Abstract
Amyloid precursor protein intracellular domain (AICD) is one of the potential candidates in deciphering the complexity of Alzheimer’s disease. It plays important roles in determining cell fate and neurodegeneration through its interactions with several adaptors. The presence or absence of phosphorylation at specific sites determines the choice of partners. In this study, we identified 20 novel AICD-interacting proteins by in vitro pull down experiments followed by 2D gel electrophoresis and MALDI-MS analysis. The identified proteins can be grouped into different functional classes including molecular chaperones, structural proteins, signaling and transport molecules, adaptors, motor proteins and apoptosis determinants. Interactions of nine proteins were further validated either by colocalization using confocal imaging or by co-immunoprecipitation followed by immunoblotting. The cellular functions of most of the proteins can be correlated with AD. Hence, illustration of their interactions with AICD may shed some light on the disease pathophysiology.
Keywords: Amyloid precursor protein intracellular domain, Adaptors, Phosphorylation, Alzheimer’s disease
Introduction
Alzheimer’s disease (AD) is a common age-related neurodegenerative disorder causing dementia among the elderly [1] and is pathologically characterized by the accumulation of amyloid plaques (called ‘senile plaques’) in the brain parenchyma. The main components of these plaques are the amyloid β (Aβ) peptides, Aβ40 and Aβ42, which are known to be neurotoxic [2]. Aβ is a proteolytic peptide fragment cleaved from the Aβ precursor protein [3] by two proteases, β- and γ-secretase [4]. In addition, dense bundles of unusual fibrils of the phosphorylated protein Tau, known as ‘neurofibrillary tangles’ (NFTs), were found in the brains of AD patients during the post-mortem examination [5]. These two molecular lesions are believed to be the primary causative factors in AD pathogenesis and their origins are explained through the “amyloid cascade” and “tau and tangle” hypotheses, respectively [6].
The role of the C-terminal domain (i.e., C-terminal Fragment, CTF) of amyloid precursor protein (APP) remains relatively obscure. However, CTF has recently been shown to be in the center of a complex protein–protein interaction network [7]. Selective phosphorylation of this domain is known to regulate the interaction with the cytosolic phosphotyrosine binding (PTB) domain or Src homology 2 (SH2) domain containing proteins involved in cell signaling [7].
Despite this emerging molecular information, a general understanding of the complexity of the disease is still lacking, which demands an ongoing search for novel candidates [8]. The APP intracellular domain (AICD), one of the CTFs, is one such potential candidate that plays important roles in biological processes including the synthesis and metabolism of APP itself [9]. The amino acid sequence of AICD is more conserved through evolution than that of Aβ [10]. Overexpression of AICD, without any change in Aβ expression, can cause age-dependent features of AD in mice [11]. But most interestingly, AICD, through its interactions with different “adaptor” proteins inside the cell, promotes transcriptional transactivation, Aβ production, APP trafficking and cellular apoptosis [9]. AICD is also involved in modulation of gene expression, cytoskeletal dynamics [8], cellular calcium homeostasis [12] and exerting neurotoxicity by inducing glycogen synthase kinase-3β expression [13]. The putative target genes for AICD include KAI1, TAGLN, GSK3β, LRP1, X11α, mDab1 among others [9].
AICD undergoes post-translational modification events, like phosphorylation of threonine (T) and tyrosine (Y) [14], [15], [16], which influence its downstream interaction with the adaptors. For example, phosphorylated Y682 in the conserved motif 682YENPTY687 of APP acts as the docking site for proteins like ShcA and Grb2 [14], [17], although the interaction of Fe65 with AICD does not require phosphorylation at this site [18].
Keeping this functional involvement of AICD in mind, our objective in this study is to find out novel interacting partners of AICD, especially those without any discernable PTB domains, and try to correlate the functions of those proteins with known AD phenotypes.
To achieve our goals, we used the tools of proteomics. We have recently shown ‘conformational switching’ [19] in AICD upon phosphorylation and in the present work we targeted novel interacting partners of AICD in its non-phosphorylated state. Several of these novel interactors might be useful in understanding the protein interaction network in AD pathogenesis.
Results
Identification of AICD interactors
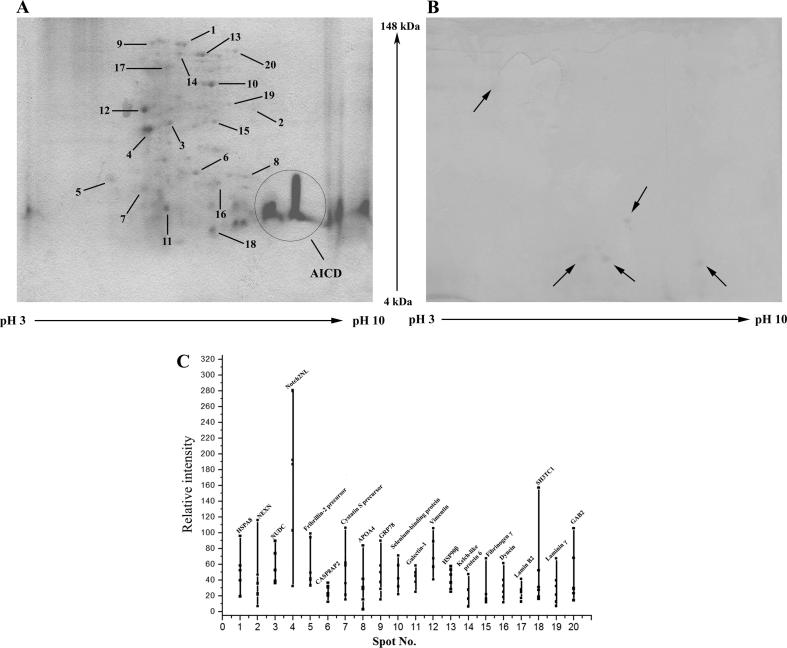
A bacterial expression system was used to express and purify His-AICD so as to minimize any untoward phosphorylation of the protein. We performed pull-down experiments with purified His-AICD as bait and whole cell extracts from regular N2A cells as prey to obtain AICD-interacting partners. Six different pull-down experiments and 2D gel runs were performed. The protein spots which were present in the experimental gel (pull down with AICD) but absent in the control gel (without AICD) were considered as potential AICD-interacting partners. On the other hand, some protein spots were present in the control gel due to non-specific binding of those proteins with the Ni-NTA resin beads and those spots were not taken into consideration. A total of 20 proteins were identified as potential AICD interacting partners. Figure 1 shows a representative 2D gel for a pull-down experiment and all the protein spots identified are marked according to the numbers given in Table 1 (the remaining five gels are included in Figure S1).
Figure 1.
