Abstract
Purpose
Zebrafish neurons regenerate from Müller glia following retinal lesions. Genes and signaling pathways important for retinal regeneration in zebrafish have been described, but our understanding of how Müller glial stem cell properties are regulated is incomplete. Mammalian Müller glia possess a latent neurogenic capacity that might be enhanced in regenerative therapies to treat degenerative retinal diseases.
Methods
To identify transcriptional changes associated with stem cell properties in zebrafish Müller glia, we performed a comparative transcriptome analysis from isolated cells at 8 and 16 hours following an acute photic lesion, prior to the asymmetric division that produces retinal progenitors.
Results
We report a rapid, dynamic response of zebrafish Müller glia, characterized by activation of pathways related to stress, nuclear factor–κB (NF-κB) signaling, cytokine signaling, immunity, prostaglandin metabolism, circadian rhythm, and pluripotency, and an initial repression of Wnt signaling. When we compared publicly available transcriptomes of isolated mouse Müller glia from two retinal degeneration models, we found that mouse Müller glia showed evidence of oxidative stress, variable responses associated with immune regulation, and repression of pathways associated with pluripotency, development, and proliferation.
Conclusions
Categories of biological processes/pathways activated following photoreceptor loss in regeneration-competent zebrafish Müller glia, which distinguished them from mouse Müller glia in retinal degeneration models, included cytokine signaling (notably NF-κB), prostaglandin E2 synthesis, expression of core clock genes, and pathways/metabolic states associated with pluripotency. These regulatory mechanisms are relatively unexplored as potential mediators of stem cell properties likely to be important in Müller glial cells for successful retinal regeneration.
Keywords: retinal regeneration, light damage, zebrafish, RNA-seq, Müller glia
In the mammalian retina, diseases and age-related biological processes result in the loss of neurons and glial scar formation.1,2 In contrast, the proliferative and neurogenic ability of Müller glial cells has been convincingly demonstrated in the teleost fish retina.3–5 The potential for neurogenesis appears to be conserved, yet dormant, in mammalian Müller glia. This latent neurogenic potential makes Müller glial cells an excellent target for regenerative therapies to promote endogenous replacement of retinal neurons.6,7
Although some aspects of Müller glial responses to injury in zebrafish resemble gliosis in mammalian Müller glia,8 the eventual outcome in the fish retina is robust regeneration of neurons. Several critical, sequential steps contribute to the overall success of retinal regeneration in zebrafish, including Müller glial activation and dedifferentiation; a self-renewing, asymmetric cell division of Müller glia; generation of multipotent retinal progenitor cells (RPCs); and RPC proliferation, migration, and differentiation.4,5,9 Much of our current understanding of Müller glial responses to retinal injury in zebrafish is based on identification of genes of interest through microarray gene profiling of RNA isolated from whole retina or laser-capture microdissected retinal layers,10–13 or RNA isolated from heterogeneous cell populations containing both Müller glia and Müller glia–derived RPCs.14,15 To date, only one gene profiling study has examined isolated, injury-activated Müller glial cells prior to the initial cell division and therefore free of contaminating Müller glia–derived progenitor cells.14 Because annotation of the microarray gene chips available at the time was very incomplete, this analysis yielded expression data for less than 15% of currently known zebrafish genes.
Our long-term goal is to identify transcriptional changes that might regulate the initial acquisition of stem cell–like properties in zebrafish Müller glia in response to injury, to facilitate future clinical therapies designed to induce Müller glial–dependent retinal regeneration in the human retina. In this study we sequenced transcripts from zebrafish Müller glia isolated with fluorescence-activated cell sorting (FACS) following an acute photic lesion. To constrain our interrogation to initial events that trigger the acquisition of stem cell–like properties, we focused the analysis on the phase during which Müller glia are dedifferentiating, prior to the first mitotic cell division at ∼24 hours post lesion (hpl) in our acute photic lesion model. These data reveal a large, rapid, and dynamic response in Müller glia in response to photoreceptor injury, and identify specific gene targets and pathways that have not been considered previously in studies of zebrafish retinal regeneration.
We next used bioinformatic analysis tools to compare the transcriptional responses of zebrafish and mouse Müller glia to photoreceptor damage, taking advantage of the most comparable, publicly available mouse data from isolated Müller glia in two retinal degeneration models.16 We identify and discuss transcriptional changes in the mouse models and compare these changes to the findings from our zebrafish data.
Methods
Transgenic zebrafish (Danio rerio) Tg(gfap:EGFP)mi200217,18 and TL strain wild-type zebrafish were maintained using standard husbandry protocols.19 All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Michigan and abided by the ARVO Statement for the Use of Animals in Ophthalmic and Visual Research.
The regenerative response in Müller glia was induced as previously described20 by exposing free-swimming, adult Tg(gfap:EGFP)mi2002 zebrafish (8–11 months old) to intense light (>120,000 lux) from an EXFO X-Cite 120W metal halide lamp (EXFO Photonic Solutions, Quebec City, QC, Canada) for 30 minutes (Fig. 1A). This acute light lesion results in the selective death of photoreceptors, with peak cell death response at ∼24 hpl. Retinas from nonlesioned and light-lesioned Tg(gfap:EGFP)mi2002 zebrafish were surgically removed and dissociated into single cells (Figs. 1B, 1C, respectively). Green fluorescent protein (GFP)-positive (GFP+) Müller glia cells were collected by FACS (Fig. 1D). Nonlesioned, wild-type zebrafish were used as GFP-negative (GFP−) controls for sorting. We collected Müller glia at 0 (nonlesioned), 8, and 16 hpl, prior to their initial asymmetric, stem cell–like division. These times were chosen to avoid including the rapidly dividing RPCs in the neurogenic clusters that begin to appear after the initial asymmetric division of the Müller glia at 20 to 36 hpl.21 Although the glial-specific promoter (gfap) is no longer active in RPCs, they retain the EGFP label through perseverance of the fluorescent protein for several days.20 Therefore, GFP+ cells isolated after 20 hpl inevitably include a heterogeneous mixture of Müller glia and RPCs.
Figure 1.
Schematic representation of the Müller glia RNA-seq experimental design. (A) Photoreceptors were ablated in free-swimming Tg(gfap:EGFP)mi2002 fish using an acute light-lesion paradigm. (B) Retinas were dissected from unlesioned controls (0 hpl) and 8 and 16 hpl Tg(gfap:EGFP)mi2002 fish. Retinas were also dissected from unlesioned, wild-type (nontransgenic, GFP−) control fish. (C) Dissected retinas from each group were pooled and dissociated. (D) Dissociated samples were subjected to fluorescence-activated cell sorting (FACS), and GFP+ cells were collected. The FACS plots shown are representative images from the final gating and collection of actual samples. (E) RNA was isolated from the sorted cells and checked by Bioanalyzer for quality and concentration. Samples with an RNA integrity number (RIN) above 7.0 were advanced to library preparation. The Bioanalyzer electropherogram shown is a representative plot from an actual sample with a RIN of 8.6. The x-axis is in seconds, which corresponds to size. The y-axis shows fluorescent units (FU), corresponding to the amount of RNA. (F) RNA-seq libraries were prepared and checked again via Bioanalyzer. The Bioanalyzer electropherogram shown is a representative plot from a sample library preparation. The x-axis shows the size in base pairs. The y-axis shows fluorescent units, corresponding to the amount of DNA. (G) The RNA-seq libraries were then sequenced on an Illumina GAIIx. (H) The sequencing data were processed with bioinformatic tools for differential expression analysis, gene ontology analysis, and pathway analysis.
RNA was extracted from the sorted cells and the quality was checked with Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) (Fig. 1E). Samples with an RNA integrity number (RIN) of acceptable quality (>7.0) were used for Illumina RNA-seq library preparation22 (Fig. 1F). RNA-seq libraries were subjected to deep sequencing on an Illumina GAIIx sequencer (Illumina, Inc., San Diego, CA, USA) (Fig. 1G). Sequencing data were analyzed for differential gene expression, gene ontology enrichment, and enriched pathways (Fig. 1H).
Additional methodological details and a complete description of the bioinformatic analysis pipeline are in the Supplementary Methods. Supplementary Data S1, S2, and S3 contain complete datasets for differential gene expression data, gene ontology enrichment, and enriched pathways, respectively.
