Abstract
Immortality and tumorigenicity are two distinct characteristics of cancers. Immortalization has been suggested to precede tumorigenesis. To understand the molecular mechanisms of tumorigenicity and cancer progression in mammary epithelium, we established a tumorigenic cell model by means of heavy-ion radiation of an immortal cell model, which was created by overexpressing the human telomerase reverse transcriptase (hTERT) in normal human mammary epithelial cells. We examined the expression profile of this tumorigenic cell line (T_hMEC) using the hTERT-overexpressing immortal cell line (I_hMEC) as a control. In-depth RNA-seq data was generated by using the next-generation sequencing (NGS) platform (Life Technologies SOLiD3). We found that house-keeping (HK) and tissue-specific (TS) genes were differentially regulated during the tumorigenic process. HK genes tended to be activated while TS genes tended to be repressed. In addition, the HK genes and TS genes tended to contribute differentially to the variation of gene expression at different RPKM (gene expression in reads per exon kilobase per million mapped sequence reads) levels. Based on transcriptome analysis of the two cell lines, we defined 7053 differentially-expressed genes (DEGs) between immortality and tumorigenicity. Differential expression of 20 manually-selected genes was further validated using qRT-PCR. Our observations may help to further our understanding of cellular mechanism(s) in the transition from immortalization to tumorigenesis.
Keywords: NGS, RNA-seq, Tumorigenicity, Immortality, Radiation, Breast cancer
Introduction
Cancers are life-threatening diseases with enormous complexities, which involve dynamic changes in the genome at both genetic and epigenetic levels [1], [2]. Although numerous types and grades of cancers have been defined on the basis of their origin, ample studies suggest that they may share certain routes to malignant transformation, such as chromosome instability, self-sufficiency, insensitivity to antigrowth signals, unlimited replicative potential, apoptosis evasion, sustained angiogenesis and tissue invasion (or metastasis) [3], [4]. In addition, many lines of evidence indicate that tumorigenesis in human cancers is a multistep process. Genetic and epigenetic alterations accumulate at each step, which drive progressive transformation of normal human cells into highly malignant derivatives [2], [5], [6]. Among these steps, there are two distinct phenomena: immortality and tumorigenicity. Immortalization occurs early during tumor progression [7], and tumorigenic cells are always transformed from immortal cells [5], [6], [8]. There are substantial differences between immortality and tumorigenicity, and many genes are differentially expressed when comparing immortal and tumorigenic cells [9], [10], [11], [12].
Immortalization does not necessarily confer tumorigenicity but tumorigenicity is the prerequisite to cancer development and remains the prime problem in cancer treatment. Therefore, elucidation of the mechanisms underlying tumorigenicity, especially identification of tumorigenicity-associated genes, is essential for facilitating early diagnosis and effective therapy. Previous studies indicated that the expression of human telomerase reverse transcriptase (hTERT) is significantly higher in certain cancerous tissues than in non-cancerous tissues [13], [14]. Moreover, overexpression of hTERT in normal and telomerase-negative cells often induces immortalization [8], [15]. We therefore took advantage of hTERT-induced immortalization to examine a specific tumorigenic effect: radiation-induced tumorigenesis of hTERT-immortalized cells.
Breast cancer is the most common cancer among women worldwide, comprising 16% of all female cancers (http://www.who.int/cancer/detection/breastcancer/en/index1.html). Development of appropriate model systems is therefore critical for understanding the molecular mechanisms of breast cancer. In this study, we induced normal human mammary epithelial cells (hMEC) to become immortal by overexpressing the hTERT gene (I_hMEC), and then performed heavy ion radiation on the immortal cell model to produce tumorigenic cells (T_hMEC). Given the sensitivity and accuracy of the next-generation sequencing (NGS) platforms [16], [17], we performed transcriptome analysis of I-hMEC and T-hMEC using RNA-seq based on NGS to identify genes and the possible molecular mechanisms involved in breast cancer tumorigenicity.
Results
RNA-seq and sequence tag mapping
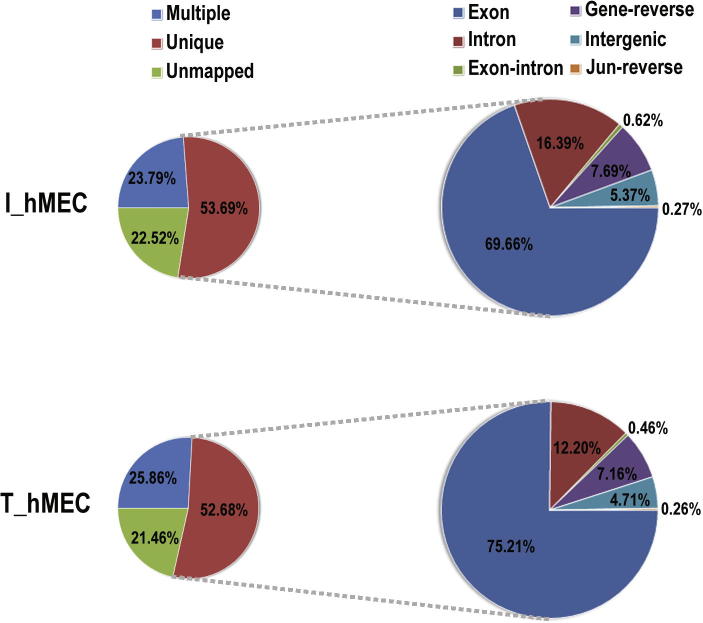
We used poly-A purified mRNAs for this study from the two human mammary epithelial cell lines: the hTERT-induced immortal cell line (I_hMEC) and tumorigenic cell line (T_hMEC). After quality filtering, we obtained 51,895,024 (92.47%) out of 56,121,440 tags from I_hMEC and 47,177,391 (93.33%) out of 50,549,359 tags from T_hMEC. We then mapped these tags to the human genome and a database of unique exon-junction sequences generated in this study (see Materials and methods), using the SOLiD sequencing system (Life Technologies, Foster City, CA). Among the high-quality tags, majority of the reads (about 80%) were mapped to the genome assembly, and about 50% of the tags in each library (about 25 million tags) were mapped to unique locations (unique tags), which are sufficient for quantitative analysis of genes covering all biologically-relevant abundance classes [18], [19]. For comparison of gene expression, we focused on the tags that are uniquely mapped to exons. In each library, about 70% of the uniquely mapped tags are confined to exons, with the remaining mapped to intronic, reverse, and intergenic sequences (Figure 1).
Figure 1.