Representative 2D gel pattern of AICD-interacting proteins A. Experimental gel with His-AICD as bait protein. The identified protein spots are marked as 1–20 as listed in Table 1. His-AICD is present as a patch in the gel (marked by a circle) due to high abundance. B. Control gel without His-AICD included in pull-down experiment. The identified protein spots in panel A are absent in the control gel. The marked spots in the control gel are present even in the absence of His-AICD and were excluded from consideration. Both gels were stained according to the blue silver staining procedure using Coomassie G. C. Relative intensity (in arbitrary units) of each spot with respect to control from six different pull-down experiments. The protein names corresponding to the spot numbers are also given. The same molecular weight markers and pH 3–10 IPG strips were used in all the experiments.
Table 1.
List of potential AICD-interacting proteins identified by MALDI-MS
| Spot No. | Protein name | Swiss-Prot accession no. | MW (kDa)/pI | MASCOT score | Molecular function |
|---|---|---|---|---|---|
| 1 | HSPA8 | P11142 | 71.1/5.37 | 247 | Molecular chaperone |
| 2 | NEXN protein (fragment) | Q7Z2X0 | 26.6/8.97 | 93 | Regulation of cell migration |
| 3 | NPD011/NUDC | Q9Y266 | 38.0/5.71 | 75 | Neurogenesis & neuronal migration |
| 4 | Notch2NL | Q7Z3S9 | 26.0/5.22 | 122 | Involved in notch signaling pathway |
| 5 | Fibrillin-2 precursor | P35556 | 314.8/4.76 | 142 | Structural components of extracellular calcium-binding microfibril |
| 6 | CASP8AP2 (fragment) | Q5T791 | 220.3/6.08 | 91 | Involved in apoptotic signaling |
| 7 | Cystatin S precursor | P01036 | 16.2/4.95 | 106 | Strongly inhibits papain and ficin |
| 8 | APOA4 | P06727 | 22.0/9.34 | 201 | May have a role in VLDL secretion and catabolism |
| 9 | GRP78/HSPA5 | P11021 | 72.1/5.03 | 226 | Facilitates multimeric protein complex assembly inside ER |
| 10 | Selenium-binding protein | Q13228 | 52.3/6.13 | 136 | May be involved in intra-Golgi protein transport |
| 11 | Galectin-1 | P09382 | 14.6/5.33 | 83 | May regulate apoptosis, cell proliferation and differentiation |
| 12 | Vimentin | P08670 | 53.5/5.06 | 139 | Class III intermediate filaments |
| 13 | HSP90β | P14625 | 62.6/6.4 | 198 | Molecular chaperone, with ATPase activity |
| 14 | Kelch-like protein 6 | Q8WZ60 | 69.1/5.83 | 53 | B-lymphocyte antigen receptor signaling |
| 15 | Fibrinogen γ | P02679 | 51.0/5.68 | 100 | Monomers that polymerize into fibrin |
| 16 | Dynein, axonemal | Q0VDD8 | 76.0/6.01 | 70 | Microtubule-associated motor protein |
| 17 | Lamin B2 | Q03252 | 67.6/5.29 | 55 | Component of nuclear lamina |
| 18 | SH3TC1 | Q8TE82 | 146.8/5.85 | 17 | Exact function is not known |
| 19 | Laminin γ | Q9Y6N6 | 171.9/6.29 | 27 | Attachment, migration and organization of cells into tissues |
| 20 | GAB2 | Q9UQC2 | 74.4/8.54 | 66 | Adaptor for transmitting various signals |
Note: Molecular functions for the identified proteins were obtained from the Uniprot database. MW, molecular weight; SH3TC1, SH3 domain and tetra-tricopeptide repeats-containing protein 1.
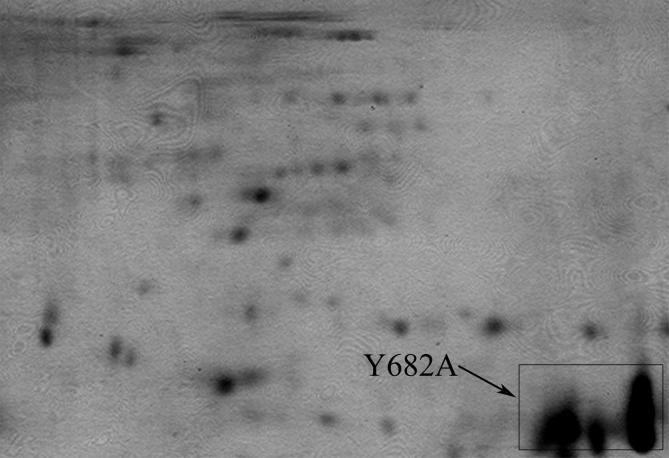
In addition, we also performed the pull-down experiment using purified mutated AICD (Y682A) as bait. As shown in Figure S2, the 2D gel pattern was indistinguishable from those using the purified wild type protein. MS analysis for these spots have identified the same proteins (data not shown). These data indicated that no PTB protein was identified as an AICD interacting partner, as expected. Since the bacterial expression system employed is this study could not guarantee any site-specific phosphorylation (e.g., at Y and T) of the overexpressed proteins, we did not recover any of the previously reported AICD-interacting proteins, especially the PTB proteins interacting specifically to the ‘NPTY’ motif. Hence, our results provided an initial idea about a subset of interacting partners of AICD which might interact with some other parts of the domain.
The function annotation of the identified proteins was obtained from Uniprot database. We found that these 20 proteins could be grouped into several functional classes including molecular chaperones, structural proteins, proteins related to signaling and transport, adaptor proteins, motor proteins and apoptosis-related proteins. The identified proteins and their molecular functions along with the MASCOT score, Swissprot accession numbers and MW/pI are listed in Table 1. The detailed MALDI-MS spectra and images of the significant hits based on MOWSE scores are available on request from the authors.
Validation of the identified AICD-interacting partners
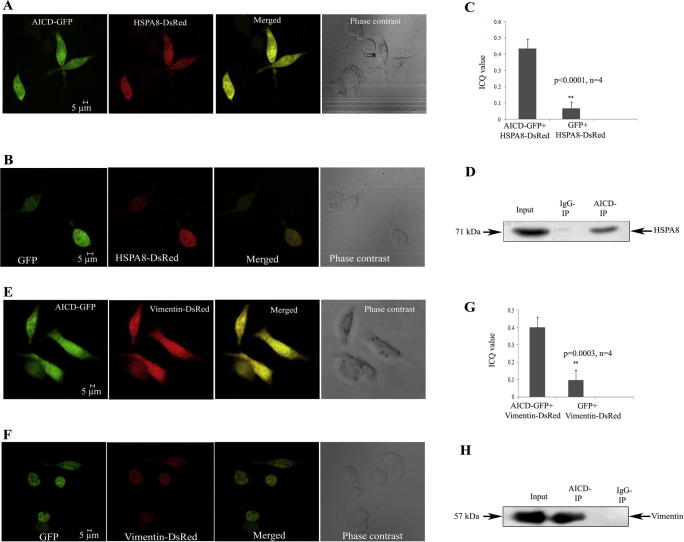
Validation for nine of the identified proteins was done by co-immunoprecipitation (co-IP) experiments. Furthermore, heat shock 70 kDa protein 8 (HSPA8) and vimentin, were also validated by co-localization studies using confocal imaging.
