Summary
The technique of challenging postmortem tissue explants with inflammation inducer such as lipopolysaccharide (LPS) followed by gene expression analysis is used widely for evaluating the immune‐suppressing effect of bioactives. Using porcine colonic tissue as an ex‐vivo model of mammalian intestinal gut, this study evaluated the effect of incubation time on the integrity of gene transcripts and activation of inflammatory immune gene cascade by LPS treatment. Post‐slaughter colon was removed surgically and explants were incubated for 0, 3, 6 and 12 h and the abundance of mRNA transcripts of a panel of 92 immune genes were evaluated using quantitative polymerase chain reaction (qPCR) arrays. The mRNA transcripts were highly intact after 0 and 3 h of incubation; however, after 6 h the degradation was clearly evident. Following 3 h incubation, 98·8% and 100% mRNA transcripts were detectable in the colonic tissue harvested from weaned and mature pigs, respectively. In the explants of weaned piglets, LPS treatment activated inflammatory signalling pathways [high mobility group B1 (HMGB1), dendritic cell maturation, interleukin (IL)‐6, IL‐8, IL‐17F], while these pathways were inhibited by dexamethasone treatment. Activations of inflammatory genes were also evident in the explants collected from the mature pigs subjected to ex‐vivo incubation for 3 h in the absence or presence of LPS. It is concluded that the colonic explant remains physiologically viable and responsive to immunological challenge for up to 3 h ex‐vivo.
Keywords: gene transcripts, immune response, inflammation, mRNA integrity
Introduction
Ex‐vivo treatment of porcine intestinal tissue is a useful tool, enabling evaluation of the immune modulatory potential of bioactive compounds 1, 2, 3. An advantage of this technique is that the tissue samples can be challenged with inflammatory inducing agents, such as lipopolysaccharide (LPS), without infecting live animals while still maintaining the cellular heterogeneity of the gut, a feature which is absent in cell lines. A number of studies have used pig intestinal tissue explants derived from both small 4, 5 and large 3, 4 intestines for ex‐vivo challenge with LPS and for the investigation of immunomodulatory effects of bioactive compounds on the gut epithelium. Immunological challenge experiments which utilize porcine intestinal tissue typically involve dissection of the intestinal tissue from animals fed previously with bioactive compounds for weeks, followed by ex‐vivo incubation of the tissue explants in presence of a proinflammatory‐inducing agent such as bacterial LPS in a near physiological environment 1, 3. The tissue explants is then processed for RNA isolation and quantitative polymerase chain reaction (qPCR) analysis. Testing the effects of bioactives by oral intake in live animals means that there are limitations to the numbers of samples that can be tested. Hence, the ex‐vivo technique is a potentially practical alternative for screening larger number of test compounds.
A successful application of qPCR requires that the postmortem tissue remains physiologically viable during the period of ex‐vivo treatment. Any treatment of the postmortem tissue is likely to cause physical and mechanical injury which can lead to inflammation and activation of cellular nucleases which, in turn, results in the loss of physiological viability and accelerates RNA degradation within the tissue 6. High‐quality intact RNA is a fundamental requirement for qPCR. Tissues such as skeletal muscle 7, 8, brain 9, 10 and connective tissues such as ligament, tendon and cartilage 11 have a slow rate of postmortem RNA degradation, as metabolic activity of these tissues is relatively slow. However, tissues with relatively higher metabolic activities such as liver 12, and intestinal tissues such as colon 13, were also reported to withstand postmortem degradation when handled carefully. Therefore, it may be expected that the postmortem intestinal tissue of pigs remains physiologically viable for some time before undergoing complete degradation. Such a time‐window is likely to be affected by the length of postmortem delay and the prevailing physicochemical environment surrounding the tissue 14.
Another requirement of immune‐related gene expression studies that utilize post‐mortem intestinal tissue ex‐vivo is that the biological response to challenge should be comparable to that exhibited by tissue in vivo. Recently, an ex‐vivo experimental set‐up has been explored which involves challenging of porcine colonic explants with LPS followed by evaluation of the expression of inflammatory immune markers such as interleukin (IL)−8, IL‐6 and tumour necrosis factor (TNF)‐α 15, 16. However, there is limited information relating to the biochemical pathways which are affected in post‐mortem tissue explants and if the responses are comparable to those of intact tissue and how these are affected by the age of the tissue donor. This information is necessary to explore the potential of ex‐vivo treatment of post‐mortem tissues in pigs and other mammalian species, including humans. Therefore, the objectives of this study were: (1) to assess the effects of incubation time and age of the donor on the integrity of a panel of inflammatory immune genes over a 12‐h period and (2) to evaluate the effect of LPS treatment on the activation of inflammatory gene cascade in post‐mortem colonic explants.
Materials and methods
Animals and collection of tissue samples
All experimental procedures were conducted under experimental licence from the Irish Department of Health in accordance with the Cruelty to Animals Act 1876 and the European Communities (Amendments of the Cruelty to Animals Act, 1876) Regulation, 1994.
Weaned piglet experiment
Male (aged 35 days) Large White × Landrace cross‐breed pigs (n = 6) were weaned at the age of 26 days and fed a weaning diet for 9 days, as described by Leonard et al. 2. The animals were assessed routinely for salmonella and health scores were taken on a daily basis. All animals were deemed healthy with no evidence of any gastrointestinal problems. Colonic samples were collected immediately post‐mortem. An area of the proximal colon (approximately 10 cm length) was dissected, and after cleaning the digesta the tissue was rinsed with sterile phosphate‐buffered saline (PBS) and processed immediately. To minimize the time gap between the collection of the tissue sample and ex‐vivo incubation, the facilities for processing of the colonic tissue and ex‐vivo incubation were set up close to the animal dissection facility. The overlying connective tissue layer was removed carefully and a section of approximately 1·5 × 1·5 cm of the colonic explants was transferred into 1 ml Dulbecco's modified Eagle's medium (DMEM) and incubated for 0, 3, 6 and 12 h in a humidified cell culture incubator with 5% CO2 at 37ºC. Tissue explants was incubated in a basal media, and no antibiotic or nutritional supplement was added to minimize any interference of these additives on the integrity of gene transcripts and/or viability of the explants. To evaluate the effect of LPS (source: Escherichia coli strain B4; Sigma‐Aldrich, St. Louis, MO, USA), tissue samples (1·5 × 1·5 cm) were subjected to incubation for 3 h in the presence or absence of LPS (10 μg/ml). The anti‐inflammatory compound dexamethasone (10 nM) was added, along with LPS and tissue explants, and were incubated for 3 h. Tissue explants were removed from the media, blotted dry and stored in RNAlater (Applied Biosystems, Foster City, CA, USA) overnight at room temperature. The RNAlater was removed prior to storing the tissue samples at −80°C.