Results
A Large, Rapid, and Dynamic Transcriptional Response in Zebrafish Müller Glia After Photoreceptor Injury With Acute Light Damage
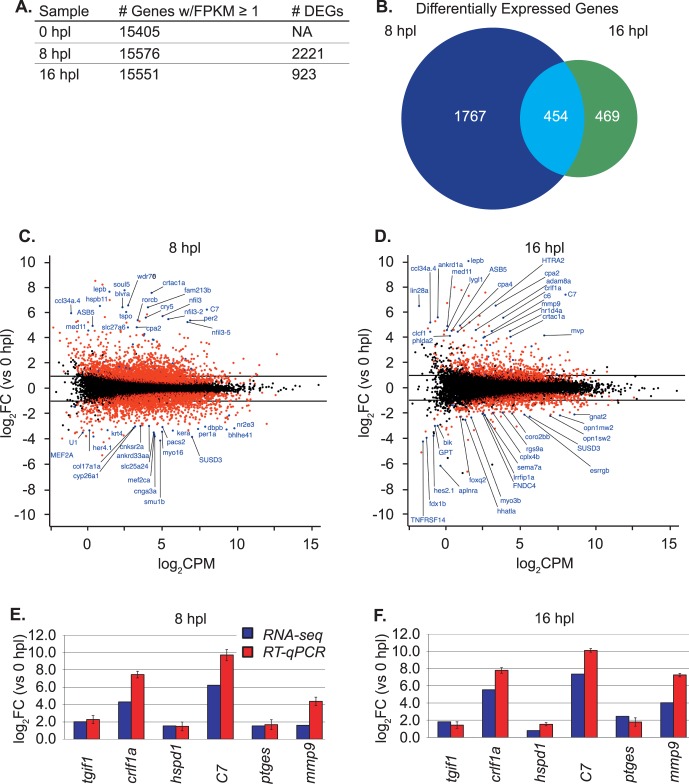
To identify genes that were differentially expressed at each time point, we used the commonly accepted threshold values22 for differentially expressed genes of |log2 fold change| (FC) ≥ 1 and a false discovery rate (FDR) ≤ 0.05. This analysis identified 2690 genes differentially expressed in Müller glia isolated from light-lesioned zebrafish retinas at 8 or 16 hpl compared with Müller glia from control (unlesioned) retinas (Fig. 2A; Supplementary Data S1). The number of genes differentially expressed at 8 hpl (n = 2221) is almost 2.5 times greater than at 16 hpl (n = 923) (Fig. 2A), although the total number of genes expressed above a threshold of 1 FPKM (fragment per kilobase of exon per million reads mapped) did not differ among the samples (Fig. 2A). A Venn diagram plot reveals 1767 differentially expressed genes specific to 8 hpl, 469 genes specific to 16 hpl, and 454 genes common to both (roughly 20% of the genes differentially expressed at 8 hpl remain differentially expressed at 16 hpl) (Fig. 2B). Figures 2C and 2D display the log2FC in expression as a function of log2 counts per million (CPM) mapped reads from RNA-seq data for all differentially expressed genes at 8 hpl and 16 hpl, respectively. The top 20 positively and negatively regulated known zebrafish genes are labeled in blue (log2FC values for 8 hpl are listed in Supplementary Table S2 and for 16 hpl in Supplementary Table S3). Other genes with a significant FDR (≤0.05) are represented by red dots.
Figure 2.
Distribution of expressed and differentially expressed genes (DEGs) in zebrafish Müller glia. (A) The total number of expressed and differentially expressed genes at each sample time point. Genes considered to be expressed were those with a fragment per kilobase of exon per million reads mapped (FPKM) value ≥ 1. (B) The number of expressed genes in each group is based on Entrez GeneID, which were used for further analyses. Differentially expressed genes are those with an absolute log2 fold change (|log2FC|) ≥ 1 and a false discovery rate (FDR) ≤ 0.05. (C, D) Transformed log ratio and mean average (MA) plot [log2FC versus log2 counts per million (CPM)] at 8 hpl and 16 hpl, respectively. Dots represent genes: red for genes with a FDR ≤ 0.05 and blue for genes of interest. Labeled genes are the top differentially expressed genes in either direction. (E, F) Validation of RNA-seq data for selected genes at 8 hpl and 16 hpl, respectively, with reverse transcription–quantitative polymerase chain reaction (RT-qPCR). Primers are listed in Supplementary Table S1. The x-axis shows the target gene and the y-axis shows the log2FC versus unlesioned control. RNA-seq log2FC data are denoted by blue bars, and RT-qPCR log2FC data are denoted by red bars. Error bars for the RT-qPCR data represent the standard error of the mean for three biological replicates.
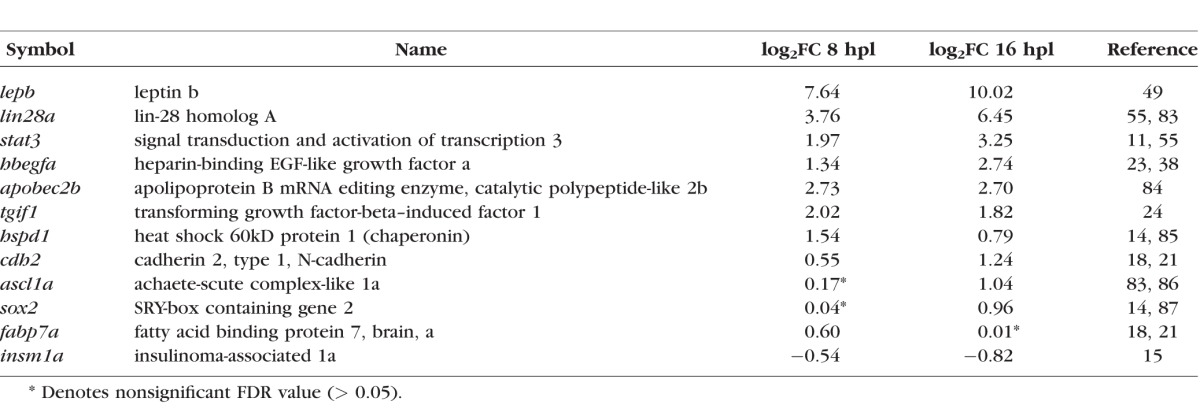
The Table lists the log2FC values for selected differentially expressed genes previously identified in microarray profiling studies and/or implicated in zebrafish retinal regeneration.4,5,9 Many of these genes belong to specific categories of biological processes, including stress response (hspd1), immune response/cytokines (stat3, lepb), secreted factors (hb-egf), Müller glial cell dedifferentiation and acquisition of stem cell properties (tgif1, lin28, apobec2b), and cell adhesion (cdh2). A few of the known regeneration genes with small or below criterion values of differential expression in this RNA-seq dataset (Table) are primarily implicated in processes that occur later than those assayed here, such as cell fate specification (ascl1a, sox2) and progenitor proliferation (fabp7a, insm1a).
Table.
Log2Fold Changes (FC) of Selected Regeneration-Associated Genes in Müller Glia at 8 and 16 Hours Post Lesion (hpl)
Validation of expression levels with RT–quantitative PCR (qPCR) confirmed the RNA-seq data for several genes of interest—TGFβ-induced factor homeobox 1 (tgif1), cytokine receptor like factor 1a (crlf1a), heat shock protein family D member 1 (hspd1), complement component 7 (c7), prostaglandin E synthase (ptges), and matrix metallopeptidase 9 (mmp9) at 8 hpl (Fig. 2E) and 16 hpl (Fig. 2F)—many of which have experimentally confirmed roles in zebrafish retinal regeneration.14,23,24
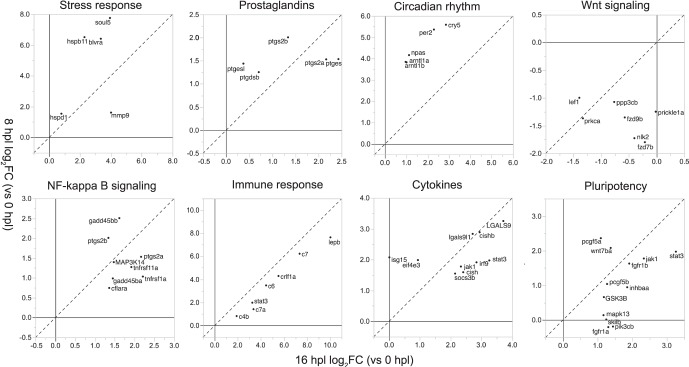
We next performed gene ontology (GO) and pathway-level analyses with KEGG and Reactome tools as described in more detail in the Supplementary Methods. Reactome data agreed with KEGG data, yet were highly redundant and thus not included in figures presented here. To provide a global overview of the rapid and dynamic transcriptional changes in Müller glia as they respond to photoreceptor loss and prepare to generate retinal progenitors, we discuss the results by classifying these data into eight general, nonexclusive, categories: stress response, prostaglandins, circadian rhythm, Wnt signaling, nuclear factor–κB (NF-κB) signaling, immune response, cytokines, and pluripotency. To illustrate dynamic transcriptional changes in Müller glia as they prepare for an asymmetric stem cell–like mitotic division, Figure 3 plots the log2FC differential expression of genes in each of these categories at 8 vs. 16 hpl.
