Abstract
Linking similar proteins structurally is a challenging task that may help in finding the novel members of a protein family. In this respect, identification of conserved sequence can facilitate understanding and classifying the exact role of proteins. However, the exact role of these conserved elements cannot be elucidated without structural and physiochemical information. In this work, we present a novel desktop application MotViz designed for searching and analyzing the conserved sequence segments within protein structure. With MotViz, the user can extract a complete list of sequence motifs from loaded 3D structures, annotate the motifs structurally and analyze their physiochemical properties. The conservation value calculated for an individual motif can be visualized graphically. To check the efficiency, predicted motifs from the data sets of 9 protein families were analyzed and MotViz algorithm was more efficient in comparison to other online motif prediction tools. Furthermore, a database was also integrated for storing, retrieving and performing the detailed functional annotation studies. In summary, MotViz effectively predicts motifs with high sensitivity and simultaneously visualizes them into 3D strucures. Moreover, MotViz is user-friendly with optimized graphical parameters and better processing speed due to the inclusion of a database at the back end. MotViz is available at http://www.fi-pk.com/motviz.html.
Key words: MotViz, sequence motif, structural visualization, algorithm, bioinformatics
Introduction
Computationally understanding and annotating the central dogma of molecular biology encompasses the success of bioinformatics. From sequence to its annotation as well as from protein sequence to structure prediction and visualization, all processes are being executed with high efficiency through in silico tools and databases. It is noteworthy that number of protein sequences grows rapidly and is far beyond the number of experimental structures. According to a recent description, SWISS-PROT (1) database contains 525, 997 protein sequence entries, while Protein Data Bank (PDB) (2) comprises only 71,794 protein structures. The tremendous amount of sequence data is in dire need of analysis in order to understand the biological meanings and to establish appropriate relationships between sequences and structures. Considering that protein structures are relatively conserved and adopt only a limited number of folds, it is possible to predict their 3D structures by knowledge-based information (3). Devising and implementing novel algorithms will largely contribute to determination of protein structure or function.
Generally, to achieve insight into structures through Cartesian coordinates, it is essential to have a visualization tool. During recent years, a variety of data visualization tools have been developed to meet increasing data complexity. These include Rasmol, VMD, Cn3D, Swiss PDB Viewer, Chimera, Jmol and PyMol 4, 5, 6, 7, 8, 9, 10. Jmol (9) is a popular Java-based freeware and standalone high-end tool for protein structural monitoring. The browser applet in Jmol supports the loading of multiple molecules with independent movement, surfaces and molecular orbitals, biological units and crystal symmetry, cavity visualization, translucency, high-quality rendering, arbitrary objects such as arrows/planes and true slabbing properties.
In the current study, a plug-in MotViz has been built and added into Jmol version 11.2.1 in order to extend its applicability. Specialized features in MotViz include sequence motif retrieval from protein 3D structure in parallel to structural analysis based on the physiochemical properties. Additionally, a local database has been created to facilitate user in re-evaluation of results with better performance. Finally, for detailed annotation of selected motifs, links of web-based tools like Prosite, MEME and Hits 11, 12, 13 have been incorporated into MotViz.
Methods
We updated Jmol protein visualization tool version 11.2.1 by incorporating new plug-in and modules. These new features include sequence annotation, motif searching, motif visualization and graphical output of their physiochemical chart, motifs conservation score calculation and their comparisons through bar graph representation. MotViz algorithm created in this study predicts motifs from the loaded 3D protein structure. This algorithm uses a dynamic approach and latest-updated data from UniProt and InterPro 14, 15 databases. MotViz algorithm works in five phases, which are described as follows.
Phase 1 Retrieving amino acid sequence from 3D coordinates
Protein sequence is dissected from 3D coordinates of determined amino acids at Phase 1. MotViz retrieves protein sequence from the structural coordinates, because the target of MotViz is to hunt structural motifs in parallel to sequence motifs, as some amino acids might be missing in 3D coordinates but likely available in the protein Fasta sequence.
