Abstract
Although strand-biased gene distribution (SGD) was described some two decades ago, the underlying molecular mechanisms and their relationship remain elusive. Its facets include, but are not limited to, the degree of biases, the strand-preference of genes, and the influence of background nucleotide composition variations. Using a dataset composed of 364 non-redundant bacterial genomes, we sought to illustrate our current understanding of SGD. First, when we divided the collection of bacterial genomes into non-polC and polC groups according to their possession of DnaE isoforms that correlate closely with taxonomy, the SGD of the polC group stood out more significantly than that of the non-polC group. Second, when examining horizontal gene transfer, coupled with gene functional conservation (essentiality) and expressivity (level of expression), we realized that they all contributed to SGD. Third, we further demonstrated a weaker G-dominance on the leading strand of the non-polC group but strong purine dominance (both G and A) on the leading strand of the polC group. We propose that strand-biased nucleotide composition plays a decisive role for SGD since the polC-bearing genomes are not only AT-rich but also have pronounced purine-rich leading strands, and we believe that a special mutation spectrum that leads to a strong purine asymmetry and a strong strand-biased nucleotide composition coupled with functional selections for genes and their functions are both at work.
Keywords: Strand-biased gene distribution, Strand-biased nucleotide composition, Horizontal gene transfer, Purine asymmetry, GC content
Introduction
The unidirectional (5′–3′) movement of the DNA replication fork divides the two strands into leading strand (LeS) and lagging strand (LaS). LeS is replicated continuously while LaS is synthesized discontinuously through a RNA priming process producing the Okazaki fragments that are subsequently joined together by a DNA ligase [1]. This is the first level of replication-associated asymmetry that is proposed to have influence on genomic organizational features, such as strand-biased gene distribution (SGD), strand-biased nucleotide composition (SNC), and mutation patterns [2], [3], [4], [5], [6], [7]. Recently, another level of replication-associated asymmetry has been realized: the dimeric DNA polymerases, which are usually encoded by the same dnaE gene in most bacterial genomes, such as Escherichia coli [8], were surprisingly found to be encoded by different genes in some other bacteria [9], [10]. Our recent studies have further clarified the diverse dnaE genes into a group of orthologous genes—dnaE1, dnaE2, dnaE3, and polC [11]. These replicative DnaE dimers were divided into three essential gene groups or enzyme isoform groups: the asymmetric polC-dnaE3 (in charge of DNA synthesis for LeS and LaS, respectively) and the two symmetric dnaE1-dnaE1 groups, including two subgroups, namely dnaE1-dnaE1|polV and dnaE1-dnaE1|dnaE2 [12]. Ample experimental evidence supports the idea that dnaE2 and polC are responsible for the relatively high and low GC contents, respectively [12], [13], [14]. However, their contributions to SGD and their relationship with other factors influential to SGD remain to be demonstrated.
To date, a dozen or so hypotheses have been put forward to explain SGD of prokaryotes [2], [15], [16], [17], [18], [19], [20], [21], [22], [23] that primarily lead into two mechanistic interpretations. The first interpretation concerns process-/function-based selection—head-on collision avoidance between components or complexes of the replication and transcription machineries. It states that the obvious adverse effect of head-on collision (for genes on LaS) between replication and transcription selectively drives more genes to be on LeS (co-orientation collision was observed) [15], [16], [17]. Consistent with this statement, highly expressed genes [18], [19] and multi-gene operons that are subjected to more intensive transcription interruptions [20] as well as essential genes [21], [22] are proposed to reside preferentially on LeS. However, it is convoluted by the following reasons. First, the lack of large-scale empirical data to define gene expressiveness and essentiality hinders accurate definitions of such genes and a comprehensive database has not yet been built. Second, although the key contribution of essential genes to SGD is not disputed, these genes only accounted for a fraction of the total among bacterial genomes—generally 500 genes per genome—according to the database of essential genes (DEG) [24]. Third, there are actually significant overlaps among highly expressed genes, multi-gene operons, and essential genes, as demonstrated based on oligonucleotide-based microarray experiments in Buchnera [25]. Therefore, there is one possibility that these three categories of genes (highly expressed genes, multi-gene operons, and essential genes) may be hard to differentiate. The second interpretation concerns polC, since most polC-harboring bacteria have much higher SGD (78% on average) as compared to those of the non-polC group (58% on average) [23], albeit a few known exceptions. SGD is arguably correlated not only with the presence of polC, but also with a mutation/selection-related force—purine asymmetry that is also unique to this group of bacteria [2].
However, these hypotheses have overlooked two major mechanisms that alter both genome composition and gene content of unicellular organisms—gene gain by horizontal gene transfer (HGT) and gene loss [26], [27], [28], [29]. For example, the LeS protein-coding genes on Mycobacerium leprae accounted for 66% of the total genes when 1116 pseudogenes are excluded and 61% if they are included. In other words, if these pseudogenes are deleted over time, gene loss is able to lead to a change in the LeS gene percentage or proportion from 61% to 66% in M. leprae. Similarly, if there is a biased gene acquisition between LeS and LaS, it also contributes to SGD in a very significant way. In summary, selection forces for SGD can be differentiated into at least two different levels. The first is background level that concerns strand-associated genes in the context of gene functionality, expressiveness, and essentiality, while the second level attributes to horizontally-transferred genes and gene loss. The latter is often observed when the bacterial host range is reduced. In this study, we investigated SGD at both function-centric (background level) and event-associated (gene gain and loss) levels. We performed a large-scale comparative analysis on 364 non-redundant bacterial genomes and provided insights into strand preference of horizontally-transferred genes and its potential roles in SGD as well as their underlying mechanisms regarding to SNC between LeS and LaS.
