Abstract
Lipid bodies store oils in the form of triacylglycerols. Oleosin, caleosin and steroleosin are unique proteins localized on the surface of lipid bodies in seed plants. This study has identified genes encoding lipid body proteins oleosin, caleosin and steroleosin in the genomes of five plants: Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Selaginella moellendorffii and Physcomitrella patens. The protein sequence alignment indicated that each oleosin protein contains a highly-conserved proline knot motif, and proline knob motif is well conserved in steroleosin proteins, while caleosin proteins possess the Dx[D/N]xDG-containing calcium-binding motifs. The identification of motifs (proline knot and knob) and conserved amino acids at active site was further supported by the sequence logos. The phylogenetic analysis revealed the presence of magnoliophyte- and bryophyte-specific subgroups. We analyzed the public microarray data for expression of oleosin, caleosin and steroleosin in Arabidopsis and rice during the vegetative and reproductive stages, or under abiotic stresses. Our results indicated that genes encoding oleosin, caleosin and steroleosin proteins were expressed predominantly in plant seeds. This work may facilitate better understanding of the members of lipid-body-membrane proteins in diverse organisms and their gene expression in model plants Arabidopsis and rice.
Keywords: Gene expression, Phylogeny, Proline knot motif, Protein alignment, Sequence logo
Introduction
Plant seeds store lipids in the form of triacylglycerols (TAGs) in specialized organelles called lipid bodies [1], [2], [3]. The lipid bodies are also referred to as oil-bodies/globules/vesicles/spherules, oil or fat droplets, or oleosomes [4], [5], [6]. The stored lipids in oilseeds provide energy for the growth of the germinated seedling [5], [7]. The TAG matrix of the lipid body is surrounded by a layer of phospholipids and proteins oleosin, caleosin and steroleosin [5], [7], [8], [9], [10]. These proteins were identified using a proteomic approach to isolated lipid bodies of plant seeds [6], [11], [12], [13], [14].
Oleosin is a reliable marker for lipid body [3], [6], [12], [15], [16], [17]. The oleosin proteins are present in two weight-forms (L and H) which coexist in all lipid bodies [18]. H-form can be distinguished from the L-form using immunological methods [6], [8], [18], due to the presence of an extra stretch of amino acids at the C-terminal domain of H-form [3], [6], [8], [18]. The L and H forms of oleosin are well conserved among diverse plant species [18], although the N- and C-terminal regions vary in length, leading to oleosin with diverse molecular weights [19].
The oleosin proteins are expressed in seed and floral anther [7]. A common feature of all oleosins is to provide stability to discrete oil-bodies during seed desiccation [2], [3], [5], [6], [12]. The Arabidopsis double mutant of oleosin1-oleosin2 showed a drastic reduction in seed germination which was attributed to extreme expansion of oil-bodies [7]. Similarly, the anther-type oleosins also provide stability to pollen lipid bodies [20], [21] and play a role in the development of pollen and the pollen coat [22]. In addition, the lipid bodies in rice embryo and aleurone layer contain two forms of oelosin, which provide stability to the oil bodies [23].
The hallmark of all oleosins is the proline (Pro) knot motif, which is Pro-hydrophobic amino acids (5x)-Ser-Pro- hydrophobic amino acids (3x)-Pro [7]. The Pro knot is located in the central anchoring domain, which is highly conserved among diverse plant species [24]. The targeting of oleosin to lipid bodies is facilitated by the Pro knot [3], [25], [26].
Caleosin is a lipid body-associated protein involved in mobilization of stored lipids during seed germination [8], [9], [10], [27]. Caleosin was found as a major protein in cycad oil-bodies without the presence of any oleosin [28]. The N-terminal domain of caleosin contains single EF-hand motif for calcium (Ca2+)-binding, and the C-terminal domain contains a few phosphorylation sites [6], [9], [17], [29]. A less conserved Pro knot motif is also found in caleosin proteins [6], [30].
Steroleosin is a minor lipid body protein that was proposed to mobilize lipid bodies during sesame seed germination [10]. All steroleosins are characterized by the presence of a sterol-binding dehydrogenase/reductase domain containing the conserved active site region [S-(12x)-Y-(3x)-K] for sterol-coupling dehydrogenase activity [10]. Steroleosin protein contains a Pro knob motif in the middle of the N-terminal hydrophobic segment [10]. It is still unclear whether the Pro knob motif plays a role in steroleosin targeting to lipid bodies [10].
It has become a challenge to assemble pertinent information from the sequencing data of a wide diversity of plant genomes. The information is spread over numerous genome databases and is presented in different formats. Much of this data originates from high-throughput studies, and several nucleotide sequences code for hypothetical proteins. Therefore, a comparative genomic approach to identify individual gene family members is essential. Moreover, characterizing the gene families is necessary to investigate the members associated with a particular gene family to study their evolutionary relationship. The main purpose of the current study was to explore the genomes of the model eudicots (Arabidopsis thaliana, Populus trichocarpa), monocot (Oryza sativa), lycophyte (Selaginella moellendorffii) and bryophyte (Physcomitrella patens) to identify genes coding for oleosin, caleosin and steroleosin. Earlier studies have identified Arabidopsis members for these gene families [5], [19]. The data presented here is an extended view to include the members of O. sativa, P. trichocarpa, S. moellendorffii, and P. patens.
Results
We aimed to characterize the gene families of oleosin, caleosin and steroleosin in eudicots, monocot, bryophyte and lycophyte. A comparative genomic approach was used for identification of genes coding for lipid body proteins oleosin, caleosin and steroleosin in A. thaliana, O. sativa, P. trichocarpa, S. moellendorffii and P. patens.