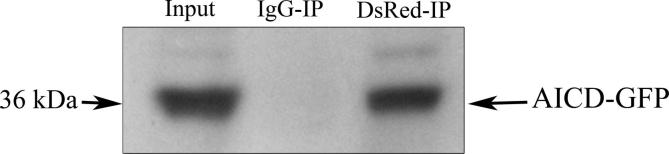
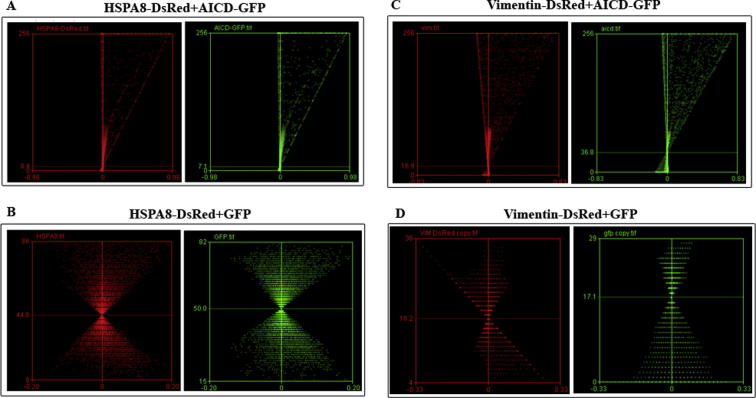
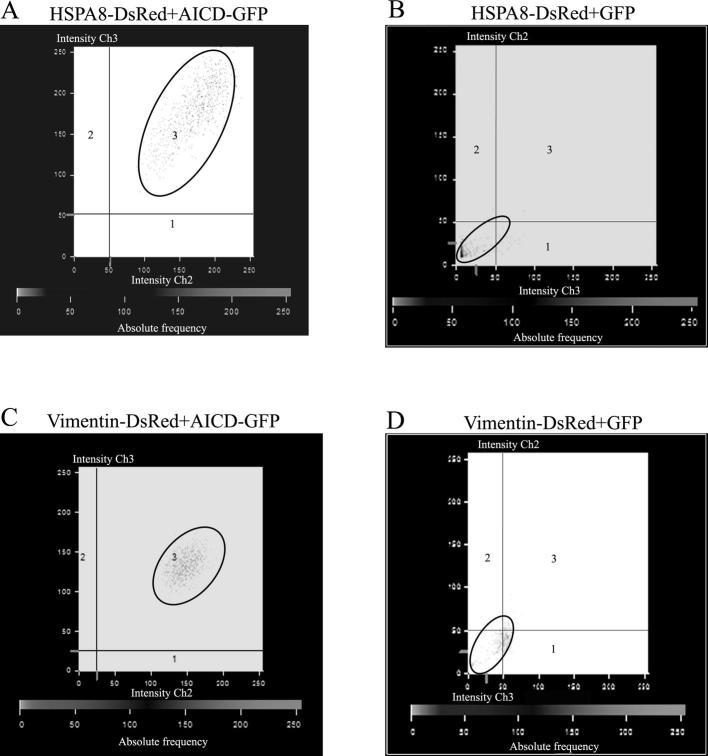
The interactions of AICD with HSPA8 and vimentin were verified by confocal microscopy as well as by co-IP (Figure 2). AICD-GFP and HSPA8-DsRed were co-transfected into Neuro2A (N2A) cells while cells co-transfected with HSPA8-DsRed and empty GFP vector served as negative control. The colocalization of AICD-GFP with HSPA8-DsRed in N2A cells (Pearson’s coefficient value, R2 = 0.82) was high (Figure 2A). On the other hand, HSPA8-DsRed did not show significant colocalization with empty GFP vector (R2 = 0.13) in the negative control experiment (Figure 2B). The difference in the averaged intensity correlation quotient (ICQ) values, which indicates the correlation of two different signals coming from the same pixel, obtained from these two experiments (0.43 ± 0.06 for the experimental set and 0.07 ± 0.03 for the control set, n = 4) was found to be significant (P < 0.0001) (Figure 2C). In the co-IP experiment, presence of HSPA8-DsRed was detected in the whole cell extract as well as in the AICD-immunoprecipitate with anti-AICD antibody, but not in the control IgG-immunoprecipitate in absence of anti-AICD antibody using western blot, which verified their interaction (Figure 2D). Moreover, a reciprocal co-IP experiment was performed and presence of AICD-GFP was verified in the HSPA8-DsRed immunoprecipitate (Figure S3).
Figure 2.
Validation of interaction of AICD with HSPA8 and vimentin A. Colocalization of AICD-GFP and HSPA8-DsRed in N2A cells. Magnification bars are included. Pearson’s correlation coefficient (R2) for colocalization is 0.82. B. Colocalization pattern of empty GFP vector with HSPA8-DsRed. No significant colocalization was obtained. Pearson’s correlation coefficient (R2) for colocalization is 0.13. C. Bar graph showing the significant difference in normalized ICQ for colocalization between AICD-GFP and HSPA8-DsRed with that between empty GFP vector and HSPA8-DsRed (n = 4, P < 0.0001). D. Western blots showing co-IP of HSPA8-DsRed with AICD-GFP. Bound proteins were immunoprecipitated with anti-AICD antibody and blotted with anti-DsRed antibody. E. Colocalization of AICD-GFP and vimentin-DsRed in N2A cells. Pearson’s correlation coefficient (R2) for colocalization is 0.91. F. Colocalization pattern of empty GFP vector with Vimentin-DsRed. No significant colocalization was obtained. Pearson’s correlation coefficient (R2) is 0.36. G. Bar graph showing the significant difference in ICQ for colocalization between AICD-GFP and Vimentin-DsRed to that between empty GFP vector and Vimentin-DsRed (n = 4, P = 0.0003). H. Western blots showing co-IP of vimentin-DsRed with AICD-GFP. Bound proteins were immunoprecipitated with anti-AICD antibody and blotted with anti-DsRed antibody.