Mapping summary of the two cell models for breast cancer Left panels show percentages of tags mapped to multiple loci and to unique loci and unmapped tags. Panels on the right show detailed mapping results of uniquely-mapped tags. Exon: tags mapped to exonic region; Intron: tags mapped to intronic region; Exon–intron: tags mapped to exon–intron junctions; Gene-reverse: tags reversely mapped to gene; Intergenic: tags mapped to intergenic region; Jun-reverse: tags reversely mapped to exon-exon junctions. I_hMEC represents immortalized human mammary epithelial cells and T_hMEC represents tumorigenic human mammary epithelial cells.
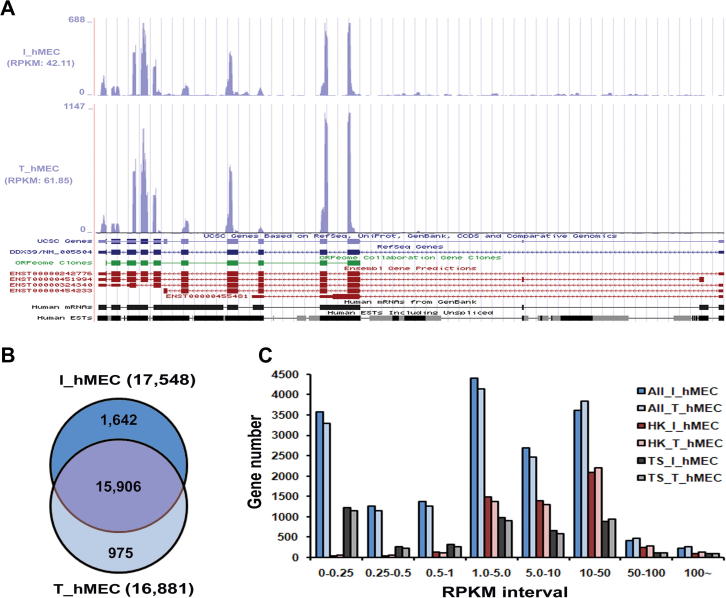
RPKM (gene expression in reads per exon kilobase per million mapped sequence reads) value was used to represent the expression level for each gene [18]. Comparison of RPKM and tag coverage (the normalized number of mapped tags in each genomic region based on the total tag count in each library) showed that RPKM exhibited results consistent with tag coverage in both libraries. Taking the gene ddx39 as an example, number of mapped tags increased greatly from I_hMEC to T_hMEC for almost all exons although tag coverage varied drastically between exons (Figure 2A). Similarly, the RPKM values of this gene increased from 42.11 (I_hMEC) to 61.85 (T_hMEC). Fragmentation of the oligo-dT primed cDNA is suggested to be more biased towards the 3′ end of the transcript [20]. We therefore fragmented oligo-dT primed RNA instead of cDNA during library construction. As shown in Figure 2A, tag coverage for exons is not obviously biased towards the 3′ end. Therefore, the improvement of experimental methods may contribute to improved accuracy of the RPKM measurement that helps to provide more reliable results.
Figure 2.
Overview of gene expression in immortal and tumorigenic cells A. Sequencing tag distribution in two cell models I_hMEC and T_hMEC using ddx39 as an example. Genes are annotated based on information from UCSC, Refseq and Ensembl. B. Venn diagram of genes expressed in I-hMEC and T_hMEC. C. RPKM distribution and variation between I-hMEC and T_hMEC. I_hMEC is an immortalized human mammary epithelial cell line and T_hMEC is a tumorigenic human mammary epithelial cell line.
We then evaluated the expression profile of each library. In I_hMEC, 27,198 and 17,548 transcripts are verified by at least one and at least five tags, respectively, and similar numbers are mapped in T_hMEC, which are 26,474 and 16,881. To avoid background noise and sequencing errors, we limited our analysis to genes with expression verified by five or more tags. According to this criterion, 18,523 transcripts were included, among which, 15,906 (85.87%) are shared by both libraries and 2617 (14.13%) are unique. Notably, more genes are uniquely expressed in I_hMEC than in T_hMEC (Figure 2B).
We also examined the expression abundance in each library (Figure 2C) and found that genes were often expressed at two obvious RPKM intervals: 0–0.25 and 1.0–50. The RPKM value of 10 appears to be a critical point because T_hMEC expresses more genes whose RPKM value is greater than 10, while I_hMEC expresses more genes whose RPKM value is smaller than 10. As house-keeping (HK) genes and tissue-specific (TS) genes are expressed at different levels and may both contribute to cancer development [21], [22], we therefore examined gene expression for these two groups separately. We identified 6003 HK and 7982 TS genes (see Materials and methods, and Table S1). Consistent with the results of the analysis on all expressed genes, RPKM value of 10 appears as a turning point for both HK genes and TS genes (Figure 2C). These results indicate that both HK genes and TS genes are expressed at different levels between immortal cell and tumorigenic cell.
Expression modulation during the tumorigenic process
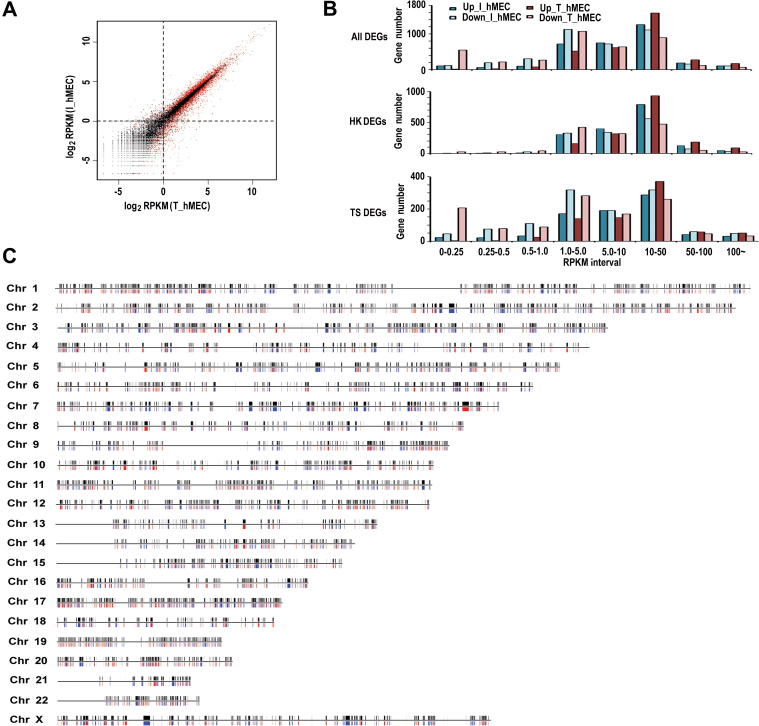
We first investigated the expression correlation between the two libraries (I_hMEC and T_hMEC), using genes detected by at least one tag. It is clearly shown that the expression of transcripts with RPKM values greater than 1.0 in both libraries is well-correlated (r2 = 0.91), while genes with RPKM values smaller than 1.0 exhibit weak correlation (r2 = 0.24) (Figure 3A). The weak correlation for poorly-expressed genes may be attributed to sampling bias.
