counts per basepair for each gene, and correlated gene expression level to the distribution of SNPs. We found a highly significant Pearson coefficient between gene expression level and SNP density (Figure 2). Our result showed that this correlation was transcript-centric, i.e., it was equally significant for exonic and intronic sequences as well as for sequences that were a sum of the two. In fact, this correlation became weaker when all 9,732 HK genes, consisting of mostly expression-variable genes (EVGs), were used for calculating the correlation (Figure S1). This may be caused by heterogeneous expressions of EVGs between germline and somatic cells, and therefore this poor correlation does not validate the relationship between transcription and genetic variations.
Figure 2.
Correlations between the expression intensity of EIGs and the density of SNPs. Gene expression levels were indicated by the average value of RPKM from all 10 tissues shown in Figure 1. The density of SNPs is measured by SNP counts per bp within each transcript or gene. The values are displayed in logarithmic scales. This correlation can be seen in exons (A), whole genes including both exons and introns (B) and introns (C) (all P<0.0001).
Transcription-associated mutations
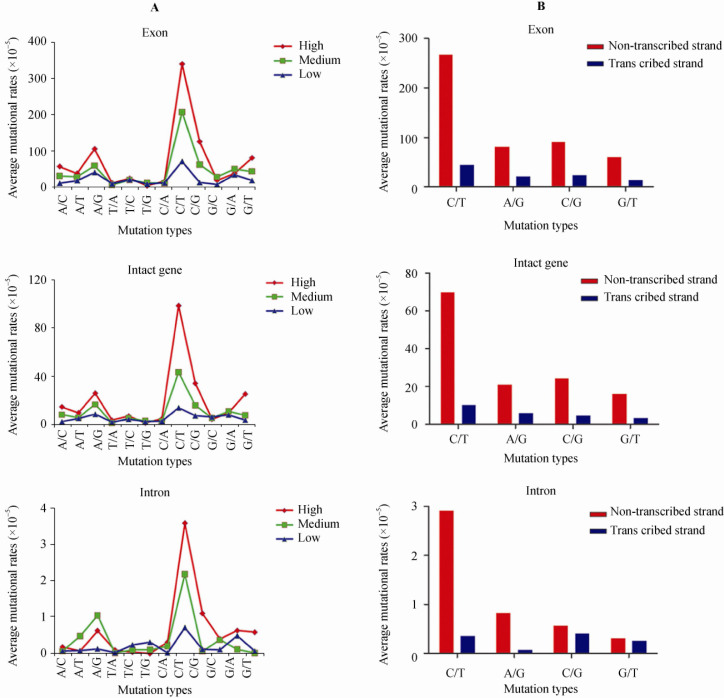
We further examined correlations between the density of each type of nucleotide substitution and gene expression level. Although each type of substitution was significantly correlated with gene expression level (Figure S2), the relative mutation rates of the four major types (C→T >> A→G = C→G > G→T) increased significantly when gene expression level was elevated (Figure 3A). This was also observed in intronic regions. Therefore, we speculated that these four mutations were mainly accumulated as a result of frequent gene transcription. Moreover, we estimated the average mutation rate for each mutation type in non-transcribed and transcribed strands of EIGs (Figure 3B) and identified significant asymmetries of mutation rates of C→T, A→G, C→G and G→T between the two strands. We found that C→T occurrence on the non-transcribed strand was about seven times higher than that on the transcribed strand (χ21df =158, P<0.0001). Similar trend was observed for the other three major mutation types including A→G, C→G and G→T, where their non-transcribed strands had about four times (χ21df =34, P<0.0001), five times (χ21df =40, P<0.0001) and four times (χ21df =30, P<0.0001) more mutations than the transcribed strand, respectively. These mutational asymmetries were also revealed in the intronic sequences, which reflects a neutral mutation rather than selection. In fact, mutational asymmetries of C→T and A→G in mammalian genes were reported by Green et al. in 2003 (5) and they suggested that these asymmetries could be caused by transcription-coupled repair (TCR) process. However, they failed to mention the asymmetries of C→G and G→T mutations, perhaps due to the weaker effects. Nonetheless, in our results, we clearly observed all four major mutations including C→T, A→G, C→G and G→T.