Similarly, validation of the interaction between AICD and vimentin was also carried out by confocal imaging as well as by co-IP. Significant colocalization was revealed between AICD-GFP and vimentin-DsRed (Figure 2E, R2 = 0.91) but not in case of vimentin-DsRed and empty GFP vector (Figure 2F, R2 = 0.36). Both AICD and vimentin were observed to be co-localized in cytosol as well as in the nucleus, which is consistent with the cellular distribution reported elsewhere [20]. A significant difference (n = 4; P = 0.0003) in the averaged ICQ values between experimental (0.40 ± 0.09) and control (0.08 ± 0.06) sets of experiments was apparent for vimentin (Figure 2G). In addition, our co-IP experiments showed that vimentin-DsRed was detected in the whole cell extract and in the AICD-immunoprecipitate but absent in the control IgG-immunoprecipitate (Figure 2H).
Representative images of the ICQ analysis for each of the confocal experiments are included in Figure S4 (A–D).
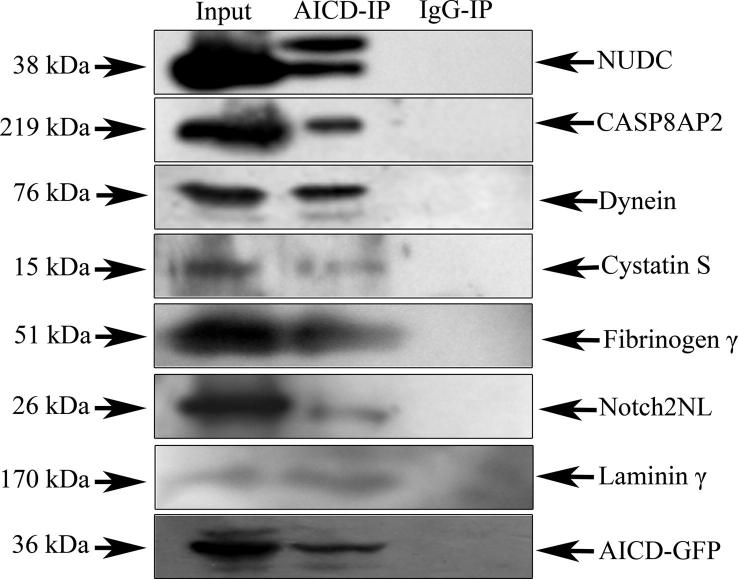
Validation of interaction with AICD for seven other proteins namely, NUDC, notch2NL, cystatin S precursor, dynein, fibrinogen γ, laminin γ and CASP8AP2, were done by co-IP experiments followed by western blotting (Figure 3). Presence of each of the above-mentioned proteins in western blot clearly verified the interactions and validated our results.
Figure 3.
Validation of interaction of AICD with seven other proteins Interactions of seven other potential interacting partners of AICD namely NUDC, CASP8AP2, dynein, cystatin S precursor, fibrinogen γ, notch2NL and laminin γ were validated by co-IP followed by western blot. Western blot of co-IP with each protein shows presence of target protein in whole cell extract (input) as well as in AICD-immunoprecipitate sample (AICD-IP) and absence in the control immunoprecipitate sample (IgG-IP). A representative blot reprobed for AICD-GFP was shown in the bottom panel.
Discussion
AICD is known to have two non-overlapping repertoires of interactors depending on its state of phosphorylation [9]. In their study with phosphorylated forms of AICD, Tamayev et al. have shown that mutually exclusive phosphorylation at Y682 or T668 changed its interactome pattern [21]. They have also reported that AICD can bind to some proteins like NUMB in a phosphorylation-independent manner [22]. For an intrinsically unstructured protein (IUP) like AICD, a “conformational switch” upon phosphorylation at Y682 is now established, ensuring different binding interfaces for two different genres of adaptors. The range of alteration of binding for non-PTB binding partners, when Y682 is mutated, needs to be checked spectroscopically in future. In a real scenario there should be a balance of phosphorylated to non-phosphorylated forms of AICD within the cell. In our study, we have emphasized the later part of the interactome and, as expected, have not found any of the known interactors of AICD which have been reported to be PTB domain-containing proteins requiring phosphorylation of AICD at either Y682 or T668. To ensure that our targets were interacting partners of AICD independent of any phosphorylation event, AICD was expressed in a bacterial expression system where site-specific phosphorylation could be ruled out, and further possibility of any enzymatic modification at specific position of resin-bound AICD should be minimal. The cholinergic hypothesis behind hippocampal vulnerability in Alzheimer’s disease has been supported very recently [23]. Keeping this in mind, we have chosen the Neuro2A cell line where the signaling was choline driven, expecting direct lines of newly identified interacting partners and AD-like condition. Since the interactions of AICD with other proteins within the cell might have a cumulative influence on the diseased condition, our goal was to identify as many numbers of interacting partners of AICD and to assess the functional implications of these interactions.
Of the 20 interactors that were deciphered (Table 1), seven proteins namely laminin [24], HSPA8, HSP90β, GRP78/HSPA5 [25], [26], GAB2 allele [27], vimentin [28], and cystatin C [29] were already described in the literature to be involved in the AD process. The fact that AICD interacted with these proteins gave a new dimension to it. Several others like selenium-binding protein, galectin-1, kelch-like protein 6 and SH3 domain and tetra-tricopeptide repeats-containing protein have never been reported in connection to AD and any guess regarding their functional significance would be premature.
A neurodegenerative disease like AD is caused by the gain of toxic function of misfolded proteins/peptides (Aβ and Tau) and the shift of balance towards misfolded proteins could be ascribed to aberrant chaperone activities [25]. Delineation of interactions of AICD with molecular chaperones may possibly illustrate its involvement in disease pathogenesis. A number of studies have recently been reported explaining the possible role of molecular chaperones such as the heat shock proteins (HSPs) in AD [26], [30]. These interactions might disrupt the protein folding machinery and increase the proportion of misfolded proteins in the cytosol. For example, cytosolic HSP70 and HSP90 were implicated in inhibiting early amyloid aggregates [31] and the equilibrium of the reaction could be disrupted due to their involvement in the interaction with AICD.
The caspase family of proteins were reported to be required for apoptotic signaling within the cell and for induction of neuronal apoptosis by β-amyloid, especially by caspase8 [32]. The interaction of AICD with caspase 8 associated protein 2 (CASP8AP2) fragment, therefore, would indicate an indirect association of AICD in neuronal apoptosis. AICD was reported to be involved in induction of neuron-specific apoptosis [10], and hence, as a component of the caspase complex, it could possibly enhance the apoptotic signal by caspase 8.
