Abstract
Background
Stem cell therapies appear promising for treating certain neurodegenerative disorders and molecular imaging methods that track these cells in vivo could answer some key questions regarding their survival and migration. Bioluminescence imaging (BLI), which relies on luciferase expression in these cells, has been used for this purpose due to its high sensitivity.
New Method
In this study, we employ BLI to track luciferase-expressing human neural progenitor cells (hNPCLuc2) in the rat striatum long term.
Results
We show that hNPCLuc2 are detectable in the rat striatum. Furthermore, we demonstrate that using this tracking method, surviving grafts can be detected in vivo for up to 12 weeks, while those that were rejected do not produce bioluminescence signal. We also demonstrate the ability to discern hNPCLuc2 contralateral migration.
Comparison with Existing Methods
Some of the advantages of BLI compared to other imaging methods used to track progenitor/stem cells include its sensitivity and specificity, low background signal and ability to distinguish surviving grafts from rejected ones over the long term while the blood-brain barrier remains intact.
Conclusions
These new findings may be useful in future preclinical applications developing cell-based treatments for neurodegenerative disorders.
Keywords: Luciferase, Bioluminescence, Rat, Human Neural Progenitor Cells
1 Introduction
Stem cell therapies have emerged as promising treatment methods for a number of neurodegenerative diseases, as these cells are capable of surviving, migrating and integrating into the central nervous system (CNS). Neural stem and progenitor cells, distinct in their ability to differentiate into cells of the neural lineage, are especially well-suited for cell-based treatment of neurodegenerative disorders. Utilizing post-mortem histological analyses, these cells have been shown to survive over 7 months in the CNS of wild type rats and have been used in several preclinical studies of disorders such as Huntington’s disease (HD) and Parkinson’s disease (PD) (Lindvall, Barker et al. 2012, Gowing, Shelley et al. 2013). Specifically, fetal brain-derived human neural progenitor cells (hNPC) have been transplanted in the striatum of HD-induced rat model, demonstrating their ability to migrate, protect the striatum, and initiate recovery (McBride, Behrstock et al. 2004). Furthermore, protection of injured dopaminergic neurons has been demonstrated following striatal transplantations of hNPC overexpressing glial cell line-derived neurotrophic factor (GDNF) in animals experiencing PD-like partial striatal lesions (Behrstock, Ebert et al. 2006). Finally, a study in PD symptomatic non-human primates showed human neural stem cell (hNSC) survival and migration as well as animals’ functional improvement following cell transplants (Redmond, Bjugstad et al. 2007). Overall, neural progenitor/stem cells appear to be a promising tool for therapy of neurodegenerative diseases.
While delaying neural degeneration with neural progenitor/stem cells seems possible, one of the major roadblocks in therapeutic efforts arises from the inability to monitor cell fate in vivo. In animal studies, cell survival and migration can be assessed using post-mortem histological analyses (Tang, Shah et al. 2003, Behrstock, Ebert et al. 2008, Riley, Federici et al. 2009). However, a method of in vivo noninvasive, longitudinal cell tracking in clinical settings would be invaluable, allowing scientists to understand cell dynamics in single subjects as well as cohorts and adapt stem/progenitor cell therapies for further studies.
Several molecular imaging techniques are used for non-invasive stem cell tracking in vivo (Gera, Steinberg et al. 2010). In order for cells to be efficiently detected, they must first be distinguished from surrounding tissues. Additionally, the ideal imaging modality must be sensitive enough to detect the appropriate cell number required for treatment and have sufficient resolution to identify their location and migration over time. Furthermore, to achieve meaningful information from a cellular imaging modality, cell signal must also be reflective of survival/viability. Currently, no one imaging technique has been shown to successfully address all of these important issues.
Bioluminescence imaging (BLI) is an optical imaging technique that relies on light emission from the cells or tissues of interest. It has been explored for stem cell tracking because of its capability of detecting small populations of cells (Kim, Tsuji et al. 2006, Daadi, Li et al. 2009). BLI exhibits low background signal due to emission of optical light without an external light source, as well as the lack of autobioluminesence in mammalian tissues. In order to be detected with BLI, stem cells must first be induced to express a luciferase protein. Among them, firefly luciferase was originally extracted from the North American firefly and then further engineered to be used for imaging purposes. For signal to be detected, stem cells must also be in the presence of ATP and O2, which in concert with luciferase allow D-luciferin to be converted into oxy-luciferin and light. Luciferase expression has been used for a variety of assays such as gene expression quantification (Lipshutz, Flebbe-Rehwaldt et al. 2000), tumor development tracking in rats (Kondo, Goldman et al. 2009), and stem cell localization in mice (Bradbury, Panagiotakos et al. 2007) showing that BLI is valuable in determining cell viability and approximate location in vivo. Until this point, luciferase overexpression has not been explored for detection of slow proliferating progenitor cells in the rat brain, particularly in a structure as deep as the striatum. Concerns about bioluminescence signal penetrating rat’s skull, brain tissue and hair have been some of the reasons behind the reservations of the scientific community.
In this study, for the first time, luciferase expression in hNPC was induced to assess and track cells in vivo in the rat striatum. We show that these cells can be visualized long-term in vivo and that their survival and location can be deduced from BL images. These methodological findings may be useful in future preclinical applications aimed at developing cell-based treatments for neurodegenerative disorders.