Figure 3.
Comparative analyses of mutation rates of EIGs. A. EIGs were equally classified into highly-, moderately- and lowly-expressed genes based on their expression levels (RPKM values). This classification scheme was also applied for the following figures. The mutation rates vary among genes expressed at different levels. The mutation rates of C→T, A→G, C→G and G→T exhibited remarkable increases when expression levels are elevated. B. The frequencies of the four major mutation types including C→T, A→G, C→G and G→T showed asymmetry between transcribed DNA strands and the non-transcribed counterparts.
Here, we propose an explanation as to why there is a correlation between gene expression level and genetic variation. Transcription-associated DNA mutations, caused mainly by the low fidelity of TCR polymerase 1, 15, 16, 17, 18, 19, 20, are also observed in other eukaryotic genomes even those of prokaryotes since TCR is actually universal. If a gene is frequently transcribed, its template (or non-transcribed) strand is repaired more often and thus accumulates more TCR-associated mutations. Therefore, highly-expressed genes, compared to lowly-expressed genes, should harbor more mutations and exhibit higher genetic diversity in human populations.
SNP gradient
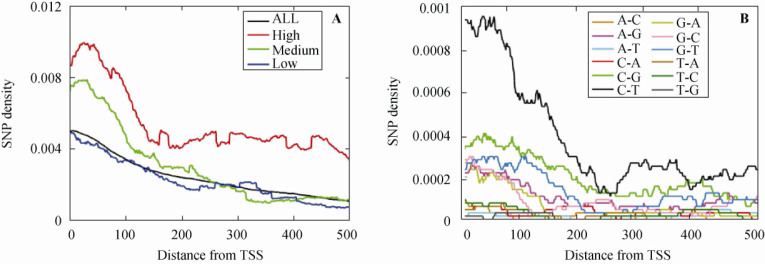
We also investigated the distribution of SNPs within the length of transcripts and found that SNPs were more abundant near transcription start sites (TSS), tapering off toward the 3′ end (Figure 4A). This gradient was more evident in highly-expressed genes. Furthermore, we examined the gradient for each type of mutations around the TSS and found that the mutations C→T, A→G, C→G and G→T had stronger gradients than other mutational types (Figure 4B). As for why SNPs are significantly enriched toward the 5′ end of EIGs, we believe that this phenomenon is also relevant to TCR mechanisms. According to our current understanding of TCR, DNA repair is triggered by the DNA damage-induced stalling of RNA polymerase II (RNAPII). Subsequently, the TCR complex displaces RNAPII at the site of DNA damage and removes the lesion. When the transcription process is disrupted by DNA damage and stalled RNAPII, genes have to be transcribed again from the start. As a result, this gives the 5′ end of a gene more opportunities to be repaired, i.e., when new damage occurs after the RNAPII and TCR complex have already passed by. Whenever mismatches occur after the completion of the repair process, variations become inherited following DNA replication. Since the mutations C→T, A→G, C→G and G→T are mainly caused by the TCR process, they are more prevalent at TSS. We had previously proposed this explanation for the existence of a compositional gradient observed in Gramineae genes (1) and similar explanations were proposed for mammalian genomic data as well (5). However, we can not rule out other possible mechanisms, such as the effect of CpG islands (21), which may enhance such a gradient effect when situated at the 5′ end of TUs.
Figure 4.
The SNP gradients at the 5′ end of EIGs. A. The expression levels of different classes of genes are color-coded. The intensity of the gradients is correlated to gene expression levels. B. The mutation types are color-coded. C→T, A→G, C→G, and G→T exhibited stronger gradient effects. TSS: transcription start site.