Mature pig experiment
The colonic tissue samples were collected from seven cross‐bred [Meatline boars × (Large White × Landrace) sows] male pigs aged 115 days. All animals received an identical ration and subjected to the same animal husbandry practices as described by Vigors et al. 17. No serological testing was performed, and therefore the pathogen load of the animals was unknown. The animals were deemed healthy and no visible symptoms of diarrhea were evident prior to euthanasia. The animals were euthanized by lethal injection using Euthatal (Pentobarbitone Sodium BP; Merial Animal Limited, Woking, UK) at a rate of 1 ml/1·4 kg body weight. Following slaughter the entire digestive tract was removed by blunt dissection and the colonic samples were collected. A defined region of the proximal colon (∼ 5 cm length) was dissected, and after removing the digesta the tissue was washed with sterile PBS and processed immediately, as described above. The overlying muscle tissue layer was removed carefully and a section of approximately 1·5 × 1·5 cm was then incubated in DMEM for 3 h in a humidified cell culture incubator with 5% CO2 at 37ºC in the presence or absence of LPS. Tissue samples were processed as described above.
The integrity of mRNA over different time‐points (0, 3, 6 and 12 h post‐mortem) was determined in the weaned piglets, while only two time‐points (0, 3 h) were evaluated in the mature pigs. The response to LPS treatment in the expression of immune genes was evaluated in both weaned piglet and mature pigs at the 3‐h time‐point.
RNA extraction
Total RNA was extracted from 25 mg tissue samples using GenElute™ Mammalian Total RNA Miniprep Kit (Sigma‐Aldrich), following the manufacturer's instructions. RNA was dissolved in 20 μl of nuclease free water and then subjected to deoxyribonuclease I (DNase I) (Sigma‐Aldrich) treatment to eliminate the genomic DNA contamination. Column purification of the RNA was performed using GenElute™ mammalian total RNA miniprep kit (Sigma‐Aldrich). Total RNA was finally dissolved in 50 μl 0·1% diethylpyrocarbonate (DEPC)‐treated water and stored at −80ºC.
Quantity and integrity of total RNA
The total RNA was quantified using a NanoDrop™‐ND 1000 (Thermo Fisher Scientific Inc. Boston, MA, USA). The integrity of total RNA was determined through analysing 1 μl (200–500 ng) of total RNA in an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) using RNA Nano LabChips (Caliper Technologies Corporation, Hopkinton, MA, USA). The RNA integrity number (RIN) value is an empirical measure of RNA integrity based on the intensities of 28S and 18S rRNA bands. The RIN values of RNA from the tissue explants of weaned piglets subjected to 0, 3, 6 and 12 h incubation were reported previously 15.
Quantitative reverse transcription (RT)–PCR analysis
The cDNA synthesis was performed with 1 μg of total RNA using the RevertAid H minus first‐strand cDNA synthesis kit (Fermentas GmbH, St Leon‐Rot, Germany) following the manufacturer's protocol. The quantitative expression of a panel of 92 target genes and four internal reference genes involved in a number of immune signalling pathways were evaluated using a PCR array. The array plate was run on a 7300 RT–PCR system (Applied Biosystems). For the PCR array experiment, 25 μl of cDNA (after 1 : 5 dilution) from six individual animals of each treatment group were pooled to generate a cDNA pool for the treatment. qPCR was performed on a 20 μl reaction mixture per well, which contained 1 μl pooled cDNA, 9 μl water and 10 μl SYBR Green Master Mix (Applied Biosystems). The thermal cycle conditions were 94°C for 30 s followed by 60°C for 1 min for 40 cycles. The mRNA abundances were expressed in CT values, the number of PCR cycles after which the PCR product crosses a threshold value. In this experiment, a CT value of 35 was considered as the cut‐off limit.
The mRNA abundance of the 92 target genes included in the PCR array was normalized to the geometric mean of three reference genes [beta actin (ACTB), hypoxanthine guanine phosphoribosyl transferase 1 (HPRT1) and beta glucuronidase (GUSB)] and following the 2–ΔΔCt method. Average ΔCt was calculated as the difference of Ct values of any target gene minus average of the Ct value of the two reference genes. Then, fold change was calculated as 2(–average ΔCt target gene)/2(–average ΔCt reference gene).
Ingenuity pathway analysis (IPA)
The fold change values (cut‐off ± 2·0‐fold) were analysed using Ingenuity Systems Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA, USA; http://www.ingenuity.com) and the relevant canonical pathways, cellular processes and upstream/downstream regulatory molecules were identified using the default setting in the software. The statistical probability (P‐value) of the observed number of genes affecting a particular biological function was calculated based on Fisher's exact test. This P‐value indicates the statistical probability of the observed number of genes affected out of the total number of genes evaluated in the PCR array for the biological function.
The correlation between the relationship direction and gene expression was determined by calculating the Z‐score following the formula Z = (N +−N –)/√N, where N + represents the number of genes whose expression follows the same direction while N – represents the number of genes whose expression follows an opposite direction of the expression of a particular gene compared to that already available in the IPA knowledge database. N indicates the total number of genes affected. A high‐stringency Z‐score between ≥+2·0 or ≤–2·0 were applied to identify the most relevant cellular functions and associated upstream/downstream regulators.
Results
Gene expression in colonic explants of weaned piglets
In the colonic explants collected immediately post‐slaughter (0 h) from the weaned piglets, 89·6% of the mRNA transcripts were detectable by real‐time qPCR (Ct values < 35·0). Following 3 h incubation, the mRNA of 98·8% genes were detectable with an increase in the mRNA abundance of ten genes (CXCL2, IL6, IL8, TNFA, IL17F, JUN, TANK, MIC2, NFKBIA and RELA) while a decrease for one gene (TLR6) (gene expression summary list in Table 1; fold change values in Table 2). No alteration in the mRNA expression was observed for the remaining 75 targets (Fig. 1a). However, following further incubation at 6 and 12 h, the mRNA expressions were reduced to 70·9 and 27·9%, respectively (Fig. 1b,c). The fact that mRNA remained mainly active following 3 h incubation, to identify the biochemical pathways of mRNA degradation and loss of viability the fold change data from 6 h incubation were analysed by Ingenuity pathway analysis.
Table 1.
Number of porcine gene transcripts [of a total 96 genes included in the polymerase chain reaction (PCR) array] up, down or unaltered due to ex‐vivo incubation of porcine colonic tissue explants in the absence or presence of lipopolysaccharide (LPS) treatment in weaned and mature pigs.
| No. of genes (cut‐off 2·0‐fold) | ||||
|---|---|---|---|---|
| Colonic tissue donor | Treatment comparisons | Up | Down | No change |
| LPS treatment: | ||||
| Weaned piglets | 3 versus 0 h | 10 | 1 | 75 |
| 6 versus 0 h | 4 | 25 | 57 | |
| 12 versus 0 h | 0 | 62 | 24 | |
| Mature pigs | 3 versus 0 h | 12 | 0 | 74 |
| LPS treatment: | ||||
| Weaned piglets | 3 (+LPS) versus 0 h | 17 | 11 | 58 |
| 3 (Dexa+LPS) versus 0 h | 0 | 23 | 63 | |
| 3 (Dexa+LPS) versus 3 h (+LPS) | 4 | 17 | 65 | |
| Mature pigs | 3 (+LPS) versus 0 h | 10 | 2 | 74 |
Dexa = dexamethasone.