Results
Strand-biased gene distribution
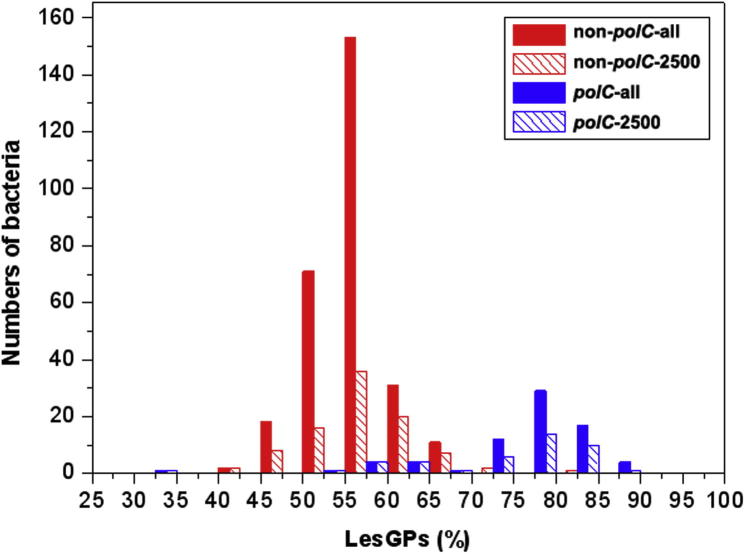
We first divided 364 non-redundant bacterial genomes into two groups based on the presence or absence of polC, resulting in 76 polC and 288 non-polC bacteria (Table S1). We subsequently calculated the LeS gene proportion (LesGP) for each bacterium (see Methods for details) to show that most of the non-polC bacteria have relatively fewer genes on LeS, with LesGPs ranging from 50% to 60%, whereas LesGPs of the polC-harboring bacteria (or simply the polC bacteria) are generally above 75% (Figure 1). In addition, to examine the contribution of gene loss to SGD, we also restricted our analysis to genomes within each group, whose gene counts are less than 2500, and again results showed that the polC group exhibits higher LesGPs than the non-polC group. In summary, we found that the polC group tends to have more genes on LeS regardless whether they once experienced drastic gene loss or not.
Figure 1.
SGD between polC and non-polC bacteria We analyzed 364 non-redundant bacterial genomes based on a simple grouping scheme: non-polC (including dnaE1-dnaE1|polV and dnaE1-dnaE1|dnaE2; red) and polC (polC-dnaE3|polV; blue), and LesGP was calculated for each bacterium. We also show results for all bacteria in each collection: (1) non-polC-all and polC-all stand for genomes that contain all bacteria in the groups and (2) non-polC-2500 and polC-2500 indicate genomes that have <2500 genes.
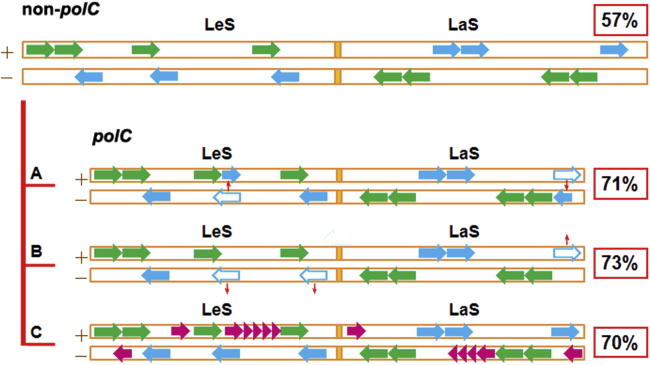
For better clarity, we describe schematically several possible gene gain-and-loss scenarios leading to SGD (Figure 2). The first scenario concerns biased gene rearrangement (without any gene loss) between LeS and LaS (Figure 2A). For instance, when biased within a genome, gene transfer often leads to gene jumping from LaS to LeS, which may result in significant increase in LesGP. The second scenario is biased gene loss (including genes that deteriorate into pseudogenes, Figure 2B), since even a limited number of gene loss events from LaS are able to result in increase in LesGP. The third scenario is biased gene acquisition, where LeS may just gain a few more genes than LaS due to process-selections, such as a high expression level of a translation machinery component (Figure 2C). Since gene loss-and-gain events occur very frequently among unicellular organisms, we believe that an analysis on highly conserved genes should be able to provide a useful hint for what may happen for other genes.
Figure 2.
Schematic illustration of factors affecting SGD under the unified grouping scheme We use a small number of genes to represent ratios (∼57% and ∼70% on average for the non-polC and polC groups, respectively) of conserved genes between LeS (solid green arrows) and LaS (solid blue arrows). There are three possible scenarios where specific changes alter LesGP: biased gene rearrangement with two genes (open blue arrows) transferred to LeS from LaS increases LesGP from 57% to 71% (A); biased gene loss with three events (open blue arrows) from LaS leads to the increase of LesGP from 57% to 73% (B); biased gene acquisition, such as adding 11 genes to LeS but adding 2 to LaS (pink arrows and arrow-heads), results in the change of LesGP from 57% to 70% (C).
Contribution of essential genes to SGD
We analyzed the contribution of essential genes to SGD based on the DEG dataset [24]. We chose a few examples including five from the non-polC and four from the polC groups to calculate their LesGPs (Table 1). The average LesGPs for essential genes is 62% for the non-polC and 89% for the polC bacteria, both of which are higher than those for total genes, 57% and 75%, respectively. Nevertheless, the most obvious finding is that the average LesGP of essential genes for the polC group (89%) is significantly higher than that of the non-polC group (62%), clearly demonstrating that gene essentiality does contribute to SGD but is not the sole and major cause.
Table 1.
LesGP of essential genes based on the DEG database
| Bacteria | Strand | LaS∗ | LeS∗ | LesGP (%) | Group |
|---|---|---|---|---|---|
| E. coli | + | 115 | 181 | 57 | non-polC(62%)† |
| − | 146 | 169 | |||
| Haemophilus influenzae | + | 195 | 238 | 57 | |
| − | 175 | 253 | |||
| Helicobacter pylori | + | 74 | 98 | 62 | |
| − | 52 | 109 | |||
| Mycobacterium tuberculosis | + | 94 | 205 | 70 | |
| − | 84 | 218 | |||
| S. typhimurium | + | 77 | 132 | 64 | |
| − | 78 | 145 | |||
| Bacillus subtilis | + | 7 | 120 | 94 | polC(89%)† |
| − | 7 | 91 | |||
| M. genitalium | + | 11 | 57 | 83 | |
| − | 10 | 47 | |||
| Staphylococcus aureus | + | 8 | 98 | 94 | |
| − | 4 | 97 | |||
| S. pneumoniae | + | 12 | 46 | 87 | |
| − | 5 | 63 |
Note: LeS, leading strand; LaS, lagging strand.
Number of genes on LeS or LaS.
Average LesGP.
Contribution of conserved genes to SGD
We also examined the strand translocation of conserved genes between LeS and LaS and detected its possible role in SGD. To characterize strand-jumping ability of conserved genes between LeS and LaS, we chose two phylogenetically well-characterized datasets, viz., Buchnera (representing non-polC group) and Mycoplasma (representing the polC group), containing 484 and 206 conserved protein-coding genes, respectively. We plotted the relative hierarchical gene distribution for genomes of the two genera (Figure 3).