2 Methods
2.1 Cell culture
Human neural progenitor cells were isolated between 10 and 15 weeks gestation using the protocols set by the National Institutes of Health (NIH) and the local ethics committees at the University of Wisconsin, Madison and University of Freiburg, Germany. All of the work was approved by the Institutional Review Board. A previously described method was used to prepare human cortical neural progenitor cells, G010 line, from fetal brains and induce their optimal cell expansion (Svendsen, Caldwell et al. 1997). These cells were grown as neurospheres in basic medium containing Dulbecco’s modified Eagle medium (DMEM, Sigma-Aldrich, St. Louis, MO) and Ham’s F12 (Sigma-Aldrich) (7:3), and penicillin/streptomycin/amphotericin B (PSA, 1% v/v, Life Technologies, Carlsbad, CA), supplemented with B27 (2% v/v, Invitrogen), epidermal growth factor (EGF, 100 ng/ml, Millipore Corp., Billerica, MA,), fibroblast growth factor-2 (FGF-2, 20 ng/ml, WiCell Research Institute, Inc.) and heparin (5 µg/ml, Sigma-Aldrich). Neurospheres were passaged approximately every 14 days by chopping with McIlwain automated tissue chopper (Mickle Engineering, Gomshall, Surrey, UK) (Svendsen, ter Borg et al. 1998). After passage 10, the cells were switched to maintenance medium: basic medium supplemented with N2, EGF, leukemia inhibitory factor (LIF, 10 ng/ml, Millipore), FGF-2 and heparin, helping to increase the rate of expansion and permitting stable growth for another 20 to 30 passages.
2.2 Transient Luciferase Expression
Transient luciferase expression was established using the Lonza Nucleofection System (Lonza Group Ltd., Basel, Switzerland). Nucleofection is a non-viral method of transfection that employs both electroporation and lipofection in order to achieve high cDNA incorporation with minimal cell death. Luciferase cDNA, pF9A-Luc2 (Promega Corp., Madison, WI), was under the control of the cytomegalovirus (CMV) promoter. Briefly, hNPC were dissociated using a Trypsin solution (TrypLE, Invitrogen) and 5×106 cells were resuspended in 100µl Basic Nucleofection Solution Primary Neurons (Kit VPI-1003) with 4 µg of pF9A-Luc2. The cells were then pulsed using the C-30 program and immediately transferred into pre-warmed fresh maintenance medium. For in vitro analysis, 3.0×104 transfected cells were plated as monolayers on glass coverslips that were pre-coated with poly-ornathine (Sigma-Aldrich, 0.1 mg/ml) and poly-L-laminin (Sigma-Aldrich, 50 µg/ml) to promote adherence. In preparation for in vivo analysis, 3×106 transfected cells were added to a well in a 6-well plate coated with polyhydroxyethylmethacrylate (p-HEMA, Sigma-Aldrich, 12 mg/ml) to minimize adherence. Luciferase activity was assessed 24–48 hours following cell plating using either immunostaining, GloMax® 96 Microplate Luminometer or In Vivo Imaging System (IVIS Spectrum, Perkin Elmer, Waltham, MA) or 48–72 hours following cell transplantation using IVIS.
2.3 Stable Luciferase Expression
PCR was used to amplify the Luc2 gene from pF9A-Luc2 using 10X Platinum Pfx Amplification Buffer (Invitrogen), 10 mM dNTP Mix (Promega), 50 mM MgSO4 (Promega), 100 ng template DNA, Platinum Pfx DNA Polymerase (Invitrogen), 10X Enhancer and primer mix containing 15 µM BamHI (5’-CTAGCGGGATCCCGTCGAGAATTAGCTTCGCCACCATGGAAGATGCCAAAAACATTAAGAAGG-3’) and 15 µM MluI (5’-GGTACCACGCGTGAATTGATCCTCACACGGCGATCTTGCCGCCCTTCTTGGCC-3’) restriction sites. The PCR cycle parameters were: 94 °C for 2 min once, 94 °C for 15 sec, 50 °C for 30 sec, 68 °C for 90 sec, the last three steps were repeated 25 times, 68 °C for 7 min once. Subsequently, the Luc2 gene was ligated into a self-inactivating lentiviral vector with posttranscriptional cis-acting regulatory element of woodchuck hepatitis virus and mouse phosphoglycerate kinase 1 promoter (SIN-W-PGK) shuttle vector. Lentivirus was produced from the Luc2-SIN-W-PGK shuttle vector and was used to infect wild type hNPC (hNPCWT) as previously described (Capowski, Schneider et al. 2007). Briefly, the cells were dissociated with TrypLE and incubated overnight with the viral titer of 100 ng p24 per million cells of luciferase lentivirus in half of the normal volume of maintenance medium. The medium was doubled 24 hours later, and half of the medium was replaced twice per week thereafter while the infected cells (hNPCLuc2) were allowed to reform spheres. hNPCLuc2 continued to be passaged by chopping (as described in section 2.1) for approximately 15 weeks, at which point they were used for further experiments.