Protein alignment and sequence logos
Table S1 shows the list of oleosin genes found in the genomes of the five plant species examined. These genes were identified as described in materials and methods. The genomic loci AT5G07530, AT5G07550, AT5G07560 and AT5G56100 in A. thaliana were not included in the list of oleosins as they were annotated as glycine-rich protein oleosin domain, oleosin-like, lipid-binding oleosin and glycine-rich protein/oleosin, respectively. Therefore, 11 oleosin genes (AtOleo1–11) were identified in A. thaliana based on a protein alignment (Figure S1). AtOleo1–4 as annotated in the TAIR database, while AtOleo5–11 were numbered according to their order of appearance on the chromosomes. The encoded proteins possessed the conserved Pro knot motif. Similarly, based on the presence of well conserved Pro knot motif, the genome-wide studies have identified oleosin-encoding genes from other species, including O. sativa (OsOleo1–6, numbered according to the order of appearance on the chromosomes), P. trichocarpa (PtOleo1–5), S. moellendorffii (SmOleo1–3) and P. patens (PpOleo1 and 2), respectively (Figure S1). We then constructed sequence logo using 319 oleosin protein sequences available in the Pfam database (PF01277), which come from a wide range of plant species, to study the conservation of amino acids (Figure S2). The data indicated the highly conserved prolines (P) of the Pro knot motif (Figure S2). Additionally, phenylalanine (F) and serine (S) were relatively conserved in the Pro knot motif too.
The identified members of caleosin gene family in the five plant species are given in Table S2. Six (AtClo1–6) and eight (OsClo1–8) caleosin genes were identified in A. thaliana and O. sativa, respectively. There were two caleosin genes in P. trichocarpa (PtClo1 and 2), P. patens (PpClo1 and 2), and one in S. moellendorffii (SmClo1) (Table S2). Protein alignment showed that the Dx[D/N]xDG motif is well conserved in caleosins (Figure S3). The sequence logo profile of 216 caleosin proteins from Pfam identified the Dx[D/N]xDG motif although the relative contribution of these amino acid residues was low, compared to histidine (H), F, P and glycine (G) (Figure S4).
Steroleosins identified in the genomes of the five plant species are listed in Table S3. There were six steroleosin or hydrosteroid dehydrogenase (HSD1–6) genes in A. thaliana (AtSlo1–6) and O. sativa (OsSlo1–6), seven in P. trichocarpa (PtSlo1–7) as well as P. patens (PpSlo1–7), and three in S. moellendorffii (SmSlo1–3) (Table S3). The protein alignment showed the Pro knob motif was present in steroleosins of all five species (Figure S5), although the Pro knob motif was absent in OsSlo1. In addition, protein alignment of steroleosins also indicated that the active site amino acids S, tyrosine (Y) and lysine (K) were conserved in Slo1–6 from A. thaliana and Slo1, 2, 4 and 5 from rice. However, S was substituted by A in Slo3 from rice and in Slo4 and Slo7 of P. trichocarpa, whereas K was replaced by asparagine (N) in Slo6 from rice and by glutamic acid (E) in Slo1 of S. moellendorffii, respectively. Nonetheless, S and Y were well conserved in a total of 54,179 steroleosin protein sequences obtained from Pfam as corroborated from the sequence logo profile (Figure S6).
Phylogenetic aspects of oleosins, caleosins and steroleosins
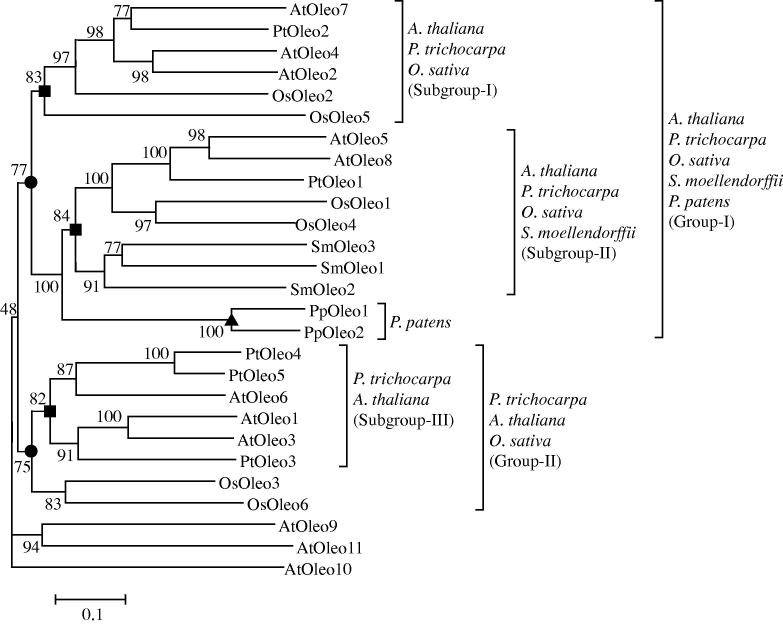
Phylogenetic trees were constructed to evaluate the evolutionary relationships among the members of oleosins, caleosins and steroleosins. The phylogenetic tree for 27 oleosin proteins of the five plant species examined is shown in Figure 1. Two major groups of oleosin proteins are present (closed circles). Group-I consisted of oleosins from all the five plant species, while group-II consisted of oleosins only from magnoliophyte such as P. trichocarpa, A. thaliana and O. sativa. Further analysis classified oleosins into three major subgroups (closed squares, Figure 1). Subgroup-I consisted of oleosins only from magnoliophyte, subgroup-II of oleosins from vascular plants including A. thaliana, P. trichocarpa, O. sativa and S. moellendorffii, and subgroup-III of oleosins from eudicots including P. trichocarpa and A. thaliana. Within group-I, the oleosins from bryophyte P. patens are clustered into separate clade (closed upward triangle, Figure 1). These data suggests that oleosins might have independently evolved at least four times (in eudicots, magnoliophytes, vascular plants and bryophyte), and with convergent evolution in vascular plants (A. thaliana, P. trichocarpa, O. sativa and S. moellendorffii).
Figure 1.