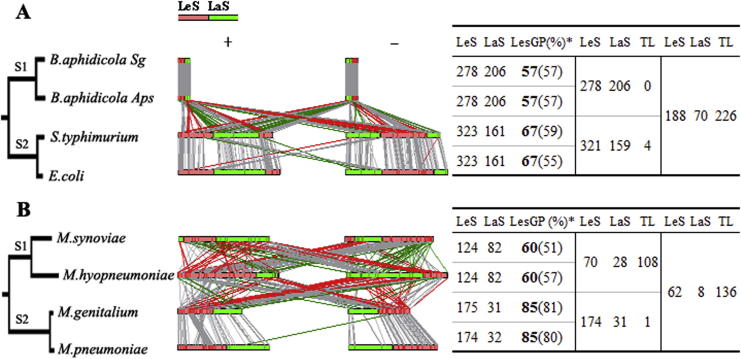
Figure 3.
A case study on strand-preference of conserved genes of the non-polC and polC bacteria We classified the strand distribution of 484 conserved genes in Buchnera (non-polC group; A) and 206 conserved genes in Mycoplasma (polC group; B). We built phylogenetic trees based on the NJ method (bootstrap value = 1000) using 16s rRNA sequences (Mega 4.0; the left panel). The trees were drawn to the scale, with branch lengths in the same units as those of the evolutionary distances used to infer phylogeny. We computed the evolutionary distance using the number of nucleotide variations per sequence as the unit. We selected two bacteria, Ehrlichia canis and Mesoplasma florum, as outgroups to root the trees in (A) and (B), respectively. All positions containing gaps and missing data were eliminated from the dataset. Genes on the positive (+) and negative (-) strands were separated into the LeS (solid red bars) and LaS (solid green bars) groups (the middle panel) and the direction of gene transfer were color-coded accordingly: LaS to LeS (red), LeS to LaS (green), no inter-strand transfer event (gray). The number of genes and their distributions were summarized in the table (the right panel). LesGP values (%) of conserved (bold) and total genes (in parentheses) were also calculated. Genes transferring between strands are classified as translocatable (TL).
The Buchnera group contains two subgroups (S1 and S2) according to its phylogenetic tree (Figure 3A). Subgroup S1 comprises B. aphidicola Sg and B. aphidicola Aps, whereas subgroup S2 includes Salmonella typhimurium and E. coli. There are 484 conserved genes within subgroup S1 with 278 on LeS and 206 on LaS, respectively, in both B. aphidicola Sg and B. aphidicola Aps. There are 278 conserved genes on LeS and 206 on LaS that are shared by the two bacteria, thus there is no translocatable (TL) gene observed between them (Figure 3A). Similar observation was found within subgroup S2. There are 484 conserved genes including 323 on LeS and 161 on LaS in both S. typhimurium and E. coli. Only four TL genes were identified. However, only 188 genes on LeS and 70 genes on LaS are shared when the two subgroups S1 and S2 are compared. We thus infer that, during the divergence of subgroups S1 and S2, their conserved genes must have experienced numerous biased gene rearrangements, which can be explained by the higher LesGP (10%) in subgroup S2. In addition, it should be noted that although subgroup S2 has an average of 10% more genes on LeS, there is no obvious biased distribution of total genes between the two subgroups (both around 57% on average).
In Mycoplasma, a genus of the polC group (Figure 3B), Mycoplasma synoviae and Mycoplasma hyopneumoniae have the same number of conserved genes on each strand, 124 on LeS and 82 on LaS. However, they only share 70 and 28 genes on LeS and LaS, respectively, which lead to 108 TL genes in subgroup S1. In contrast to subgroup S1, the synteny or gene order is well conserved in subgroup S2 where only a single TL gene was classified. In addition, we also found that the longer branch length of the ancestor of Mycoplasma genitalium and Mycoplasma pneumoniae (in subgroup S2) correlated well with the higher LesGPs of this group. In contrast, although greater divergence and substantial gene transfer events were observed in the two bacteria of subgroup S1, there were no obvious differences in their LesGPs. However, we observed obvious disparities between the two subgroups on LesGPs of conserved genes and total genes (Figure 3B).
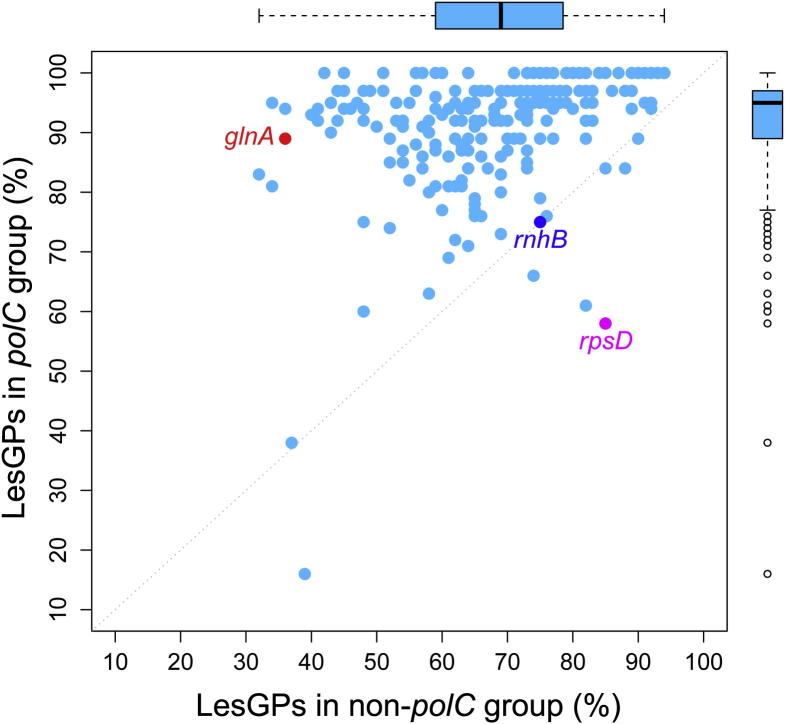
We also identified 239 conserved genes from 249 well-annotated bacterial genomes (Figure 4), and our results demonstrate that there are higher chances for the same genes to reside on LeS of the polC bacteria than the non-polC bacteria, with an average proportion of 92% for the former but only 68% for the latter. Furthermore, KEGG orthology (KO) function analysis on these genes revealed that most of them are involved in essential cellular processes, such as translation and replication, as well as amino acid and nucleotide metabolisms (Figure S1).