Figure 3.
DEGs between immortal cell and tumorigenic cell A. Scatter-plot of gene expression abundance. DEGs are highlighted in red to differentiate from the background (black). Genes that are mapped by at least one tag in its exonic region were evaluated. Genes with different RPKM values exhibit different correlations between I_hMEC and T_hMEC. The coefficient – r2 is 0.91 (P < 0.001) for genes with RPKM >1.0 but only 0.24 (P < 0.001) for genes with RPKM <1.0 in both libraries. B. Gene expression abundance for different DEGs in each library. Up-regulated DEGs during the tumorigenic process are referred as Up_I_hMEC in I-hMEC and Up_T_hMEC in T_hMEC; Down-regulated DEGs during the tumorigenic process are referred as Down_I_hMEC in I-hMEC and Down_T_hMEC in T_hMEC, respectively. C. DEG distribution in human chromosomes. Black bars above the horizontal line represent all DEGs; red and blue bars represent up-regulated and down-regulated DEGs, respectively.
We identified 7053 DEGs using the software DEGseq with P < 0.001 (Figure 3A). In particular, expression of 3804 (53.93%) genes was repressed or down-regulated, while expression of the remaining 3249 (46.07%) genes was activated or up-regulated in T_hMEC (Table S2). In I_hMEC, more genes with RPKM >10.0 are up-regulated, while more genes with RPKM <5.0 are down-regulated during tumorigenic process. As a result, more genes are expressed at RPKM <0.25 and >10 in T_hMEC (Figure 3B). This may help to explain the previous observation that RPKM 10 acts as a turning point between I_hMEC and T_hMEC (Figure 2C). Therefore, the turning point, RPKM 10, is mostly due to different regulation of genes at different RPKM intervals.
To further investigate whether HK genes and TS genes influence the transcription landscape between the two cell lines, we compared DEGs among different RPKM intervals and found that HK genes and TS genes contributed differently to the transcriptome landscape variation during the tumorigenic process. For example, 55.02% of HK genes are up-regulated, while only 40.52% TS genes are up-regulated. Moreover, expression of more HK genes with RPKM >5.0 was up-regulated such that there are more HK genes with RPKM >10 in T_hMEC. However, expression of more TS genes was repressed at all RPKM intervals indicated, and in particular, we found that the 0–0.25 RPKM interval in T_hMEC contains mostly down-regulated TS genes in the tumorigenic process (Figure 3B). This result indicates that HK genes and TS genes are differentially regulated in tumorigenesis: HK genes contribute more to the pool of up-regulated genes, whereas TS genes contribute more to the pool of down-regulated genes. Such different tendency between HK genes and TS genes may have significant impact on the transcription landscape and is also help to explain the turning point of RPKM 10.
As genes from different chromosomes may contribute differentially to tumorigenesis, we further examined the distribution of DEGs in each chromosome (Figure 3C and Table S3). For instance, chromosome 18 contains the highest percentage of down-regulated genes. Conversely, chromosome 17 contains the highest percentage of up-regulated genes and all DEGs, respectively. High percentage of all DEGs was also observed on chromosomes 12, 19, 18, 16, 5, 11, and 8 (range from high to low). Further examination revealed that the three breast cancer-associated genes—BRCA1 [23], ERBB2 [24] and P53 [25]—are all located on chromosome 17, and another important gene BRCA2 is on chromosome 13 [26]. According to our data, these genes are differentially regulated during tumorigenesis: BRCA1 is up-regulated, as opposed to ERBB2 and P53, which are down-regulated. However, we did not find a significant difference in BRCA2 expression between I_hMEC and T_hMEC. Additionally, we found many other cancer-associated genes among DEGs on chromosome 17 (Table S4). Our results agree with previous studies that genetic alterations of chromosome 17 are the most frequent changes identified in breast cancers and more tumor-associated genes may reside on this chromosome [25], [27], [28]. Therefore, genes on chromosome 17 may play an important role in tumorigenesis.
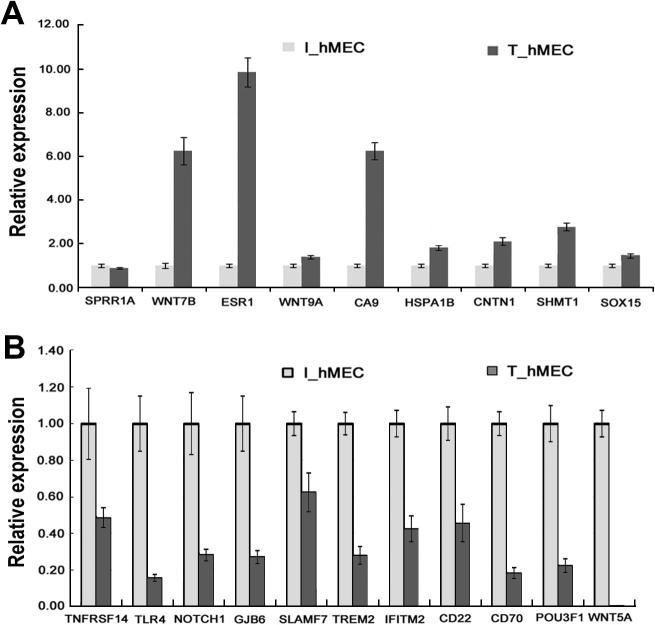
To validate the RNA-seq results, we manually selected 20 significantly-varied DEGs according to fold change and expression level and performed quantitative real-time PCR on these genes (Figure 4). In particular, we selected 6 lowly-expressed genes with a fold change of more than 5 (RPKM <2) (the higher RPKM in the two libraries was used to represent the expression level of a particular gene) such as ESR1 and POU3F1, 6 genes expressed in the medium range with a fold change of more than 3 (2< RPKM <10) such as TNFRSF14 and WNT7B, and 8 highly-expressed genes with a fold change of more than 2 (RPKM >10) such as NOTCH1 and SOX15. Our qRT-PCR results indicated that expression of most genes (19 out of 20) was consistent with that shown by RNA-seq. Therefore, significantly-varied DEGs can be validated even at relatively low expression levels. However, a single gene (SPRR1A) failed the test, although the expression of SPRR1A is not very low (RPKM is 0.31 for I_hMEC and 3.48 for T_hMEC). It has been reported that a large number of both non-protein-coding regions and protein-coding sequences are transcribed into long non-coding RNAs [29], which may affect our mapping results and the way differentially expressed genes are defined. However, long non-coding RNAs are supposed to be mostly expressed at low levels [30], implying that significant influences may only be confined to limited number of genes.