The phylogeny of oleosin proteins The unrooted phylogeny was bootstrapped to 100 and scaled to 0.1 substitutions. The bootstrap values are given at each node. At, Arabidopsis thaliana; Os, Oryza sativa; Pt, Populus trichocarpa; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
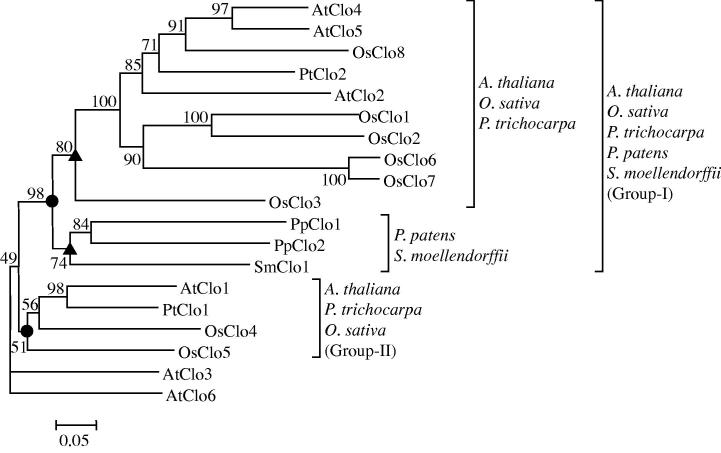
The phylogeny for 19 caleosin protein sequences of the five plant species is shown in Figure 2. The caleosin proteins were phylogenetically classified into two major groups, I and II (closed circles, Figure 2). The group-I consisted of caleosins from all the five species; and the caleosins from magnoliophyte formed group-II. The caleosins from bryophyte P. patens and lycophyte S. moellendorffii emerged as separate clades within group-I. Another separate clade within group-I was of caleosins from magnoliophyte (closed upward triangles, Figure 2). The data indicates divergent evolution of caleosins in magnoliophyte, lycophyte and bryophyte.
Figure 2.
The phylogeny of caleosin proteins The unrooted phylogeny was bootstrapped to 100 and scaled to 0.05 substitutions. The bootstrap values are given at each node. At, Arabidopsis thaliana; Os, Oryza sativa; Pt, Populus trichocarpa; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
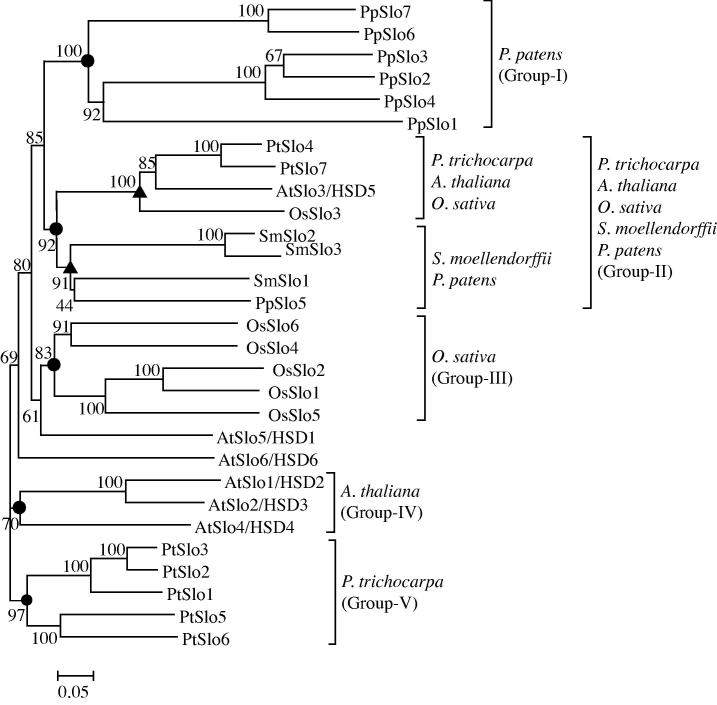
The phylogeny constructed from steroleosin proteins of the five species is shown in Figure 3. The steroleosin proteins were classified into five major groups, I–V (closed circles). The steroleosin proteins in group-I were from P. patens, group-II from all the five species, group-III from O. sativa, group-IV from A. thaliana, and group-V from P. trichocarpa. Within group-II, two separate clades were formed with one from magnoliophyte and the other from lycophyte S. moellendorffii and bryophyte P. patens, respectively (closed upward triangles). The results indicate that steroleosins are divergent class of lipid body proteins which might have independently evolved within the members of magnoliophyte and bryophyte.
Figure 3.
The phylogeny of steroleosin proteins The unrooted phylogeny was bootstrapped to 100 and scaled to 0.05 substitutions. The bootstrap values are given at each node. At, Arabidopsis thaliana; Os, Oryza sativa; Pt, Populus trichocarpa; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
Expression profiles of oleosins, caleosins and steroleosins in Arabidopsis and rice
The publicly available gene expression microarray datasets were explored to study expression patterns of oleosins, caleosins and steroleosins in A. thaliana and O. sativa. The gene expression of Arabidopsis oleosins, caleosins and steroleosins were studied in 12 different samples (callus, cell culture, cotyledons, hypocotyl, stem, rosette, shoot apical meristem (SAM), roots, flower, silique, seed and imbibed seed). According to Levene’s test (0.001 < P < 0.054), an increased expression of Arabidopsis oleosin genes (Oleo1–4 and 6–8) was detected in seeds and imbibed seeds (Figure S7A). The expression of Oleo3 was upregulated in SAM as compared to other oleosin genes (Figure S7A). The expression of Oleo5 and 9–11 was not studied due to the unavailability of microarray probes for these genes in the TAIR database. Similarly, we examined the expression of caleosin-coding genes in Arabidopsis. Expression of Clo3 and Clo4 was upregulated in cell culture, cotyledons, stem, rosette, SAM, flower and silique as compared to Clo1 and Clo2 (Figure S7B). An enhanced expression of Arabidopsis Clo4 was detected in hypocotyl and root tissue as compared to other Clo genes (Figure S7B). The expression data of Arabidopsis Clo5 was found to be identical to Clo4 in the TAIR database. Expression data was not available for Arabidopsis Clo6 in the TAIR database. The Arabidopsis steroleosin genes Slo3, Slo5 and Slo6 showed a higher level of expression in seed and imbibed seed as compared to Slo4 (Figure S7C). The data for microarray probes of Slo1 and Slo2 were not available in the TAIR database.