Figure 4.
Probability of conserved genes residing on LeS between the non-polC and polC bacteria We used 239 conserved genes from 249 well-annotated bacterial genomes for this analysis. The corresponding values for the non-polC (top) and polC (right) groups are labeled outside the frame. Points mapped on the diagonal line indicate genes that have equal chance to stay on the LeS between the non-polC and polC bacterial groups, such as rnhB. Most of the genes, such as glnA, are found preferentially residing on the LeS of the polC (above the diagonal) group. Only a small number of genes remain below the diagonal, such as rpsD, primarily residing on the LeS of the non-polC group.
Contribution of biased horizontal gene transfer to SGD
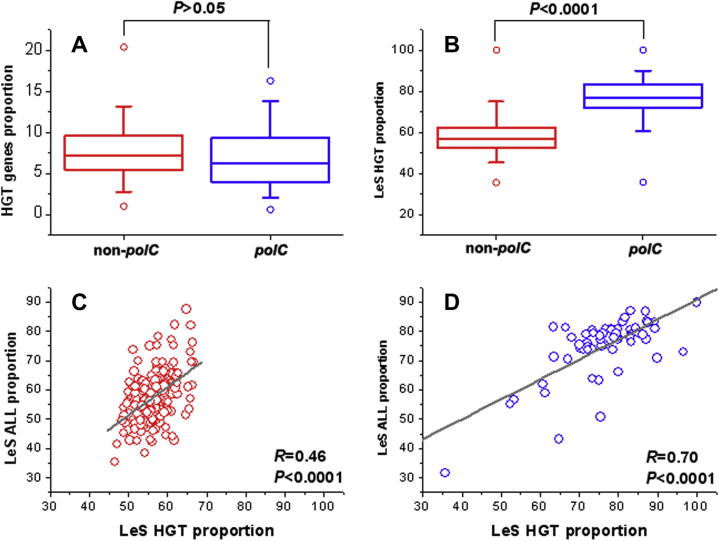
HGT occurs frequently among unicellular organisms, especially bacteria, and plays an essential role in bacterial genome evolution. We propose that it also contributes to SGD. We estimated the proportion of horizontally-transferred genes for both polC and non-polC groups, which was around 5% to 10% in general. There is no statistically significant difference in the proportion of horizontally-transferred genes (unpaired two-tailed t test, P > 0.05) between genomes of the two groups if all horizontally-transferred genes are considered (Figure 5A). A similar analysis with a special consideration of strand preference reveals that the preferences of horizontally-transferred genes on the LeS are 75% and 55% on average for polC and non-polC bacteria, respectively. Such strand preferences are statistically significant (unpaired one-tailed t test, P<0.0001; Figure 5B). We also examined the linear correlations between LesGPs of horizontally-transferred genes and those of total genes in the two groups (Figure 5C and D). Although both groups exhibit significant positive correlations (P < 0.0001), the correlation coefficient of the polC bacteria is much greater (R = 0.70) than that of the non-polC bacteria (R = 0.46).
Figure 5.
Contribution of horizontally-transferred genes to SGD We show the proportion of horizontally-transferred genes (A) over all genes in each bacterium and the strand preference of horizontally-transferred genes of the non-polC (red) and polC (blue) bacterial groups (B) as box plots. The horizontal lines in the boxes, the open circles over and bellow the boxes, and the vertical scale bars indicate median, maximum, minimum, and quartiles, respectively. The LesGP between horizontally transferred genes and total genes of the non-polC (C) and polC bacterial groups (D) are linearly correlated.
Relationship between SGD and SNC
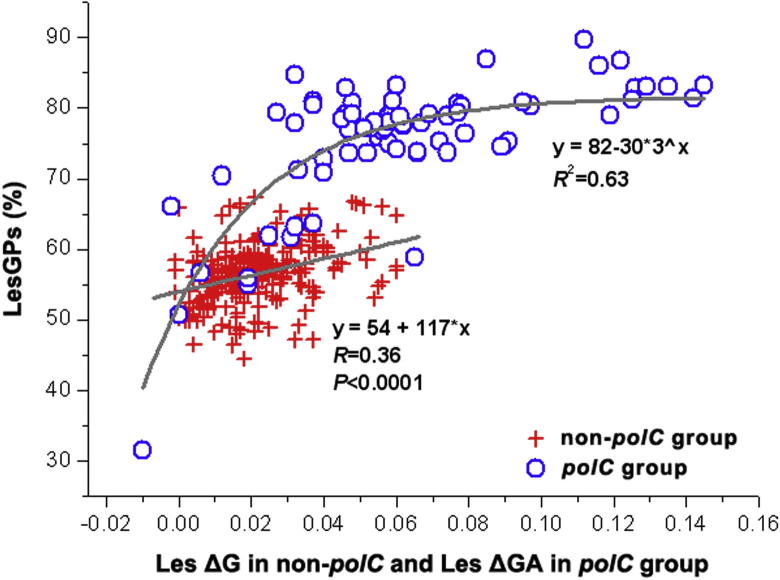
We further tested the correlation of LesGP and SNC, by using the differences in nucleotide composition between LeS and LaS: ΔA, ΔT, ΔC, and ΔG in the non-polC (Figure S2) and polC groups (Figure S3). It is clear that only the LeS G-dominance (ΔG or lagging-strand ΔC) (R = 0.36, P < 0.0001) of the non-polC bacteria contributes to SGD, whereas both G-dominance (ΔG; R2 = 0.49) and A-dominance (ΔA; R2 = 0.26) of the polC bacteria are positively correlated to LesGP. We further summarized this correlation by plotting ΔA in the non-polC group and ΔA + ΔG (x axis) against LesGP (y axis) in Figure 6.
Figure 6.
Correlation between SGD and SNC Correlations between LesGP and SNC are illustrated. Only ΔG (red crosses) in the non-polC group but both ΔG and ΔA (ΔGA; blue circles) in the polC group are positively correlated with LesGP. ΔG and ΔA stand for the nucleotide frequency differences between Les and LaS.