Phase 2 Launching BLASTP and fetching Fasta sequences
At the second phase MotViz retrieves similar protein sequences related to the visualized protein sequence to hunt for conserved motifs. UniProt BlastP is launched by calling the EBI web service to obtain IDs of the 50 most conserved sequences (with a maximum difference of 10−50 E-value). If this E-value difference is found in about topmost 30 BLASTP sequences, then only those sequences are picked. Fasta sequences of selected proteins are then fetched from the UniProt server and saved for further analysis.
Phase 3 Performing multiple sequence alignment
At the third phase multiple sequence alignment (MSA) of closely-related protein sequences (from phase 2) is performed to locate the conservation points using ClustalW.
Phase 4 Locating conservations and determining motifs
This step identifies conservation points by reading ClustalW “*, :, .”, where “*”, “:” and “.” represents identical, conserved and semi-conserved residue, respectively. MotViz algorithms will identify ---*--:--.--- as the worst case and ---***--- as the best case. Furthermore MotViz also locates the positions of the determined motifs in the protein sequence and stores the information in the database.
MotViz Algorithm Steps
-
I.
Input: PDB ID of protein
-
II.Search: motifs of PDB ID in database
-
a.If(motifs)
-
i.store: motifs in array m of length k
-
ii.For i
 1 to k
1 to k-
1.Calculate_physiochemical_properties(m(i))
-
2.Input: loc of m(i) location
-
3.Output: m(i) and loc
-
4.Output: m(i) C value graph
-
1.
-
i.
-
b.Else
-
i.Input Amino Acid sequence from 3D coordinates
-
ii.Input UniProt Psiblast parameters
-
iii.Launch UniProt_Psiblast()
-
iv.getResult in Array results
-
v.diff
 Calculate E-value difference (e−50)location from results
Calculate E-value difference (e−50)location from results -
vi.For j
 1 to diff do sequence(j)
1 to diff do sequence(j)  results(j)
results(j) -
vii.Store sequence in MotViz database
-
viii.Launch ClustalW MSA(sequence)
-
ix.OutputRes
 Read ClustalW .output file
Read ClustalW .output file -
x.Input
 array new_motifs of dynamic length
array new_motifs of dynamic length -
xi.Input
 cont_gap, cons_leng, motif_No, start to 0
cont_gap, cons_leng, motif_No, start to 0 -
xii.for k
 1 to OutputRes
1 to OutputRes  lengthif (cont_gap< 3)
lengthif (cont_gap< 3)-
1.If (OutputRes(k) == * || OutputRes(k) ==: || OutputRes(k) == .)If (cons_leng = 0)start
 kcons_leng++
kcons_leng++ -
2.Else
-
a.cont_gap++
-
a.
Else if (cont_gap > 2 && cons_leng >2)-
3.Calculate C_Value (start to k)
-
4.new_motifs(motif_No) = OutputRes(start to k)
-
5.start =0, cons-leng =0, cont_gap=0
-
1.
-
xiii.Store new_motifs array in MotViz database
-
i.
-
a.
Phase 5 Calculating conservation value
At the final phase, conservation of each predicted motifs is calculated using the MotViz algorithm according to the following formula.
Where Cv is conservation value; ‘i’ is from 0 to 3, x1 is number of identical residues; w1 is identity score (in this case 1 is optimal score); x2 is number of completely-conserved residues; w2 is score for completely-conserved residues (maximum value is 0.85); x3 is number of semi-conserved residues; w3 is score for semi-conserved residues (maximum value is 0.70) and is the total length of motif.
PSI-BLAST was used during the algorithmic development of MotViz (16). Dependency of PSIBLAST (16) is on activation.jar, commons-cli-1.1.jar, commons-logging-1.0.2.jar, mail.jar, servlet.jar, xercesimpl.jar, axis.jar, commons-discovery-0.2.jar, jaxrpx. jar, saaj.jar and wsdl4j-1.4.1.jar, which are available at http://www.ebi.ac.uk/Tools/webservices/services/dbfetch (17). ClustalW (18) was used for performing MSA of protein sequences retrieved from UniProt.