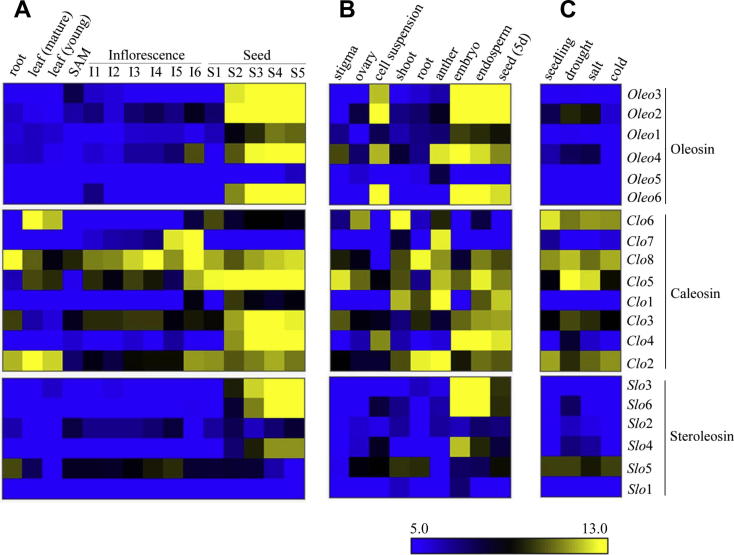
The rice oligonucleotide array data was explored for oleosins, caleosins and steroleosins at different vegetative (cell suspension, shoot, root, leaf, SAM) and reproductive (inflorescence, anther, stigma, ovary, embryo, endosperm, seed) stages (Figure 4). The expression of rice Oleo1 was detected through stages S2–S5 (Figure 4A). The rice Oleo4 was expressed in inflorescence stage-I6, stigma and anther (Figure 4A and B). The expression of most of the rice oleosin genes (Oleo2, Oleo3, Oleo4 and Oleo6) was upregulated in cell suspension, embryo, endosperm and seed (Figure 4B).
Figure 4.
The heat map showing gene expression of oleosins, caleosins and steroleosins in rice A. Gene expression of oleosin, caleosin and steroleosin in vegetative (root, leaf and SAM) and reproductive (inflorescence and seed) stages of plant development. The microarray was performed using tissue samples from O. sativa L. cv. IR64. ROAD experiment ID for this panel is GSE6893. B. Gene expression of oleosin, caleosin and steroleosin in cell suspension, shoot, root and reproductive (stigma, ovary, anther, embryo, endosperm and seed) stages. The microarray was performed using tissue samples from O. sativa L. cv. Nipponbare. ROAD experiment ID for this panel is GSE7951. C. Expression of oleosin, caleosin and steroleosin genes under abiotic stresses. The control seedlings were 7-day-old. The microarray was performed using tissue samples from O. sativa L. cv. IR64. ROAD experiment ID for this panel is GSE6901. The microarray probe set IDs are given in Table S4. The corresponding gene names are given on the right of the heat map. The tissue samples or stress conditions for which microarray was performed are given on top of the heat map. The gene expression scale is from 5.0 (downregulation of genes) to 13.0 (upregulation of genes).
The dynamic expression pattern of rice caleosin genes (Clo1–8) in vegetative and reproductive stages is shown in Figure 4A. The genes Clo2–5 and Clo8 were upregulated in seed stages S2–S5; the rice Clo6 was upregulated in young and mature leaf tissue, and was expressed in seed stage-S1; the gene Clo7 was upregulated only in inflorescence stages I5 and I6 (Figure 4A). The expression level of rice Clo1, Clo5, Clo7 and Clo8 was upregulated in anther tissue (Figure 4B). In seed stages-S4 and S5, the expression of rice steroleosin genes Slo3, Slo4 and Slo6 was upregulated (Figure 4A). The steroleosin genes Slo3 and Slo6 were upregulated in embryo and endosperm tissue (Figure 4B).
The expression of rice Oleo, Clo and Slo was analyzed in abiotic conditions of drought, salt and cold stress (Figure 4C). The expression of Oleo2 was enhanced in drought and salt stresses; the expression of Clo3 and Clo5 was upregulated also in drought and salt stress treatments. Conversely, the expression of Clo2 was down-regulated by drought and, to a lesser extent, by salt stress; expression of Clo6 and Clo8 was slightly down-regulated by drought and salt stresses, respectively. Additionally, expression of Slo5 was down-regulated by salt stress, while expression of the remaining steroleosin genes (Slo1–4 and Slo6) was consistently low under abiotic stress treatments (Figure 4C).
Discussion
In this study, the members of gene families of lipid body proteins oleosin, caleosin and sterolesoin in A. thaliana, O. sativa, P. trichocarpa, S. moellendorffii and P. patens were evaluated by protein alignment, phylogenetic analyses, sequence logos and expression profiles. The current study reveals genes encoding lipid body proteins in model plants. The phylogenetic classification of these three gene families has identified several independent clades for categories of eudicots, monocot, bryophyte and lycophyte which led to evolutionary comparisons.
High-throughput nucleotide sequencing has led to the identification of numerous novel protein-coding genes, some of which remain to be characterized and functionally assigned. The current study has focused on the in silico identification of plant lipid-body–membrane proteins for which the physiological functions remain elusive.
Non-storage plant tissues like floral tapetum, pollen, and vegetative cells are known to contain oleosin [3], [20], [31], which is in agreement with the present study. Sixteen oleosin genes of Arabidopsis were categorized into five seed-type oleosins, eight anther-type oleosins, and three seed-and-anther-type oleosins [5], [19]. The present study has identified 11 genes in the genome of Arabidopsis which encode oelosin proteins. The loci AT5G07530, AT5G07550, AT5G07560 and AT5G56100 were not included as these loci were annotated as glycine-rich proteins although their protein sequences show the conserved Pro knot motif. The loci AT5G07571 (Oleo9) and AT5G56100 (glycine-rich protein) from the current study were not identified by Kim et al. [19]. The loci AT5G07510, AT5G07520 and AT5G07540 which were identified as oleosins by Kim et al. [19] were not included in the current study as they were annotated as glycine-rich protein 14, 18 and 16, respectively. In Arabidopsis, Oleo1 was the most abundant seed oleosin isoforms [11], [32] and its knockdown resulted in the formation of large lipid bodies in seeds [5]. The present study showed increased expression of Oleo1 in seeds and imbibed seeds of Arabidopsis. The deficiency of oleosin causes loss of freezing tolerance in seeds of Arabidopsis [33]. RNA interference of soybean oleosin resulted in giant lipid body formation and impaired seed germination [34]. The expression of oleosin was spatially and developmentally regulated [5] and controlled by the transactivator ABI3 [5], [35]. The current study also provides evidence of the overall increased expression of oleosins (Oleo1–4 and 6–8) in seeds and imbibed seeds of Arabidopsis. The exclusive upregulation of Arabidopsis Oleo3 in SAM suggests its vital role in the developing plant meristem. The lack of differential expression of Oleo5, and the differential expression of Oleo1 indicate the developmental-selectivity of oleosin gene expression in rice.