Mechanisms of SNC: selection vs. mutation
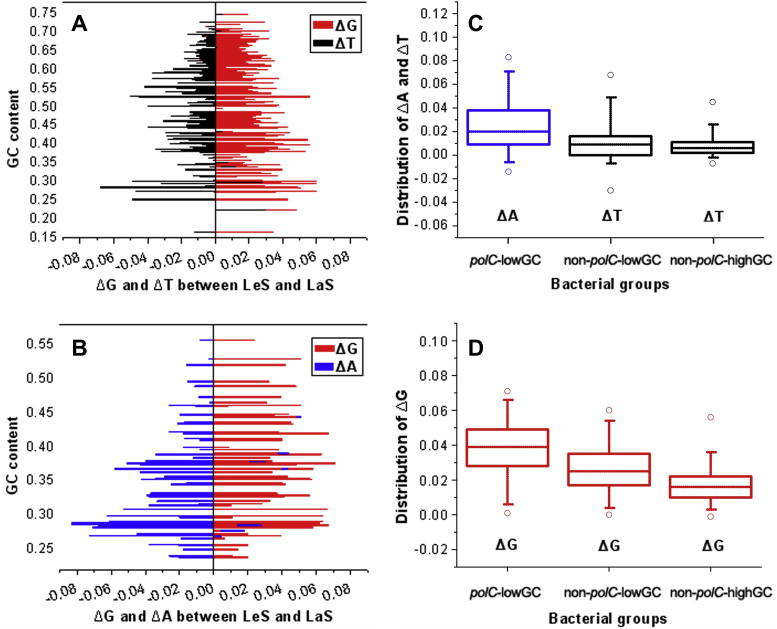
We further explored the relationship between genomic GC (gGC) content and SNC, aiming to reveal the relative roles of selection and mutation in SGD (Figure 7). In the non-polC group, the LeS ΔT ranges approximately from 0 to 0.02, and no obvious difference was found between the “non-polC-lowGC” and the “non-polC-highGC” groups (unpaired two tailed t-test, P > 0.05) (Figure 7C). The insensitivity of the LeS ΔT to gGC variation implies that LeS ΔT is independent of gGC and possibly caused by mutation. As to the LeS ΔG, the situation appears more complex. First, the LeS ΔG of the “non-polC-highGC” bacteria represents a basal level of ΔG around 0.01–0.03, which was elevated to 0.02–0.04 in the “non-polC-lowGC” bacteria (Figure 7D). Second, LeS ΔG further increases to 0.03–0.05 in the polC bacteria and such increases indicate that selection becomes more pronounced since a greater majority of bacteria in this group are extremely GC-poor. In addition, the LeS A-dominance only exists in the polC bacteria and correlates negatively with gGC content (R = −0.53, P < 0.0001; Figure 7B). Therefore, the basal level of LeS ΔG and ΔT (both around 0.02) is possibly shaped by mutation but not by selection. The effect of selection on ΔG and ΔA becomes more pronounced as gGC content decreases.
Figure 7.
Correlation between gGC content and SNC We show correlations between nucleotide dominance (LeS G- and T-dominances in Panel A and LeS G- and A-dominances in Panel B) and gGC content variations. ΔG (red), ΔT (black), and ΔA (blue) values are plotted as horizontal lines. The distribution of positive values of ΔT (C), ΔA (C), and ΔG (D) are shown in different grouping schemes. Results from both high gGC- (>50%) and low gGC-content (⩽50%) genomes in non-polC group as well as low gGC-content genomes only in the polC group are summarized.
Discussion
SGD: conservation vs. essentiality
What contributes to SGD of prokaryotic genomes is not straightforward but a combined effect of mutational and selective forces on genes; some are quantitative and some may be simply qualitative. That is to say, if there is no biased gene distribution on the two strands at the background level, SGD should be primarily affected by gene gain-and-loss regardless whether they are results of HGT or simple gene loss. Alternatively, if there is already a biased gene distribution, gene gain-and-loss may further balance this bias in non-polC bacteria, whereas it maintains or even intensifies the bias in polC bacteria. But how do bacterial genes translocate between the LeS and LaS and how does HGT affect SGD? To address these questions, we performed a detailed case study on conserved gene distributions of four phylogenetically related bacterial clades, two from each group, based on the fact that conserved genes tend to be essential [30].
In the non-polC group, the top 10% LesGPs of conserved genes in clade or subgroup S2 resulted from biased gene rearrangement (more genes tend to stay on the LeS in subgroup S2, as compared with S1), and such strand preference implies that inter-strand gene exchange is very frequent in bacterial genome evolution. Such unusual gene rearrangement may be replication-directed [31] and related to a rapid evolution process of the E. coli subgroup S2 as compared with the extreme genomic stasis of Buchnera subgroup S1 [32], which is reported to evolve from an enterobacterium-like ancestor some 200–250 million years ago [33], [34]. However, there is no obvious difference observed between LesGPs of total genes in these two subgroups (both averagely around 57%), suggesting that there must be other factors, such as HGT that is responsible for further stabilizing or balancing their SGD, around 50%–60%.
In the polC group, greater disparities in the LesGPs were observed not only when compared the conserved genes between the two subgroups (the LesGPs of 60% and 85% on average in S1 and S2, respectively) but also the total genes between the two subgroups (54% vs. 80.5%). We infer that it is the inter-subgroup divergences (the ancestor of S1 group bacteria vs. the ancestor of S2 group bacteria) not the intra-subgroup divergences that underly the obvious disparities between the LesGPs of these two subgroups. To be more specific, the longer branch length of the ancestor of the two δ-mycoplasma species suggests a longer evolutionary distance, and therefore there must have been a stronger selective pressure leading to the biased inter-strand gene arrangements and “strand jumping” where most LaS (20/28) and TL genes (92/108) in the subgroup S1 stay on the LeS in subgroup S2, resulting in the current higher LesGP. We can now draw a tentative conclusion that no matter what mechanism, either selection-based (rearrangement of conserved gene) or mutation-related (gene gain-and-loss), underlies a more biased gene distribution, they are all modulated to echo with the more biased gene distribution in the polC bacteria.