Table 2.
Differential expression of inflammatory genes in porcine colonic explants from weaned piglets in the presence or absence of lipopolysaccharide (LPS) and dexamethasone (Dexa) (fold change values ≥ 2·0‐ and ≤ −2.0‐fold are shown).
| Genes | Fold change | |||
|---|---|---|---|---|
| 3/0 h | 3 LPS/0 h | 3 h Dexa+LPS/3 h LPS | 3 h Dexa+LPS/0 h | |
| Chemokine (C‐X‐C motif) ligand 10 (CXCL10) | +11·53 | +37·58 | +2·42 | – |
| Interleukin 6 (IL6) | +5·05 | +31·17 | −3·82 | −19·33 |
| Tumour necrosis factor alpha (TNFA) | +4·55 | +23·62 | +2·65 | – |
| Interleukin 8 (IL8) | +3·50 | +41·99 | – | −2·42 |
| Jun proto‐oncogene (JUN) | +2·29 | +15·05 | +2·29 | – |
| Interleukin 17F (IL17F) | +2·18 | +3·23 | – | – |
| TRAF family member‐associated NF‐κB activator (TANK) | +2·08 | – | – | −3·61 |
| MHC class I‐related antigen 2 (MIC2) | +2·07 | – | – | – |
| Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha (NFKBIA) | +2·07 | +7·79 | – | – |
| v‐rel avian reticuloendotheliosis viral oncogene homologue A (RELA) | +2·01 | – | – | – |
| Toll‐like receptor 6 (TLR6) | −3·14 | −2·05 | −3·77 | – |
| Interleukin 1 beta (IL1B) | – | +5·78 | – | – |
| Chemokine (C‐C motif) ligand 19 (CCL19) | – | +4·29 | – | – |
| Chemokine (C‐C motif) ligand 5 (CCL5) | – | +3·82 | – | – |
| Toll‐like receptor 9 (TLR9) | – | +3·14 | – | – |
| Toll‐like receptor 8 (TLR8) | – | +2·70 | – | – |
| Interferon (alpha, beta and omega) receptor 1(IFNAR1) | – | +2·66 | −3·00 | −3·95 |
| Intercellular adhesion molecule 1 (ICAM1) | – | +2·59 | – | – |
| Prostaglandin–endoperoxide synthase 2 (PTGS2) | – | +2·43 | −2·24 | −3·32 |
| Chemokine (C‐X‐C motif) ligand 4 (CCL4) | – | +2·27 | – | – |
| Interleukin 17A (IL17A) | – | +2·13 | +2·65 | – |
| Mitogen‐activated protein kinase kinase kinase 8‐like (MAP3K8) | – | −2·08 | −2·12 | – |
| Interleukin 21 (IL21) | – | −2·26 | – | −2·15 |
| Toll‐like receptor 5 (TLR5) | – | −2·38 | – | – |
| Toll‐like receptor 1 (TLR1) | – | −2·41 | – | – |
| Tumour necrosis factor receptor superfamily, member 1A (TNFRSF1A) | – | −2·41 | −2·02 | – |
| Chemokine (C‐X‐C motif) ligand 9 (CXCL9) | – | −2·88 | – | – |
| S100 calcium binding protein A3 (S100A3) | – | −3·20 | −5·30 | −3·49 |
| Lysozyme (LYZ) | – | −3·41 | – | – |
| Chemokine (C‐X‐C motif) ligand 11 (CXCL11) | – | −8·16 | – | – |
| Toll‐like receptor 2 (TLR2) | – | – | −2·05 | – |
| Nuclear factor of kappa B light polypeptide gene enhancer in B‐cells 1 (NFKB1) | – | – | −2·06 | – |
| Interleukin 15 (IL15) | – | – | −2·12 | – |
| Interleukin 18 (IL18) | – | – | −2·37 | – |
| Interleukin 1 receptor antagonist (IL1RN) | – | – | −2·40 | −2·43 |
| Mitogen‐activated protein kinase 8 (MAPK8) | – | – | −2·51 | −2·50 |
| v‐akt murine thymoma viral oncogene homologue 1 (AKT1) | – | – | −2·52 | −3·32 |
| Mitogen‐activated protein kinase 9 (MAPK9) | – | – | −2·84 | −4·29 |
| Interleukin 5 (IL5) | – | – | −8·91 | −14·65 |
| Nitric oxide synthase 2, inducible (NOS2) | – | – | −13·79 | −23·96 |
| Complement component 5 (C5) | – | – | – | −2·54 |
| Chemokine (C‐X‐C motif) ligand 2 (CXCL2) | – | – | – | −4·77 |
| Interleukin 4 receptor (IL4R) | – | – | – | −2·00 |
| Myeloid differentiation primary response 88 (MYD88) | – | – | – | −2·03 |
| Peroxisome proliferator‐activated receptor gamma (PPARG) | – | – | – | −2·24 |
| Signal transducer and activator of transcription‐3 (STAT3) | – | – | – | −2·55 |
| Toll‐like receptor 4 (TLR4) | – | – | – | −2·47 |
| Tumour necrosis factor (ligand) superfamily, member 10 (TNFSF10) | – | – | – | −3·74 |
| TNF receptor‐associated factor 4 (TRAF4) | – | – | – | −2·25 |
| TNF receptor‐associated factor 6, E3 ubiquitin protein ligase (TRAF6) | – | – | – | −2·07 |
NF‐κB = nuclear factor kappa B; MHC = major histocompatibility complex
Figure 1.
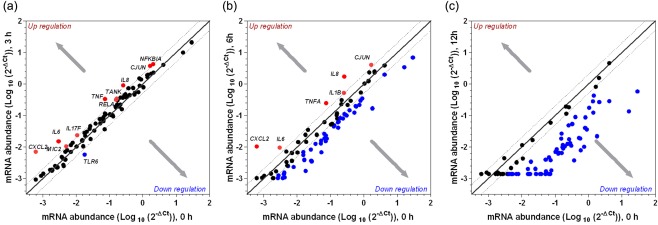
Differential expression of porcine inflammation‐related genes in the postmortem colonic explants harvested from the weaned piglets and subjected to incubation for 3 (a), 6 (b), and 12 (c) h in comparison to the most intact mRNA transcript profile (0 h). Sections of postmortem colonic tissue were processed and incubated in 1 ml Dulbecco's modified Eagle's medium (DMEM) in a humidified cell culture incubator maintained at 37°C with 5% CO2 for different time‐periods (0, 3, 6 and 12 h postmortem for weaned piglets and 0 and 3 h postmortem for mature pigs). The abundance of gene transcripts in the colonic explants at 0, 3, 6 and 12 h were measured using a customized quantitative real‐time polymerase chain reaction (PCR) array and the gene expression value is presented relative to the most intact tissue (0 h).