SNP periodicity
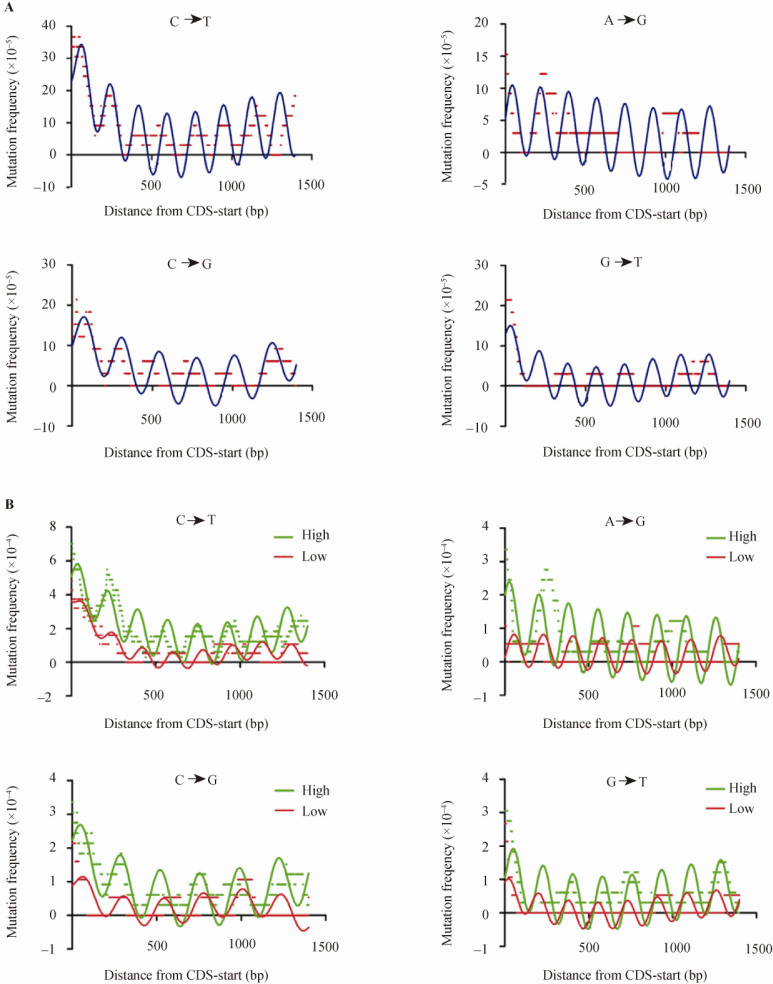
Periodicity of SNPs was found around TSSs in human genomes (22). We plotted the frequency for each of the 12 mutation types (measured as SNP counts) over aligned EIGs at coding sequences (CDS)-start in a 100 bp window and 1 bp step (Figure S3) and found that C→T, A→G, C→G and G→T displayed significant periodicities (Figure 5A). The C→T periodicity appears to be the strongest. We further performed power spectrum analysis for each mutation type for the gene sets, demonstrating that the length range for the major peaks was 170-250 nt (Figure S4). We curve-fitted the periodical changes to show that the periodicities from mutation types C→T, A→G, and G→T fitted better to the 176 nt periodicity, but C→G preferred the 233 nt periodicity. Other mutation types, such as A→T and A→C, also exhibit weaker periodicities (Figure S5). However, we noticed that the mutations appeared to have irregular periodicities or multiple periodicities. We speculate that this irregular pattern may reflect the result of selection that often eliminates deleterious mutations and renders the periodicity obscure. Furthermore, when correlating the periodicity to gene expression level, we found that these mutation periodicities became more pronounced among highly-expressed EIGs (Figure 5B), indicating that transcription-associated mutations contribute significantly to the formation of genetic variation periodicities.
Figure 5.
The periodicities of SNPs and its relationship with gene expression. A. Average mutation frequencies in a 100 bp window and 1 bp step plotted as a function of transcript length (from CDS-start) for EIGs. The periodical changes of mutations C→T (upper left), A→G (upper right), C→G (lower left) and G→T (lower right) can be equally observed in the TUs. The red plots indicate the observed SNP density. The periodicities were curve-fitted (marked by blue line) using a nonlinear regression equation as described in Methods. All the parameters and statistical tests were listed in Table S5. B. The relationship between SNP periodicity and gene expression level. The highly-expressed EIGs show stronger periodicity. The parameters and statistical tests for fitting were listed in Table S5.