2.4 Immunocytochemistry
Immunocytochemistry was performed to ensure that the Luc2 protein was expressed after transfection and lentiviral infection and to assess cell differentiation compared to hNPCWT controls. Cells were fixed using 4% paraformaldehyde (PFA) for 20 minutes and rinsed in phosphate buffered saline (PBS). Cells were blocked in PBS with 5% normal donkey serum (NDS) and 0.2% Triton X-100 (Sigma-Aldrich) for 30 minutes. For luciferase staining, cells were blocked in 1% bovine serum albumin (BSA) and 0.2% Triton X-100 in PBS. Primary antibodies against luciferase (Millipore, goat polyclonal 1:500), βIII-tubulin (Sigma-Aldrich, mouse monoclonal 1:1000), or glial fibrillary acidic protein (GFAP, Dako, Carpinteria, CA, polyclonal rabbit 1:1000) were used. Primary antibody incubation for 1 hour at RT was followed by PBS washes, a 30 minute incubation with the appropriate secondary antibody and nuclear labeling with Hoechst 33258 (Sigma-Aldrich, 0.5 µg/ml). All double labels were performed simultaneously. Images were acquired using the Nikon Intensilight C-HGFI camera and Nikon Eclipse (80i) fluorescence microscope. The quantification of fluorescence was done using NIS Element D Software by counting the ratio of Hoechst-positive nuclei to immunostained cells and converting it to percent in three to six independent fields from at least three coverslips. The data counted was plotted as mean ± SEM.
2.5 BrdU Labeling
To observe cell proliferation rates, hNPCWT and hNPCLuc2 were pulsed with bromodeoxyuridien (BrdU) as previously described (Wright, Li et al. 2003, Suzuki, Wright et al. 2004). Briefly, cells were incubated with 0.2 µM BrdU (Sigma-Aldrich) for 16 hours and subsequently dissociated into single cell suspensions using TrypLE. Dissociated cells were then plated as monolayers on glass coverslips, allowed to attach for two hours in maintenance medium and then fixed with 4% PFA for 20 minutes. Alternatively, BrdU-pulsed hNPCWT and hNPCLuc2 were plated onto glass coverslips and maintained for 14 days in plating media (basic medium supplemented with 2% B27) with half medium replacement twice per week before fixation.
Fixed cells were incubated with 2N HCl in PBS for 20 minutes at 37 °C, then quenched with 0.1M Na-Borate buffer (pH 8.5) for 10 minutes at RT. Cells were blocked in PBS with 5% NDS and Triton X-100 for 30 minutes. The primary antibody against BrdU (Accurate Chemical, Westbury, NY, monoclonal rat 1:500), was incubated for 1 hour at RT and then overnight at 4 °C, followed by PBS washes and a 30 minute incubation with the appropriate secondary antibody and Hoechst 33258 nuclear labeling. When double labeling with BrdU and luciferase was required, luciferase staining was completed prior to commencing the BrdU stain.
2.6 Western Blot
hNPCLuc2 and hNPCWT were washed in PBS, lysed by shaking for 15 minutes at 4 °C in RIPA buffer (Sigma-Aldrich) and protease inhibitor (1:20), centrifuged for 10 minutes at 2000 RPM and protein lysates were stored at −80 °C. Protein concentration was determined by the DC Protein Assay Kit (Bio-Rad Laboratories, Inc., Hercules, CA) and BSA as a standard curve. Sample concentrations were calculated using FLUOstar OPTIMA (BMG Labtech GmbH, Ortenberg, Germany). Approximately 15 µg of protein was loaded into Mini-PROTEAN® TGX Precast gels (Bio-Rad Labs), separated with 110 V for approximately 1.5 hours then electro-transferred onto Immobilon transfer membrane (Millipore) for 70 minutes at 250 mA. The membrane was blocked in 5% milk in Tris-buffered saline with 0.5% Tween 20 (TBST) for 1 hour at RT and then exposed to the primary antibody against luciferase (Promega Corp, 1:1000, goat,) in blocking buffer overnight at 4 °C. Anti-goat secondary antibody conjugated to peroxidases (Dako, Promega Corp., 1:1000) was applied in blocking buffer for one hour at RT, followed by exposure by chemiluminescence kit (Pierce ECL Western Blot Substrate, Thermo Fisher Scientific, Inc. Waltham, MA). Equal protein loading was confirmed by incubating the membrane in the Coommassie blue stain for 15–20 minutes.
2.7 In Vitro Imaging
In vitro imaging of Luc2-transfected cells was performed using the GloMax 96 Microplate Luminometer (Promega Corp.) and IVIS. Luc2-transfected cells were plated at a density of 1×105 cells/well in a 96-well plate coated with poly-L-laminin to permit cell adherence. The next day, 0.2, 2, or 20 mM luciferin (VivoGlo™) was added to triplicate wells for 1 hour, with no luciferin addition as a negative control. The cells were scanned in the GloMax® 96 Microplate Luminometer using the luminescence protocol.
Luc2-transfected hNPC or hNPCLuc2 were also imaged using IVIS. Luciferase-expressing and wild type cells were plated at a density of 1×105 cells/well in a 24-well plate. The next day, 0–2 mM luciferin was added for 1 hour. The cells were imaged with IVIS bioluminescence protocol using 620 nm emission, 30 seconds exposure time and 1 cm camera height.
2.8 Animals
Sprague Dawley rats were obtained from Taconic (Hudson, NY). The animals were housed under controlled temperature (20–26°C) and illumination (12 hours on, 12 hours off) with unlimited access to laboratory chow and water. The University of Wisconsin-Madison and NIH guidelines were followed for all animal experiments.