In addition, we found that among 125 essential genes of M. genitalium, all of which are verified experimentally, 98 (78%) are in our conserved gene dataset, which further consolidate the conclusion that essential genes tend to be conserved and there are significant overlaps between essential (conserved) and highly expressed genes, especially those organized as multi-gene operons [25]. A systematic effort is necessary to experimentally define an exhaustive list of highly expressed genes and operons, which are shared by most known bacteria. On the one hand, our analysis on the distribution of essential genes based on the DEG database supports that gene essentiality does have a significant influence on SGD, driving it up from 57% to 62% (5% increase) for the non-polC bacteria and from 75% to 89% (14% increase) for the polC bacteria. On the other hand, a higher level (27%) of average LesGP in the polC bacteria than in the non-polC bacteria demonstrates that gene essentiality is not the only contributor to SGD. Nevertheless, a stronger selection mechanism, process-based or function-based, may also contribute to this major effect among the polC group bacteria.
We also looked into conserved genes individually for well-annotated genomes and noticed that it is more likely to find conserved genes on LeS in the polC bacteria than in the non-polC bacteria. For instance, glnA has only 36% of the possibility to reside on LeS of non-polC bacteria but such possibility increases to as high as 89% in the polC bacteria. There are only a few exceptional cases, such as rpsD that has higher chance to be on LeS of non-polC bacteria and rnhB that has equal chance to stay on both strands. Therefore, we infer that there is indeed strand preference for conserved genes in the polC group bacteria, which contributes directly to SGD. Our KO function analysis on these genes verifies that there are also functional constraints on SGD [35], consistent with the contribution of gene conservation and essentiality.
SGD at dynamic level: new insights into HGT and gene loss
HGT and gene loss are two major forces that constantly alter gene repertoires of individual prokaryotic genomes [26], [27], [28], [29]. Site-specific recombination under the assistance of uptaking signal sequences has been reported to affect both orientation and efficiency of HGT [36], [37], [38]. Yet, no obvious evidence has been provided to describe whether SGD is related to HGT and gene loss. Fortunately, a large-scale identification effort for putative horizontally-transferred genes and a related database (HGT-DB) [39] offer us an opportunity to estimate their strand distributions. Our results indicate that although no significant statistical difference in the total proportion of horizontally-transferred genes between the non-polC and polC group bacteria (both ranging from 5% to 10%, even more than 20% in some bacteria), there is a strong strand preference for these genes. For example, the LesGP of horizontally-transferred genes in the non-polC bacteria is averaged as 58%, whereas it is as high as 78% for the polC bacteria. Therefore, unlike in the non-polC group, there are more genes that tend to be transferred to LeS in the polC bacteria, consistent with a previous study performed on Bacillaceae species (belong to the polC group) [40]. In addition, this can also be inferred from the fact that horizontally-transferred genes, different from its balancing role in the non-polC bacteria, can further maintain or even intensify LesGP in the polC bacteria. Nevertheless, it remains to be investigated whether the LeS preference of horizontally-transferred genes is related to the excess of uptaking signal sequences [41], [42]. In summary, horizontally-transferred genes correlate positively with SGD of both non-polC and polC bacteria but such correlation is stronger in the latter (R = 0.70) than in the former. We believe that this new observation will provide better insights into bacterial genome evolution.
As to the influence of gene loss to SGD, it is hard to measure quantitatively because of lacking systematical data acquisition methods and data curation. However, preliminary analysis in bacteria with gene numbers less than a cutoff value of 2500, which are intuitively regarded as bacteria once experienced dramatic genome reduction (or gene loss), may provide a window to look into the relative extent of gene losses in different strands. Our results demonstrate that although all these genomes have experienced extensive gene losses, the polC bacteria still exhibit a more biased gene distribution, such as B. aphidicola (non-polC group, with a LesGP about 57%) vs. M. genitalium (polC group, with a LesGP about 81%). Thus, a strand-biased gene loss is quite obvious.
Mutation vs. selection
Although it has been known that there is a LeS T-dominance [43] among non-polC bacteria, our analyses revealed that it has little to do with SGD and is independent of genomic GC content, suggesting that the LeS T-dominance originates most likely from mutations. However, a G-dominance in the same group is observed to be positively correlated with SGD and increases as gGC decreases from a basal content level similar to that of the LeS ΔT of the non-polC bacteria. We believe that ΔG is influenced by both mutation and selection but the effect of selection exerting on ΔG is more obvious in the polC bacteria that are generally GC-poor [12]. In addition, in the polC group bacteria, the LeS ΔA can be inferred to be largely introduced by selection since it only stands out in the GC-poor bacteria and correlates negatively with gGC variation. In addition, LeS ΔA is also found to underlie SGD of this bacterial group. It seems that the LeS A-dominance can explain the more pronounced LesGPs of this group.
A unified explanation for the underlying mechanisms of SGD
SGD is a complicated phenomenon that involves not only mutation-related forces, such as HGT, gene loss, purine asymmetry, and SNC, but also selection-related forces, such as process- and function-selection mechanisms, which either enhance or reduce the effect of mutation-related forces (Table 2). Our large-scale comparative analysis on 364 non-redundant bacterial genomes leads to a new stratification in addition to dnaE-based grouping: SGD distribution at both background and dynamic levels.
Table 2.
Common mechanisms underlying SGD
| Mechanisms | non-polC group | polC Group | |
|---|---|---|---|
| LesGP of total genes⁎ | 58% | 78% | |
| LesGP of essential genes⁎ | 62% | 89% | |
| LesGP of conserved genes⁎ | 68% | 92% | |
| LesGP of horizontally-transferred genes⁎ | 55% | 75% | |
| SNC vs. SGD | LeS ΔT | No correlation | – |
| LeS ΔA | – | R2 = 0.26 | |
| LeS ΔG | R = 0.36, P < 0.0001 | R2 = 0.49 | |
| LeS ΔG + ΔA | – | R2 = 0.63 | |
Note: “–” indicates that no nucleotide dominance was observed in the corresponding group.
Average LesGP.
At the background level, both process- and function-selections contribute to SGD, including essentiality and expressivity of genes, coupled with rearrangement and conservation. The replication-transcription collision avoidance hypothesis is an example to explain the effect of process-selections. Selection may work at three levels at least. First, selection can act at the gene level where functional and beneficial genes are acquired through HGT. Second, selection can act at the gene variation level where constant threat of loss-of-function mutations and positive selections on function-improving or advantageous mutations force genes to shift from one strand to the other. Third, selection can also act at the population level where fitness selects for a combined effect of advantageous genes and their variations. Essentially, genes are selected by their functions, regardless where they come from and how they change.