Biochemical pathways affected in colonic explants of weaned piglets at 6 h
At 6 h, a total of 12 most relevant canonical pathways were identified (Table 3). All these biochemical pathways except the phosphatidylinositol‐4,5‐bisphosphate 3‐kinase (PI3K) signalling pathway were inhibited, indicating a possible loss of physiological viability of the colonic explants at 6 h. The diseases or functions annotation analysis (Table 4) indicated further that cellular activities including activation, differentiation, migration and aggregation of cells decreased with an increase in cell death and mortality. The ten most relevant upstream regulatory molecules underlying the gene expression at 6 h incubation are shown in Table 5. Activation of one kinase [interleukin 1 receptor‐associated kinase 4 (IRAK4)], one transcription factor [interferon (IFN) regulatory factor 2 (IRF2)] and three trans‐membrane receptors [C‐type lectin domain family 7 member A (CLEC7A), Toll‐like receptor (TLR)‐10 and TLR‐11] and/or inhibition of three cytokines [tumour necrosis factor (TNF)‐α, IFN‐α, IFN‐γ, IFN‐β1] and one trans‐membrane receptor (CD5) were predicted to underlie the gene expression pattern observed after 6 h incubation of the colonic explants.
Table 3.
Predicted canonical pathways affected in the colonic explants harvested from the weaned piglets and subjected to ex‐vivo incubation for 6 h.
| Canonical pathway | Genes affected | ‐log (P‐value) | Z‐score |
|---|---|---|---|
| NF‐κB signalling | 21/172 | 3·07E01 | −3·578 |
| Dendritic cell maturation | 19/178 | 2·65E01 | −2·982 |
| Toll like receptor signalling | 18/74 | 1·09E32 | −1·941 |
| NGF signalling | 8/113 | 9·66E00 | −2·646 |
| TREM1 signalling | 19/76 | 3·41E01 | −2·524 |
| Production of nitric oxide and reactive oxygen species in macrophages | 14/186 | 1·72E01 | −2·496 |
| Lymphotoxin β receptor signalling | 7/56 | 1·03E01 | −2·449 |
| IL‐17A signalling | 9/66 | 1·35E01 | −2·333 |
| RANK signalling | 9/89 | 1·22E01 | −2·333 |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 20/125 | 3·16E01 | −2·324 |
| MIF regulation of innate immunity | 8/44 | 1·31E01 | −2·121 |
| PI3K signalling | 7/132 | 7·62E00 | +1·890 |
NF‐κB = nuclear factor kappa B; IL = interleukin; RANK = receptor activator of nuclear factor κB; PI3K = phosphatidylinositol 3‐kinase; MIF = migration inhibitory factor; TREM = triggering receptor expressed on myeloid cells 1; NGF = nerve growth factor.
Table 4.
The most conspicuous diseases or functions annotation underlying the loss of viability in the colonic explants harvested from the weaned piglets and subjected to ex‐vivo incubation for 6 h.
| Diseases or functions annotation | Predicted activation state | Z‐score | P‐value |
|---|---|---|---|
| Activation of cells | Decreased | −3·742 | 9·47E‐43 |
| Differentiation of cells | Decreased | −3·288 | 1·59E‐22 |
| Proliferation of cells | Decreased | −3·059 | 6·60E‐23 |
| Migration of cells | Decreased | −3·021 | 3·90E‐26 |
| Cell movement | Decreased | −2·992 | 3·68E‐27 |
| Aggregation of cells | Decreased | −2·680 | 1·60E‐15 |
| Organization of cytoskeleton | Decreased | −2·362 | 5·17E‐15 |
| Synthesis of eicosanoid | Decreased | −2·361 | 6·53E‐33 |
| Generation of cells | Decreased | −2·341 | 5·70E‐29 |
| Production of protein | Decreased | −2·300 | 6·21E‐17 |
| Growth of plasma membrane projections | Decreased | −2·165 | 2·04E‐15 |
| Expression of RNA | Decreased | −2·063 | 2·14E‐23 |
| Infection | Increased | 2·743 | 2·36E‐25 |
| Bacterial infections | Increased | 2·415 | 1·18E‐40 |
| Morbidity or mortality | Increased | 2·189 | 1·12E‐35 |
| Organismal death | Increased | 2·112 | 4.37E‐34 |
Table 5.
Predicted upstream regulatory molecules linked to the loss of physiological viability and cell death in the colonic explants harvested from the weaned piglets and subjected to ex‐vivo incubation for 6 h.
| Upstream regulator | Molecule type | Predicted state | Z‐score | P‐value of overlap |
|---|---|---|---|---|
| IRAK4 | Kinase | Activated | 2·382 | 3·52E‐18 |
| IRF2 | Transcription regulator | Activated | 2·186 | 5·13E‐11 |
| CLEC7A | Trans‐membrane receptor | Activated | 2·171 | 1·16E‐09 |
| TLR‐10 | Trans‐membrane receptor | Activated | 2·000 | 9·04E‐11 |
| TLR‐11 | Trans‐membrane receptor | Activated | 2·000 | 6·31E‐10 |
| Interferon alpha | Interferon family | Inhibited | −2·768 | 7·18E‐31 |
| TNF | Cytokine | Inhibited | −2·745 | 3·67E‐32 |
| IFN‐γ | Cytokine | Inhibited | −2·708 | 1·17E‐34 |
| IFN‐Β1 | Cytokine | Inhibited | −2·429 | 4·37E‐21 |
| CD5 | Trans‐membrane receptor | Inhibited | −2·414 | 1·45E‐09 |
IRAK4 = interleukin 1 receptor‐associated kinase 4; IRF2 = interferon (IFN) regulatory factor 2; CLEC7A = C‐type lectin domain family 7 member A; TLR = Toll‐like receptor; TNF = tumour necrosis factor.