Figure 3.
Differential regulation of genes in selected categories that characterize the Müller glial injury response. For each category (each part of figure), log2FC at 8 hpl (y-axis) is plotted versus log2FC at 16 hpl (x-axis) for top differentially expressed genes (|log2FC| ≥ 1 and FDR ≤ 0.05) within each category. Dashed line represents equivalent values of differential expression of a given gene at both sample times (slope = 1 and y-intercept = 0).
Stress Response, Prostaglandin Metabolism, and Circadian Rhythm Pathways Are Enriched and Wnt Signaling Is Repressed in Zebrafish Müller Glia at 8 hpl
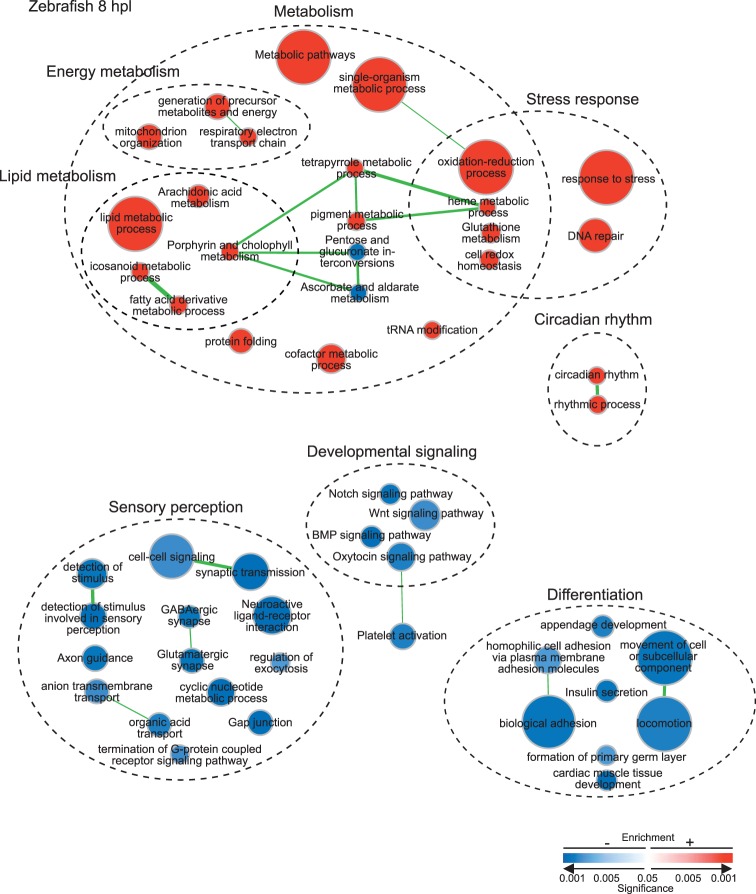
The top positively regulated annotated gene at 8 hpl with 7.75 log2FC is heme-binding protein soul 5 (soul5), an oxidative stress–induced protein (Figs. 2C, 3; Supplementary Table S2). Other evidence of a strong oxidative stress response includes elevated expression of biliverdin reductase A (blvra), heat shock protein alpha-crystallin-related b11 (hspb11), and heat shock 60-kDa protein 1 (hspd1) (Figs. 2C, 3; Supplementary Table S2). The enriched GO terms and KEGG pathways in Figure 4 similarly show strong positive regulation of redox and biological processes and pathways indicative of a stress response.
Figure 4.
Pathways and biological processes enriched at 8 hpl in zebrafish Müller glia. Enrichment map of enriched KEGG pathways and biological process GO terms (with redundancy reduced) with FDR ≤ 0.05. Circular node color reflects positive (red) or negative enrichment (blue); color intensity reflects the significance of the enrichment; and area reflects the size of the gene set. Lines (green) represent significant overlap in genes within linked sets, and line thickness represents degree of overlap.
Two categories of biological responses that are rapidly activated in Müller glia, but have not previously been associated with retinal regeneration, are prostaglandin metabolism and circadian rhythm (Figs. 2C, 3, 4). Metabolic processes and pathways, in general, are very highly enriched at 8 hpl (Fig. 4); especially prominent are pathways associated with lipid metabolism, including icosanoids (also called eicosanoids), fatty acid derivatives, and arachidonic acid. These metabolic changes are reflected in the increased expression of zebrafish orthologs of prostaglandin E synthase (ptges, ptges1), prostaglandin endoperoxide synthase (ptges, ptges1), and prostaglandin D2 synthase (ptgdsb) genes in the biosynthetic pathway for the eicosanoid prostaglandin E2 (PGE2) (Fig. 3, Supplementary Fig. S1A).
Somewhat unexpectedly, several core circadian clock genes are included in the top 20 positively regulated genes at 8 hpl (Figs. 2C, 3, Supplementary Fig. S2A; Supplementary Table S2), and the GO terms associated with circadian and biological rhythms are positively enriched (Fig. 4). With the exception of period 1a (per1a), which is negatively regulated (Fig. 2C; Supplementary Table S2), several core clock genes are positively regulated (Figs. 2C, 3; Supplementary Table S2; Supplementary Fig. S2A), including period 2 (per2), cryptochrome 5 (cry5), neuronal PAS domain protein 2 (npas2), and orthologs (arntl1a, arntl1b) of aryl hydrocarbon receptor nuclear translocator-like (Arntl1), also known as Bmal1.
Enrichment analyses reveal that Wnt signaling is negatively regulated at 8 hpl and remains repressed at 16 hpl (Figs. 3, 4) with reduced expression of Wnt target genes such as lymphoid enhancer binding factor 1 (lef1) and frizzled class receptors (fzd7b, fzd9b) (Fig. 3). Activity is likewise dampened in the Notch and bone morphogenetic protein (BMP) signaling pathways (Fig. 4), and the Notch target hairy-related 4, tandem duplicate 1 (her4.1) is among the top negatively regulated genes (Fig. 2C; Supplementary Table S2).
Negatively expressed genes and negatively enriched GO terms and KEGG pathways at both 8 and 16 hpl include several indicative of photoreceptors and neural functions, including sensory transduction and synaptic mechanisms (Figs. 2C, 2D, 3, 4, 5; Supplementary Tables S2, S3). Some of the negatively regulated photoreceptor-specific genes have relatively high log2CPM, such as the rod photoreceptor gene nuclear subfamily 2 group E member 3 (nr2e3) at 8 hpl (Fig. 2C) and cone photoreceptor genes G protein subunit alpha 2 (gnat2) and cone opsins (opn1mw2, opn1sw2) at 16 hpl (Fig. 2D). These results suggest that the preparations of FACS-isolated Müller glia are contaminated with photoreceptor transcripts, as has been noted in previous microarray profiling studies of sorted Müller glia from zebrafish (Supplementary Figs. S3B, S3C; Supplementary Tables S4, S5),14,15 as well as microarray and RNA-seq expression profiles of FACS-isolated GFP+ Müller glia from adult mouse retina (Supplementary Figs. S3D, S3E, S3F; Supplementary Tables S4, S5).25,26 Even single Müller glial cells hand-picked from young and adult mouse retinas can show significant levels of photoreceptor-specific transcripts (Supplementary Figs. S4A–S4I; Supplementary Tables S4, S5).16 This unavoidable photoreceptor contamination is likely attributed to the dense network of Müller glial processes that enwrap the photoreceptor cells. As this tight physical association would predict, the inverse is also true—Müller glial–specific transcripts (retinaldehyde binding protein 1; Supplementary Table S5) are found in FACS-isolated rod and cone photoreceptors from adult mouse (Supplementary Figs. S5A–S5F; Supplementary Table S4).27,28 Furthermore, rod photoreceptor–specific transcripts such as rhodopsin and Gnat1 (Supplementary Table S5) are abundantly present in 97.5% pure cone photoreceptor preparations (Supplementary Fig. S5B; Supplementary Table S4).28 Contaminating transcripts are avoided only when the offending cell type is not present; for example, rods are absent in the Nrl−/− mouse (Supplementary Fig. S5E).27 The negative enrichment values of transcripts related to photoreceptors at 8 or 16 hpl versus unlesioned samples in our dataset (Figs. 4, 5, sensory perception) are consistent with a reduction in cross-contamination caused by concurrent degeneration of light-damaged photoreceptors.