2.9 Cell Transplantation
Transiently or stably luciferase-expressing and hNPCWT were transplanted into the rat brain. Transfected cells were transplanted two days after transfection while stably expressing hNPCLuc2 were transplanted over four months after lentiviral infection. Cells were dissociated into single cells using TrypLE and resuspended at a density of 1.5×105 cells/ul in Leibowitz transplantation medium (L 15 Leibowitz, Invitrogen) / 0.6% Glucose (D-(+)-Glucose, Sigma-Aldrich) in PBS (1:1) and B27 (2% v/v). Transplants were done using a 30g sharp tip needle connected to a 10 µl Hamilton syringe secured by a stereotaxic frame (David Kopf Instruments, Tujunga, CA) in isoflurane-anesthetized adult female rats. Either three striatal sites (AP+1.0, −1.0 and −1.2; ML ±3.0, ±3.7 and ±4.5; DV −4.5 from Bregma) were used on their own or in addition to three cortical sites (AP+1.0, −1.0 and −1.2; ML ±3.0, ±3.7 and ±4.5; DV −2.0). Alternatively, cells were transplanted in only two striatal sites (AP +0.5 and −0.1; ML ±3.3 and ±2.8; DV −4.5). In all instances, 3×105 cells were transplanted per site. hNPCWT were injected contralaterally as a negative control. For striatal and cortical transplants (N=5), the needle was first lowered into the striatum for 2 minutes. Using a pump, 3.0×105 cells were injected at the rate of 1 ul/min for 2 minutes, injecting 2 µl of cells. The needle was left in place for 2 minutes, then slowly lifted to the cortex for transplantation of 3.0×105 cells using the same procedure. This process was repeated for each transplantation site for a total of 1.8×106 cells. For striatal only transplants, animals received either 9.0×105 cells in three sites (N=7) or 6×105 cells in two sites (N=10). In addition, serial dilutions (1.5×105, 1.5×104 and 1.5×103 cells/µl) of hNPCLuc2 were injected into the striatum to determine the minimum cell number that can be detected in vivo. By injecting 1 µl/min for 2 minutes, injecting 2 µl of cells per site, into three sites, the first animal received 9×105 cells, the second 9×104 and the third 9×103, and each animal received non-luciferase-expressing hNPC contralaterally as a negative control.
For cell migration experiments, quinolinic acid (QA) was used to promote hNPCLuc2 migration. Ipsilateral cell migration in the rostral direction was induced by injecting 100 nM QA at AP 0.48, ML −3.0 and DV −4.0 followed after a week by hNPCLuc2 cell transplantation into the striatum (AP −1.1, ML −2.8, −3.4, and −4.0, and DV −4.0) (N=3). Contralateral cell migration along the corpus callosum was induced by injecting QA at AP 0.48, ML 3.0 and DV −2.5 followed after a week by hNPCLuc2 cell transplantation into the cortex (AP 1.0, 0.48 and −0.04, ML −3.0 and DV −2.0) (N=3).
Intramuscular transplantations of hNPCLuc2 in the tibialis anterior (TA) hind limb muscle were performed as described previously (Suzuki, McHugh et al. 2008). Briefly, partial muscular lesion was induced using bupivacaine (BVC) hydrochloride (0.35mg per muscle; Sensorcaine-MPF, AstraZeneca, London, UK) injected unilaterally into TA one day before hNPCLuc2 transplantations using a 30-gauge needle connected to a 1 ml syringe. hNPCLuc2 (10,000 cells/µl in 50 µl) were injected 24 hours later into the muscle using a 33-gauge needle connected to a 100 µl Hamilton syringe.
All animals received daily intraperitoneal (IP) cyclosporine injections (10 mg/kg, Sandimmun, Novartis, Basel, Switzerland) starting the day before transplantation. After the final scan, animals were perfused with chilled 0.9% saline and subsequently with 4% PFA. The brains were removed, post-fixed in 4% PFA overnight and then cryopreserved in 30% sucrose before sectioning at 30–40 µm with a sliding microtome (Leica Microsystems, Bannockburn, IL).
2.10 In Vivo Imaging and Migration Analysis
One to three days after hNPCLuc2 transplantation, animals were imaged using IVIS. Animals imaged long-term were scanned after 7 days and at least three more times thereafter for up to 12 weeks. Finally, animals used in the study to determine the minimal number of detectable cells were imaged 7 and 21 days following surgeries. Only hNPC stably expressing luciferase were used for imaging studies longer than three days. Rats were anesthetized in a holding chamber with 4% isoflurane in compressed air, injected with VivoGlo™ luciferin (150 mg/kg, IP) and moved into the scanner after 15 minutes, where they were maintained on 2–3.5% isoflurane. The scans were performed using the bioluminescence protocol with open emission, 60 seconds exposure and 3.0 cm camera height.
Image analysis assessing cell migration was performed at each imaging time point. In each image a rectangular region of interest (ROI) was selected based on anatomical landmarks such as the ears and eyes. The ROI limits were defined as the most medial parts of the eyes in the medial-lateral direction and most caudal part of the eyes and the most rostral part of the ears in the rostral-caudal direction. For detecting ipsilateral migration, the ROI was split in half horizontally and the mean luminescence signal level was measured in the rostral (R) and caudal (C) halves of the ROI. The ratio R/C was calculated for each time point and the change in this ratio was noted over time. For detecting contralateral migration, the ROI was split in half vertically and the mean luminescence signal level was measured on the right (R) and left (L) side of the ROI. The ratio R/L was calculated for each time point and the change in this ratio was observed over time. Imaging parameters, ROI size and measurement procedure were kept consistent within and between subjects to minimize human errors in migration analysis measurements.