Caleosin and steroleosin are lipid body proteins that are involved in cytoplasmic signaling [6]. The abundance of caleosin and steroleosin transcripts was similar in sesame seed lipid bodies [10], which is consistent with the present findings on their expression levels in Arabidopsis seeds. Caleosins were found in plant seed lipid body, pollen, shoot, and root tip, in accordance with the current data of gene expression in Arabidopsis and rice [30], [36], [37]. Caleosin was expressed in tapetal and germ line cells of olive anthers [36]. The current study has identified expression of rice Clo7 to be differentially upregulated in late stages (I5 and I6) of inflorescence development and anther tissue. Expression of caleosin is upregulated in seeds and promotes seed dormancy in abscisic acid (ABA) and osmotic stress conditions [17]. The present study has revealed differential upregulation of rice Clo4 in seed stages S2–S5. The study showed high expression levels of Clo8 in roots as compared to Clo2 and Clo3, implicating a role of Clo2 in modulating root related traits.
Arabidopsis Clo1 and Clo2 show Ca2+-dependent peroxygenase activity [37], [38], [39]. Arabidopsis Clo3, also named as Responsive to Dehydration 20 (RD20), was biochemically identified as a peroxygenase [40]. RD20 is induced by various abiotic stress factors [39], [40] and gets involved in water stress responses and plant growth [40]. Expression of Arabidopsis Clo3 was upregulated by ABA treatment and under stress conditions [38], [39], [40], [41]. The current study has identified upregulation of rice Clo3 by drought and salt stresses implicating its vital role in response to environmental cues. The stress-responsive Arabidopsis Clo4 is a negative regulator of ABA responses [42]. The current study has shown an increased expression of both Clo3 and Clo4 in some of the vegetative (cotyledons, stem, rosette, and SAM) and reproductive (flower and silique) stages as compared to Clo1 and Clo2 in Arabidopsis. The current study demonstrates that some members of caleosin gene family may be involved in the vegetative to reproductive stage transition.
The caleosins are found in higher plants and fungi [16], [27]. Earlier studies have reported seven caleosin genes in Arabidopsis (AtClo1–7) [38], [39]. The locus AT1G23250, although annotated as a caleosin-related family protein, lacks the characteristic Dx[D/N]xDG motif for Ca2+-binding [43] and therefore was excluded from the current study.
The sterol-binding dehydrogenases/reductases have been identified as signal transduction components in microorganisms and mammals [10]. As reported earlier, Arabidopsis genome encodes for eight steroleosin genes [6], [10]. The current study identified six steroleosin genes in Arabidopsis genome. Sequence comparison shows that the loci AT5G50690 (HSD7/HSD4) and AT5G50700 (HSD1) are identical to AT5G50590 (HSD4) and AT5G50600 (HSD1), respectively. Steroleosin is expressed in maturing sesame seeds [10]. The present findings also show an increase in expression of steroleosins (Slo3, Slo5 and Slo6) in Arabidopsis seeds and imbibed seeds. The current study in O. sativa shows an increased expression of Slo3, Slo4 and Slo6 in seeds of Indica cultivar but reduced expression of Slo4 in seeds of Japonica cultivar. An increase in expression of steroleosins (Slo3, Slo4 and Slo6) is also revealed in rice embryo and endosperm. The current findings demonstrate the upregulation of rice Slo5 in root, shoot and inflorescence implying its crucial role in a variety of plant functions.
The evolutionary lineage of a particular gene family can be traced by comparison of gene families with organisms from different branches of evolution studied in the context of their phylogeny. The phylogenetic trees were constructed to examine evolutionary relationship of gene families for oleosins, caleosins and steroleosins among eudicots, monocot, bryophyte and lycophyte based on a comparative genomic approach. Our results showed that a group of oelosins as well as caleosins in A. thaliana, P. trichocarpa and O. sativa (magnoliophyte) was evolved along with S. moellendorffii (lycophyte) and P. patens (bryophyte). The other group of oleosins and caleosins was evolved specifically in magnoliophyte. Some members of the gene family of steroleosin have evolved independently in A. thaliana, P. trichocarpa, O. sativa and P. patens.
This study explores gene families of oleosin, caleosin and steroleosin from diverse organisms. The dense sampling of members of these gene families provides wide gene coverage for phylogenetic analysis. The study describes well-resolved phylogenies of oleosins, caleosins and steroleosins from eudicots, monocot, bryophyte and lycophyte. This article also provides an in-depth transcriptome profiles of lipid body proteins in Arabidopsis and rice. The gene expression patterns revealed that the oleosin genes are expressed predominantly in seeds of Arabidopsis and rice. A more dynamic expression pattern for caleosin genes was detected which was regulated both spatially and temporally in rice. This article may provide a better understanding of the lipid-body-membrane proteins in magnoliophyte, bryophyte and lycophyte.
Materials and methods
Retrieval of sequences
Arabidopsis protein sequences of oleosin, caleosin and steroleosin were downloaded from the Arabidopsis information resource (TAIR) (http://www.arabidopsis.org) [44]. The full-length protein sequences of Oryza sativa subsp. Japonica (rice) were downloaded from the rice genome annotation project database (http://rice.plantbiology.msu.edu) [45]. The protein sequences of P. trichocarpa, P. patens and S. moellendorffii were retrieved from the Department of Energy Joint Genome Institute database (http://www.jgi.doe.gov) [46]. Arabidopsis protein sequences of steroleosin were used as query against the NCBI protein database to retrieve corresponding gene family members of P. trichocarpa and P. patens.