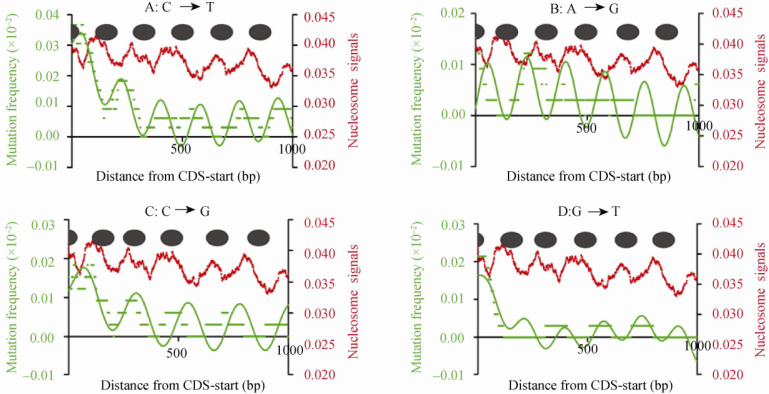
Genetic variation periodicities (170-250 nt) have been reported to be associated with nucleosome positioning in killifish (23) and yeast 24, 25, 26 genomes. We therefore further investigated the relationship between this periodicity and nucleosome positioning. Mutation rates of the major mutation types were aligned along nucleosome arrays of TUs, determined by plotting nucleosome positioning information using the publicly-available MNase-digested chromatin sequences (27). We observed that the linker DNA showed higher mutation rates than nucleosome-protected DNA, and the local mutation rates varied according to a periodicity corresponding to the nucleosome-protected length (Figure 6). However, we noticed that there was some inconsistency between SNP periodicity and nucleosome positioning, especially for the last several nucleosomes. The effect is more obvious in the C→G mutation type. Here, we proposed that there are two reasons responsible for it. On the one hand, as mentioned above, SNP periodicities and the underlying SNPs have been disturbed by selection, and thus it cannot match perfectly to nucleosome positioning. On the other hand, nucleosome positioning shows variability along TUs, especially for highly-expressed TUs. Nucleosomes are generally aligned around transcription starts at a significant positioning periodicity of ~185 bp in the human genome 27, 28. However, the distances between the two adjacent nucleosomes are rather variable among different TUs. Among the highly-transcribed genes, nucleosomes often display a reduction in content and a sharp increase in fuzziness, which often leads to a larger gap between two nucleosomes. In addition, nucleosome positioning in our case was based on data from human T cells, and thus there was a possibility that some of the nucleosome positioning data may be different from data obtained from germline cells where SNP periodicities were expected to better match nucleosome positioning.
Figure 6.
The oscillation of mutation frequencies along nucleosome arrays. The nucleosome arrays were aligned from CDS-start of the highly-expressed EIGs. The Y axis on the left shows the normalized number of sequence tags from the sense strand at each position. The inferred nucleosomes are shown by the filled ovals. The mutation frequencies, indicated along the Y axis on the right side, C→T(A), A→G (B), C→G (C) and G→T (D), were calculated based on the sequence covered by nucleosome arrays. Note that higher mutation rates were present in the linker DNA regions.
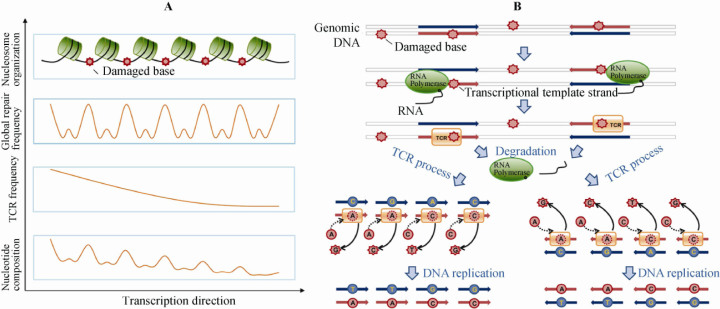
This result suggests a common molecular mechanism that explains the causes of mutation periodicities. Histones and the DNA double helix are packaged into compact forms to prevent DNA from damage in contrast to the linker region between nucleosome folds, which is exposed to the ionic environment more often than the protected fractions, and thus is vulnerable to mutagenesis 29, 30. Therefore, the mutagen-susceptible linker DNA is believed to be damaged more frequently, repaired more often, and leaves more mutations caused by low-fidelity of the DNA polymerases of TCR 4, 31. Since nucleosomes are not randomly positioned along DNA sequences and their regularity appears to coincide with transcriptional units, the different damage-repair frequency between linker DNA and wrapper DNA finally causes oscillation of TCR-associated mutations along nucleosome arrays (Figure 7A). Moreover, highly-expressed EIGs are transcribed more frequently and repaired more often, thus leading to more pronounced periodicity.