The protein cystatin C, on the other hand, was reported to inhibit Aβ deposition in the AD mouse model [29]. Both cystatin C and cystatin S are members of the cystatin superfamily [33], the interaction of AICD with cystatin S precursor, as we have shown, might show a distant line between the proteolytic generation of these toxic fragments and their subsequent deposition.
Apart from an aggregation issue, loss of structural integrity is a deleterious feature of AD [34]. The structural proteins fibrillin-2 precursor, fibrinogen γ and vimentin, which were identified as AICD interactors in this study, should have important roles in cellular disintegration. Fibrinogen γ-A chain precursor was previously reported as a candidate CSF biomarker for AD [35], which is in agreement with our result. The interaction of protein fibrillin2, a structural component of extracellular Ca2+-binding microfibril, with AICD could relate AICD to cellular calcium homeostasis. In addition, we also revealed two important traffic components as AICD interactors. These include dynein, which is a microtubule based molecular motor involved in vesicular transport, mitosis and ciliary and flagellar beating [36], and NudC, which is a dynein-associated nuclear movement protein involved in mitosis and cytokinesis [37]. In an AD-like cellular model, AICD is transported via endocytic vesicles in association with growth factor receptor-bound protein 2 (Grb2) in the cytosol [38]. Grb2 is known to interact with dynein via its N-terminal SH2 domain [39]. As AICD is also transported into the nucleus and influences cytoskeletal dynamics [8], its interaction with a dynein complex therefore is significant. Additionally, NUDC plays roles in neurogenesis and neuronal migration and AICD may affect these processes by interacting with NUDC. Two other novel interactors including LaminB2, a component of the inner nuclear membrane and the nuclear envelope framework [40], and NEXN, a filamentous actin-binding protein involved in cell migration [41], imply the possibility of AICD’s involvement in cell motility and division. Together with our observations regarding the dynein complex, these data suggests the involvement of AICD in the loss of cellular integrity, which is a novel aspect that should be explored further.
In addition to cellular disintegration, generation of pathologically important proteolytic fragments of APP would destabilize cellular homeostasis and dynamics. Major disruptions in lipogenic pathways in AD are well established. Apolipoprotein A-IV (ApoA4), a cholesterol-binding lipoprotein, is reported to be associated with AD pathology [42] and its interaction with AICD signifies similar disruption possibilities. Another finding, laminin, has been implicated in a wide spectrum of biological processes including cell adhesion, differentiation, migration, signaling, neurite outgrowth and metastasis [43]. Interestingly, laminin was reported to exert anti-amyloidogenic effects in rat primary hippocampal neurons, by interacting with Aβ and inhibiting amyloid fibril formation [24]. The interaction of AICD with laminin γ may have far-reaching effects on the disease process.
Notch, which is well known in cell proliferation, is processed similarly to APP and its intracellular domain (NICD) goes directly into the nucleus [44]. NICD competitively inhibits AICD-Tip60-Fe65 complex to affect neuronal cell death [45]. Our observation of notch 2 N-terminal like (Notch2NL) as an interacting partner of AICD precludes the possibility of a similar inhibitory effect on intracellular AICD dynamics.
There are reports that Grb2-associated binding protein 2 (GAB2) influences AD neuropathology in ApoEε4 carrier individuals [27] and is an AD susceptibility gene [46]. Understandably, it is an important protein in respect of AD pathogenesis which we have obtained as an AICD-interacting partner.
Despite the lack of direct biophysical examinations like fluorescence resonance energy transfer (FRET) or isothermal titration calorimetry (ITC) to support interactions of each pair, the list of identified novel interactors of AICD presented in this work enriches the repertoire of functions that a natively-unfolded protein could achieve post generation. Starting from early survival events like prevention of protein aggregates, altered cellular motility, vesicle trafficking to cell death, via cytoskeletal restructuring, AICD holds promise as an important modulator downstream of intramembrane proteolysis. Despite initial scepticism regarding the stability of this CTF [3], [47], recent results support its role in creating AD-like phenotypes in mouse model [11]. It is possible that the initial scepticism regarding the stability of AICD came out of experiments which followed non-amyloidogenic processing of APP on the plasma membrane. Recent reports, on the contrary, focussed on amyloidogenic lysosome membrane centric events resulting in the generation of a relatively stable AICD having downstream cellular consequences. Expanding the AICD interactome and delineating its newer functionalities would help resolve this apparent ambiguity.
Materials and methods
Molecular cloning of AICD
The nucleotide sequence coding the human AICD fragment (NCBI Accession number NP_000475.1; amino acid 724-770 of the whole APP protein) was cloned into the bacterial expression vector pET28a+ and also into mammalian expression vector pEGFP (C-terminal fusion) as described earlier [19], [38]. The mutant His-AICD (Y682A) in pET28a+ was generated using site-directed mutagenesis. HSPA8-DsRed-C1 and Vimentin-DsRed-C1 constructs were obtained as generous gifts from Prof. Nitai P. Bhattacharyya of Saha Institute of Nuclear Physics, Kolkata. All constructs were confirmed by sequencing.
Expression and purification of His-AICD
The plasmid containing wild type AICD or mutant AICD (Y682A) in pET28a+ was transformed using the CaCl2 transformation method into the Escherichia coli BL21-DE3 for protein expression. As described earlier, His-tagged proteins were purified using Ni-NTA resin column [19]. The concentration of the His-AICD proteins (wild type and mutant) was measured using the Bradford protein assay. This purified AICD and the mutant proteins were used as the “bait” protein in the pull-down assay.
Cell culture and extraction of Neuro2A proteins
N2A cells were grown at 37 °C in a humidified 5% CO2 incubator in DMEM media supplemented with 10% fetal bovine serum (FBS). For confocal imaging or co-IP experiments, log phase-N2A cells were transfected with constructs of interest using Lipofectamine Reagent (Invitrogen, USA) (ratio 1:3 for DNA:lipofectamine) as instructed by the manufacturer to obtain transient overexpression of the target proteins.
After confluency, the cells were pellet down and washed with cold 1 × phosphate buffered saline (PBS) on ice. The cell pellets were resuspended in lysis buffer (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 150 mM NaCl, 2 mM PMSF, 0.5% NP-40 and 1 × protease inhibitor cocktail, Pierce) and rapid freezing and thawing was carried out to rupture the cell walls. After centrifugation at 13,000 rpm for 15 min at 4 °C, the supernatant containing the whole cell proteins was collected and used as the “prey” protein in the pull-down assay or as input for the co-IP experiments.