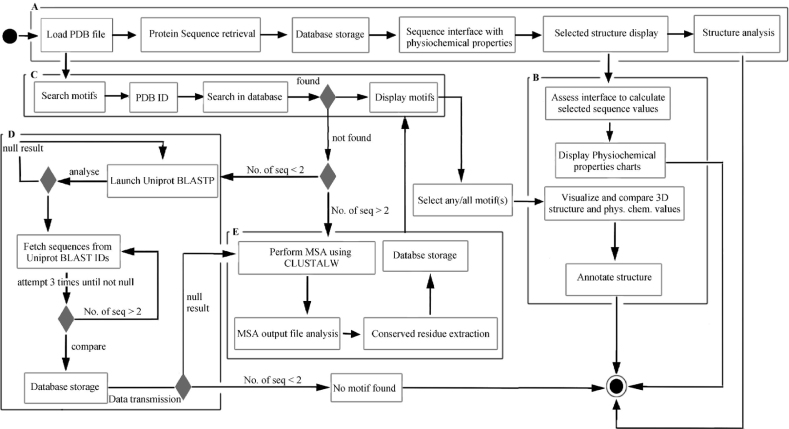
The conserved segments across all aligned sequences are marked by numerical values and subsequently positioned into the structural coordinates. Figure 1 shows the schematics flow of the MotViz algorithm. Predicted conserved areas are considered to be of functional relevance and are stored in a separate database for easy retrieval.
Figure 1.
Schematic overview of motif searching and visualization algorithm (MSVA). A. AA sequence is retrieved from the PDB, visualized and stored into local database. User may examine the respective sequence properties. B. Calculation and displaying the AA physiochemical properties of selected sequence region or motif(s). The corresponding structure is rendered with distinct color. C. Searching in database and displaying the list of motifs present in the loaded PDB file. D. If motifs are not found in the MotViz database, UniProt PSI-BLAST (16) is launched and results are stored in MotViz database for MSA. E. MSA is performed using ClustalW (18) and conserved regions are identified and stored in MotViz database to carry out step В and step C. The circle at the lower right corner represents inspection to approve the quality of product.
Results
Sequence retrieval and analysis
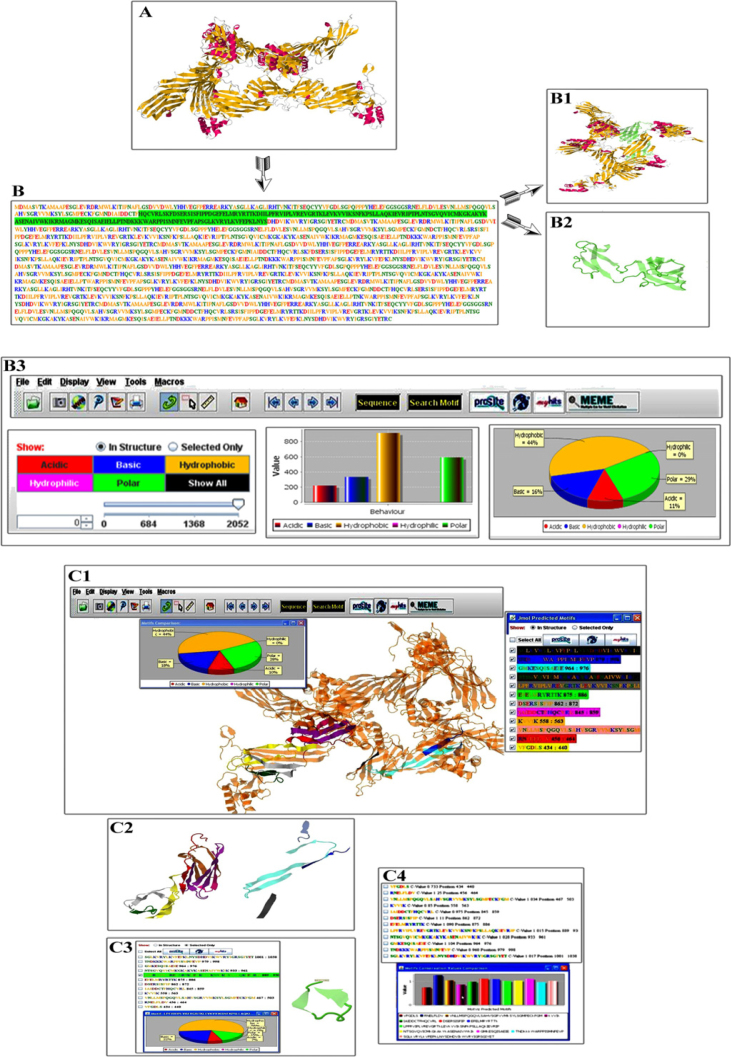
Protein structure files were obtained from PDB (2). Here, we took 3ML6, the structure of complex between dishevlled2 and clathrin adaptor AP-2, as example (Figure 2A). Upon clicking the sequence button, there displays a new panel at the left bottom corner bearing the protein sequence (Figure 2B). The sequence segments are highlighted within protein structure with same color representation (Figure 2B1, 2B2). In the same panel, there are sub-panels for bar and pie charts, button to display the physiochemical properties (hydrophobic, acidic, basic and polar residues) together with slider and input spinner for selecting the protein sequence or to visualize the selected amino acids properties. In addition, two radio-buttons are enabled for selecting the visualization option of either whole structure or selected sequence only. For selected sequence segments, pie and bar chart features are also enabled together with slider selection (Figure 2B3). Multiple visualization aspects that have already been embedded in Jmol (9), including balls-sticks, cartoon, space-filled (Vander Waals), ribbon model and wireframe-trace, are enabled for detailed analysis.
Figure 2.