2.11 Histological Analysis
Brain sections were blocked with 3% NDS, 0.3% Triton X-100 in PBS for 1 hour then incubated with primary antibodies against either human cytoplasmic marker (hCyto, Stem Cells Inc, Newark CA, mouse, 1:200), luciferase (Promega, goat, 1:100) or GFAP (Dako, mouse, 1:200) overnight at RT. They were then washed and incubated with appropriate fluorescent-conjugated secondary antibodies for 1 hour. Sections were mounted and coversliped using DAPI mounting medium (Vectashield Hard Set, Mounting Medium with DAPI, Vector Labs).
2.12 Statistical Analysis
Unpaired Student’s two-tailed t tests were used to compare data between various time points or different groups for all experiments requiring cell counts. Migration data comparing cell signal shift over time between histologically determined migration and non-migration groups was analyzed using two-way analysis of variance (ANOVA) for migration, time and their interaction. A Bonferroni test was applied to account for multiple comparisons. All analysis was done using GraphPad Prism 5 software. Data are shown as mean ± SEM. P values < 0.05 were considered statistically significant.
3 Results
3.1 hNPC Transiently Express Luciferase
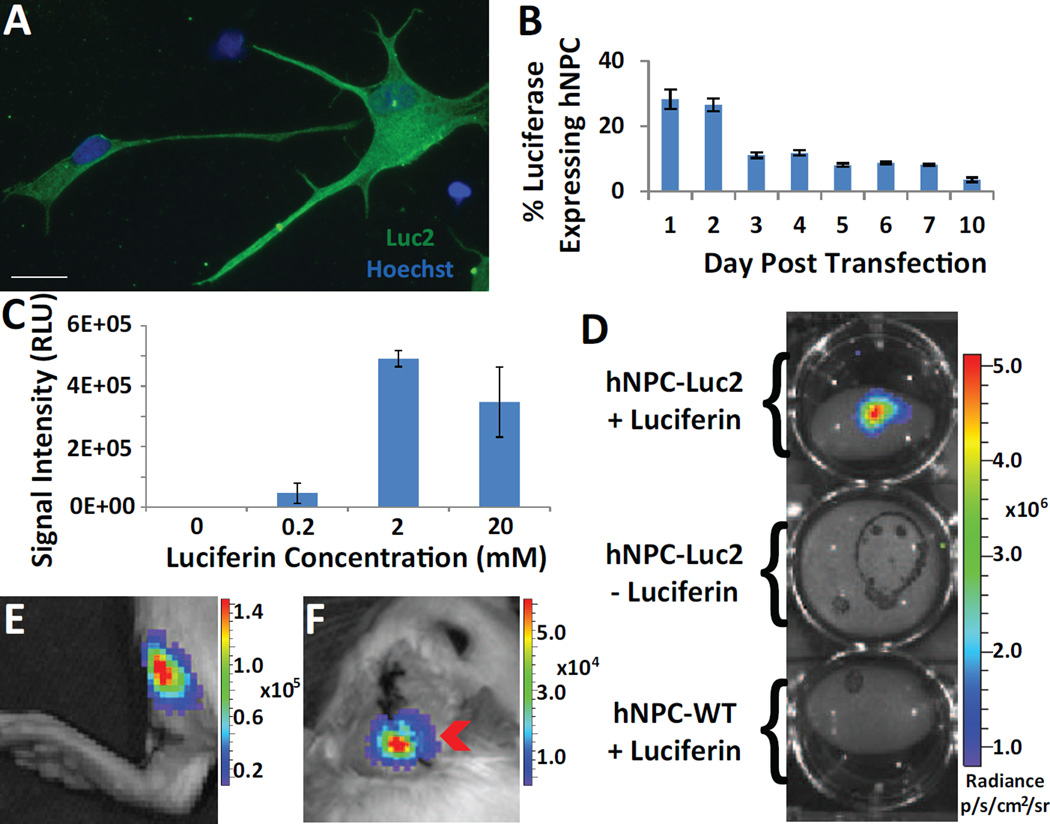
We used nucleofection to induce transient luciferase expression in hNPC in vitro, for ultimate in vivo cell detection. Immunocytochemistry with a luciferase antibody confirmed that hNPC transiently express robust levels of luciferase (Fig. 1A). To determine the percent of hNPC that express luciferase and the time course of luciferase expression following nucleofection, transiently expressing hNPC were plated as single cells and immunostained over ten days. The results show that for up to 2 days following nucleofection, at least 28% of total hNPC express luciferase. The luciferase expression then greatly decreases on day three following transfection, reducing to less than 5% of total hNPC expressing luciferase by day ten (Fig. 1B).
Fig. 1.
Transient luciferase expression in hNPC. (A) Immunocytochemical image of hNPC with robust transient expression of luciferase. (B) Quantification of immunocytochemical analysis determines the percent of hNPC transiently expressing luciferase. (C) Comparison of signal intensity (relative light units, RLU) of hNPC exposed to different concentrations of luciferin. (D) In vitro image of Luc2-transfected hNPC incubated with luciferin, compared with those not incubated with luciferin and WT hNPC incubated with luciferin. (E) Detection of 5.0×105 Luc2-transfected hNPC in rat left hind limb muscle. (F) Detection of Luc2-transfected hNPC in right cortex (9.0×105) and striatum (9×105) of rat. All data is given as mean ± SEM. Scale bar 20 µm.