Protein alignments and phylogenetic studies
The full-length protein sequences of oleosin, caleosin and steroleosin were imported to the ClustalX multiple sequence alignment program [47]. The neighbor-joining (NJ) method was adopted to construct unrooted phylogenies of oleosins, caleosins and steroleosins. The phylogeny was constructed on alignment file using the molecular evolutionary genetics analysis (MEGA) 4 software program [48].
Building sequence logos
The sequence logos were built for protein sequences of oleosins (Pfam ID – PF01277), caleosins (Pfam ID – PF05042) and steroleosins (Pfam ID – PF00106) based on the profile Hidden Markow Model (pHMM) [49] of the Pfam database (http://pfam.janelia.org) [50]. A total of 319, 216 and 54,179 protein sequences of oleosin, caleosin and steroleosin, respectively, available in the Pfam database were utilized to generate sequence logos.
Gene expression analysis
The gene expression status of Arabidopsis oleosins, caleosins and steroleosins at different stages was referred to the Genevestigator response viewer (https://www.genevestigator.com) [51] of the TAIR database. The gene expression data of oleosins, caleosins and steroleosins in rice was downloaded from the rice oligonucleotide array database (ROAD) (http://www.ricearray.org) [52]. For Figure 4A, the rice inflorescence represents following stages of development: floral organ initiation (I1), meiotic stage (I2 and I3), microspore stage (I4), vacuolated pollen stage (I5) and mature pollen stage (I6). The rice seed development represents following stages: early globular embryo (0–2 days after pollination) (DAP) (S1), middle and late globular embryo (3–4 DAP) (S2), embryo morphogenesis (5–10 DAP) (S3), embryo maturation (11–20 DAP) (S4), dormancy and desiccation tolerance (21–29 DAP) (S5).
Competing interests
The author has no competing interests to declare.
Acknowledgements
The author is grateful to the reviewers for their helpful suggestions.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.08.006.
Appendix Supplementary. material
The amino acid alignment of oleosin proteins The gene names are given on left and the numbers of amino acids are shown on right. The conserved proline knot motif is indicated on top of the alignment. The section of alignment containing the proline knot motif is shown. The figure is made as described in materials and methods. At, Arabidopsis thaliana; Os, Oryza sativa; Pt, Populus trichocarpa; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
The sequence logo of the proline knot motif in oleosin proteins The conserved proline residues are indicated with an asterisk on top of the logo.
The amino acid alignment of caleosin proteins The gene names are given on left and the numbers of amino acids are shown on right. The conserved Dx[D/N]xDG motif is indicated on top of the alignment. The part of alignment with the Dx[D/N]xDG is shown. The figure is made as described in materials and methods. At, Arabidopsis thaliana; Os, Oryza sativa; Pt, Populus trichocarpa; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
The sequence logo of the Dx[D/N]xDG motif in caleosin proteins The Dx[D/N]xDG motif is shown on top of the alignment. The conserved amino acids flanking the Dx[D/N]xDG motif are indicated with an asterisk on top of the alignment.
The amino acid alignment of steroleosin proteins of A. thaliana (At), O. sativa (Os), P. trichocarpa (Pt), P. patens (Pp) and S. moellendorffii (Sm) The gene names are given on left and the numbers of amino acids are shown on right. The conserved proline knob motif is indicated on top of the alignment. The active site amino acids (S, Y and K) are indicated with down-arrows on top of the alignment. The figure is made as described in materials and methods. The parts of alignment containing the proline knob motif and active site amino acids are shown.
The sequence logo of the active site amino acids S, Y and K in steroleosins The active site amino acids are represented with an asterisk on top.
The gene expression pattern of oleosin (A), caleosin (B), and steroleosin (C) in Arabidopsis The tissue source is given on the X-axis and the relative expression of genes on 22k microarray on the Y-axis. The gene names of oleosins (Oleo), caleosins (Clo) and steroleosins (Slo/HSD) are given on the right. The error bars are for standard error (S.E.). The microarray data were normalized using Affymetrix MAS5 software [44]. The expression data from different sources were scaled up or down using a Target intensity value (TGT) of 200 [44].
The locus IDs and nomenclature for oleosin (Oleo) encoding genes.
The locus IDs and nomenclature for caleosin (Clo) encoding genes.
The locus IDs and nomenclature for steroleosin (Slo) encoding genes.
The microarray probe set IDs, TIGR locus IDs and gene names of oleosins, caleosins and steroleosins in Oryza sativa.