MotViz working interface encompassing feature’s track individually. A. Loaded 3ML6 (complex between Dishevlled2 and Clathrin adaptor AP-2) structure. B. Sequence retrieved from PDB structure. B1. Selected amino acids in the sequence are visualized in structure. B2. Relative amino acids highlighted in structure only. B3. Individual amino acid physiochemical properties charts (pie and bar), buttons and slider panel to select the region(s) of choice are indicated. Detailed view of amino acid physiochemical properties charts. Green color represents the polar amino acids (G, S, T, Y, C, Q and N); orange color represents the hydrophobic amino acids (A, V, L, I, P, W, F and M); red color indicates acidic amino acids (D and E); blue color is for basic residues (K, R and H); while other amino acids are represented by Tan color. C1. List of predicted motifs in multi-color panel. Pie chart indicates the AA physiochemical properties of predicted motifs in parallel to their structure representation. C2. Predicted motifs in structure. C3. Selected motifs in motif panel as well as in structure. C4. Bar chart representing the predicted motif conservation rate analysis encompassing the whole protein. Moreover, conservation value (Cv) along with individual motif location for each predicted motif is also indicated.
Motif prediction and visualization
Sequence retrieved from the 3D model of loaded protein structure is stored in a local database (MotViz-Database). Individual motifs are retrieved by clicking the search motif button. Motifs are predicted from conserved sites generated using the MotViz algorithm against the query sequence and stored in the local database (MotViz-Database). Later, a motif panel is displayed after retrieving motifs in the right upper corner (Figure 2C1). The motif panel contains a comprehensive list of all predicted motifs. To facilitate the user, motif positions in retrieved sequence are made available. Also, the individual motifs can be selected by clicking check box (Figure 2C2). Radio-buttons help in selecting either the visualization option or select motifs only within structure. In order to select all motifs, a select all check box is added to display all motifs simultaneously. By selecting this checkbox, all motifs can be visualized in 3D structure accordingly containing the same color scheme to that of corresponding motif sequences. By clicking the selected only button, selected motifs are displayed in the structure (Figure 2C3). The conservation value calculated for each predicted motif along with its location in the respective sequence is displayed. Moreover, a comprehensive bar chart helps in categorizing related predicted motif(s) on the basis of conservation rate (Figure 2C4).