Following confirmation of luciferase expression in fixed cells, we next used optical imaging to observe bioluminescence signal from luciferase-expressing live cells both in vitro and in vivo. Luciferin substrate is an essential component in bioluminescence imaging of firefly luciferase. To determine the optimal concentration of the luciferin substrate for in vitro detection of hNPC-Luc2, we incubated cells in three different concentrations of luciferin and assessed the BLI signal intensity using a GloMax® 96 MicroplateLuminometer. Two mM of luciferin provided the highest signal intensity and hence was used in all subsequent in vitro experiments (Fig. 1C). An additional imaging system, IVIS, confirmed high levels of luciferase activity in live hNPC-Luc2 following exposure to 2 mM luciferin (Fig. 1D). Importantly, the IVIS imaging data also showed that neither hNPCWT exposed to luciferin nor hNPC-Luc2 not incubated with luciferin produced signal above background (Fig. 1D), confirming that hNPCWT show no endogenous levels of luciferase activity and that luciferin must be catalyzed by luciferase for optical light emission.
Having confirmed that luciferase-expressing hNPC can be detected in vitro using IVIS, we next investigated their detectability in vivo. Two days following transfection with Luc2 cDNA, a total of 1.8×106 cells were injected in the cortex (9.0×105) and striatum (9.0×105) of adult rats. The cortex, a dorsal structure compared to the striatum, was injected to increase the likelihood of in vivo cell detection. Additionally, 5×105 of the same cells were injected in the hind limb muscle (tibialis anterior) as a positive control. Three days following transplantation, the animals received IP injections of luciferin and were imaged 15 minutes later. In vitro, cells still showed transient expression at 5 days post-transfection, which was reinforced in vivo by the continued expression of luciferase that permitted cell detection in the muscle (Fig. 1E) and the brain (Fig. 1F), although the signal from the muscle was significantly higher despite a lower number of transplanted cells. These proof-of-concept experiments were important to confirm that luciferase-expressing hNPC can indeed be detected both in vitro and in vivo.
3.2 hNPC Stably Express Luciferase
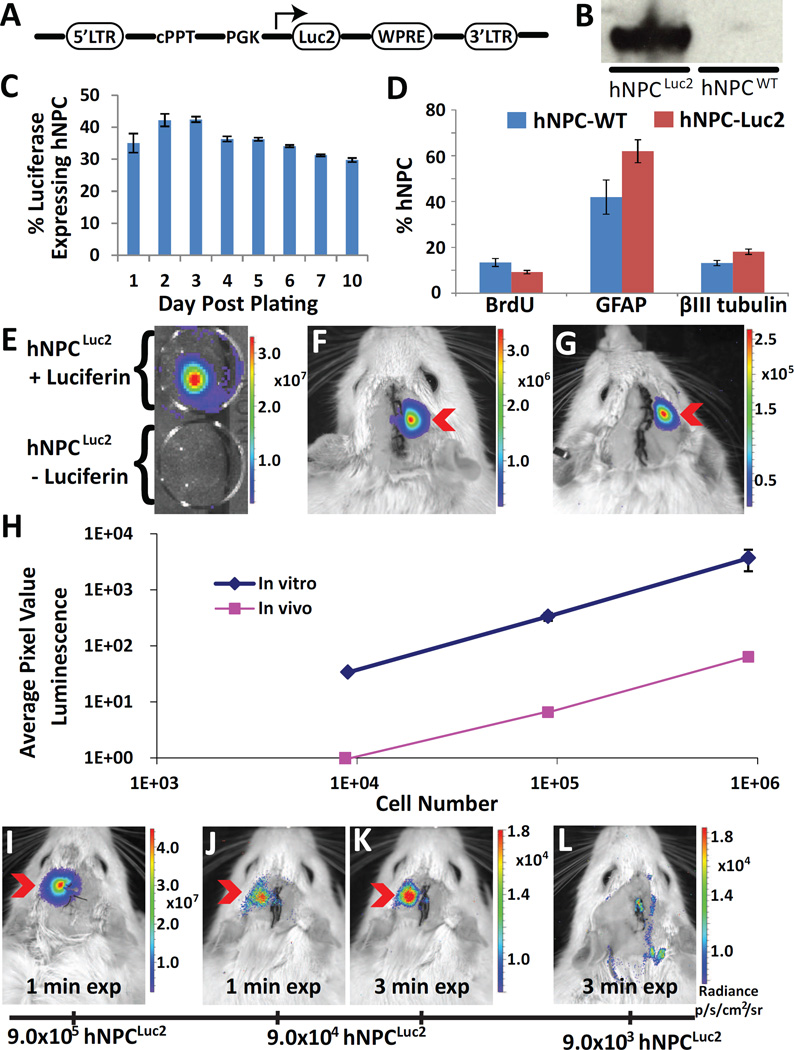
Our transient expression results show promise in that the luciferase reporter protein can be used for in vitro and in vivo imaging of hNPC. However, the short duration of reporter protein expression indicates that stable protein expression in hNPC is required for cell tracking over the course of an in vivo cell therapy experiment. Several of our previous studies have demonstrated successful stable protein expression in hNPC using lentiviral infection (Behrstock, Ebert et al. 2006, Capowski, Schneider et al. 2007, Suzuki, McHugh et al. 2007, Behrstock, Ebert et al. 2008). This method relies on insertion of a potentially sizeable gene into the genome in both dividing and non-dividing cells and provides high infection efficiency. The Luc2 lentiviral shuttle vector consisted of Luc2 cDNA under the control of mouse PGK promoter to provide constitutive protein expression (Fig. 2A). The lentivirus generated from the construct was used to infect hNPCWT. Three months following infection, stable expression of the Luc2 protein in hNPCLuc2 was verified in two ways. First, Western blot analysis of protein extracted from both hNPCLuc2 and hNPCWT and stained against luciferase revealed that luciferase was produced by hNPCLuc2 but not by hNPCWT (Fig. 2B). In addition, hNPCLuc2 were plated as single cells and immunostained for luciferase over ten days. Luciferase expression was maintained for at least 10 days post-plating and quantification showed that approximately 40% of total hNPC stably expressed luciferase (Fig. 2C). The stable expression of luciferase was confirmed for up to four months post-infection in vitro and additional three months post-transplants using histology. This suggests that hNPC expressing the luciferase reporter protein can be used for long-term cell tracking. To increase the percentage of hNPC stably expressing luciferase, we did a subsequent infection on hNPCWT that yielded 53.5±1.5% of total hNPC expressing luciferase (data given as mean±SEM). Given that we achieved an infection level over 50%, it was unnecessary to stress the hNPCLuc2 with cell sorting. This hNPCLuc2 population had a similar long-term expression profile to the initial hNPCLuc2 stable line (Fig. 2B and C) and was used for subsequent transplantation experiments.