Figure 5.
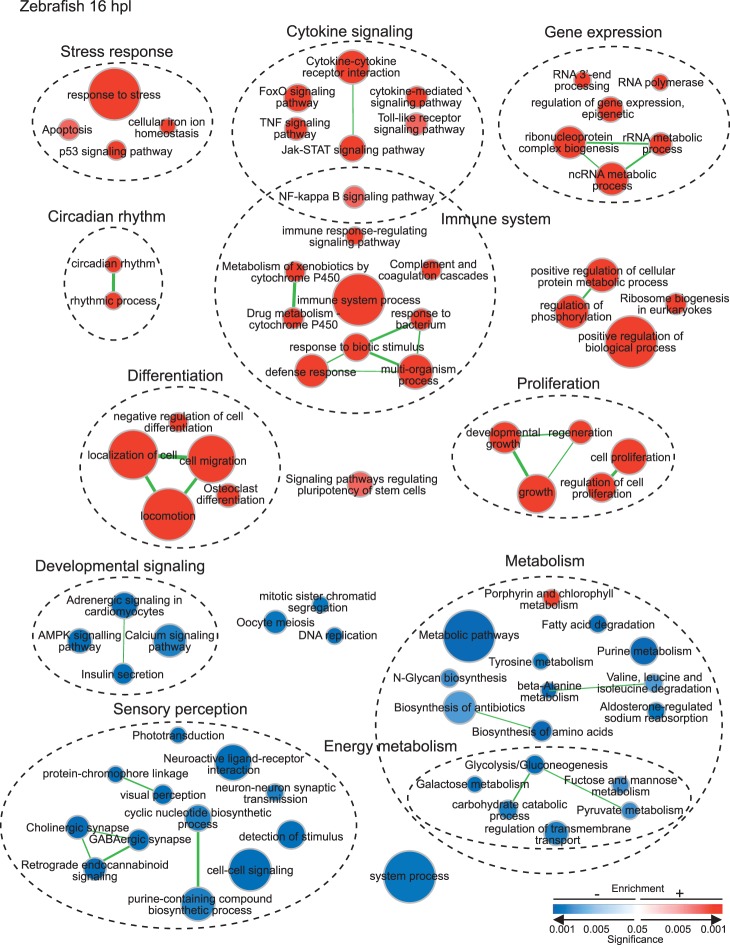
Pathways and biological processes enriched at 16 hpl in zebrafish Müller glia. Enrichment map of enriched KEGG pathways and redundancy-reduced biological process GO terms (with redundancy reduced) with FDR ≤ 0.05. Circular node color reflects positive (red) or negative enrichment (blue), color intensity reflects the significance of the enrichment, and area of the circle reflects the size of the gene set. Lines (green) represent significant overlap in genes within linked sets, and thickness represents degree of overlap.
NF-κB Signaling, Immune Responses, Cytokine Signaling, and Pluripotency Processes and Pathways Are Enriched in Zebrafish Müller Glia at 16 hpl
Activation of biological processes and pathways indicative of oxidative stress and the regulation of redox homeostasis, which were already evident at 8 hpl (Figs. 3, 4), continue to be enriched at 16 hpl, but with a clear shift toward more prominent activation of signaling pathways related to inflammation, including both innate and adaptive immune responses (Figs. 3, 5).
The NF-κB signaling pathway is activated by both oxidative stress and proinflammatory cytokines,29,30 and it regulates cell survival, cell proliferation, and inflammation.31–33 Positive enrichment of this pathway (Fig. 5) is reflected by upregulation of several genes (Fig. 3), including tumor necrosis factor receptor superfamily orthologs (tnfrsf1a, tnfrsf11a), mitogen-activated protein kinase kinase kinase 14 (map3k14), and growth arrest and DNA-damage-inducible 45 beta orthologs (gadd45ba, gadd45bb). Notably, two of the upregulated prostaglandin synthetases (ptgs2a and ptgs2b) mentioned previously are also in the NF-κB signaling pathway (Fig. 3).
A number of pathways associated with cytokine-mediated signaling and immune processes, including Janus kinase-Signal Transducer and Activator of Transcription (Jak-STAT) and tumor necrosis factor (TNF) signaling pathways (Fig. 5), are positively enriched at 16 hpl, as reflected by upregulation of genes (Fig. 3) such as janus kinase 1 (jak1), signal transducer and activator of transcription 3 (stat3), suppressor of cytokine signaling 3b (socs3b), cytokine inducible SH protein orthologs (cish, cishb), and interferon regulator factor 9 (irf9). Activation of immune responses mediated by the complement cascade is also very prominent at 16 hpl: Several complement component genes (c4a, c6, c7a, c7) show a log2FC > 2.0 (Figs. 2D, 3; Supplementary Table S3). The most highly enriched gene at 16 hpl is leptin b (lepb), a hormone that signals via Jak-STAT and modulates the immune system.34
In addition to cytokine signaling and immune responses, the Jak-STAT signaling pathway is also implicated in regulation of pluripotency in stem cells (Fig. 3). Other pluripotency-related enriched genes are associated with several growth factor signaling pathways: fibroblast growth factor receptor 1 orthologs (fgfr1a, fgfr1b), wingless-type MMTV integration site, family member 7a (wnt7a), glycogen synthase kinase 3b (gsk3b), inhibin beta Aa (inhbaa), and chromatin modifier polycomb group ring finger 5 orthologs (pcgf5a, pcgf5b).
Whereas biological processes associated with cell growth and differentiation (e.g., cell movement, biological adhesion, tissue development, insulin secretion) were negatively regulated at 8 hpl (Fig. 4), by 16 hpl evidence for positive enrichment of proliferation processes and pathways (e.g., cell proliferation, negative regulation of cell differentiation, regulation of epigenetic gene expression, locomotion and cell migration, regeneration, growth) was very strong (Fig. 5), suggestive of the transition to cell cycle entry and production of RPCs. Consistent with this interpretation is that signaling pathways regulating pluripotency of stem cells were positively enriched at 16 hpl (Fig. 5).
Mouse Retinal Degeneration Model Analysis
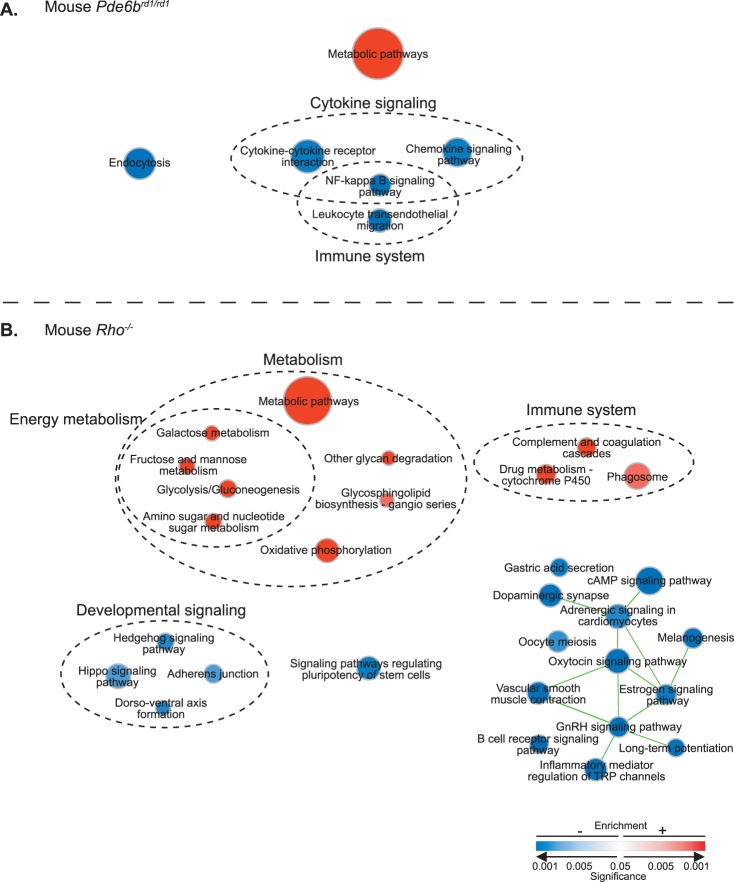
To identify transcriptional changes that distinguish regeneration-competent zebrafish Müller glia from mammalian Müller glia, which lack the ability to regenerate retinal neurons, we analyzed a publicly available microarray dataset of single isolated mouse Müller glia16 from two different mouse models of retinal degeneration, Pde6brd1/rd1 and Rho−/−, which vary in their degeneration kinetics.35,36 We used data only from the initial “major rod death phase” (postnatal day 13 and postnatal week 8 for Pde6brd1/rd1 and Rho−/−, respectively) as these were most comparable to our zebrafish dataset, which focused on the initial response of Müller glia to photoreceptor loss. We performed a parallel differential expression analysis of biological processes and pathways, as illustrated in Figures 6A and 6B and described in more detail in Supplementary Methods.