Performance results
Evaluation of the motif search result was carried out by querying MotViz predicted motifs using multiple online motif finding tools. To validate MotViz predicted motifs, the performance of these predicted motifs was tested by comparing the MotViz predicted motifs with other online motif prediction tools including Prosite, MEME, PATTERN, BLOCKS, Pfam, ProDom, PRINTS 11, 12, 19, 20, 21, 22, 23. Table 1 summarized the verified sensitivity, specificity, precision and accuracy of results predicted by MotViz and other motif prediction tools 11, 12, 19, 20, 21, 22, 23 for 9 protein families randomly selected. These protein families include Wnt protein family, Gli protein family, histone protein family, ribosomal protein family, HIV protein family, PHD-finger protein family, Hox protein family, HSP100 protein family and Frizzled protein family. These selected protein families were analyzed (four proteins in average for each family) for the presence of motifs using MotViz. Highly conserved motifs which were predicted only by MotViz were listed in Table 2 along with their conservation value score (Cv). MotViz specificity was calculated by averaging Cv greater than 0.60 and an E-value greater than 1009 for MotViz predicted motifs 11, 12, 19, 20, 21, 22, 23. MotViz sensitivity was measured by taking the averages of Cv and E-values of the predicted motifs.
Table 1.
Comparative performance of motif searching tools
| Database/Method | Time (s) | Sensitivity | Precision | Specificity | Accuracy | No. of predicted motifs |
|---|---|---|---|---|---|---|
| Prosite | 230.12 | 84.43 | 94.31 | 88.92 | 92.65 | 37 |
| MEME | 348.48 | 92.11 | 93.52 | 95.362 | 92.02 | 139 |
| PATTERN | 165.63 | 80.78 | 96.24 | 98.43 | 95.13 | 13 |
| BLOCKS | 1330.73 | 90.59 | 90.10 | 93.62 | 88.73 | 99 |
| Pfam | 219.12 | 88.34 | 89.37 | 95.10 | 87.15 | 78 |
| ProDom | 198.34 | 81.23 | 94.83 | 98.32 | 92.45 | 15 |
| PRINTS | 165.63 | 82.93 | 94.67 | 98.10 | 91.89 | 27 |
| MotViz | 0.0699 | 96.18 | 95.82 | 97.43 | 93.45 | 233 |
Table 2.
List of highly conserved motifs predicted by MotViz only
| PDB ID | Motif sequence | Cv | Location |
|---|---|---|---|
| 3ML6 | VFGDLS | 0.733 | 434-440 |
| 3ML6 | RNELFLDV | 0.9875 | 456-464 |
| 3ML6 | KVVIK | 0.85 | 558-563 |
| 3ML6 | GMKESQISAEIE | 0.99104 | 964-976 |
| 2XQL | AGNAARDNK | 0.883 | 232-241 |
| 2XQL | IIPRHLQLA | 0.9013 | 244-253 |
| 2XQL | LGKVTIAQG | 0.677 | 263-272 |
| 2WOM | KIEELRQH | 0.9893 | 200-208 |
| 2WOM | GIRK | 0.925 | 554-558 |
| 1KHY | PSQD | 0.812 | 203-207 |
| 1KHY | AGQLRTDIN | 0.772 | 315-324 |
| 1KHY | ISS | 0.85 | 359-362 |
| 1KHY | LSALLN | 0.733 | 426-432 |
| 1KHY | GSVS | 0.675 | 435-439 |
| 1KHY | NITQAIEQ | 0.656 | 526-534 |
| 3MDF | KHRGFAF | 0.871 | 127-134 |
| 3MDF | RTIRV | 0.679 | 157-162 |
| 3MEY | TEALRFPPVM | 0.655 | 44-54 |
| 3MEY | YLKSFPNL | 0.9832 | 63-71 |
| 3MEY | CFRREPSKHL | 0.825 | 139-149 |
| 3MEY | VDYASDPFF | 0.95 | 192-201 |
| 3MEY | LKFELLIPL | 0.838 | 216-225 |
Note: Highly-conserved motifs predicted from 9 protein families by MotViz were verified by Cv.