Gene expression in the colonic explants of weaned piglets by LPS treatment
In the colonic explants of weaned piglets, the LPS treatment for 3 h resulted in an increase in the mRNA abundance of inflammatory genes when compared to the 0‐h time‐point (Table 2). As expected, the mRNA expression of a total of 17 genes, mainly inflammatory cytokines and chemokines, were activated while 11 genes were inhibited due to LPS treatment. An increased mRNA expression of proinflammatory transcription factors (NFKBIA, CJUN), chemokines (CXCL2, CCL19, CCL4 and CCL5), proinflammatory cytokines (IL1B, IL6, IL8, IL17F and TNFA) and TLR receptors (TLR8, TLR9) were evident. Ingenuity canonical pathway analysis revealed that the LPS‐mediated response in weaned piglets occurred through activation of major inflammatory pathways, including high mobility group box 1 (HMGB1), dendritic cell maturation, IL‐17F signalling, TLRs and IL‐6 signalling (Table 6). Based on the expression of majority of the inflammatory markers and their sensitivity to LPS treatment, it was evident that the post‐mortem colonic explants remain physiologically viable up to the 3‐h time‐point.
Table 6.
Predicted canonical pathways affected in the colonic explants harvested from the weaned piglets and subjected to ex‐vivo incubation for 3 h in the presence of lipopolysaccharide (LPS) or LPS and dexamethasone combined.
| LPS | LPS+ dexamethasone | |||||
|---|---|---|---|---|---|---|
| Canonical pathway | Genes affected | ‐log (P‐value) | Z‐score | Genes affected | ‐log (P‐value) | Z‐score |
| HMGB1 signalling | 9/120 | 1·35E01 | +2·333 | 11/120 | 1·75E01 | −3·317 |
| IL‐17F in inflammatory diseases | 6/44 | 1·06E01 | +2·236 | 7/44 | 1·29E01 | −2·449 |
| Dendritic cell maturation | 8/177 | 1·02E01 | +2·121 | 12/177 | 1·76E01 | −3·464 |
| IL‐8 signalling | 4/184 | 3·99E00 | +2·000 | 7/184 | 1·56E01 | −3·162 |
| IL‐6 signalling | 7/116 | 9·83E00 | +1·890 | 10/116 | 8·42E00 | −2·449 |
| Acute‐phase response signalling | 6/169 | 7·07E00 | +1·633 | 9/169 | 1·21E01 | −3·000 |
| Activation of IRF by cytosolic pattern recognition receptors | 5/62 | 7·73E00 | +1·342 | 8/62 | 1·39E01 | −1·414 |
| TREM1 signalling | 10/75 | 1·76E01 | +1·265 | 8/75 | 1·33E01 | −2·828 |
| TNFR1 signalling | 4/49 | 6·28E00 | +1·000 | 5/49 | 8·26E00 | −2·236 |
| Toll‐like receptor signalling | 9/74 | 1·55E01 | +1·342 | 10/74 | 1·77E01 | −1·890 |
| LPS/IL‐1 mediated inhibition of RXR function | 4/221 | 3·69E00 | +1·000 | 6/221 | 6·39E00 | −2·236 |
| Pattern recognition receptors | 12/125 | 1·95E01 | +0·707 | 12/125 | 1·95E01 | −2·828 |
| Production of nitric oxide and reactive oxygen species | 6/180 | 6·91E00 | ‐ | 8/180 | 1·01E01 | −2·121 |
| PPAR signalling | 6/93 | 8·63E00 | −1·633 | 7/93 | 1·05E01 | −2·646 |
| NF‐κB signalling | 10/172 | 1·39E01 | −0·632 | 8/172 | 1·03E01 | −2·121 |
| LXR/RXR activation | 6/121 | 7·94E01 | −0·816 | 8/121 | 1·15E01 | +2·121 |
HMGB1 = high mobility group box 1 protein; IL = interleukin; TREM1 = triggering receptor expressed on myeloid cells 1; TNFR1 = tumour necrosis factor receptor 1; PPAR = peroxisome proliferator‐activated receptors; NF‐κB = nuclear factor kappa B.
Attenuation of LPS‐induced response by anti‐inflammatory drug dexamethasone
Compared to LPS induction of colonic explants for 3 h or the explants harvested immediately post‐mortem (0 h), treatment with dexamethasone resulted in inhibition of the mRNA expression of a number of proinflammatory marker genes, including IL6, IFNAR1, PTGS2, MAPK8, MAPK9, AKT1, NOS2 and IL5 (Table 2). The expression of NOS2 genes were down‐regulated due to dexamethasone treatment by −13·8‐ and −24·0‐fold when compared to LPS treatment for 3 h and colonic tissue collected immediately after slaughter (0 h), respectively. It was evident that five of the inflammatory signalling pathways (HMGB1 signalling, IL‐17F in inflammatory disease, dendritic cell maturation, IL‐8 signalling and IL‐6 signalling) activated by LPS treatments were inhibited by dexamethasone treatment (Table 6). Dexamethasone treatment (in the presence of LPS) inhibited a number of other inflammatory pathways, including acute‐phase response signalling, triggering receptor expressed on myeloid cells 1 (TREM1) signalling, TNF‐R1 signalling, LPS/IL‐1‐mediated inhibition of RXR function, pattern recognition receptors, production of nitric oxide and reactive oxygen species and nuclear factor kappa B (NF‐κB) signalling pathways. The peroxisome proliferator‐activated receptor gamma (PPAR) and LXR/RXR activation pathways were the major canonical pathways activated by the dexamethasone treatment.
Gene expression in the colonic explants of mature pigs
In the colonic explants collected from the mature pigs, 89·6% mRNA transcripts were detectable (CT values less than 35) in the 0‐h tissue samples. Incubation of the colonic explants for 3 h caused no reduction in the relative abundance (compared to the 0‐h time‐point) of the gene transcripts (Table 1). While the mRNA of all 86 gene transcripts were detectable and were mainly unchanged at 3 h (Fig. 2), the abundance of 12 genes (AKT1, TNFA, IL1B, IL6, IL8, IL12B, IL17A, CCL2, CXCL2, JUN, LYZ and NFKBIA) were increased by more than 2·0‐fold (Table 7), indicating that at 3 h, the mRNA derived from mature pigs was still predominantly undegraded. Following an LPS challenge of the colonic explants for 3 h, the mRNA abundance of inflammatory markers (TNFA, IL1B, IL6, IL8, IL4, IL12B, MIC2, CCL2, JUN and NFKBIA) increased by > 2·0‐fold while that of two genes (CSF1 and LYZ) decreased by < −2·0‐fold (Fig. 2; Table 7). A closely similar activation profile of the inflammatory genes was evident in the explants from the mature pigs following 3 h incubation in the presence or absence of LPS.
Figure 2.