Pull-down experiment
Pull-down experiments were performed using the ProFound™ Pull-Down PolyHis Protein: Protein Interaction Kit (Thermo Scientific, Prod No. 21277). Purified His-AICD was used as bait and non-transfected N2A cell extract was used as prey in this experiment. Purified His-AICD was bound to the Ni-NTA based resin column for 4 h. After washing thrice with lysis buffer to remove unbound AICD, AICD-bound resin was incubated overnight with N2A whole cell extract. For 200 μg of bait protein, 800 μg of prey protein was used. This experiment was repeated several times to obtain a reproducible pattern of spots on 2D gel. Every time a control experiment was done by following the same pull-down protocol in absence of bait protein (His-AICD). After discarding the flow-through and subsequent washing with lysis buffer, the interaction complex as well as the control fraction was eluted using 300 mM imidazole. In addition, we also performed the same pull-down experiment using mutated His-AICD protein as bait where a point mutation, Y682A, was introduced in the ‘NPTY’ motif of AICD.
Two-dimensional gel electrophoresis
The eluted protein complex from the pull-down experiment was acetone precipitated and the proteins were pelleted by centrifugation at 12,000 rpm for 10 min. The protein pellet was dissolved in 2D rehydration buffer (8 M urea, 4% CHAPS, 2% DTT, 2% ampholyte pH 3–10 and 30 mM Tris–HCl pH 8.5) and IPG strips were rehydrated with the experimental sample and the control. After the first dimension (isoelectric focusing), IPG strips were equilibrated for 15 min in reducing equilibration buffer (50 mM Tris–HCl pH 8.8, 6 M urea, 30% glycerol, 2% SDS and 50 mM DTT) followed by 15 min in alkylating equilibration buffer containing 2.5% (w/v) iodoacetamide instead of DTT.
After gel electrophoresis, the 2D gels were stained with coomassie brilliant blue or silver or blue silver staining [48]. Different staining procedures were used in different experiments depending on the amount of protein present in the eluted fraction after pull-down. After staining, the protein spots were compared between the experimental and the control gel. The proteins spots which were present in the experimental gel but absent in the control gel were considered to be potential AICD-interacting partners. We compared the relative intensity (in arbitrary units) of each unique spot in the experimental gel to that in the control gel by selecting the same spot volume. The ratios of intensity for each spot obtained in at least five among the total six different pull-down experiments were calculated and the relative intensities were plotted (Figure 1C). For some of the proteins (e.g., spot 9), the corresponding spot was detected in all the six gels, which were all included in the plot. The spots were excised from the gel and processed for the subsequent MALDI-MS identification.
MALDI-MS analysis and protein identification
The excised spots were destained and then tryptic digestion was performed overnight In-Gel tryptic Digestion Kit (Pierce, USA). The digestion mixture was lyophilized in a Heto vacuum centrifuge (Thermo) and resuspended in 50% Acetonitrile (ACN) and 0.1% Trifluoroacetic acid (TFA) solution. The dissolved samples were mixed 1:1 (v/v) with α-Cyano-4-hydroxycinnamic acid (CHCA) matrix and spotted on the MALDI plate and air dried. MALDI analyses were performed with a 4700 Proteomic Analyzer MALDI-TOF/TOF (Applied Biosystems) for identification of the protein spots. Peptide mass fingerprinting data was acquired in positive MS reflector mode using a fixed laser intensity of 5500 with 2000–3000 laser spots in the range 800–4000 Da, signal to noise ratio 10 and mass exclusion tolerance of 150 ppm. Internal calibration was done in the instrument with minimum signal to noise of 20 and mass tolerance of ±300 ppm taking monoisotopic peaks only. The isolation of peptides of interest was carried out at a relative resolution of 50 (full width at half maximum) and data from 3000 to 5000 laser shots were collected. Spectral data was analyzed with GPS Explorer™ version 3.6 (Applied Biosystems). Peptide identification was based on the MASCOT database scoring algorithm from Swiss-Prot and NCBI protein databases using search settings of: single missed tryptic cut, fixed carbamidomethylation, variable methionine oxidation and N-terminal acetylation and 150 ppm mass accuracy. Autolytic tryptic peaks were excluded in the MASCOT search parameter and P < 0.05 was considered significant during identification. Since we had taken a gel-based proteomics approach, we could identify specific proteins from a particular spot according to the molecular weight and pI from the protein list given by MASCOT server.
Confocal microscopy
N2A cells were transfected with AICD-GFP in combination with HSPA8-DsRed or vimentin-DsRed. A vector control was set by overexpressing empty GFP vector with either of HSPA8-DsRed and vimentin-DsRed in N2A cells to nullify the contribution of GFP in the colocalization studies.
An LSM 510 META confocal laser scanning microscope (Carl Zeiss, Jena, Germany) was used for the imaging studies and images were captured with either 40× or 63× objectives. The extent of colocalization was determined by the square of the Pearson correlation coefficient value (Rp), where this squared value, R2, was defined as the percentage of variance of the first channel which could be explained by or which was dependent on the variance of the second channel. Only when this percentage of overlap between the two channels exceeded 0.50, did we consider those to have significant colocalization. As Pearson’s coefficient might not be sensitive to the intensity of the background, we performed intensity correlation analysis (ICA) as described by Li et al. [49] and obtained the intensity correlation quotient (ICQ) to test whether the color overlap was dependent (0 < ICQ ⩽ 0.5) or random (ICQ∼0) using ImageJ open-source software (http://www.rsb.info.nih.gov/ij). No thresholding was applied for the ICQ analyses. For statistical analysis, unpaired t-test was performed to compare the normalized ICQ values of the negative controls and experimental sets for both HSPA8 and vimentin using the online software GraphPad QuickCalcs.
Co-immunoprecipitation
For the interaction validation of HSPA8 or vimentin with AICD, N2A cells were transiently co-transfected with AICD-GFP and either of vimentin-DsRed or HSPA8-DsRed constructs. After harvesting, soluble cell extracts were prepared similarly as described previously (see “Cell culture and extraction of Neuro2A proteins”) from these co-transfected N2A cells. Sixty micrograms of the whole cell extract was used as input in each experiment and 200 μg extract was incubated overnight with anti-mouse AICD antibody (monoclonal, Chemicon, Cat. No. mab343) and Protein A/G beads. After SDS–PAGE and western transfer, presence of particular protein in the AICD interaction complex was revealed by immunoblotting with anti-DsRed primary antibody (Clontech, rabbit polyclonal, Cat. No. 632496). In addition, a reciprocal co-immunoprecipitation was also performed for interaction between HSPA8 and AICD. Whole cell extract from N2A cells expressing HSPA8-DsRed and AICD-GFP was incubated with anti-DsRed antibody and the complex was blotted with anti-GFP antibody (Clontech, mouse monoclonal, Cat. No. 632375).