Fig. 2.
Stable luciferase expression in hNPC (hNPCLuc2). (A) Schematic of lentiviral construct. LTR, the long terminal repeat; PGK, the mouse phosphoglycerate kinase 1 promoter; WPRE, the post transcriptional regulatory element of woodchuck hepatitis virus, cPPT, the central polypurine tract; SIN, self-inactivating. (B) Western blot of hNPCLuc2 and hNPCWT lysates stained against luciferase. (C) Percentage of hNPC stably expressing luciferase more than seven months post infection. (D) Comparison of hNPCLuc2 and hNPCWT in their capacity to proliferate (BrdU marker) and differentiate (βIII tubulin and GFAP markers). (E) In vitro image of hNPCLuc2 with and without luciferin incubation. (F) Detection of hNPCLuc2 in right cortex (9.0×105) and striatum (9.0×105) of WT rats. (G) Detection of 9×105 hNPCLuc2 in right striatum only of WT rats. (H) Plot of average pixel intensity compared to cell number (9×105, 9×104, 9×103) for in vitro and in vivo conditions. Minimal cell number study confirms the ability to detect (I) 9×105 hNPCLuc2 with autoexposure (approximately 60 sec) and (J) 9×104 hNPCLuc2 in the left striatum of WT rats using both auto exposure (approximately 60 sec) and (K) 3 min exposure. (L) Minimal cell number study shows that 9×103 cannot be distinguished from background even using longer exposure time. All data is given as mean ± SEM.
Before using lentivirus-infected hNPC stably expressing luciferase in further experiments, we needed to ensure that neither the lentivirus nor stable luciferase expression altered cell proliferation and differentiation capacity. hNPCLuc2 and hNPCWT proliferation was compared by pulse-labeling cells with BrdU, a synthetic nucleotide which incorporates into the DNA of replicating cells. After labeling, single cells were plated down for one to three hours then fixed and fluorescently immunostained with an anti-BrdU antibody to detect newly dividing cells. Approximately 10% of hNPCWT and hNPCLuc2 stained positive for BrdU, showing that neither lentiviral infection nor stable luciferase expression significantly change cell proliferation rates (Fig. 2D). Furthermore, our previous studies have shown that hNPC differentiate mainly into astrocytes and a limited number of neurons (Svendsen, ter Borg et al. 1998). To examine the differentiation capacities of hNPCLuc2 compared with hNPCWT, both cell types were plated as single cells and allowed to differentiate for 14 days without any growth factors in the media, to promote differentiation into astrocytes and neurons. Immunostaining with a glial fibriallary acidic protein (GFAP) antibody or βIII tubulin antibody was used to assess the number of astrocytes and neurons, respectively. Both hNPCWT and hNPCLuc2 differentiated into approximately 50% astrocytes and 15% neurons (Fig. 2D). The remaining cells are presumably undifferentiated, nestin-postive cells (data not shown). This data demonstrated no significant differences in differentiation rates between hNPCLuc2 and hNPCWT suggests that, along with proliferation rate, differentiation potential is not altered following lentiviral infection or stable luciferase expression.
3.3 Detection of hNPCLuc2 in Optical Imaging System
After verifying that hNPC could stably express luciferase without changes in their proliferation or differentiation, we examined whether stably-expressing hNPCLuc2 could be detected in vitro and in vivo with IVIS. To confirm the ability of hNPCLuc2 detection in vitro, hNPCLuc2 were plated as single cells for 24 hours and then incubated with luciferin for one hour prior to imaging with IVIS. As a negative control, hNPCLuc2 not exposed to luciferin were also imaged. The acquired images demonstrated that hNPCLuc2 incubated with luciferin could be detected, while the hNPCLuc2 not incubated with luciferin produced no signal (Fig. 2E).