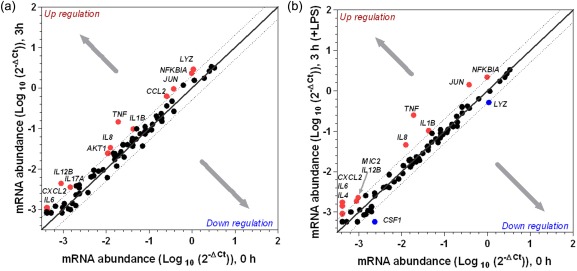
Differential expression of porcine inflammation‐related genes in the postmortem colonic explants harvested from the mature pigs and subjected to incubation at 3 h in the absence (a) or presence of lipopolysaccharide (LPS) (b). Sections of postmortem colonic tissue from mature pigs were processed and incubated in 1 ml Dulbecco's modified Eagle's medium (DMEM) containing 10 μg/ml LPS in a humidified cell culture incubator maintained at 37°C with 5% CO2 for 3 h. The abundance of gene transcripts was measured using a customized quantitative real‐time polymerase chain reaction (PCR) array and the gene expression value is presented relative to the tissue harvested at 0 h time‐point.
Table 7.
Differential expression of inflammatory genes in porcine colonic explants from mature pigs in presence or absence of lipopolysaccharide (fold change values ≥ 2.0‐ and ≤ −2.0‐fold are shown).
| Genes | Fold change | |
|---|---|---|
| 3/0 h | 3 (+LPS)/0 h | |
| Tumour necrosis factor alpha (TNFA) | +7·41 | +13·16 |
| Interleukin 1 beta (IL1B) | +2·17 | +2·41 |
| Interleukin 17A (IL17A) | +2·36 | – |
| Interleukin 6 (IL6) | +2·20 | +3·38 |
| Jun proto‐oncogene (JUN) | +2·44 | +3·78 |
| Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha (NFKBIA) | +2·28 | +2·22 |
| Interleukin 12B (IL12B) | +4·79 | +2·10 |
| Chemokine (C‐C motif) ligand 2 (CCL2) | +2·33 | +4·16 |
| Chemokine (C‐X‐C motif) ligand 2 (CXCL2) | +2·55 | – |
| Interleukin 8 (IL8) | +2·58 | +3·65 |
| Lysozyme (LYZ) | +2·58 | −2·10 |
| v‐akt murine thymoma viral oncogene homologue 1 (AKT1) | +2·14 | – |
| Major histocompatibility complex (MHC) class I‐related antigen 2 (MIC2) | – | +2·33 |
| Interleukin 4 (IL4) | – | +2·19 |
| Colony‐stimulating factor (CSF1) | – | −4·12 |
Discussion
The present study was conducted to identify the post‐mortem time‐window during which porcine colonic explants remain physiologically viable and responsive to the LPS challenge. Colonic explants subjected to ex‐vivo incubation for up to 3 h have generated highly intact gene transcripts comparable to those from the tissue explant collected immediately after euthanasia. In contrast, incubation beyond 6 h resulted in degradation of gene transcripts and an onset of cellular mechanisms leading to cell death. Ex‐vivo treatment with LPS for 3 h resulted in activation of inflammatory immune pathways that can be inhibited by treatment with the anti‐inflammatory drug dexamethasone. Our results demonstrate that a post‐mortem time‐window of 0–3 h is valid for immunological challenge/treatment of the pig colonic tissue due to the fact that the integrity of gene transcripts remains unaffected and the tissue responded to LPS and dexamethasone treatments.
In this experiment, following 3 h incubation of the colonic explants, the gene expression profile of 86 genes and activation of a number of inflammatory immune pathways indicated that the tissues remain physiologically viable during 0–3 h. However, the viability of the tissue explants was compromised at 6 h and beyond, as the degradation of large number of mRNA transcripts occurred and activation of biochemical pathways leading to cell death was evident. Degradation of mRNA transcripts could be due to a number of reasons. First, intestinal tissues have a high level of RNAse activity 18, which might be activated further in response to mechanical disruption and removal of overlying muscle layer during processing of the tissue explants. Secondly, the colonic tissue has a high metabolic activity, and due to lack of oxygen supply 19 and inadequate buffering of the culture media the explants will inevitably suffer hypoxia beyond the 3‐h incubation period. Such a hypoxic condition 20 is expected to increase the acidity (this was visible by a change in the colour of the media to yellow at 6 and 12 h post‐incubation). Thirdly, the colonic tissue harbours large numbers of micro‐organisms, which may contribute to a rapid degradation of the post‐mortem tissue beyond 3 h. The fact that all weaned piglets had ad‐libitum access to the same diet and water and all animals were maintained in an identical husbandry environment it is unlikely that, within this group of animals, diet or husbandry conditions can cause any major difference that could have a confounding effect on the mRNA integrity.
The extended physiological viability of the colonic explants during the post‐mortem time‐window of 0–3 h was also evident from the fact that there was no decrease in the abundance of mRNA transcripts all 86 genes in the mature pigs. In the colonic explants of weaned piglets and mature pigs, at 0–3 h, up‐regulated common inflammatory genes were TNFA, IL6, IL8, NFKBIA and JUN, even in the absence of any exogenous inflammatory stimulus. An activation of these genes can be expected due to an induction of the inflammatory biochemical pathways in the post‐mortem tissue. Inflammation and associated biochemical pathways were inhibited following incubation of the explants 6 h and beyond. The activation of PI3K signalling at 6 h indicated potential activation of cellular autophagy mechanisms leading to cell death 21. This was supported further by the predicted diseases or functions annotation analysis, where the markers of morbidity or mortality of the cells/tissue/organisms were increased. A reduction in the cellular activities and production of RNA and protein synthesis also indicated loss of cell viability at 6 h.
To understand further the role of upstream regulatory molecules underlying the molecular/biochemical processes activated at 6 h, the most potent regulators were identified as activation of IRAK4, IRF2 and inhibition of IFN‐α and TNF‐α. IRAK4 is a serine/threonine kinase that plays a key role in the IL‐1/TLR‐mediated activation of NF‐κB inflammatory pathway 22. In the absence of any inflammatory stimuli, activation of IRAK4 might be highly critical to the induction of proinflammatory response that was evident in the post‐mortem tissue at 3 h and 6 h. IRF2 is a transcription factor involved, along with IRF1, in the transcription of the IFN system 23. IRF2, as such, causes transcriptional repression of IFN‐responsive genes, including IFN‐α. Repression of IFN‐α was also predicted to be another upstream regulatory phenomenon at 6 h. TNF‐α is a proinflammatory cytokine reported to be involved in regulating cell proliferation in mouse colonic cells 24. At high concentrations (100–1000 ng/ml) TNF‐α was found to inhibit cell proliferation, while at lower concentrations (0·1–1·0 ng/ml) the same cytokine stimulated cell proliferation 24. In the present experiment the fact that the mRNA abundance of the TNFA gene remained elevated at 3 h and 6 h, a higher activity of TNF‐α in the post‐mortem tissue explants, might contribute to the inhibition of cell proliferation and an increase of subsequent cell death beyond 6 h.