For validating the interaction of AICD with other 7 proteins, N2A cells were transfected with AICD-GFP and co-IP was performed using anti-AICD antibody to pull the AICD-interaction complex and the immunoblots were probed with antibody against the endogenous protein of interest (AICD-interacting protein). A negative control was set for each of the interaction validations using mouse anti-IgG primary antibody instead of anti-AICD antibody to rule out non specific binding of the proteins with the A/G beads. The immunoblot was reprobed with anti-AICD antibody to verify the presence of AICD in the complex. Mouse monoclonal antibodies to dynein (Cat. No. ab23905), notch2NL (Cat. No. ab57806) and NUDC (Cat. No. ab56999) were purchased from Abcam. Antibodies to cystatin S (mouse monoclonal, Cat. No. sc133496), fibrinogen γ (mouse monoclonal, Cat. No. sc133157), laminin γ 3 (rabbit polyclonal, Cat. No. sc25719) and CASP8AP2 (rabbit polyclonal, Cat. No. sc9088) were purchased from Santa Cruz Biotechnology.
Authors’ contributions
DM conceived the idea and planned the project. AC designed and carried out the experiments. DM and AC prepared the manuscript. Both authors read and approved the final manuscript.
Competing interests
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the Structural Proteomics and Genomics of Human Genetic Disorders Project of Department of Atomic Energy, Govt. of India. AC acknowledges Council of Scientific and Industrial Research, Govt. of India for his fellowship.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.07.002.
Supplementary material
Supplementary Figure 1.
Supplementary Figure S1
Supplementary Figure 2.
Supplementary Figure S2
Supplementary Figure 3.
Supplementary Figure S3
Supplementary Figure 4.
Supplementary Figure S4
Supplementary Figure 5.
Supplementary Figure S5
Supplementary material
References
- 1.Selkoe D.J. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 2.Yankner B.A. Mechanisms of neuronal degeneration in Alzheimer’s disease. Neuron. 1996;16:921–932. doi: 10.1016/s0896-6273(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 3.Nunan J., Shearman M.S., Checler F., Cappai R., Evin G., Beyreuther K. The C-terminal fragment of the Alzheimer’s disease amyloid protein precursor is degraded by a proteasome-dependent mechanism distinct from gamma-secretase. Eur J Biochem. 2001;268:5329–5336. doi: 10.1046/j.0014-2956.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- 4.Wilquet V., Strooper B.D. Amyloid-beta precursor protein processing in neurodegeneration. Curr Opin Neurobiol. 2004;14:582–588. doi: 10.1016/j.conb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Chen F., David D., Ferrari A., Götz J. Posttranslational modifications of tau-role in human tauopathies and modeling in transgenic animals. Curr Drug Targets. 2004;5:503–515. doi: 10.2174/1389450043345236. [DOI] [PubMed] [Google Scholar]
- 6.Mudher A., Lovestone S. Alzheimer’s disease-do tauists and baptists finally shake hands? Trends Neurosci. 2002;25:22–26. doi: 10.1016/s0166-2236(00)02031-2. [DOI] [PubMed] [Google Scholar]
- 7.Russo C., Venezia V., Repetto E., Nizzari M., Violani E., Carlo P. The amyloid precursor protein and its network of interacting proteins: physiological and pathological implications. Brain Res Brain Res Rev. 2005;48:257–264. doi: 10.1016/j.brainresrev.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Müller T., Meyer H.E., Egensperger R., Marcus K. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer’s disease. Prog Neurobiol. 2008;85:393–406. doi: 10.1016/j.pneurobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Raychaudhuri M., Mukhopadhyay D. AICD and its adaptors-in search of new players. J Alzheimers Dis. 2007;11:343–358. doi: 10.3233/jad-2007-11311. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama K., Ohkawara T., Hiratochi M., Koh C.S., Nagase H. The intracellular domain of amyloid precursor protein induces neuron-specific apoptosis. Neurosci Lett. 2008;444:127–131. doi: 10.1016/j.neulet.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 11.Ghosal K., Vogt D.L., Liang M., Shen Y., Lamb B.T., Pimplikar S.W. Alzheimer’s disease-like pathological features in transgenic mice expressing the APP intracellular domain. Proc Natl Acad Sci U S A. 2009;106:18367–18372. doi: 10.1073/pnas.0907652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamid R., Kilger E., Willem M., Vassallo N., Kostka M., Bornhövd C. Amyloid precursor protein intracellular domain modulates cellular calcium homeostasis and ATP content. J Neurochem. 2007;102:1264–1275. doi: 10.1111/j.1471-4159.2007.04627.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim H.S., Kim E.M., Lee J.P., Park C.H., Kim S., Seo J.H. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J. 2003;17:1951–1953. doi: 10.1096/fj.03-0106fje. [DOI] [PubMed] [Google Scholar]
- 14.Tarr P.E., Roncarati R., Pelicci G., Pelicci P.G., D’Adamio L. Tyrosine phosphorylation of the beta-amyloid precursor protein cytoplasmic tail promotes interaction with Shc. J Biol Chem. 2002;277:16798–16804. doi: 10.1074/jbc.M110286200. [DOI] [PubMed] [Google Scholar]
- 15.Ando K., Iijima K.I., Elliott J.I., Kirino Y., Suzuki T. Phosphorylation-dependent regulation of the interaction of amyloid precursor protein with Fe65 affects the production of beta-amyloid. J Biol Chem. 2001;276:40353–40361. doi: 10.1074/jbc.M104059200. [DOI] [PubMed] [Google Scholar]
- 16.Iijima K., Ando K., Takeda S., Satoh Y., Seki T., Itohara S. Neuron-specific phosphorylation of Alzheimer’s beta-amyloid precursor protein by cyclin-dependent kinase 5. J Neurochem. 2000;75:1085–1091. doi: 10.1046/j.1471-4159.2000.0751085.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou D., Noviello C., D’Ambrosio C., Scaloni A., D’Adamio L. Growth factor receptor-bound protein 2 interaction with the tyrosine-phosphorylated tail of amyloid beta precursor protein is mediated by its Src homology 2 domain. J Biol Chem. 2004;279:25374–25380. doi: 10.1074/jbc.M400488200. [DOI] [PubMed] [Google Scholar]
- 18.Borg J.P., Ooi J., Levy E., Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S., Raychaudhuri M., Sen U., Mukhopadhyay D. Functional implications of the conformational switch in AICD peptide upon binding to Grb2-SH2 domain. J Mol Biol. 2011;414:217–230. doi: 10.1016/j.jmb.2011.09.046. [DOI] [PubMed] [Google Scholar]