Figure 7.
Schematic illustration for the proposed mechanism of TCR-associated mutations and sequence variation-defined periodicity. A. Mutation-derived periodicity is a result of nucleosome positioning. Nucleosomes are regularly positioned near the transcription starts along genes. Since the linker DNA is exposed and more vulnerable to mutagenesis, it is damaged more frequently, repaired more often, and therefore leaves more mutations resulting from TCR. In addition, the mutation frequencies are higher at the 5′ end of highly expressed EIGs, leading to a descending mutation gradient toward the 3′ end. B. After transcription initiation, DNA damage occurring on the template (transcribed) strand results in stall and degradation of RNA polymerase II (RNAPII) when TCR displaces RNAPII and initiates the DNA repair process. In the process of repair, due to the low-fidelity of DNA polymerase, A and C are often added regardless of the damaged bases, leading to A or C mismatches at the damaged loci. After one round of DNA replication, mutations are fixed in the daughter cells. Since TCR is asymmetric, only the template strand is repaired, and the resulting distribution of mutations would also be asymmetric. Therefore, the strong asymmetry of the four major mutation types (C→T, A→G, C→G and G→T) is easily explained in such a way; C→T is most prevalent as it represents a transition rather than a transversion like C→G and G→T, while the size difference between purine and pyrimidine may explain why the frequency of A→G is much lower than C→T.
Discussion
In this study, we characterized transcript-centric mutations in human genomes. Although the gradient and periodicity indeed exist in the human genome, we cannot rule out the possibility that natural selection also contributes to both because a significant fraction of mutations are believed to be random 32, 33. There are several lines of evidence supporting the idea that the mutation-defined periodicity should reflect the intrinsic mutation process in the human genome rather than the result of natural selection. First, the gradient and periodicity extend from CDS to introns (Figure S6). Since most introns are thought to be non-functional, this pattern should reflect a neutral mutation process. Second, most mutations (60%) occurred often in the most upstream 1.5 kb regions among the studied TUs and associated with, by and large, minor alleles (<0.1) (Figure S7), which were not subjected to strong population-based selection yet, as they were at most neutral or weakly deleterious. Third, the fact that the genetic variations, gradient and periodicity correlate strongly with gene expression levels suggests a possibly neutral accumulation of mutations that are not yet strongly selected for as proposed previously by many other authors 1, 3, 5, 31, 34. In the mean time, this biased mutational process provides an excellent explanation for the non-random distribution of SNPs in human genomes 6, 35, 36, 37, 38, 39.
Although TCR has been thought as the causative reason of transcript-centric mutations 1, 5, 16, alternative hypotheses exist. For instance, the deamination of cytosine and adenine in the exposed single DNA strand when a gene is transcribed can be used to explain the gradient formation 1, 3, 5, 40, 41. At present, although it is difficult to distinguish between the two contributing factors as to which one is the major or minor, we argue that TCR makes the decisive contribution to this mutation asymmetry for at least highly-expressed genes. There are three reasons. First, the higher rates of C/G→T and A/C→G are consistent with the error spectrum of DNA polymerase ŋ involved in the DNA repair process, showing an error-prone outcome in adding A and C to the target DNA (Figure 7B). Second, deaminations (cytosine and adenine) alone do not explain the mutation asymmetry for C→G and G→T. We nevertheless demonstrated that they are also transcript-centric and strand-biased albeit weaker than the other two types, C→T and A→G, which are transitions rather than transversions. Third, TCR is the only transcript-centric mechanism that allows us to explain why the periodicity of genetic mutations is correlated with the expression level of genes. If the deamination of cytosine and adenine in the linker of nucleosome spaces was a major contributing factor, we should be able to observe the periodicity over a much longer range in entire genes.