In the colonic explants from the weaned piglets, as well as mature pigs, LPS treatment resulted in the up‐regulation of a number of genes, including transcription factors (NFKBIA, JUN) driving key inflammatory pathways and proinflammatory cytokines (TNFA, IL6 and IL8). LPS is generally secreted by gram‐negative microbes in the intestinal gut and is a potent inducer of the proinflammatory response 25. The up‐regulation of proinflammatory cytokine genes including IL1A, IL6, IL8 and TNFA following treatment of colonic explants with LPS was reported previously 1, 3, 16. In the intestinal epithelium, LPS is expected to mediate the proinflammatory response through TLRs, in particular TLR‐4 26, 27. In the present experiment, LPS treatment did not increase the mRNA abundance of the TLR4 gene in colonic explants ex vivo, which is consistent with our previous observation 3. Of the nine TLR genes evaluated (TLR‐1, ‐2, ‐4, ‐5, ‐6, ‐7, ‐8, ‐9 and ‐10), only the mRNA abundance of TLR8 and TLR9 were increased while there was a decrease in the TLR1, TLR5 and TLR6 genes in the colonic explants from weaned piglets following the LPS treatment. None of the TLR genes responded to LPS treatment in the explants from the mature pigs.
As evident in this experiment, a proinflammatory response at 3 h in the explants from weaning and mature pigs even in the absence of LPS treatment could be due to mechanical injury 28, tissue hypoxia 29 and/or microbial contamination 30. Cellular responses to microbial invasion or a hypoxic cell environment are generally mediated by transmembrane receptors, including the TLRs 31. However, in this study, in the colonic explants derived from weaned piglets at 3 h post‐mortem, TLR6 was down‐regulated. The TLR6 gene codes a transmembrane protein that recognizes a Mycoplasma hyponeumoni‐derived pathogen recognition antigen pattern 32, 33. Therefore, the observed up‐regulation of the inflammatory immune markers caused by LPS treatment may be due to other stimuli including mechanical injury, tissue hypoxia and microbial contamination. Furthermore, co‐treatment of the tissue explants with LPS and dexamethasone, an anti‐inflammatory compound 34, resulted in the down‐regulation of most of the LPS‐induced proinflammatory genes, thereby nullifying the proinflammatory effect of LPS in the tissue explants, further supporting the fact that the tissue explants remained physiologically responsive for up to 3 h.
An evaluation of the abundance of mRNA transcripts in mature pigs and weaned piglets revealed that the age of the animals had only a minor effect on the mRNA profile. Nevertheless, the explants from mature pigs had only a few proinflammatory cytokine genes expressed differentially as a result of ex‐vivo incubation or in response to LPS treatment for 3 h post‐mortem. This could indicate that the colonic tissue of the weaned pigs is more responsive to inflammatory stimuli, possibly because the colonic tissue of weaning piglets is undergoing a period of adaptation to microbial challenge and nutritional stresses 35, 36, 37. In contrast, the mature pigs are expected to have a more stably developed immune system and are more resistant to a wider range of inflammatory stimuli 37, 38. This is in agreement with previous reports on LPS‐mediated induction of proinflammatory cytokine gene expression (IL6 and IL8) in 49‐day‐old pigs 3 and 56‐day‐old pigs 1.
In the present experiment, screening of mRNA expression of genes was based on the pooling of equal amount of cDNA from six (weaned piglets) or seven (mature pig) animals. Pooling of cDNA from individual animals could result in a reduction of biological variability which is otherwise present in each animal. However, in the present experiment, any reduction of biological variability would have only a minimal affect on the interpretation of biological mechanisms based on the differential gene expression pattern, which was obvious and consistent. In fact, the pooling of cDNA may even result in a greater accuracy of gene expression measurement where pooled cDNA is evaluated in only one array/treatment, as in the present experiment 39.
Conclusions
The exploitation of time‐window of 0–3 h during which the post‐mortem tissue explants remains physiologically viable provides an excellent opportunity to study the interaction of these tissue explants with drugs and bioactive compounds ex‐vivo. This has already been explored for the evaluation of the immune modulatory potential of bioactive neutraceuticals derived from milk and seaweed 16, 40. One distinct advantage of challenging tissue explants ex‐vivo is that the experiment needs only a small section (1·5 × 1·5 cm) of the tissue and a small quantity (1 mg) of the test sample, and therefore several test samples can be evaluated simultaneously on samples harvested from the same donor. Taken together, this study supports the validity of using ex‐vivo mammalian intestinal tissue explants as a tool in the exploration of bioactivity in the gut of both drugs and bioactive compounds.
Disclosures
The authors declare no disclosures relating to this publication.
Acknowledgements
This project (Grant‐Aid Agreement no. MFFRI/07/01) is carried out under the Sea Change Strategy with the support of the Marine Institute and the Department of Agriculture, Food and the Marine, funded under the National Development Plan 2007–2013.
References
- 1. Smith AG, O'Doherty JV, Reilly P et al The effects of laminarin derived from Laminaria digitata on measurements of gut health: selected bacterial populations, intestinal fermentation, mucin gene expression and cytokine gene expression in the pig. Br J Nutr 2011; 105:669–77. [DOI] [PubMed] [Google Scholar]
- 2. Leonard SG, Sweeney T, Bahar B et al Effect of dietary seaweed extracts and fish oil supplementation in sows on performance, intestinal micro‐flora, intestinal morphology, volatile fatty acid concentrations and immune status of weaned pigs. Br J Nutr 2011; 105:549–60. [DOI] [PubMed] [Google Scholar]
- 3. Sweeney T, Collins CB, Reilly P et al Effect of purified beta‐glucans derived from Laminaria digitata, Laminaria hyperborean and Saccharomyces cerevisiae on piglet performance, selected bacterial populations, volatile fatty acids and pro‐inflammatory cytokines in the gastrointestinal tract of pigs. Br J Nutr 2012; 108:1226–34. [DOI] [PubMed] [Google Scholar]
- 4. Danielsen EM, Sjostrom H, Noren O et al Biosynthesis of intestinal microvillar proteins. Biochem J 1982; 202:647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kik MJL, Koninkx JFJG, van den Muysenberg A et al Pathological effects of Phaseolus vulgaris isolectins on pig jejunal mucosa in organ culture. Gut 1991; 32:886–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee JH, Fitzgerald JB, Dimicco MA et al Mechanical injury of cartilage explants causes specific time‐dependent changes in chondrocyte gene expression. Arthritis Rheum 2005; 52:2386–95. [DOI] [PubMed] [Google Scholar]