Figure 4.
qPCR validation of up-regulated and down-regulated DEGs Relative expression was validated for up-regulated DEGs (A) and down-regulated DEGs (B), respectively, using qPCR. Expression of selected genes was normalized using GSK3A as the internal control. Data was shown as mean ± SD of three independent experiments.
Function categorization of DEGs
We further compared gene function categories between up-regulated and down-regulated genes using enrichment analysis of GO through WebGestalt [31]. The GO enrichment analysis helps to find which GO terms are overrepresented in a large gene list. According to the results, DNA replication, RNA transcription, and translation are significantly and also specifically enriched in up-regulated genes, together with nucleotide metabolism and ribonucleo protein biogenesis, all of which are characteristic of cell proliferation. In contrast, the down-regulated genes are significantly and specifically enriched in cell communication, signal transduction, cell adhesion and migration. In addition, most cell cycle arrest genes and positive regulators of apoptosis are down-regulated (Table 1). These results indicate that tumorigenic cells are heavily tuned into a proliferating mode, ignoring cell communication and adhesion.
Table 1.
Enriched biological processes in GO terms involving up- and down-regulated genes during tumorigenesis
| Category | GO ID | Statistics |
|---|---|---|
| Up-regulated | ||
| DNA replication | GO:0006260 | C = 226; O = 85; E = 38.56; R = 2.20; rawP = 8.67e−14; adjP = 6.42e−12 |
| Gene expression | GO:0010467 | C = 3672; O = 867; E = 626.57; R = 1.38; rawP = 8.43e−33; adjP = 1.62e−30 |
| RNA processing | GO:0006396 | C = 556; O = 261; E = 94.87; R = 2.75; rawP = 1.12e−62; adjP = 3.23e−59 |
| RNA splicing | GO:0008380 | C = 292; O = 144; E = 49.82; R = 2.89; rawP = 1.20e−37; adjP = 5.78e−35 |
| Translation | GO:0006412 | C = 410; O = 152; E = 69.96; R = 2.17; rawP = 5.81e−23; adjP = 7.30e−21 |
| Ribonucleoprotein complex biogenesis | GO:0022613 | C = 180; O = 113; E = 30.71; R = 3.68; rawP = 3.13e−43; adjP = 2.26e−40 |
| Macromolecule metabolic process | GO:0043170 | C = 6304; O = 1368; E = 1075.67; R = 1.27; rawP = 6.80e−39; adjP = 3.93e−36 |
| Ribosome biogenesis | GO:0042254 | C = 122; O = 80; E = 20.82; R = 3.84; rawP = 6.58e−33; adjP = 1.36e−30 |
| Ribonucleoprotein complex assembly | GO:0022618 | C = 69; O = 40; E = 11.77; R = 3.40; rawP = 1.93e−14; adjP = 1.80e−12 |
| Cellular metabolic process | GO:0044237 | C = 7309; O = 1566; E = 1247.16; R = 1.26; rawP = 4.22e−46; adjP = 4.77e−43 |
| Nitrogen compound metabolic process | GO:0006807 | C = 4378; O = 971; E = 747.03; R = 1.30; rawP = 2.03e−26; adjP = 2.79e−24 |
| DNA metabolic process | GO:0006259 | C = 552; O = 169; E = 94.19; R = 1.79; rawP = 9.52e−16; adjP = 9.16e−14 |
| RNA metabolic process | GO:0016070 | C = 2486; O = 580; E = 424.19; R = 1.37; rawP = 6.05e−19; adjP = 6.99e−17 |
| Cell cycle | GO:0007049 | C = 909; O = 268; E = 155.11; R = 1.73; rawP = 6.14e−22; adjP = 7.39e−20 |
| Regulation of ligase activity | GO:0051340 | C = 78; O = 42; E = 13.31; R = 3.16; rawP = 1.42e−13; adjP = 1.00e−11 |
| Down-regulated | ||
| Regulation of cell communication | GO:0010646 | C = 1003; O = 250; E = 185.59; R = 1.35; rawP = 9.74e−08; adjP = 4.77e−05 |
| Intracellular signaling cascade | GO:0007242 | C = 1565; O = 365; E = 289.58; R = 1.26; rawP = 2.37e−07; adjP = 9.03e−05 |
| Small GTPase mediated signal transduction | GO:0007264 | C = 502; O = 138; E = 92.89; R = 1.49; rawP = 3.27e−07; adjP = 0.0001 |
| Anatomical structure morphogenesis | GO:0009653 | C = 1223; O = 285; E = 226.30; R = 1.26; rawP = 6.31e−06; adjP = 0.0006 |
| Cellular response to chemical stimulus | GO:0070887 | C = 285; O = 83; E = 52.74; R = 1.57; rawP = 7.13e−06; adjP = 0.0006 |
| Cell migration | GO:0016477 | C = 371; O = 104; E = 68.65; R = 1.51; rawP = 3.58e−06; adjP = 0.0004 |
| Cell-matrix adhesion | GO:0007160 | C = 108; O = 43; E = 19.98; R = 2.15; rawP = 1.83e−07; adjP = 7.84e−05 |
| Positive regulation of apoptosis | GO:0043065 | C = 420; O = 118; E = 77.72; R = 1.52; rawP = 7.11e−07; adjP = 0.0001 |
| Protein localization | GO:0008104 | C = 962; O = 232; E = 178.01; R = 1.30; rawP = 4.09e−06; adjP = 0.0004 |
| Protein modification process | GO:0006464 | C = 1529; O = 380; E = 282.92; R = 1.34; rawP = 3.78e−11; adjP = 1.30e−07 |
| Phosphate metabolic process | GO:0006796 | C = 1225; O = 289; E = 226.67; R = 1.27; rawP = 1.84e−06; adjP = 0.0002 |
| Cell cycle arrest | GO:0007050 | C = 104; O = 40; E = 19.24; R = 2.08; rawP = 1.39e−06; adjP = 0.0002 |
| Positive regulation of biological process | GO:0048518 | C = 1865; O = 417; E = 345.10; R = 1.21; rawP = 3.82e−06; adjP = 0.0004 |
Note: C, the number of reference genes in the category; O, the number of genes in the gene set and also in the category; E, the expected number in the category; R, ratio of enrichment; rawP, the P value from hypergeometric test; and adjP, the P value adjusted by multiple test adjustment.