References
- 1.Yatsu L.Y., Jacks T.J. Spherosome membranes: half unit-membranes. Plant Physiol. 1972;49:937–943. doi: 10.1104/pp.49.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy D.J. Structure, function and biogenesis of storage lipid bodies and oleosins in plants. Prog Lipid Res. 1993;32:247–280. doi: 10.1016/0163-7827(93)90009-l. [DOI] [PubMed] [Google Scholar]
- 3.Huang A.H.C. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996;110:1055–1061. doi: 10.1104/pp.110.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slack C.R., Bertaud W.S., Shaw B.D., Holland R., Browse J., Wright H. Some studies of the composition and surface of oil bodies from the seed cotyledons of safflower and linseed. Biochem J. 1980;190:551–561. doi: 10.1042/bj1900551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siloto R.M., Findlay K., Lopez-Villalobos A., Yeung E.C., Nykiforuk C.L., Moloney M.M. The accumulation of oleosins determines the size of seed oil-bodies in Arabidopsis. Plant Cell. 2006;18:1961–1974. doi: 10.1105/tpc.106.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Schoot C., Paul L.K., Paul S.B., Rinne P.L. Plant lipid bodies and cell–cell signaling: a new role for an old organelle? Plant Signal Behav. 2011;6:1732–1738. doi: 10.4161/psb.6.11.17639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimada T.L., Hara-Nishimura I. Oil-body-membrane proteins and their physiological functions in plants. Biol Pharm Bull. 2010;33:360–363. doi: 10.1248/bpb.33.360. [DOI] [PubMed] [Google Scholar]
- 8.Tzen J.T., Peng C.C., Cheng D.J., Chen E.C., Chiu J.M. A new method for seed oil body purification and examination of oil body integrity following germination. J Biochem. 1997;121:762–768. doi: 10.1093/oxfordjournals.jbchem.a021651. [DOI] [PubMed] [Google Scholar]
- 9.Chen J.C., Tsai C.C., Tzen J.T. Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil-bodies of plant seeds. Plant Cell Physiol. 1999;40:1079–1086. doi: 10.1093/oxfordjournals.pcp.a029490. [DOI] [PubMed] [Google Scholar]
- 10.Lin L.J., Tai S.S., Peng C.C., Tzen J.T. Steroleosin, a sterol-binding dehydrogenase in seed oil-bodies. Plant Physiol. 2002;128:1200–1211. doi: 10.1104/pp.010982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolivet P., Roux E., D’Andrea S., Davanture M., Negroni L., Zivy M. Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem. 2004;42:501–509. doi: 10.1016/j.plaphy.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Huang A.H.C. Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:177–200. [Google Scholar]
- 13.Qu R., Wang S.M., Lin Y.H., Vance V.B., Huang A.H. Characteristics and biosynthesis of membrane proteins of lipid bodies in the scutella of maize (Zea mays L.) Biochem J. 1986;235:57–65. doi: 10.1042/bj2350057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen E.C., Tai S.S., Peng C.C., Tzen J.T. Identification of three novel unique proteins in seed oil bodies of sesame. Plant Cell Physiol. 1998;39:935–941. doi: 10.1093/oxfordjournals.pcp.a029457. [DOI] [PubMed] [Google Scholar]
- 15.Napier J.A., Stobart A.K., Shewry P.R. The structure and biogenesis of plant oil bodies: the role of the ER membrane and the oleosin class of proteins. Plant Mol Biol. 1996;31:945–956. doi: 10.1007/BF00040714. [DOI] [PubMed] [Google Scholar]
- 16.Murphy D.J. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 17.Frandsen G.I., Mundy J., Tzen J.T. Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant. 2001;112:301–307. doi: 10.1034/j.1399-3054.2001.1120301.x. [DOI] [PubMed] [Google Scholar]
- 18.Tzen J.T., Lai Y.K., Chan K.L., Huang A.H. Oleosin isoforms of high and low molecular weights are present in the oil bodies of diverse seed species. Plant Physiol. 1990;94:1282–1289. doi: 10.1104/pp.94.3.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H.U., Hsieh K., Ratnayake C., Huang A.H. A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem. 2002;277:22677–22684. doi: 10.1074/jbc.M109298200. [DOI] [PubMed] [Google Scholar]
- 20.Mayfield J.A., Fiebig A., Johnstone S.E., Preuss D. Gene families from the Arabidopsis thaliana pollen coat proteome. Science. 2001;292:2482–2485. doi: 10.1126/science.1060972. [DOI] [PubMed] [Google Scholar]
- 21.Jiang P.L., Wang C.S., Hsu C.M., Jauh G.Y., Tzen J.T. Stable oil bodies sheltered by a unique oleosin in lily pollen. Plant Cell Physiol. 2007;48:812–821. doi: 10.1093/pcp/pcm051. [DOI] [PubMed] [Google Scholar]
- 22.Kawanabe T., Ariizumi T., Kawai-Yamada M., Uchimiya H., Toriyama K. Abolition of the tapetum suicide program ruins microsporogenesis. Plant Cell Physiol. 2006;47:784–787. doi: 10.1093/pcp/pcj039. [DOI] [PubMed] [Google Scholar]
- 23.Wu L.S., Wang L.D., Chen P.W., Chen L.J., Tzen J.T. Genomic cloning of 18 kDa oleosin and detection of triacylglycerols and oleosin isoforms in maturing rice and post-germinative seedlings. J Biochem. 1998;123:386–391. doi: 10.1093/oxfordjournals.jbchem.a021949. [DOI] [PubMed] [Google Scholar]
- 24.Tzen J.T., Huang A.H. Surface structure and properties of plant seed oil bodies. J Cell Biol. 1992;117:327–335. doi: 10.1083/jcb.117.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abell B.M., Holbrook L.A., Abenes M., Murphy D.J., Hills M.J., Moloney M.M. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9:1481–1493. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzen J.T., Lie G.C., Huang A.H. Characterization of the charged components and their topology on the surface of plant seed oil bodies. J Biol Chem. 1992;267:15626–15634. [PubMed] [Google Scholar]
- 27.Zienkiewicz K., Zienkiewicz A., Rodríguez-García M.I., Castro A.J. Characterization of a caleosin expressed during olive (Olea europaea L.) pollen ontogeny. BMC Plant Biol. 2011;11:122. doi: 10.1186/1471-2229-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang P.L., Chen J.C., Chiu S.T., Tzen J.T. Stable oil bodies sheltered by a unique caleosin in cycad megagametophytes. Plant Physiol Biochem. 2009;47:1009–1016. doi: 10.1016/j.plaphy.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Naested H., Frandsen G.I., Jauh G.Y., Hernandez-Pinzon I., Nielsen H.B., Murphy D.J. Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol. 2000;44:463–476. doi: 10.1023/a:1026564411918. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh K., Huang A.H. Endoplasmic reticulum, oleosins and oils in seeds and tapetum cells. Plant Physiol. 2004;136:3427–3434. doi: 10.1104/pp.104.051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang P.L., Jauh G.Y., Wang C.S., Tzen J.T. A unique caleosin in oil bodies of lily pollen. Plant Cell Physiol. 2008;49:1390–1395. doi: 10.1093/pcp/pcn103. [DOI] [PubMed] [Google Scholar]