Figure 6.
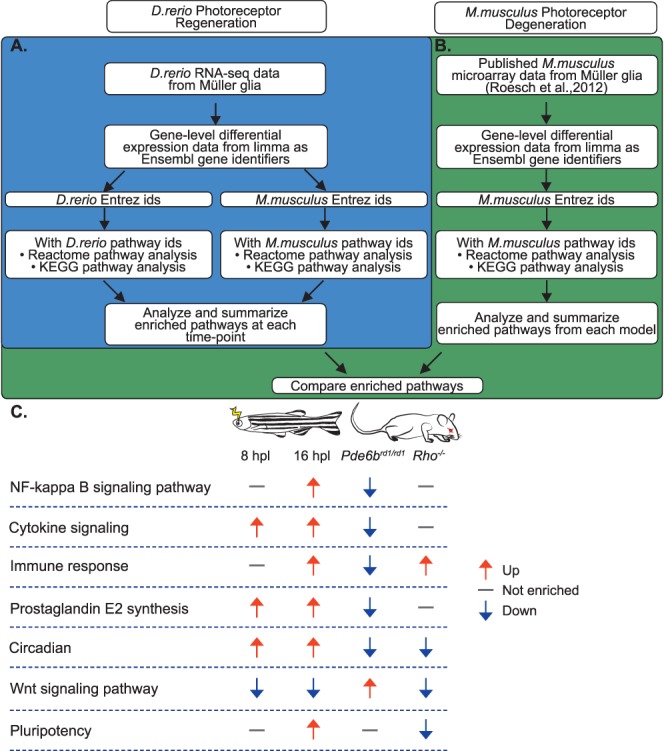
Comparison between zebrafish and mouse Müller glial responses to photoreceptor loss. (A) Schematic showing the methodology for the pathway analysis based on RNA-seq data for the initial stages of Müller glia–dependent photoreceptor regeneration in zebrafish (D. rerio). (B) Schematic showing the methodology for the pathway analysis of microarray data from single Müller glia isolated from mouse (Mus musculus) photoreceptor degeneration models, and the comparison between D. rerio and M. musculus. (C) Summary of key differences revealed by pathway enrichment analysis. Columns from left to right: signaling pathway or biological process, zebrafish Müller glia at 8 hpl, zebrafish Müller glia at 16 hpl, mouse Müller glia from Pde6brd1/rd1 retinal degeneration model at postnatal day 13, mouse Müller glia from Rho−/− retinal degeneration model at 8 weeks postnatal. Arrows and corresponding colors represent the overall direction of regulation according to the legend at the right; gray dashes represent pathways or processes that were not enriched compared with controls. Direction of regulation for 8 and 16 hpl zebrafish Müller glia is based on data from Figures 4 and 5, respectively. Direction of regulation for Pde6brd1/rd1 and Rho−/− Müller glia is based on data from Figures 6A and 6B, respectively, with the exception of Wnt signaling. Pde6brd1/rd1 is based on results from Reactome analysis (positive enrichment of “formation of the beta-catenin:TCF transactivating complex,” Supplementary Data S3) and biological process GO terms (positive enrichment of “regulation of canonical Wnt signaling pathway,” Supplementary Data S2). Rho−/− is based on biological process GO term analysis (negative enrichment of “regulation of canonical Wnt signaling pathway,” Supplementary Data S2).
Pathway enrichment analysis in the Pde6brd1/rd1 retinal degeneration model revealed strong positive regulation of metabolic responses in mouse Müller glia (Fig. 7A), but in contrast to zebrafish Müller glia, the NF-κB signaling pathway, cytokine signaling and immune system responses were negatively enriched (Figs. 6C, 7A). In the Rho−/− mouse retinal degeneration model, a strong metabolic response was also seen the Müller glia (Fig. 7B); but in contrast to the Pde6brd1/rd1 model, the complement cascade pathway was positively enriched (Fig. 7B) similarly to zebrafish at 16 hpl (Fig. 5). Genes in the biosynthetic pathway leading to PGE2, which were strongly upregulated in fish Müller glia at both 8 and 16 hpl (Figs. 3, 6C, Supplementary Fig. S1A) do not exhibit this effect in mouse Müller glia (Fig. 6C, Supplementary Fig. S1B), although several genes in the arachidonic acid metabolism pathway are differentially but inconsistently regulated in both mouse degeneration models (Supplementary Fig. S1B). Similarly, the positive regulation of core circadian clock genes seen in zebrafish Müller glia, including per2, clock, and arntl (Fig. 6C, Supplementary Fig. S2A), is not replicated in mouse Müller glia; instead, negative regulators of Per1 (beta-transducin repeat containing E3 ubiquitin protein ligase, Btrc, and S-phase kinase-associated protein 1a, Skp1a) are strongly upregulated, and Per1 and Arntl (Bmal1) are downregulated (Fig. 6C, Supplementary Fig. S2B). Wnt signaling is repressed in zebrafish Müller glia and variably regulated in the degeneration models (Fig. 6C; Supplementary Material S2, S3). Finally, unlike in zebrafish, in the Rho−/− model, signaling pathways regulating pluripotency in stem cells were negatively enriched (Figs. 6C, 7B).
Figure 7.
Pathways enriched in mouse degeneration models. Enrichment map of KEGG pathways with FDR ≤ 0.05 in (A) Pde6brd1/rd1 and (B) Rho−/− degeneration models. Circular node color reflects positive (red) or negative enrichment (blue); color intensity reflects the significance of the enrichment; and area reflects the size of the gene set. Lines (green) represent significant overlap in genes within linked sets, and line thickness represents degree of overlap.
Discussion
The inability of mammals to endogenously regenerate neurons is in marked contrast to the highly regeneration-competent zebrafish.7,37 The objectives of the current study were to determine a more complete catalog of transcript expression changes during the early steps of Müller glia response to neuronal loss and to compare gene regulatory responses of regeneration-competent zebrafish Müller glia with those of Müller glia in mouse models of retinal degeneration. To help identify regeneration-relevant factors that are missing from mammalian Müller glia, which fail to regenerate retinal neurons, we compared transcriptional changes in zebrafish Müller glia to publicly available transcriptome data from single mouse Müller glia isolated from two genetic models of retinitis pigmentosa in which rod photoreceptors degenerate.16 This cross-species analysis was admittedly less than ideal for several reasons. (1) The photoreceptor injury models were not equivalent (acute photic damage of cones and rods in zebrafish versus genetically induced rod degeneration in mouse). (2) The methods for isolating Müller glia were different (FACS isolation of GFP-labeled zebrafish Müller glial cells versus single mouse Müller glial cells selected based on morphology). (3) The methods for transcriptome analysis were different (RNA-seq for zebrafish Müller glia versus microarray analysis for mouse Müller glia). Despite these limitations, the comparison highlighted several potentially important differences between regeneration-competent Müller glia in zebrafish and Müller glial responding to retinal degeneration in mice.