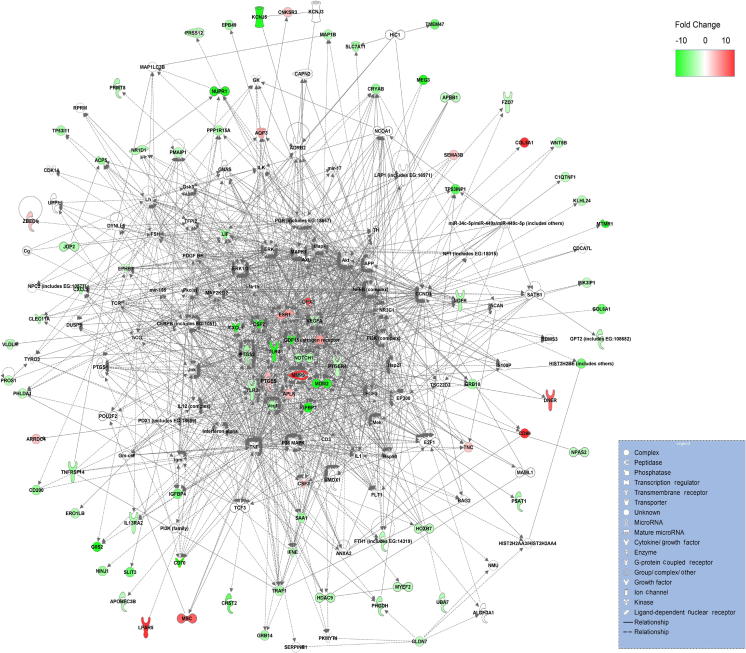
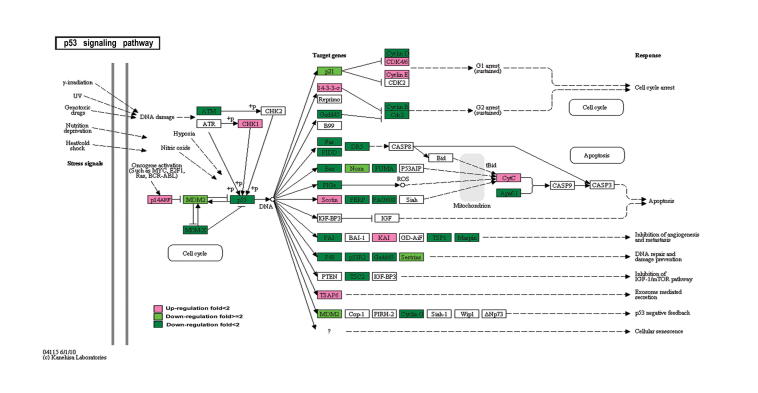
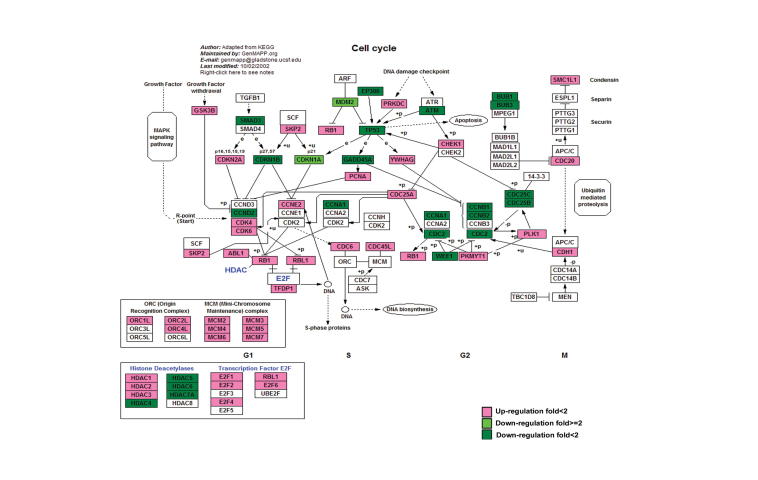
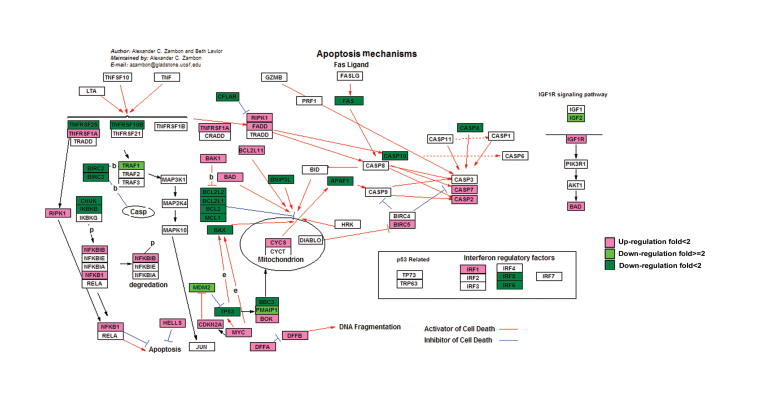
Many DEGs identified in this study are involved in the important processes in cancer development such as cell cycle, apoptosis, and p53 signaling pathway (Figures S1–S3). Deterministic mechanisms between immortality and tumorigenicity may lie in the significantly-regulated genes and these genes would have great potential as useful biomarkers. We thus further investigated the significantly-regulated DEGs (fold change ⩾2, and the sum of RPKM values of the two libraries is greater than 1.5). Accordingly, 364 genes were selected, including 270 down-regulated and 94 up-regulated genes (Table S5). KEGG enrichment analysis showed that 12 significantly-regulated genes are involved in “pathways in cancer” (Table S6). Since our cell models are established to simulate the progression of breast cancer, these data suggest that these 364 genes may play an important function in tumorigenicity. In addition, we performed gene interaction network analysis using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) on the 364 significantly regulated genes (Figure 5). We placed genes that always interact with other genes in the core modules of the network, and found that 16 significantly-regulated genes are in the core modules. In particular, expression of ESR1, GPX3, PTGE3, MMP9, and APLN was significantly up-regulated, while expression of CXCL5, CSF2, VEGFA, PTGS2, TLR4, NOTCH1, PTGER4, TLR2, MDM2, IGFBP2, and GDF15 was significantly down-regulated. Although ESR1 is expressed at a low level, its differential expression has been verified using qRT-PCR experiments (Figure 4).