In conclusion, the possibility of a transcript-centric mechanism which causes a distinct pattern of genetic mutations is intriguing. If most of the human genome—as well as all animal genomes—are transcribed (42), then most genetic variations should reveal sequence signatures that reflect gene expression levels as well as the mechanisms that regulate expression. Our results indicate that gene expression levels are strongly correlated with the density of SNPs in TUs, and the gradient of genetic variations is found at the 5′ end of TUs. These observations have strong implications for our understanding of natural selection and evolution. That is, mutations may not actually be generated randomly, but rather tend to accumulate more in genes that are expressed more often, or at higher levels, in germline cells to be inheritable. Such mutations may also be prevalent in somatic cells, especially stem or progenitor cells, resulting in a rapid accumulation of mutations in the differentiated cell lineages when they are not terminally differentiated. This also implies that mutation pressure can be reduced if exons are away from the 5′ end of transcripts, or gene expression level is reduced. In addition, one can imagine that a gene, either functional or non-functional, is able to mutate faster just by being highly expressed in germline cells and that gene variants are certainly selected at different levels, including cell, tissue, organ, individual and even population, where gene actually plays functional role. A Pandora’s Box is now open.
Materials and Methods
Data source
We collected RNA-sequencing data from 10 human tissues (9) and mapped them onto the human genome sequence (hg18) using MAQ (43). Uniquely-mapped sequence reads were annotated according to Refseq defined genes (44). To analyze mRNA expression quantitatively, we calculated RPKM (8) as the expression parameter. Since the 5′ portion of mRNAs is frequently truncated in the process of RNA-seq library construction (8), the RPKM value of the last exon is often preferred as a measure of gene expression levels. When there are no reads mapped to the last exon, expression levels are defined by averaging RPKM values from the entire gene. A RPKM threshold value of 0.3 was used to filter out background noise.
In order to identify and select expression-invariable genes, we first isolated 9,732 shared or ubiquitously-expressed genes from 10 human tissues and divided them into 1,001 groups according to their expression levels to estimate the relative expression levels. We subsequently performed a hierarchical clustering analysis based on the relative expression levels. Data on human genetic variations were obtained from NCBI dbSNP database (snp128) (14), and the sequences for nucleosome occupancy mapping were obtained from SRA (SRA 000234) database (45).
Power spectrum analysis
Power spectrum analysis (31) was used for detecting mutational periodicity along coding sequences. To accelerate calculation, we used the Fast Fourier Transform algorithm to compute power spectrum. For a sequence χk of length N (N is a positive integer), its power spectrum is expressed as:
Where i2 = −1, and fj = j / N(j = 0,1,2…,N −1).
To identify periodicities that are in phase from CDS-start, we aligned the begining of CDS and calculated the mutation rate for each nucleotide position in a 100 bp window to generate a binary sequence xk(k = 0,1,2…,1400). We also applied power spectrum analysis to mutational frequencies.
Regression analyses
The third order polynomial equation, coupled with a sine wave, was used for fitting the curve for periodical changes of mutational rates. The equation is:
Where Y represents average mutation rate at a 100 bp window in a 1 bp step, and x represents position away from CDS-start. The right side of this equation can be split into two parts: the first
describes a baseline for mutational rate, and the second
yields a sine undulation with a period length estimated by power spectrum analysis. The parameters used for the curve-fitting are listed in Table S5.
Authors’ contributions
JY and SH designed the experiments. PC, QL and FD analyzed the data. PC, QL and JY wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have no competing interests to declare.
Acknowledgements
This work was supported by grants from the National Basic Research Program (973 Program; 2011CB944100 and 2011CB944101), National Natural Science Foundation of China (90919024) awarded to JY, and Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-R-01-04) to SH.
Contributor Information
Songnian Hu, Email: husn@big.ac.cn.
Jun Yu, Email: junyu@big.ac.cn.