Previous gene expression studies of the zebrafish retinal response to injury have shown a rapid induction of stress response transcripts and proteins,11,12,14,38 and many of the genes identified in those studies were also present in our dataset. Although the transcriptional responses of Müller glia from Pde6brd1/rd1 and Rho−/− mice show some evidence of gene expression changes associated with gliosis and stress responses,16 these pathways were not among those identified by the KEGG enrichment analyses of these data. However, single-cell transcriptomic data on this heterogeneous population of Müller glia39 result in a lack of statistical power, which might account for this observation. Another potential explanation for this apparent difference in activation of stress response pathways is that the mouse models represent chronic photoreceptor degeneration, whereas the zebrafish model is an acute, traumatic injury.
Stress response genes may also activate other pathways that are key to regeneration. For example, hspb11 is a top upregulated gene at 8 hpl in zebrafish Müller glia; hspb11 is required for hedgehog signaling in mice,40 and this pathway has been shown to promote the stem cell potential of Müller glia in mammalian and chick retinas.41,42 Hedgehog signaling also stimulates aerobic glycolysis, a metabolic characteristic of pluripotent stem cells.43 Constitutive hedgehog signaling in developing zebrafish retinas results in an increase in the number of reactive Müller glia,44 although its role has not been studied in the context of zebrafish photoreceptor regeneration. In the mouse Rho−/− retinal degeneration model, both Hedgehog and Hippo signaling pathways are negatively regulated. Hippo signaling also induces Drosophila neural stem cell quiescence,45 inhibits zebrafish RPC proliferation, and promotes differentiation,46 but again, this pathway has not been examined in retinal regeneration.
Another signaling pathway responsive to stress is NF-κB signaling pathway, which was strongly activated in zebrafish Müller glia, but repressed in Pde6brd1/rd1 mouse Müller glia. Inhibition of NF-κB signaling in neural stem cells results in symmetric division and accumulation of Nestin/Sox2/GFAP-expressing cells, reminiscent of gliosis.47 Nuclear factor–κB signaling is reactive oxygen species (ROS) responsive and stimulates an immune response by activating transcription of cytokines and ligands of known importance in retinal regeneration, such as interleukin-6 (IL-6),48,49 TNF-alpha,38 activin A,24,50 and in zebrafish tail fin regeneration, such as mmp9.51 Several genes in the complement cascade (c4b, c6, c7, c7a) are also upregulated in zebrafish Müller glia, and related complement fragments (C3a and C5a) can directly induce retinal regeneration in the embryonic chick model through transdifferentiation of retinal pigmented epithelial cells.52 Complement activation can also induce IL-6 and TNF-alpha, resulting in NF-κB and STAT-3 activation,53,54 and stat3 activation is critical for retinal regeneration in zebrafish.49,55 The role of the immune system in regeneration is increasingly recognized, and an interaction between stem cells and immune cell–derived factors is necessary for effective tissue regeneration.56 In the zebrafish brain, inflammation is both necessary and sufficient to trigger neurogenesis that resembles a regenerative response,57 but additional studies are needed to understand the role of the immune system in retinal regeneration.
Several other signaling pathways whose activity is altered in zebrafish Müller glia and that have previously been implicated in regulating retinal regeneration include Wnt, Notch, and TGFβ/BMP.4,5,9 Both Wnt and Notch signaling are necessary for zebrafish retinal regeneration,58,59 so the early repression of these signaling pathways that we observed is somewhat puzzling, although the requirement for activation of Wnt signaling is at a later stage, to promote proliferation of RPCs and regeneration of neurons. Activation of Notch signaling in the mature retina maintains differentiated Müller glial fate, inhibits stat3 and ascl1 expression, and prevents progenitor proliferation23; hence the initial rapid downregulation of Notch signaling might promote Müller glial dedifferentiation and cell cycle reentry.4 Signaling through the TGFβ/BMP pathway also promotes Müller glia differentiation,60,61 and zebrafish mutants with impaired capacity to negatively regulate TGFβ signaling show reduced RPC proliferation and diminished regenerative capacity.24 Interestingly, inhibitors of TGFβ can replace two of the classic pluripotency factors, c-Myc and Sox2, in reprogramming fibroblasts to iPSCs.62–64
Surprisingly, circadian rhythm–related terms and pathways were among the most positively enriched in our dataset. Disruption of the circadian-regulated, heparin-binding cytokine, midkine a (mdka), in the injured zebrafish retina results in altered cell cycle kinetics, RPC proliferation, and diminished rod photoreceptor regeneration,65 but no investigations have directly addressed the role of core clock genes. Recent studies of stem cell division and differentiation implicate clock genes in functions that have no clear link to the circadian rhythm itself.66 For instance, Per2, which is upregulated in our dataset, is nonrhythmically expressed in neural stem cells (NSC) in the adult hippocampus, and regulates their proliferation and neurogenesis in mice.67,68 Recent work showed that mouse and human Müller glial cells in vitro have an endogenous circadian clock,69 but in the microarray dataset from mouse degeneration models that we analyzed, the core clock genes were downregulated.
A strong metabolic response to photoreceptor degeneration was evident in both fish and mouse Müller glia pathway enrichment analyses. The injury-induced metabolic responses in zebrafish Müller glia associated with regeneration are largely unexplored, in contrast to extensive investigations of metabolic profiles associated with reactive gliosis in mammalian Müller glia.1,70,71 The essential role of metabolism in directing cell proliferation, differentiation, and cell fate determination is increasingly recognized, and levels of glycolysis, oxidative phosphorylation, and oxidative stress provide a metabolic signature that distinguishes pluripotent stem cells, rapidly dividing progenitor cells, and differentiated cells.43,72 The initial metabolic state and energy metabolism of zebrafish Müller glia in response to photoreceptor injury includes enhanced oxidative phosphorylation and lipid metabolism, similar to neural progenitors, which are characterized by high levels of aerobic glycolysis, oxidative phosphorylation, ROS, and eicosanoid and fatty acid biosynthesis.72 Within a few hours, the profile of zebrafish Müller glia shifts to negative regulation of metabolic pathways, decreased glycolysis, and enrichment of the FoxO signaling pathway, which more resembles NSC.72 Fox-O signaling mediates ROS suppression, and inhibiting FoxO-mediated ROS suppression results in a depleted NSC population in mice.73 The rapidly shifting profile of energy and metabolism in zebrafish Müller glia toward a “stemness-like” profile may reflect preparation for the asymmetric, self-renewing division that initiates the regenerative process. An in-depth, comparative metabolomic study of zebrafish Müller glia and mammalian Müller glia could reveal metabolic pathways that regulate the regenerative response in zebrafish retina, and the relevant enzymes might be excellent therapeutic targets for pharmacologic treatments to enhance endogenous regeneration.
Of particular interest are changes in fatty acid metabolism, which play an important role in the regulation of neural stem cell proliferation.74–76 Icosanoid (also called eicosanoid)-derived metabolites, including PGE2, can regulate NF-κB, differentiation, and inflammation in stem cell populations,76 and inhibitors of PGE2-degrading enzymes potentiate tissue regeneration in mice.77 The addition of PGE2 to rat Müller glia in vitro enhances their dedifferentiation, stem cell–like properties, and proliferative abilities.78 Our transcriptome analysis suggests that PGE2 levels in zebrafish, but not in mouse, Müller glia are increased after loss of photoreceptors. Whether PGE2 promotes Müller glia–dependent retinal regeneration is unknown, but if so, this may be a promising therapeutic target.
A final and critical difference is that zebrafish Müller glia demonstrate positive enrichment of pluripotency factors and cell proliferation, whereas mouse Müller glia show reduced pluripotency factors and no evidence of cell proliferation in response to photoreceptor loss. Although mammalian Müller glia express some genes associated with RPCs and have latent neurogenic capabilities,7,26,79–82 these attributes are not enhanced in response to neuronal loss.
In conclusion, a comparison of transcriptional profiles from regeneration-competent zebrafish Müller glia and gliotic mouse Müller glia has revealed several novel, previously unexplored, categories of biological responses that could act to promote endogenous retinal regeneration: NF-κB signaling, PGE2 synthesis, expression of core clock genes, and signaling pathways and metabolic signatures associated with stem cells. These and other candidates can be explored in future investigations and may provide potential therapeutic targets that could enhance endogenous Müller glial–dependent regeneration of retinal neurons in humans.
Availability of Data and Material
The datasets supporting the conclusions of this article are available in the National Center for Biotechnology Information Sequence Read Archive and Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE86872), and are included within the article or the Supplementary Material.
Supplementary Material
Acknowledgments
The authors thank Linn Gieser (National Eye Institute) for technical help and Matthew Brooks (National Eye Institute) and Rich McEachin (University of Michigan) for insightful discussions on bioinformatic principles and practices.