- 20.Hartig R., Shoeman R.L., Janetzko A., Tolstonog G., Traub P. DNA-mediated transport of the intermediate filament protein vimentin into the nucleus of cultured cells. J Cell Sci. 1998;111:3573–3584. doi: 10.1242/jcs.111.24.3573. [DOI] [PubMed] [Google Scholar]
- 21.Tamayev R., Zhou D., D’Adamio L. The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol Neurodegener. 2009;4:28. doi: 10.1186/1750-1326-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamayev R., Zhou D., D’Adamio L. The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling. Proc Natl Acad Sci U S A. 2002;99:7102–7107. doi: 10.1073/pnas.102192599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig L.A., Hong N.S., McDonald R.J. Revisiting the cholinergic hypothesis in the development of Alzheimer’s disease. Neurosci Biobehav Rev. 2011;35:1397–1409. doi: 10.1016/j.neubiorev.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Morgan C., Inestrosa N.C. Interactions of laminin with the amyloid beta peptide. Implications for Alzheimer’s disease. Braz J Med Biol Res. 2001;34:597–601. doi: 10.1590/s0100-879x2001000500006. [DOI] [PubMed] [Google Scholar]
- 25.Barral J.M., Broadley S.A., Schaffar G., Hartl F.U. Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Koren J., III, Jinwal U.K., Lee D.C., Jones J.R., Shults C.L., Johnson A.G. Chaperone signalling complexes in Alzheimer’s disease. J Cell Mol Med. 2009;13:619–630. doi: 10.1111/j.1582-4934.2008.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiman E.M., Webster J.A., Myers A.J., Hardy J., Dunckley T., Zismann V.L. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin E.C., Acharya N.K., Sedeyn J.C., Venkataraman V., D’Andrea M.R., Wang H.Y. Neuronal expression of vimentin in the Alzheimer’s disease brain may be part of a generalized dendritic damage-response mechanism. Brain Res. 2009;17:194–207. doi: 10.1016/j.brainres.2009.08.072. [DOI] [PubMed] [Google Scholar]
- 29.Mi W., Pawlik M., Sastre M., Jung S.S., Radvinsky D.S., Klein A.M. Cystatin C inhibits amyloid-beta deposition in Alzheimer’s disease mouse models. Nat Genet. 2007;39:1440–1442. doi: 10.1038/ng.2007.29. [DOI] [PubMed] [Google Scholar]
- 30.Dou F., Netzer W.J., Tanemura K., Li F., Hartl F.U., Takashima A. Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci U S A. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans C.G., Wisén S., Gestwicki J.E. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1–42) aggregation in vitro. J Biol Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 32.Ivins K.J., Thornton P.L., Rohn T.T., Cotman C.W. Neuronal apoptosis induced by beta-amyloid is mediated by caspase-8. Neurobiol Dis. 1999;6:440–449. doi: 10.1006/nbdi.1999.0268. [DOI] [PubMed] [Google Scholar]
- 33.Barka T., van der Noen H. Expressions of the genes for cysteine proteinase inhibitors cystatin C and cystatin S in rat submandibular salivary gland. Arch Oral Biol. 1994;39:307–314. doi: 10.1016/0003-9969(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 34.Fellgiebel A., Schermuly I., Gerhard A., Keller I., Albrecht J., Weibrich C. Functional relevant loss of long association fibre tracts integrity in early Alzheimer’s disease. Neuropsychologia. 2008;46:1698–1706. doi: 10.1016/j.neuropsychologia.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Lee J.W., Namkoong H., Kim H.K., Kim S., Hwang D.W., Na H.R. Fibrinogen gamma-A chain precursor in CSF: a candidate biomarker for Alzheimer’s disease. BMC Neurol. 2007;7:14. doi: 10.1186/1471-2377-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King S.M. The dynein microtubule motor. Biochim Biophys Acta. 2000;1496:60–75. doi: 10.1016/s0167-4889(00)00009-4. [DOI] [PubMed] [Google Scholar]
- 37.Aumais J.P., Williams S.N., Luo W., Nishino M., Caldwell K.A., Caldwell G.A. Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J Cell Sci. 2003;116:1991–2003. doi: 10.1242/jcs.00412. [DOI] [PubMed] [Google Scholar]
- 38.Raychaudhuri M., Mukhopadhyay D. Grb2-mediated alteration in the trafficking of AbetaPP: insights from Grb2-AICD interaction. J Alzheimers Dis. 2010;20:275–292. doi: 10.3233/JAD-2010-1371. [DOI] [PubMed] [Google Scholar]
- 39.Giubellino A., Burke T.R., Jr., Bottaro D.P. Grb2 signaling in cell motility and cancer. Expert Opin Ther Targets. 2008;12:1021–1033. doi: 10.1517/14728222.12.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson K.L. Nuclear envelope and lamin B2 function in the central nervous system. Proc Natl Acad Sci U S A. 2010;107:6121–6122. doi: 10.1073/pnas.1000863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W., Zhang W., Han Y., Chen J., Wang Y., Zhang Z. NELIN, a new F-actin associated protein, stimulates HeLa cell migration and adhesion. Biochem Biophys Res Commun. 2005;330:1127–1131. doi: 10.1016/j.bbrc.2005.03.082. [DOI] [PubMed] [Google Scholar]
- 42.Carter C.J. Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem Int. 2007;50:12–38. doi: 10.1016/j.neuint.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Koch M., Olson P.F., Albus A., Jin W., Hunter D.D., Brunken W.J. Characterization and expression of the laminin gamma3 chain: a novel, non-basement membrane-associated, laminin chain. J Cell Biol. 1999;145:605–618. doi: 10.1083/jcb.145.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selkoe D.J. Presenilin, notch, and the genesis and treatment of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2001;98:11039–11041. doi: 10.1073/pnas.211352598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S.Y., Kim M.Y., Mo J.S., Park H.S. Notch1 intracellular domain suppresses APP intracellular domain-Tip60-Fe65 complex mediated signaling through physical interaction. Biochim Biophys Acta. 2007;1773:736–746. doi: 10.1016/j.bbamcr.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Schjeide B.M., Hooli B., Parkinson M., Hogan M.F., DiVito J., Mullin K. GAB2 as an Alzheimer disease susceptibility gene: follow-up of genomewide association results. Arch Neurol. 2009;66:250–254. doi: 10.1001/archneurol.2008.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oddo S. The ubiquitin-proteasome system in Alzheimer’s disease. J Cell Mol Med. 2008;12:363–373. doi: 10.1111/j.1582-4934.2008.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westermeier R. Sensitive, quantitative, and fast modifications for Coomassie Blue staining of polyacrylamide gels. Proteomics. 2006;6:61–64. doi: 10.1002/pmic.200690121. [DOI] [PubMed] [Google Scholar]
- 49.Li Q., Lau A., Morris T.J., Guo L., Fordyce C.B., Stanley E.F. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material