- 32.Richmond S.W., VanRooijen G.J.H., Holbrook L.A., Abenes L., Moloney M.M. Regulation of expression of oleosin isoform genes in Arabidopsis thaliana. Plant Physiol. 1997;114:256–261. [Google Scholar]
- 33.Shimada T.L., Shimada T., Takahashi H., Fukao Y., Hara-Nishimura I. A novel role of oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 2008;55:798–809. doi: 10.1111/j.1365-313X.2008.03553.x. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt M.A., Herman E.M. Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant. 2008;1:910–924. doi: 10.1093/mp/ssn049. [DOI] [PubMed] [Google Scholar]
- 35.Crowe A.J., Abenes M., Plant A., Moloney M.M. The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci. 2000;151:171–181. doi: 10.1016/s0168-9452(99)00214-9. [DOI] [PubMed] [Google Scholar]
- 36.Zienkiewicz K., Castro A.J., Alché Jde D., Zienkiewicz A., Suárez C., Rodríguez-García M.I. Identification and localization of a caleosin in olive (Olea europea L.) pollen during in vitro germination. J Exp Bot. 2010;61:1537–1546. doi: 10.1093/jxb/erq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanano A., Burcklen M., Flenet M., Ivancich A., Louwagie M., Garin J. Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J Biol Chem. 2006;281:33140–33151. doi: 10.1074/jbc.M605395200. [DOI] [PubMed] [Google Scholar]
- 38.Partridge M., Murphy D.J. Role of membrane-bound caleosin and putative peroxygenase in biotic and abiotic stress responses in Arabidopsis. Plant Physiol Biochem. 2009;47:796–806. doi: 10.1016/j.plaphy.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 39.Aubert Y., Leba L.J., Cheval C., Ranty B., Vavasseur A., Aldon D. Involvement of RD20, a member of caleosin family, in ABA-mediated regulation of germination in Arabidopsis thaliana. Plant Signal Behav. 2011;6:538–540. doi: 10.4161/psb.6.4.14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aubert Y., Vile D., Pervent M., Aldon D., Ranty B., Simonneau T. RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1975–1987. doi: 10.1093/pcp/pcq155. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi S., Katagiri T., Yamaguchi-Shinozaki K., Shinozaki K. An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol. 2000;41:898–903. doi: 10.1093/pcp/pcd010. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y.Y., Jung K.W., Yoo K.S., Jeung J.U., Shin J.S. A stress-responsive caleosin-like protein, AtCLO4, acts as a negative regulator of ABA responses in Arabidopsis. Plant Cell Physiol. 2011;52:874–884. doi: 10.1093/pcp/pcr039. [DOI] [PubMed] [Google Scholar]
- 43.Rigden D.J., Woodhead D.D., Wong P.W., Galperin M.Y. New structural and functional contexts of the Dx[DN]xDG linear motif: insights into evolution of calcium-binding proteins. PLoS One. 2011;6:e21507. doi: 10.1371/journal.pone.0021507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole R.L. The TAIR database. Methods Mol Biol. 2007;406:179–212. doi: 10.1007/978-1-59745-535-0_8. [DOI] [PubMed] [Google Scholar]
- 45.Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K. The TIGR rice genome annotation resource. improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grigoriev I.V., Nordberg H., Shabalov I., Aerts A., Cantor M., Goodstein D. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The ClustalX windows interface. flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S., Nei M., Dudley J., Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuster-Böckler B., Schultz J., Rahmann S. HMM logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finn R.D., Tate J., Mistry J., Coggill P.C., Sammut S.J., Hotz H.R. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008;2008:420747. [DOI] [PMC free article] [PubMed]
- 52.Jung K.H., Dardick C., Bartley L.E., Cao P., Phetsom J., Canlas P. Refinement of light-responsive transcript lists using rice oligonucleotide arrays: evaluation of gene-redundancy. PLoS One. 2008;3:e3337. doi: 10.1371/journal.pone.0003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The amino acid alignment of oleosin proteins The gene names are given on left and the numbers of amino acids are shown on right. The conserved proline knot motif is indicated on top of the alignment. The section of alignment containing the proline knot motif is shown. The figure is made as described in materials and methods. At, Arabidopsis thaliana; Os, Oryza sativa; Pt, Populus trichocarpa; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
The sequence logo of the proline knot motif in oleosin proteins The conserved proline residues are indicated with an asterisk on top of the logo.
The amino acid alignment of caleosin proteins The gene names are given on left and the numbers of amino acids are shown on right. The conserved Dx[D/N]xDG motif is indicated on top of the alignment. The part of alignment with the Dx[D/N]xDG is shown. The figure is made as described in materials and methods. At, Arabidopsis thaliana; Os, Oryza sativa; Pt, Populus trichocarpa; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii.
The sequence logo of the Dx[D/N]xDG motif in caleosin proteins The Dx[D/N]xDG motif is shown on top of the alignment. The conserved amino acids flanking the Dx[D/N]xDG motif are indicated with an asterisk on top of the alignment.
The amino acid alignment of steroleosin proteins of A. thaliana (At), O. sativa (Os), P. trichocarpa (Pt), P. patens (Pp) and S. moellendorffii (Sm) The gene names are given on left and the numbers of amino acids are shown on right. The conserved proline knob motif is indicated on top of the alignment. The active site amino acids (S, Y and K) are indicated with down-arrows on top of the alignment. The figure is made as described in materials and methods. The parts of alignment containing the proline knob motif and active site amino acids are shown.
The sequence logo of the active site amino acids S, Y and K in steroleosins The active site amino acids are represented with an asterisk on top.
The gene expression pattern of oleosin (A), caleosin (B), and steroleosin (C) in Arabidopsis The tissue source is given on the X-axis and the relative expression of genes on 22k microarray on the Y-axis. The gene names of oleosins (Oleo), caleosins (Clo) and steroleosins (Slo/HSD) are given on the right. The error bars are for standard error (S.E.). The microarray data were normalized using Affymetrix MAS5 software [44]. The expression data from different sources were scaled up or down using a Target intensity value (TGT) of 200 [44].
The locus IDs and nomenclature for oleosin (Oleo) encoding genes.
The locus IDs and nomenclature for caleosin (Clo) encoding genes.
The locus IDs and nomenclature for steroleosin (Slo) encoding genes.
The microarray probe set IDs, TIGR locus IDs and gene names of oleosins, caleosins and steroleosins in Oryza sativa.