At the dynamic level, HGT and gene gain-and-loss are both biased between LeS and LaS, and the influences of their accountable events on SGD are more pronounced in the polC group than in the non-polC group. And we propose that SNC, being exhibited as a mutation spectrum, is responsible for placing pressure on protein composition changes. This hypothesis demonstrates that SGD of the non-polC bacteria is weakly selected, associated with a weak LeS G-dominance, whereas it involves much stronger purine dominance, as contributed by LeS G- and A-dominances, in the polC bacteria. Therefore, SGD among prokaryotic genomes is most likely subjected directly to both mutation and selection; some are weaker and others are stronger, and the degree measurement of SGD, SNC, and HGT in such a context is of importance for functional studies of both genes and genomes.
Materials and methods
The data
We retrieved genome sequences and annotations of 539 prokaryotes from NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/; July 17, 2007). After reducing redundancy by a random selection of a single representative isolate for a given species, we obtained 364 non-redundant bacterial genomes and classified them into non-polC (288) and polC groups (76) based on the absence or presence of polC in these genomes (Table S1). To examine the contribution of gene loss to SGD, we randomly selected 42 and 92 genomes, whose gene counts are less than 2500, from the polC and non-polC groups, respectively.
The definition of LeS gene proportion
The origin and terminus of replication, used to distinguish LeS and LaS, are determined based on two methods: extracting information from DoriC database (http://www.tubic.tju.edu.cn/doric/) [44] and re-annotating genome sequences by using Z-curve [45] when positional information is not available in DoriC.
For convenience, we define the two strands of bacterial chromosomes as positive (in 5′–3′ direction) and negative strands (in 3′–5′ direction) relatively to their replication origins, respectively. The number of genes on LeS of the positive strand is defined as N+les, and that of LaS of the positive strand is denoted as N+las. Similarly, the negative-strand associated parameter are N−les and N−las, and thus the total number of genes, Ntotal = N+les+N+las+N−les+N−las. Consequently, the leading strand gene proportion (LesGP) can be described as:
Defining conserved strand-associated genes
We constructed two datasets for analyzing conserved strand-associated gene distribution from two genera Buchnera and Mycoplasma as representatives of the non-polC and polC groups, respectively. The two datasets contain 690 protein-coding genes (484 of non-polC group and 206 of polC group) that are conserved and strand-associated, confirmed based on BlastP [46] with E value <1 × 10−5, identity >30%, and coverage >80%. Duplicated genes are excluded from this analysis. We chose these two genera for two reasons. First, bacteria of both groups are experiencing a massive gene loss but have different LesGPs. Second, they show reliable divergence time based on phylogenetic analysis of 16s rRNA sequences.
We also performed a genome-wide screening for conserved genes in bacteria. We selected 249 well-annotated genomes (for which >70% genes are annotated) as a dataset for the definition of conserved genes. And we obtained 239 genes that are present in at least 225 (∼90%) genomes. We annotated these genes using KEGG database (http://www.genome.jp/kegg/) [47].
Essential gene and horizontally-transferred gene distribution
All essential genes are retrieved from the DEG database (July 17, 2007) [24] and horizontally-transferred genes are retrieved from the HGT-DB database (July 17, 2007) [39]. Their distributions are calculated based on above-described methods.
Nucleotide compositional disparities
To examine SNC, we only chose the positive strand for calculation. The total number for each nucleotide i is denoted as LeSi for LeS, and LaSi for LaS, where i represents A, T, C, or G, and their corresponding total numbers of all nucleotides are labeled as LeStotal and LaStotal, respectively. Thus, for any given nucleotide i, LeS nucleotide composition can be calculated as Cles,i = LeSi/LeStotal, and that of LaS as Clas,i = LaSi/LaStotal. The relative bias of nucleotide i between LeS and LaS is formulated as Δi = Cles,i − Clas,i.
Authors’ contributions
HW carried out comparative genome analysis, calculation of leading strand gene proportion, and drafted the manuscript. HQ was involved in the leading and lagging strand annotation and KO gene function analysis. NW contributed to the visualization of inter-strand gene distribution. ZZ, SH, and JY designed and supervised the project and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Acknowledgements
We thank Dr. Tongwu Zhang and Dr. Yongjun Fang for helpful discussions. This study is supported by grants from Knowledge Innovation Program of the Chinese Academy of Sciences (Grant No. KSCX2-EW-R-01-04), Natural Science Foundation of China (Grant No. 90919024 and 30900831), the Ministry of Science and Technology of China as the National Science and Technology Key Project (Grant No. 2008ZX10004-013), the Special Foundation Work Program (Grant No. 2009FY120100), and the National Basic Research Program (Grant No. 2011CB944100).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.08.001.
Supplementary material
KO function analysis of 239 conserved genes. The Y axis shows gene abundance (measured by the number of genes) of metabolic pathways. The four gene-rich pathways are highlighted in red.
Correlation between LesGP of all genes and SNC in the non-polC group. The SNC is measured by calculating the nucleotide frequency differences (Δi) between LeS and LaS.
Correlation between LesGP of all genes and SNC in the polC group. We used similar methods for measuring SNC as what are illustrated in Figure S2.
GC contents and LesGPs of all 364 bacteria.
References
- 1.Lewin B. Genes IX. 9th ed. Sudbury, Mass, USA: Jones and Bartlett Publishers; 2008.