Supported by National Institutes of Health (NIH) R01 EY004318 (PAR), NIH T32 EY013934 (CJS), NIH P30 EY007003, and Intramural Research Program of the National Eye Institute (EY000474 and EY000546 to AS).
Disclosure: C.J. Sifuentes, None; J.-W. Kim, None; A. Swaroop, None; P.A. Raymond None
References
- 1. Bringmann A,, Iandiev I,, Pannicke T,, et al. Cellular signaling and factors involved in Müller cell gliosis: neuroprotective and detrimental effects. Prog Retin Eye Res. 2009; 28: 423–451. [DOI] [PubMed] [Google Scholar]
- 2. Dyer MA,, Cepko CL. Control of Müller glial cell proliferation and activation following retinal injury. Nat Neurosci. 2000; 3: 873–880. [DOI] [PubMed] [Google Scholar]
- 3. Hitchcock PF,, Raymond PA. The teleost retina as a model for developmental and regeneration biology. Zebrafish. 2004; 1: 257–271. [DOI] [PubMed] [Google Scholar]
- 4. Lenkowski JR,, Raymond PA. Müller glia: stem cells for generation and regeneration of retinal neurons in teleost fish. Prog Retin Eye Res. 2014; 40: 94–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gorsuch RA,, Hyde DR. Regulation of Müller glial dependent neuronal regeneration in the damaged adult zebrafish retina. Exp Eye Res. 2014; 123: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jeon S,, Oh I-H. Regeneration of the retina: toward stem cell therapy for degenerative retinal diseases. BMB Rep. 2015; 48: 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hamon A,, Roger JE,, Yang X-J,, Perron M. Müller glial cell-dependent regeneration of the neural retina: an overview across vertebrate model systems: Müller cells and retinal regeneration. Dev Dyn. 2016; 245: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomas JL,, Ranski AH,, Morgan GW,, Thummel R. Reactive gliosis in the adult zebrafish retina. Exp Eye Res. 2016; 143: 98–109. [DOI] [PubMed] [Google Scholar]
- 9. Goldman D. Müller glial cell reprogramming and retina regeneration. Nat Rev Neurosci. 2014; 15: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cameron DA,, Gentile KL,, Middleton FA,, Yurco P. Gene expression profiles of intact and regenerating zebrafish retina. Mol Vis. 2005; 11: 775–791. [PubMed] [Google Scholar]
- 11. Kassen SC,, Ramanan V,, Montgomery JE,, et al. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007; 67: 1009–1031. [DOI] [PubMed] [Google Scholar]
- 12. Craig SEL,, Calinescu A-A,, Hitchcock PF. Identification of the molecular signatures integral to regenerating photoreceptors in the retina of the zebra fish. J Ocul Biol Dis Infor. 2008; 1: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morris AC,, Forbes-Osborne MA,, Pillai LS,, Fadool JM. Microarray analysis of XOPS-mCFP zebrafish retina identifies genes associated with rod photoreceptor degeneration and regeneration. Invest Ophthalmol Vis Sci. 2011; 52: 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qin Z,, Barthel LK,, Raymond PA. Genetic evidence for shared mechanisms of epimorphic regeneration in zebrafish. Proc Natl Acad Sci U S A. 2009; 106: 9310–9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramachandran R,, Zhao X-F,, Goldman D. Insm1a-mediated gene repression is essential for the formation and differentiation of Müller glia-derived progenitors in the injured retina. Nat Cell Biol. 2012; 14: 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roesch K,, Stadler MB,, Cepko CL. Gene expression changes within Müller glial cells in retinitis pigmentosa. Mol Vis. 2012; 18: 1197–1214. [PMC free article] [PubMed] [Google Scholar]
- 17. Bernardos RL,, Raymond PA. GFAP transgenic zebrafish. Gene Expr Patterns. 2006; 6: 1007–1013. [DOI] [PubMed] [Google Scholar]
- 18. Raymond PA,, Barthel LK,, Bernardos RL,, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006; 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio). 4th ed. Eugene OR: University of Oregon Press; 2000. [Google Scholar]
- 20. Bernardos RL,, Barthel LK,, Meyers JR,, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007; 27: 7028–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagashima M,, Barthel LK,, Raymond PA. A self-renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013; 140: 4510–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brooks MJ,, Rajasimha HK,, Swaroop A. Retinal transcriptome profiling by directional next-generation sequencing using 100 ng of total RNA. Methods Mol Biol. 2012; 884: 319–334. [DOI] [PubMed] [Google Scholar]
- 23. Wan J,, Ramachandran R,, Goldman D. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev Cell. 2012; 22: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lenkowski JR,, Qin Z,, Sifuentes CJ,, et al. Retinal regeneration in adult zebrafish requires regulation of TGFβ signaling. Glia. 2013; 61: 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xue W,, Cojocaru RI,, Dudley VJ,, Brooks M,, Swaroop A,, Sarthy VP. Ciliary neurotrophic factor induces genes associated with inflammation and gliosis in the retina: a gene profiling study of flow-sorted Müller cells. PLoS One. 2011; 6: e20326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pollak J,, Wilken MS,, Ueki Y,, et al. ASCL1 reprograms mouse Müller glia into neurogenic retinal progenitors. Development. 2013; 140: 2619–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim J-W,, Yang H-J,, Oel AP,, et al. Recruitment of rod photoreceptors from short-wavelength-sensitive cones during the evolution of nocturnal vision in mammals. Dev Cell. 2016; 37: 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mo A,, Luo C,, Davis FP,, et al. Epigenomic landscapes of retinal rods and cones. Elife. 2016; 5: e11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schreck R,, Rieber P,, Baeuerle PA. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 1991; 10: 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawai T,, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007; 13: 460–469. [DOI] [PubMed] [Google Scholar]
- 31. Widera D,, Kaus A,, Kaltschmidt C,, Kaltschmidt B. Neural stem cells, inflammation and NF-kappaB: basic principle of maintenance and repair or origin of brain tumours? J Cell Mol Med. 2008; 12: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009; 1: a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Young KM,, Bartlett PF,, Coulson EJ. Neural progenitor number is regulated by nuclear factor-κB p65 and p50 subunit-dependent proliferation rather than cell survival. J Neurosci Res. 2006; 83: 39–49. [DOI] [PubMed] [Google Scholar]
- 34. Frühbeck G. Intracellular signalling pathways activated by leptin. Biochem J. 2006; 393 (pt 1): 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Punzo C,, Kornacker K,, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009; 12: 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hippert C,, Graca AB,, Barber AC,, et al. Müller glia activation in response to inherited retinal degeneration is highly varied and disease-specific. PLoS One. 2015; 10: e0120415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gallina D,, Todd L,, Fischer AJ. A comparative analysis of Müller glia-mediated regeneration in the vertebrate retina. Exp Eye Res. 2014; 123: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nelson CM,, Ackerman KM,, O'Hayer P,, Bailey TJ,, Gorsuch RA,, Hyde DR. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Muller glia proliferation during zebrafish retinal regeneration. J Neurosci. 2013; 33: 6524–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roesch K,, Jadhav AP,, Trimarchi JM,, et al. The transcriptome of retinal Müller glial cells. J Comp Neurol. 2008; 509: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang N,, Li L,, Eguether T,, Sundberg JP,, Pazour GJ,, Chen J. Intraflagellar transport 27 is essential for hedgehog signaling but dispensable for ciliogenesis during hair follicle morphogenesis. Development. 2015; 142: 2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wan J,, Zheng H,, Xiao H-L,, She Z-J,, Zhou G-M. Sonic hedgehog promotes stem-cell potential of Müller glia in the mammalian retina. Biochem Biophys Res Commun. 2007; 363: 347–354. [DOI] [PubMed] [Google Scholar]
- 42. Sirko S,, Behrendt G,, Johansson PA,, et al. Reactive glia in the injured brain acquire stem cell properties in response to sonic hedgehog. Cell Stem Cell. 2013; 12: 426–439. [DOI] [PubMed] [Google Scholar]
- 43. Agathocleous M,, Harris WA. Metabolism in physiological cell proliferation and differentiation. Trends Cell Biol. 2013; 23: 484–492. [DOI] [PubMed] [Google Scholar]