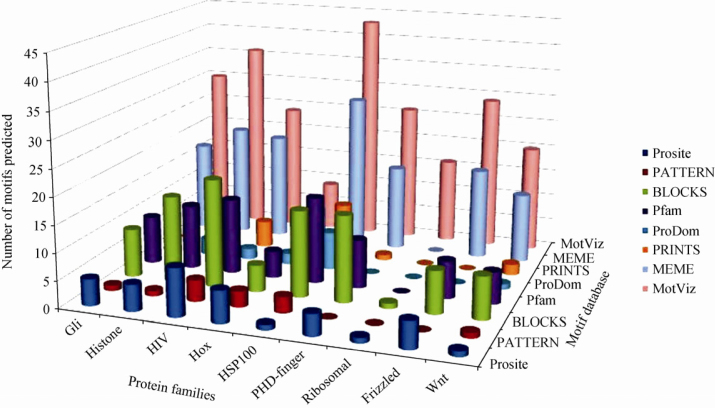
A comprehensive comparison of MotViz predicted motifs was made by combining the motif-prediction data of listed protein families. Individual motifs were predicted by afore-mentioned tools, respectively and compared to those predicted by MotViz. Subsequently, the representative sequence motifs were plotted graphically (Figure S1). The pattern clearly indicate that majority of motifs predicted by MotViz are missed by the listed tools. Furthermore, we plotted the number of motifs predicted and found that more motifs were predicted by MotViz than any listed tools for all 9 protein families. For example, MotViz predicted 9 motifs in Hox protein family, and 5-6 motifs were also predicted by MEME, Prosite, BLOCKS, PRINTS and Pfam, while PATTERN predicted only 3 motifs. In addition, MotViz predicted 36 motifs in histone protein family, out of which MEME, BLOCKS and Pfam predicted 21, 16 and 12 motifs, respectively, while the remaining tools predicted less than 10 motifs (5 by Prosite, 4 by PRINTS, 3 by ProDom and 1 by PATTERN). Furthermore, MotViz predicted 16 motifs in ribosomal family proteins, while 7 motifs were predicted by MEME. Prosite and BLOCKS only predicted 1 motif while PATTERN, Pfam, ProDom and PRINTS did not predict any motif (Figure 3). These data illustrate the MotViz algorithm efficiency is significantly high, compared to other tools.
Figure 3.
Number of motifs predicted by MotViz for protein families selected randomly. X-axis indicates protein families examined, Y-axis represents number of motifs predicted using different tools while Z-axis represents the motif databases. The performance of MotViz is superior to other tools examined, although the performance of MEME (12) is very close to that of MotViz.
Discussion
Combination of conserved sequence segments in protein with visualization in a single click offers a remarkable addition that may result in gathering the in-depth details about functional elaboration of protein structure elements. These details would largely help in dissecting functional information of short segments in parallel to their structural localization. Motifs are important for predicting protein structure or behavior and to categorize anonymous proteins into appropriate families (24). Presence of similar motifs can be annotated by analyzing multiple structures, which may provide a unique mode for determining the novel links and characterizing the functional relevance of unknown proteins.