- 7. Bahar B, Monahan FJ, Moloney AP et al Long‐term stability of RNA in post‐mortem bovine skeletal muscle, liver and subcutaneous adipose tissues. BMC Mol Biol 2007; 8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fontanesi L, Colombo M, Beretti F et al Evaluation of post mortem stability of porcine skeletal muscle RNA. Meat Sci 2008; 80:1345–51. [DOI] [PubMed] [Google Scholar]
- 9. Johnson SA, Morgan DG, Finch CE. Extensive post‐mortem stability of RNA from rat and human brain. J Neurosci Res 1986; 16:267–80. [DOI] [PubMed] [Google Scholar]
- 10. Inoue H, Kimura A, Tuji T. Degradation profile of mRNA in a dead rat body: basic semi‐quantification study. Forensic Sci Int 2002; 130:127–32. [DOI] [PubMed] [Google Scholar]
- 11. Marchuk L, Sciore P, Reno C et al Post‐mortem stability of total RNA isolated from rabbit ligament, tendon and cartilage. Biochim Biophys Acta 1998; 1379:171–7. [DOI] [PubMed] [Google Scholar]
- 12. Almeida A, Paul TJ, Magdelenat H et al Gene expression analysis by real‐time reverse transcription polymerase chain reaction: influence of tissue handling. Anal Biochem 2004; 328:101–8. [DOI] [PubMed] [Google Scholar]
- 13. Micke P, Ohshima M, Tahmasebpoor S et al Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest 2006; 86:202–11. [DOI] [PubMed] [Google Scholar]
- 14. Lee SML, Schelcher C, Gashi S et al RNA stability in human liver: comparison of different processing times, temperatures and methods. Mol Biotechnol 2013; 53:1–8. [DOI] [PubMed] [Google Scholar]
- 15. Bahar B, O'Doherty JV, Sweeney T. Assessment of RNA integrity in the post‐mortem pig colonic tissue ex‐vivo . J Anim Sci 2012; 90:22–4. [DOI] [PubMed] [Google Scholar]
- 16. Mukhopadhya A, Noronha N, Bahar B et al Anti‐inflammatory effects of a casein hydrolysate and its peptide enriched fractions on TNFa challenged Caco‐2 cells and LPS challenged porcine colonic explants. Food Sci Nutr 2014; 2:712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vigors S, Sweeney T, O'Shea CJ et al Pigs that are divergent in feed efficiency, differ in intestinal enzyme and nutrient transporter gene expression, nutrient digestibility and microbial activity. Animal 2016; doi:10.1017/S1751731116000847. [DOI] [PubMed] [Google Scholar]
- 18. Ibberson D, Benes V, Muckenthaler MU et al RNA degradation compromises the reliability of microRNA expression profiling. BMC Biotechnol 2009; 9:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Randall KJ, Turton J, Foster JR. Explant culture of gastrointestinal tissue: a review of methods and applications. Cell Biol Toxicol 2011; 27:267–84. [DOI] [PubMed] [Google Scholar]
- 20. Lenihan CR, Taylor CT. The impact of hypoxia on cell death pathways. Biochem Soc Trans 2013; 41:657–63. [DOI] [PubMed] [Google Scholar]
- 21. Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ 2005; 12:1509–18. [DOI] [PubMed] [Google Scholar]
- 22. Wang Z, Wesche H, Stevens T et al IRAK‐4 inhibitors for inflammation. Curr Top Med Chem 2009; 9:724–37.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harada H, Fujita T, Miyamoto M et al Structurally similar but functionally distinct factors, IRF‐1 and IRF‐2, bind to the same regulatory elements of IFN and IFN‐inducible genes. Cell 1989; 58:729–39. [DOI] [PubMed] [Google Scholar]
- 24. Kaiser GC, Polk DB. Tumor necrosis factor alpha regulates proliferation in a mouse intestinal cell line. Gastroenterology 1997; 112:1231–40. [DOI] [PubMed] [Google Scholar]
- 25. Sacco RE, Nibbelink SK, Baarsch MJ et al Induction of interleukin (IL)−1beta and IL‐8 mRNA expression in porcine macrophages by lipopolysaccharide from Serpulina hyodysenteriae . Infect Immun 1996; 64:4369–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yuk JM, Jo EK. Toll‐like receptors and innate immunity. J Bacteriol Virol 2011; 41:225–35. [Google Scholar]
- 27. Moue M, Tohno M, Shimazu T et al Toll‐like receptor 4 and cytokine expression involved in functional immune response in an originally established porcine intestinal epitheliocyte cell line. Biochim Biophys Acta 2008; 1780:134–44. [DOI] [PubMed] [Google Scholar]
- 28. Wehner S, Buchholz BM, Schuchtrup S et al Mechanical strain and TLR4 synergistically induce cell‐specific inflammatory gene expression in intestinal smooth muscle cells and peritoneal macrophages. Am J Physiol Gastrointest Liver Physiol 2010; 299:G1187–97. [DOI] [PubMed] [Google Scholar]
- 29. Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med 2011; 364:656–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grivel JC, Margolis L. Use of human tissue explants to study human infectious agents. Nat Protoc 2009; 4:256–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takeda K, Kaisho T, Akira S. Toll‐like receptors. Ann Rev Immunol 2003; 21:335–76. [DOI] [PubMed] [Google Scholar]
- 32. Régia Silva Sousa K, Mauric Frossard Ribeiro A, Roberto Nunes Goes P et al Toll‐like receptor 6 differential expression in two pig genetic groups vaccinated against Mycoplasma hyopneumoniae . BMC Proc 2011; 5:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar H, Kawai T, Akira S. Toll‐like receptors and innate immunity. Biochem Biophys Res Commun 2009; 388:621–5. [DOI] [PubMed] [Google Scholar]
- 34. Moynagh PN. Toll‐like receptor signaling pathways as key targets for mediating the anti‐inflammatory and immunosuppressive effects of glucocorticoids. J Endocrinol 2003; 179:139–44. [DOI] [PubMed] [Google Scholar]
- 35. Blecha F, Pollman DS, Nichols DA. Weaning pigs at an early age decreases cellular immunity. J Anim Sci 1983; 56:396–400. [DOI] [PubMed] [Google Scholar]
- 36. Wijtten PJ, van der Meulen J, Verstegen MW. Intestinal barrier function and absorption in pigs after weaning: a review. Br J Nutr 2011; 105:967–81. [DOI] [PubMed] [Google Scholar]
- 37. Stokes CR, Bailey M, Haverson K et al Postnatal development of intestinal immune system in piglets: implications for the process of weaning. Anim Res 2004; 53:325–34. [Google Scholar]
- 38. Williams NH, Stahly TS, Zimmerman DR. Effect of level of chronic immune system activation on the growth and dietary lysine. J Anim Sci 1997; 75:2481–96. [DOI] [PubMed] [Google Scholar]
- 39. Kendziorski C, Irizarry RA, Chen KS et al On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA 2005; 102:4252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bahar B, O'Doherty JV, Smyth TJ et al A cold water extract of Fucus vesiculosus inhibits lipopolysaccharide (LPS) induced pro‐inflammatory responses in the porcine colon ex‐vivo model. Innov Food Sci Emerg Technol 2016; http://dx.doi.org/10.1016/j.ifset.2016.04.014 [Google Scholar]