Supplementary Material
Figures S1-S7; Tables S1-S5
References
- 1.Wong G.K. Compositional gradients in Gramineae genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes D.E., Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 3.Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am. J. Hum. Genet. 2003;73:688–692. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green P. Transcription-associated mutational asymmetry in mammalian evolution. Nat. Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 6.Rogozin I.B., Pavlov Y.I. Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutat. Res. 2003;544:65–85. doi: 10.1016/s1383-5742(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 7.Weiss K.M. In search of human variation. Genome Res. 1998;8:691–697. doi: 10.1101/gr.8.7.691. [DOI] [PubMed] [Google Scholar]
- 8.Ramskold D. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang E.T. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marioni J. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortazavi A. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J. How many human genes can be defined as housekeeping with current expression data? BMC Genomics. 2008;9:172. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Sherry S. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J. Minimal introns are not junk? Genome Res. 2002;12:1185–1189. doi: 10.1101/gr.224602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am. J. Hum. Genet. 2003;73:688–692. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svejstrup J. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y. Error-free and error-prone lesion bypass by human DNA polymerase к in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mugal C. Transcription-induced mutational strand bias and its effect on substitution rates in human genes. Mol. Biol. Evol. 2009;26:131–142. doi: 10.1093/molbev/msn245. [DOI] [PubMed] [Google Scholar]
- 20.Mugal C. Conservation of neutral substitution rate and substitutional asymmetries in mammalian genes. Genome Biol. Evol. 2010;2:19–28. doi: 10.1093/gbe/evp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polak P., Arndt P.F. Transcription induces strand-specific mutations at the 5′ end of human genes. Genome Res. 2008;18:1216–1223. doi: 10.1101/gr.076570.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higasa K., Hayashi K. Periodicity of SNP distribution around transcription start sites. BMC Genomics. 2006;7:66. doi: 10.1186/1471-2164-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki S. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellinger R.E., Thoma F. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 1997;16:5046–5056. doi: 10.1093/emboj/16.16.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suter B. Chromatin structure modulates DNA repair by photolyase in vivo. EMBO J. 1997;16:2150–2160. doi: 10.1093/emboj/16.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Washietl S. Evolutionary footprints of nucleosome positions in yeast. Trends Genet. 2008;24:583–587. doi: 10.1016/j.tig.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Schones D.E. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang C., Pugh B.F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu Rev Biophys Biomol Struct. 1997;26:83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- 30.Tuteja N. Molecular mechanisms of DNA damage and repair: progress in plants. Crit. Rev. Biochem. Mol. Biol. 2001;36:337–397. doi: 10.1080/20014091074219. [DOI] [PubMed] [Google Scholar]
- 31.Chen K. A novel DNA sequence periodicity decodes nucleosome positioning. Nucleic Acids Res. 2008;36:6228–6236. doi: 10.1093/nar/gkn626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 33.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 34.Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 35.Amos W. Even small SNP clusters are non-randomly distributed: is this evidence of mutational non-independence? Proc. Biol. Sci. 2010;277:1443–1449. doi: 10.1098/rspb.2009.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lercher M.J., Hurst L.D. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18:337–340. doi: 10.1016/s0168-9525(02)02669-0. [DOI] [PubMed] [Google Scholar]
- 37.Tenaillon M.I. Apparent mutational hotspots and long distance linkage disequilibrium resulting from a bottleneck. J. Evol. Biol. 2008;21:541–550. doi: 10.1111/j.1420-9101.2007.01490.x. [DOI] [PubMed] [Google Scholar]
- 38.Tian D. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. 2008;455:105–108. doi: 10.1038/nature07175. [DOI] [PubMed] [Google Scholar]
- 39.Winckler W. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- 40.Duret L. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 2002;12:640–649. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 41.Hendriks G. Transcription-dependent cytosine deamination is a novel mechanism in ultraviolet light-induced mutagenesis. Curr. Biol. 2010;26:170–175. doi: 10.1016/j.cub.2009.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guttman M. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruitt K. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2006;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shumway M. Archiving next generation sequencing data. Nucleic Acids Res. 2010;38:D870–D871. doi: 10.1093/nar/gkp1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S7; Tables S1-S5