Figure 5.
Gene interaction networks of the highly-regulated DEGs We chose the top five networks based on their scores (generated using the software of Ingenuity Pathways Analysis) and merged the networks into one figure. The degree of DEG modulation (we calculated fold changes based on RPKM values) is indicated with color intensity. Solid lines indicate direct regulations and dashed lines indicate indirect regulations. Arrows point to downstream genes.
Discussion
Here we describe a cell model for identification of tumorigenicity-associated genes in breast cancers in terms of immortalization by using a combined action of over-expression of hTERT and heavy-ion radiation. Radiation of immortal cells can induce phenotypical alterations—the abilities to grow in soft agar and to form fast-growing carcinomas in nude mice [5], [6]. Therefore, this cell model becomes instrumental in studying cancer development at different molecular levels and in greater depth, and our RNA-seq-based transcriptomics analysis is the first step to a systematic study.
The RNA-seq sequencing method and the improvement in experimental technologies both facilitate to obtain more accurate results. The RNA-seq sequencing method correlates well with the microarray method and can detect more genes when compared with microarray data [16], [20]. Additionally, low background noise enables people to obtain relatively accurate results without performing replicates [16], [32]. Also, oligo-dT primed RNA instead of oligo-dT primed cDNA was fragmented to avoid sequencing bias in the 3′ end of the transcript [20]. In this study, we identified 7053 DEGs with DEGseq at a P value of <0.001, and validated the reliability of SOLiD-based RNA-seq results using quantitative real-time PCR (19 out of 20 candidate genes were validated for their differential expression, Figure 4).
In general, differential expression between TS genes and HK genes and the gene location on chromosomes, both contribute to the progression of breast cancer. According to our results, more TS genes are repressed (although most TS genes are already lowly expressed) while more HK genes are activated (especially for those that are highly expressed) during the tumorigenic process. Previous study suggested that molecular signature of disease across tissues is overall more prominent than the signature of tissue expression across diseases [33]. During cancer progression, specialization in cancerous tissues dropped due to a decrease in expression of genes that are highly specific to the normal organ [21]. In addition, we found that certain human chromosomes (or genes residing therein) may play critical roles, compared to other chromosomes during tumorigenic transformation. For example, chromosome 17 is the most frequently changed chromosome in breast cancers and more tumor-associated genes are found in this chromosome [25], [27], [28]. Consistently, we found chromosome 17 contains the highest number of DEGs during tumorigenic process.
As knowledge of mechanisms underlying breast cancer is quite limited, the search for new molecular markers is currently ongoing. Our results are consistent with previously detected markers, such as thrombospondin 1 (TSP-1, down-regulated during tumorigenicity process) [9], [10] and keratin 5 (K5, up-regulated during tumorigenicity process) [9]. Therefore, the modulation pattern of TSP-1 and K5 may be used as more reliable biomarkers for tumorigenicity. Nonetheless, there are exceptions too. For example, FN1 (fibronectin) is down-regulated in our cell model instead of up-regulated [9]. According to the existing evidence, carcinogenesis is a multistep process [2], [5], [6], however, the whole process may be constantly changing and it may be impossible to ensure that all the tumorigenicity associated genes change synchronously. On the other hand, different DEGs in tumorigenic process may be identified due to different origins of tumorigenic cell systems. In addition, we identified 145 cancer-related DEGs (Table S4) and 364 highly regulated genes (Table S5). Although more validation experiments are required for these genes, the current study may help to facilitate further investigations and provide new insights into breast cancer progression mechanisms. Future studies will address how these genes are up-regulated or down-regulated through regulatory pathways and the possible epigenetic mechanisms involved.
Materials and methods
Sample preparation and sequencing
Normal human mammary epithelial cells (hMECs) were purchased from Clonetics BioWhittaker (Walkersville, MD) and maintained in serum-free mammary epithelial basal medium supplemented with growth factors. hMEC were immortalized as described in our previous study [8]. Briefly, we shuffled hTERT cDNA from pZeoSV2-hTERT construct into pLNCX2-neo retroviral vector (BD Biosciences Clontech) at HindIII and NotI sites. Afterwards, hMECs were transduced with retroviral vector containing hTERT cDNA at the second passage, and the resultant cells were cultured for over 100 population doublings (PDs) to establish hTERT immortalized hMEC cell line (I_hMEC). We subsequently irradiated the I_hMEC cells with a single dose of 60 cGy heavy ions, which were produced at Brook Heaven National Laboratory, USA. After passaged continuously for 3–4 months, the irradiated cells were injected subcutaneously into the left flank of 4–6 week old male Nu/Nu mice (Harlan Sprague–Dawley, Indianapolis, IN) at 5 × 106 cells/site. Non-irradiated I_hMEC cells with the same number of passages were used as control. We found that only heavy ion-irradiated cells can form progressively growing tumors at four weeks post injection in nude mice. We then established tumorigenic cell lines (T_hMEC) from the above tumor nodules.
Total RNA was extracted using Trizol reagent (Invitrogen) and 1 μg total RNA from each pool was used to isolate poly(A)+ mRNA by using Oligotex (QIAGEN). We prepared cDNA libraries for RNA-seq as instructed by the manufacturer (updated 4/29/09; http://solid.appliedbiosystems.com; Life Technologies, Foster City, CA). The acquired sequence tags from SOLiD-3 were 50 bp in length. Our data were submitted to Gene Expression Omnibus Database (Accession number: GSE31310; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=nrsnvioqscecszk&acc=GSE31310).
Sequence alignment to the human genome
We aligned the sequence tags using the software of Corona_lite_v4.0r2.0 (SOLiD™ System Analysis Pipeline Tool; http://solidsoftwaretools.com/gf/project/corona/). Since some mature mRNAs consists of diverse exons due to alternative splicing, we established a database containing splice junctions for tag mapping, where the 3′ 50 bp of each exon is joined to 5′ 50 bp of its 3′ adjacent exon for each gene locus according to Ensembl database.
We mapped the sequencing tags in full-length (50 bp) to the human genome assembly, allowing for five color base mismatches. Unmapped tags were further mapped to splice junctions with the same criterion. After the first round of mapping, we trimmed tags that do not match to 45 bp for the second round of mapping with 4 mismatches allowed. We performed a series of stringency-reducing recursive mapping for salvaging more data: 40 bp with 4 mismatches, 35 bp with 3 mismatches, 30 bp with 3 mismatches, and 25 bp with 3 mismatches, respectively. We sorted matched tags into multiple and unique hits and annotated uniquely-mapped tags to genes in Ensembl database (Homo sapiens. DNA. GRCh37).