- 2.Hu J., Zhao X., Yu J. Replication-associated purine asymmetry may contribute to strand-biased gene distribution. Genomics. 2007;90:186–194. doi: 10.1016/j.ygeno.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Qu H., Wu H., Zhang T., Zhang Z., Hu S., Yu J. Nucleotide compositional asymmetry between the leading and lagging strands of eubacterial genomes. Res Microbiol. 2010;161:838–846. doi: 10.1016/j.resmic.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Rocha E.P. The organization of the bacterial genome. Annu Rev Genet. 2008;42:211–233. doi: 10.1146/annurev.genet.42.110807.091653. [DOI] [PubMed] [Google Scholar]
- 5.Trinh T.Q., Sinden R.R. Secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 6.Veaute X., Fuchs R.P. Greater susceptibility to mutations in lagging strand of DNA replication in Escherichia coli than in leading strand. Science. 1993;261:598–600. doi: 10.1126/science.8342022. [DOI] [PubMed] [Google Scholar]
- 7.V. Khrustalev V, V. Barkovsky E. A Blueprint for a Mutationist Theory of Replicative Strand Asymmetries Formation. Current Genomics 2012; 13: 55–64. [DOI] [PMC free article] [PubMed]
- 8.McHenry C.S. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem. 1988;57:519–550. doi: 10.1146/annurev.bi.57.070188.002511. [DOI] [PubMed] [Google Scholar]
- 9.Koonin E.V., Bork P. Ancient duplication of DNA polymerase inferred from analysis of complete bacterial genomes. Trends Biochem Sci. 1996;21:128–129. [PubMed] [Google Scholar]
- 10.Dervyn E., Suski C., Daniel R., Bruand C., Chapuis J., Errington J. Two essential DNA polymerases at the bacterial replication fork. Science. 2001;294:1716–1719. doi: 10.1126/science.1066351. [DOI] [PubMed] [Google Scholar]
- 11.Zhao X., Hu J., Yu J. Comparative analysis of eubacterial DNA polymerase III alpha subunits. Genomics Proteomics Bioinformatics. 2006;4:203–211. doi: 10.1016/S1672-0229(07)60001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H., Zhang Z., Hu S., Yu J. On the Molecular Mechanism of GC Content Variation among Eubacterial Genomes. Biology Direct. 2012;7:2. doi: 10.1186/1745-6150-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J., Zhao X., Zhang Z., Yu J. Compositional dynamics of guanine and cytosine content in prokaryotic genomes. Res Microbiol. 2007;158:363–370. doi: 10.1016/j.resmic.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X., Zhang Z., Yan J., Yu J. GC content variability of eubacteria is governed by the pol III alpha subunit. Biochem Biophys Res Commun. 2007;356:20–25. doi: 10.1016/j.bbrc.2007.02.109. [DOI] [PubMed] [Google Scholar]
- 15.Omont N., Kepes F. Transcription/replication collisions cause bacterial transcription units to be longer on the leading strand of replication. Bioinformatics. 2004;20:2719–2725. doi: 10.1093/bioinformatics/bth317. [DOI] [PubMed] [Google Scholar]
- 16.Mirkin E.V., Mirkin S.M. Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol. 2005;25:888–895. doi: 10.1128/MCB.25.3.888-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J.D., Berkmen M.B., Grossman A.D. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:5608–5613. doi: 10.1073/pnas.0608999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brewer B.J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 19.McLean M.J., Wolfe K.H., Devine K.M. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- 20.Price M.N., Alm E.J., Arkin A.P. Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res. 2005;33:3224–3234. doi: 10.1093/nar/gki638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocha E.P., Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 22.Rocha E.P., Danchin A. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 2003;31:6570–6577. doi: 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocha E. Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes? Trends Microbiol. 2002;10:393–395. doi: 10.1016/s0966-842x(02)02420-4. [DOI] [PubMed] [Google Scholar]
- 24.Zhang R., Ou H.Y., Zhang C.T. DEG: a database of essential genes. Nucleic Acids Res. 2004;32:D271–D272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinuelas J., Calevro F., Remond D., Bernillon J., Rahbe Y., Febvay G. Conservation of the links between gene transcription and chromosomal organization in the highly reduced genome of Buchnera aphidicola. BMC Genomics. 2007;8:143. doi: 10.1186/1471-2164-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran N.A. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 27.Ochman H., Lawrence J.G., Groisman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 28.Koonin E.V., Wolf Y.I. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunin V., Ouzounis C.A. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13:1589–1594. doi: 10.1101/gr.1092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan I.K., Rogozin I.B., Wolf Y.I., Koonin E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tillier E.R., Collins R.A. Genome rearrangement by replication-directed translocation. Nat Genet. 2000;26:195–197. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- 32.Tamas I., Klasson L., Canback B., Naslund A.K., Eriksson A.S., Wernegreen J.J. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 33.Moran N.A., Munson M.A., Baumann P., Ishikawa H. A Molecular Clock in Endosymbiotic Bacteria is Calibrated Using the Insect Hosts. Proceedings of the Royal Society of London Series B: Biological Sciences. 1993;253:167–171. [Google Scholar]
- 34.van Ham R.C., Kamerbeek J., Palacios C., Rausell C., Abascal F., Bastolla U. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci U S A. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y., Gao F., Zhang C.T. Functionality of essential genes drives gene strand-bias in bacterial genomes. Biochem Biophys Res Commun. 2010;396:472–476. doi: 10.1016/j.bbrc.2010.04.119. [DOI] [PubMed] [Google Scholar]
- 36.Smith H.O., Tomb J.F., Dougherty B.A., Fleischmann R.D., Venter J.C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 37.de Vries J., Wackernagel W. Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc Natl Acad Sci U S A. 2002;99:2094–2099. doi: 10.1073/pnas.042263399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidsen T., Rodland E.A., Lagesen K., Seeberg E., Rognes T., Tonjum T. Biased distribution of DNA uptake sequences towards genome maintenance genes. Nucleic Acids Res. 2004;32:1050–1058. doi: 10.1093/nar/gkh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Vallve S., Guzman E., Montero M.A., Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 2003;31:187–189. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao W., Golding G.B. Does gene translocation accelerate the evolution of laterally transferred genes? Genetics. 2009;182:1365–1375. doi: 10.1534/genetics.109.104216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Orvis J., Dyer D., Chen C. Genomic distribution and functions of uptake signal sequences in Actinobacillus actinomycetemcomitans. Microbiology. 2006;152:3319–3325. doi: 10.1099/mic.0.29018-0. [DOI] [PubMed] [Google Scholar]
- 42.Treangen T.J., Ambur O.H., Tonjum T., Rocha E.P. The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome Biol. 2008;9:R60. doi: 10.1186/gb-2008-9-3-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 44.Gao F., Zhang C.T. DoriC: a database of oriC regions in bacterial genomes. Bioinformatics. 2007;23:1866–1867. doi: 10.1093/bioinformatics/btm255. [DOI] [PubMed] [Google Scholar]
- 45.Guo F.B., Ou H.Y., Zhang C.T. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 2003;31:1780–1789. doi: 10.1093/nar/gkg254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
KO function analysis of 239 conserved genes. The Y axis shows gene abundance (measured by the number of genes) of metabolic pathways. The four gene-rich pathways are highlighted in red.
Correlation between LesGP of all genes and SNC in the non-polC group. The SNC is measured by calculating the nucleotide frequency differences (Δi) between LeS and LaS.
Correlation between LesGP of all genes and SNC in the polC group. We used similar methods for measuring SNC as what are illustrated in Figure S2.
GC contents and LesGPs of all 364 bacteria.