- 44. Bibliowicz J,, Gross JM. Expanded progenitor populations, vitreo-retinal abnormalities, and Müller glial reactivity in the zebrafish leprechaun/patched2 retina. BMC Dev Biol. 2009; 9: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ding R,, Weynans K,, Bossing T,, Barros CS,, Berger C. The Hippo signalling pathway maintains quiescence in Drosophila neural stem cells. Nat Commun. 2016; 7: 10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Asaoka Y,, Hata S,, Namae M,, Furutani-Seiki M,, Nishina H. The Hippo pathway controls a switch between retinal progenitor cell proliferation and photoreceptor cell differentiation in zebrafish. PLoS One. 2014; 9: e97365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y,, Liu J,, Yao S,, et al. Nuclear factor kappa B signaling initiates early differentiation of neural stem cells. Stem Cells. 2012; 30: 510–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Libermann TA,, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol. 1990; 10: 2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao X-F,, Wan J,, Powell C,, Ramachandran R,, Myers MG, Jr,, Goldman D. Leptin and IL-6 family cytokines synergize to stimulate Müller glia reprogramming and retina regeneration. Cell Rep. 2014; 9: 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jaźwińska A,, Badakov R,, Keating MT. Activin-βA signaling is required for zebrafish fin regeneration. Curr Biol. 2007; 17: 1390–1395. [DOI] [PubMed] [Google Scholar]
- 51. LeBert DC,, Squirrell JM,, Rindy J,, et al. Matrix metalloproteinase 9 modulates collagen matrices and wound repair. Development. 2015; 142: 2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Haynes T,, Luz-Madrigal A,, Reis ES,, et al. Complement anaphylatoxin C3a is a potent inducer of embryonic chick retina regeneration. Nat Commun. 2013; 4: 2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strey CW,, Markiewski M,, Mastellos D,, et al. The proinflammatory mediators C3a and C5a are essential for liver regeneration. J Exp Med. 2003; 198: 913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Viedt C,, Hänsch GM,, Brandes RP,, Kübler W,, Kreuzer J. The terminal complement complex C5b-9 stimulates interleukin-6 production in human smooth muscle cells through activation of transcription factors NF-kappa B and AP-1. FASEB J. 2000; 14: 2370–2372. [DOI] [PubMed] [Google Scholar]
- 55. Nelson CM,, Gorsuch RA,, Bailey TJ,, Ackerman KM,, Kassen SC,, Hyde DR. Stat3 defines three populations of Müller glia and is required for initiating maximal müller glia proliferation in the regenerating zebrafish retina. J Comp Neurol. 2012; 520: 4294–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eming SA,, Hammerschmidt M,, Krieg T,, Roers A. Interrelation of immunity and tissue repair or regeneration. Semin Cell Dev Biol. 2009; 20: 517–527. [DOI] [PubMed] [Google Scholar]
- 57. Kyritsis N,, Kizil C,, Zocher S,, et al. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science. 2012; 338: 1353–1356. [DOI] [PubMed] [Google Scholar]
- 58. Ramachandran R,, Zhao X-F,, Goldman D. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci U S A. 2011; 108: 15858–15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meyers JR,, Hu L,, Moses A,, Kaboli K,, Papandrea A,, Raymond PA. β-catenin/Wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neural Dev. 2012; 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ueki Y,, Wilken MS,, Cox KE,, Chipman LB,, Bermingham-McDonogh O,, Reh TA. A transient wave of BMP signaling in the retina is necessary for Müller glial differentiation. Development. 2015; 142: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stipursky J,, Gomes FCA. TGF-beta1/SMAD signaling induces astrocyte fate commitment in vitro: implications for radial glia development. Glia. 2007; 55: 1023–1033. [DOI] [PubMed] [Google Scholar]
- 62. Maherali N,, Hochedlinger K. Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol. 2009; 19: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lin T,, Wu S. Reprogramming with small molecules instead of exogenous transcription factors. Stem Cells Int. 2015; 2015: 794632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ichida JK,, Blanchard J,, Lam K,, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009; 5: 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gramage E,, D'Cruz T,, Taylor S,, Thummel R,, Hitchcock PF. Midkine-a protein localization in the developing and adult retina of the zebrafish and its function during photoreceptor regeneration. PLoS One. 2015; 10: e0121789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brown SA. Circadian clock-mediated control of stem cell division and differentiation: beyond night and day. Development. 2014; 141: 3105–3111. [DOI] [PubMed] [Google Scholar]
- 67. Borgs L,, Beukelaers P,, Vandenbosch R,, et al. Period 2 regulates neural stem/progenitor cell proliferation in the adult hippocampus. BMC Neurosci. 2009; 10: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bouchard-Cannon P,, Mendoza-Viveros L,, Yuen A,, Kærn M,, Cheng H-YM. The circadian molecular clock regulates adult hippocampal neurogenesis by controlling the timing of cell-cycle entry and exit. Cell Rep. 2013; 5: 961–973. [DOI] [PubMed] [Google Scholar]
- 69. Xu L,, Ruan G,, Dai H,, Liu AC,, Penn J,, McMahon DG. Mammalian retinal Müller cells have circadian clock function. Mol Vis. 2016; 22: 275–283. [PMC free article] [PubMed] [Google Scholar]
- 70. Bringmann A,, Pannicke T,, Grosche J,, et al. Müller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006; 25: 397–424. [DOI] [PubMed] [Google Scholar]
- 71. Bringmann A,, Wiedemann P. Müller glial cells in retinal disease. Ophthalmologica. 2012; 227: 1–19. [DOI] [PubMed] [Google Scholar]
- 72. Shyh-Chang N,, Daley GQ,, Cantley LC. Stem cell metabolism in tissue development and aging. Development. 2013; 140: 2535–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Renault VM,, Rafalski VA,, Morgan AA,, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009; 5: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Knobloch M,, Braun SMG,, Zurkirchen L,, et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 2013; 493: 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hamilton LK,, Dufresne M,, Joppé SE,, et al. Aberrant lipid metabolism in the forebrain niche suppresses adult neural stem cell proliferation in an animal model of Alzheimer's disease. Cell Stem Cell. 2015; 17: 397–411. [DOI] [PubMed] [Google Scholar]
- 76. Kang JX,, Wan J-B,, He C. Concise review: regulation of stem cell proliferation and differentiation by essential fatty acids and their metabolites. Stem Cells. 2014; 32: 1092–1098. [DOI] [PubMed] [Google Scholar]
- 77. Zhang Y,, Desai A,, Yang SY,, et al. Inhibition of the prostaglandin-degrading enzyme 15-PGDH potentiates tissue regeneration. Science. 2015; 348: aaa2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang L,, Lang L-L,, Wang Y,, Shi S,, Liu L. Prostaglandin E(2) enhances proliferation, dedifferentiation and stem-like properties of rat retinal Müller glial cells in vitro. Ophthalmic Res. 2013; 49: 100–107. [DOI] [PubMed] [Google Scholar]
- 79. Karl MO,, Reh TA. Regenerative medicine for retinal diseases: activating endogenous repair mechanisms. Trends Mol Med. 2010; 16: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jadhav AP,, Roesch K,, Cepko CL. Development and neurogenic potential of Müller glial cells in the vertebrate retina. Prog Retin Eye Res. 2009; 28: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jayaram H,, Jones MF,, Eastlake K,, et al. Transplantation of photoreceptors derived from human Muller glia restore rod function in the P23H rat. Stem Cells Transl Med. 2014; 3: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Singhal S,, Bhatia B,, Jayaram H,, et al. Human Müller glia with stem cell characteristics differentiate into retinal ganglion cell (RGC) precursors in vitro and partially restore RGC function in vivo following transplantation. Stem Cells Transl Med. 2012; 1: 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ramachandran R,, Fausett BV,, Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent let-7 microRNA signalling pathway. Nat Cell Biol. 2010; 12: 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Powell C,, Elsaeidi F,, Goldman D. Injury-dependent Müller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J Neurosci. 2012; 32: 1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Makino S,, Whitehead GG,, Lien C-L,, et al. Heat-shock protein 60 is required for blastema formation and maintenance during regeneration. Proc Natl Acad Sci U S A. 2005; 102: 14599–14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fausett BV,, Gumerson JD,, Goldman D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J Neurosci. 2008; 28: 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Millimaki BB,, Sweet EM,, Riley BB. Sox2 is required for maintenance and regeneration, but not initial development, of hair cells in the zebrafish inner ear. Dev Biol. 2010; 338: 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.