MotViz plug-in preferably isolates the conserved motifs by aligning the query sequence to a similar dataset by performing the PSI-BLAST followed by MSA using ClustalW in order to dissect the narrow regions of sequence similarities. MotViz motif prediction efficiency is much higher than other tools due to the implementation of increased conservation rate and accuracy in picking by applying filters in two consecutive steps. In the first step, PSI-BLAST at UniProt (17) is launched resulting in retrieving the 50 most related sequences and later in the second step, these sequences are further narrowed down by performing MSA using the ClustalW (18) algorithm. The subsequent identification of conserved sites was further enhanced by selecting the alignment regions of consecutive amino acids with less than three gaps.
Conclusion
MotViz tool provides better usability due to many reasons. Firstly, it generates the query sequence from 3D coordinates, searches similar proteins (with maximum E-value of 10−50) and then aligns them to find the conserved residues in a single effort. Secondly, these residues are further annotated by online motif prediction tools and are verified before appearing for visualization in protein structure. Thirdly, MotViz keeps features that have already been embedded in Jmol intact (9). Lastly, addition of database, incorporation of conservation score calculation, graphical representations and individual sequence to structural compatibility differentiate MotViz. The user may visualize each motif individually within the corresponding structure and examine structural details of the highlighted motif. The graphical output of physiochemical properties and conservation score in parallel to motif prediction make it a unique platform for user.
Authors’ contributions
MSN designed the tool and carried out the data analysis and comparison assays. SR conceived and supervised the project, developed algorithm and methods, and prepared the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have no competing interests to declare.
Acknowledgements
We thank Zahida Parveen and Nousheen Bibi for critical reading and editing of the manuscript. We thank Soft Ideas, Pvt. for technological logistics. This research has been supported by Higher Education Commission, Pakistan (Grants No. 20-1493/R&D/09).
Supplementary Material
Figure S1
References
- 1.Boeckmann B. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose P.W. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Illergard K. Structure is three to ten times more conserved than sequence--a study of structural response in protein cores. Proteins. 2009;77:499–508. doi: 10.1002/prot.22458. [DOI] [PubMed] [Google Scholar]
- 4.Sayle R.A., Milner-White E.J. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 5.Humphrey W. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27-28. [DOI] [PubMed] [Google Scholar]
- 6.Hogue C.W. Cn3D: a new generation of three-dimensional molecular structure viewer. Trends Biochem. Sci. 1997;22:314–316. doi: 10.1016/s0968-0004(97)01093-1. [DOI] [PubMed] [Google Scholar]
- 7.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 8.Pettersen E.F. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 9.Hanson R.M. Jmol – a paradigm shift in crystallographic visualization. J. Appl. Cryst. 2010;43:1250–1260. [Google Scholar]
- 10.Lill M.A., Danielson M.L. Computer-aided drug design platform using PyMOL. J. Comput. Aided Mol. Des. 2010;25:13–19. doi: 10.1007/s10822-010-9395-8. [DOI] [PubMed] [Google Scholar]
- 11.Falquet L. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey T.L. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagni M. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:W433–W437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donovan C., Apweiler R. A guide to UniProt for protein scientists. Methods Mol. Biol. 2011;694:25–35. doi: 10.1007/978-1-60761-977-2_2. [DOI] [PubMed] [Google Scholar]
- 15.Hunter S. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y. A performance enhanced PSI-BLAST based on hybrid alignment. Bioinformatics. 2011;27:31–37. doi: 10.1093/bioinformatics/btq621. [DOI] [PubMed] [Google Scholar]
- 17.Labarga A. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin M.A. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Sigrist C.J. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 20.Henikoff S., Henikoff J.G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn R.D. The Pfam protein families database. Nucleic Acids Res. 2009;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bru C. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attwood T.K. PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res. 2000;28:225–227. doi: 10.1093/nar/28.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rost B., Valencia A. Pitfalls of protein sequence analysis. Curr. Opin. Biotechnol. 1996;7:457–461. doi: 10.1016/s0958-1669(96)80124-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1