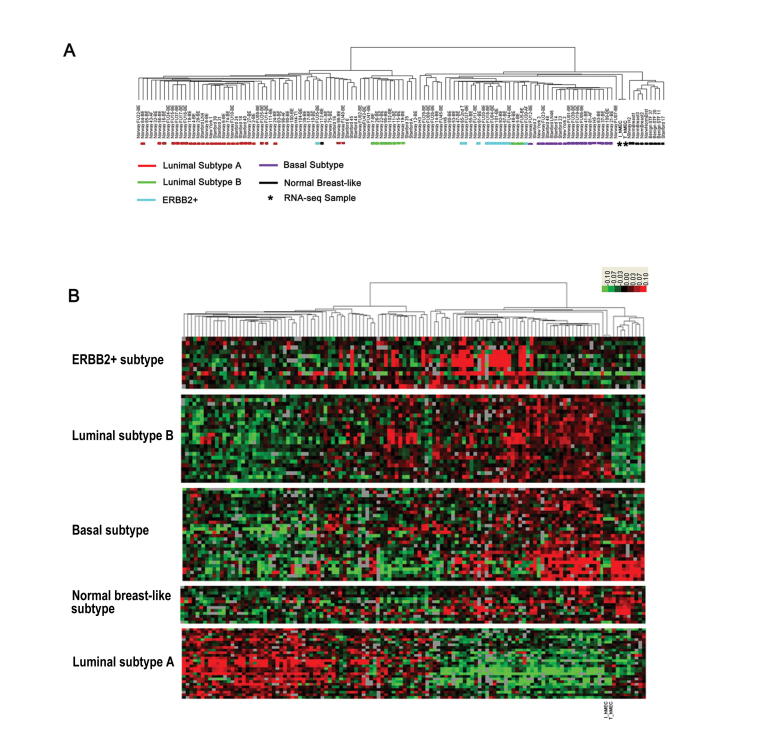
To test the accuracy of the sequencing results, we compared our RNA-seq results with a published microarray dataset [34], and our cell models are most similar to the basal-like group according to the cluster results (Figure S4). As the hMECs we used in this study are of basal epithelial origin, this result suggests that our RNA-seq data show excellent correlation with those of the microarray platform.
Analysis of DEGs
We used DEGseq (an R-based package) for DEG identification from RNA-seq data at a P value of 0.001 [32]. We performed enrichment analysis of GO terms and KEGG pathways using WebGestalt2 (http://bioinfo.vanderbilt.edu/webgestalt/) with significance at a P value of 0.001, and performed network analyses using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). We used different colors to represent different modulation patterns of DEGs in each KEGG pathway based on software package GenMAPP 2.0 (http://www.genmapp.org/).
Annotation of HK genes and TS genes
TS genes were defined as those that are preferentially expressed in a particular tissue. To annotate TS genes, we obtained two sets of TS genes with one based on EST data [35] and the other based on microarray data [22]. The combined datasets includes 7982 genes, which were named as TS genes in this study (Table S1). We defined HK genes according to the result of Ramskold et al., who determined and annotated 7882 ubiquitously expressed (UB) genes based on RNA-seq data [36]. After excluding genes that may be preferentially expressed in a particular tissue (also named as TS genes) from UB genes, we obtained 6003 genes, which were named as HK genes (Table S1).
Quantitative real-time PCR analysis
Some DEGs were selected for validation using qRT-PCR. Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA). After treated with DNAase I (Promega, Madison, WI), total RNA was reversely transcribed into cDNA (random priming) according to a standard protocol (SuperScript II reverse-transcriptase, Invitrogen, Carlsbad, CA). We performed PCR in AB7500 quantitative Real-Time PCR System with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). We analyzed each sample in duplicate, using GSK3A gene as internal reference.
Competing interests
The authors declare no competing interests.
Authors’ contributions
JY and YZ conceived the idea of sequencing, designed the whole study, supervised data analysis, and revised the manuscript. YZ and LN provided cell samples for sequencing. SH, BZ, MS, and JY (JinYang) sequenced these samples. SS mapped sequencing tags to the genome. XF and YY participated in data analysis work. JL performed qRT-PCR experiments. LM participated in sequencing work, analyzed the data, and drafted this manuscript. JY and YZ revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by grants from the National Basic Research Program (973 Program; Grant No. 2011CB944100 and 2011CB944101), National Natural Science Foundation of China (Grant No. 90919024) awarded to Jun Yu and Knowledge Innovation Program of the Chinese Academy of Sciences (Grant No. KSCX2-EW-R-01-04) to Songnian Hu. This work was also supported by the NIH National Cancer Institute (Grant No. CA127120) to Yongliang Zhao. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to acknowledge Ms. Meili Chen for constructive discussions on data analysis.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.11.001.
Contributor Information
Yongliang Zhao, Email: zhaoyongliang@big.ac.cn.
Jun Yu, Email: junyu@big.ac.cn.
Appendix A. Supplementary material
Supplementary Figure S1.
DEGs in p53 signaling pathway Up-regulated and down-regulated genes are color-coded in red and green, respectively. Color intensity represents the level of gene expression. “+p” means phosphorylation.
Supplementary Figure S2.
DEGs in cell cycle pathway Up-regulated and down-regulated DEGs are highlighted in red and green, respectively. Color intensity indicates the level of gene expression. “+p” means phosphorylation. “−p” means dephosphorylation. “+u” means ubiquitination. “e” means expression.
Supplementary Figure S3.
Comparisons between RNA-seq samples and published microarray data A. Cluster analysis of our cell lines with published microarray data from breast cancer samples. B. Gene clusters associated with each breast cancer subtype. Our RNA-seq samples are marked. I_hMEC is an immortalized human mammary epithelial cell line and T_hMEC is a tumorigenic human mammary epithelial cell line.
Supplementary Figure S4.
DEGs in apoptosis pathway Up-regulated and down-regulated DEGs are highlighted in red and green, respectively. Color intensity indicates the level of gene expression. “p” means phosphorylation. “b” means binding. “e” means expression.
House-keeping (HK) genes and tissue-specific (TS) genes.
Tumorigenicity-associated DEGs.
Distribution of tumorigenicity-associated genes on chromosomes.
DEGs involved in “pathways in cancer”.
Highly regulated DEGs.
Enriched KEGG pathways of the 364 highly regulated DEGs.
References
- 1.Kan Z., Jaiswal B.S., Stinson J., Janakiraman V., Bhatt D., Stern H.M. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 2.Momparler R.L. Cancer epigenetics. Oncogene. 2003;22:6479–6483. doi: 10.1038/sj.onc.1206774. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Rhim J.S. Neoplastic transformation of human cells in vitro. Crit Rev Oncog. 1993;4:313–335. [PubMed] [Google Scholar]
- 6.Rhim J.S., Yoo J.H., Park J.H., Thraves P., Salehi Z., Dritschilo A. Evidence for the multistep nature of in vitro human epithelial cell carcinogenesis. Cancer Res. 1990;50:5653S–5657S. [PubMed] [Google Scholar]
- 7.Kolquist K.A., Ellisen L.W., Counter C.M., Meyerson M., Tan L.K., Weinberg R.A. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat Genet. 1998;19:182–186. doi: 10.1038/554. [DOI] [PubMed] [Google Scholar]
- 8.Shao G., Balajee A.S., Hei T.K., Zhao Y. P16INK4a downregulation is involved in immortalization of primary human prostate epithelial cells induced by telomerase. Mol Carcinog. 2008;47:775–783. doi: 10.1002/mc.20434. [DOI] [PubMed] [Google Scholar]
- 9.Zajchowski D.A., Band V., Trask D.K., Kling D., Connolly J.L., Sager R. Suppression of tumor-forming ability and related traits in MCF-7 human breast cancer cells by fusion with immortal mammary epithelial cells. Proc Natl Acad Sci U S A. 1990;87:2314–2318. doi: 10.1073/pnas.87.6.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fusenig N.E., Boukamp P. Multiple stages and genetic alterations in immortalization, malignant transformation, and tumor progression of human skin keratinocytes. Mol Carcinog. 1998;23:144–158. doi: 10.1002/(sici)1098-2744(199811)23:3<144::aid-mc3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 11.Rio M.C., Bellocq J.P., Gairard B., Rasmussen U.B., Krust A., Koehl C. Specific expression of the pS2 gene in subclasses of breast cancers in comparison with expression of the estrogen and progesterone receptors and the oncogene ERBB2. Proc Natl Acad Sci U S A. 1987;84:9243–9247. doi: 10.1073/pnas.84.24.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zajchowski D.A., Sager R. Induction of estrogen-regulated genes differs in immortal and tumorigenic human mammary epithelial cells expressing a recombinant estrogen receptor. Mol Endocrinol. 1991;5:1613–1623. doi: 10.1210/mend-5-11-1613. [DOI] [PubMed] [Google Scholar]
- 13.Avilion A.A., Piatyszek M.A., Gupta J., Shay J.W., Bacchetti S., Greider C.W. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56:645–650. [PubMed] [Google Scholar]
- 14.Kirkpatrick K.L., Ogunkolade W., Elkak A.E., Bustin S., Jenkins P., Ghilchick M. HTERT expression in human breast cancer and non-cancerous breast tissue: correlation with tumour stage and c-Myc expression. Breast Cancer Res Treat. 2003;77:277–284. doi: 10.1023/a:1021849217054. [DOI] [PubMed] [Google Scholar]
- 15.Hahn W.C. Immortalization and transformation of human cells. Mol Cells. 2002;13:351–361. [PubMed] [Google Scholar]
- 16.Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu X., Fu N., Guo S., Yan Z., Xu Y., Hu H. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 19.Zenoni S., Ferrarini A., Giacomelli E., Xumerle L., Fasoli M., Malerba G. Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol. 2010;152:1787–1795. doi: 10.1104/pp.109.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez O., Reyes-Valdes M.H., Herrera-Estrella L. Cancer reduces transcriptome specialization. PLoS One. 2010;5:e10398. doi: 10.1371/journal.pone.0010398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axelsen J.B., Lotem J., Sachs L., Domany E. Genes overexpressed in different human solid cancers exhibit different tissue-specific expression profiles. Proc Natl Acad Sci USA. 2007;104:13122–13127. doi: 10.1073/pnas.0705824104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Black D.M., Nicolai H., Borrow J., Solomon E. A somatic cell hybrid map of the long arm of human chromosome 17, containing the familial breast cancer locus (BRCA1) Am J Hum Genet. 1993;52:702–710. [PMC free article] [PubMed] [Google Scholar]
- 24.Watatani M., Nagayama K., Imanishi Y., Kurooka K., Wada T., Inui H. Genetic alterations on chromosome 17 in human breast cancer: relationships to clinical features and DNA ploidy. Breast Cancer Res Treat. 1993;28:231–239. doi: 10.1007/BF00666584. [DOI] [PubMed] [Google Scholar]
- 25.Negrini M., Sabbioni S., Haldar S., Possati L., Castagnoli A., Corallini A. Tumor and growth suppression of breast cancer cells by chromosome 17-associated functions. Cancer Res. 1994;54:1818–1824. [PubMed] [Google Scholar]
- 26.Fischer S.G., Cayanis E., de Fatima Bonaldo M., Bowcock A.M., Deaven L.L., Edelman I.S. A high-resolution annotated physical map of the human chromosome 13q12-13 region containing the breast cancer susceptibility locus BRCA2. Proc Natl Acad Sci U S A. 1996;93:690–694. doi: 10.1073/pnas.93.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casey G., Plummer S., Hoeltge G., Scanlon D., Fasching C., Stanbridge E.J. Functional evidence for a breast cancer growth suppressor gene on chromosome 17. Hum Mol Genet. 1993;2:1921–1927. doi: 10.1093/hmg/2.11.1921. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W., Yu Y. The important molecular markers on chromosome 17 and their clinical impact in breast cancer. Int J Mol Sci. 2011;12:5672–5683. doi: 10.3390/ijms12095672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 30.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 33.Dudley J.T., Tibshirani R., Deshpande T., Butte A.J. Disease signatures are robust across tissues and experiments. Mol Syst Biol. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorlie T., Tibshirani R., Parker J., Hastie T., Marron J.S., Nobel A. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X., Yu X., Zack D.J., Zhu H., Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramskold D., Wang E.T., Burge C.B., Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
House-keeping (HK) genes and tissue-specific (TS) genes.
Tumorigenicity-associated DEGs.
Distribution of tumorigenicity-associated genes on chromosomes.
DEGs involved in “pathways in cancer”.
Highly regulated DEGs.
Enriched KEGG pathways of the 364 highly regulated DEGs.