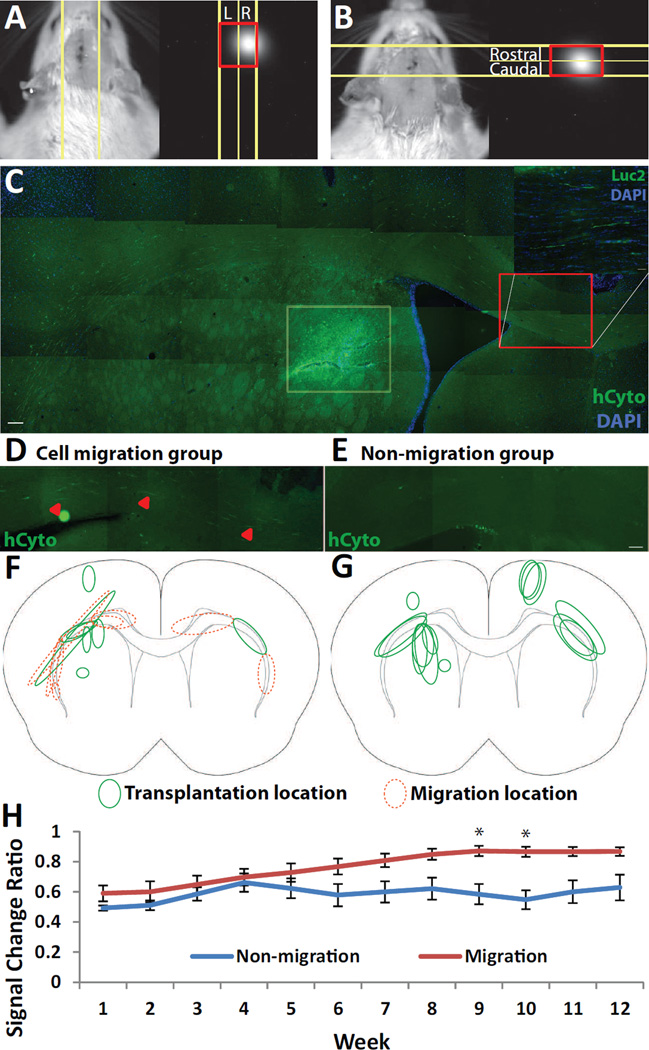
To test whether hNPCLuc2 can be imaged in vivo with IVIS, we injected the cells unilaterally into both the cortex (9.0×105) and the striatum (9.0×105) of WT rats, and into the hind limb muscle as a positive control. The animals were imaged three days following transplantation. hNPCLuc2 in both the muscle and the brain could be detected, suggesting that stable expression of luciferase can be used for identifying hNPC in vivo (data not shown). Since neurological diseases such as Parkinson’s and Huntington’s disease may benefit from cell transplants into the striatum, it was important for our study to ensure that hNPCLuc2 can be detected if injected into the striatum. We injected 9.0×105 hNPCLuc2 unilaterally into the striatum and, as the positive control, we injected hNPCLuc2 into both the cortex and the striatum as before. Indeed, signal could be detected in the positive control (Fig. 2F) and it also remained detectable when cells were injected into the striatum only (Fig. 2G). We confirmed that signal originated from the striatum, rather than from along the injection tract, using three-dimensional diffuse luminescence tomography (3D DLIT, Fig. S1) and histology (Fig. 4C). This is the first time that stable luciferase-expressing slow proliferating progenitor cells have been visualized in the rat brain, specifically in a deep structure such as the striatum.
Fig. 4.
Assessment of hNPCLuc2 migration using IVIS. Example of the method used to calculate signal movement for (A) contralateral and (B) ipsilateral migration. (C) Histology using human cytoplasmic marker (hCyto) shows hNPCLuc2 striatal graft (highlighted in light yellow box) and hNPCLuc2 migration along the corpus callosum (highlighted by red box). A zoomed medial region confirms luciferase expression. Histological comparison of two grafts which (D) did and (E) did not result in migrating hNPCLuc2. Schematic of transplant sites (green, solid line) and migration paths (orange, dotted line) for animals whose (F) hNPCLuc2 migrated contralaterally and (G) those whose cells did not. (H) Graph comparing signal change ratio (opposite side vs. original side) and time of the migration and non-migration groups. While ANOVA 2-way test determined a significant difference between the groups, asterisks suggest significant difference in the particular time points. All of the data is given as mean ± SEM, *p<0.05.Scale bar 100 µm (hCyto), 50 µm (Luc2) (C), 100 µm (D–E).
The successful visualization of cells transplanted into a more ventral brain region led us to next investigate the minimum number of hNPCLuc2 that can be detected using IVIS both in vitro and in vivo in the rat striatum. This is a fundamental question as subsequent studies may require a different number of transplanted hNPCLuc2. To determine the range of detectable cells permitted by the IVIS, we plated a monolayer of cells in serially decreasing numbers (9×105, 9×104, and 9×103). The day after plating, images of cells incubated with 2 mM luciferin showed that each cell amount could be serially detected (Fig. 2H). To determine the range of detectable cells following striatal transplantation, animals were transplanted with the same serially decreasing number of cells (9×105, 9×104 and 9×103 cells) that was shown to be detectable by IVIS. In vivo imaging at one and three weeks following surgeries showed that BLI signal could be detected from 9×105 and 9×104 hNPCLuc2 (Fig. 2I and 2J, respectively and Fig. 2H). In the animal transplanted with 9×104 hNPCLuc2, cell signal was visualized better after exposure time was increased from 1 to 3 minutes (Fig. 2K). On the other hand, the signal from the animal injected with 9×103 cells could not be distinguished from background even following exposure adjustments (Fig. 2L and H). This suggests that the lowest detectable number of hNPCLuc2 in the striatum using our infection and transplantation protocols is between 9×104 and 9×103. Furthermore, a comparison of in vitro and in vivo signal intensities revealed that the average signal intensity in vitro is approximately two orders of magnitude higher than in vivo (Fig. 2H). Overall, through these studies we were able to determine the approximate hNPCLuc2 number that can be visualized in the rat striatum and assess the magnitude of signal difference between in vitro and in vivo imaging.
3.4 Long Term Detection of hNPCLuc2 in Optical Imaging System
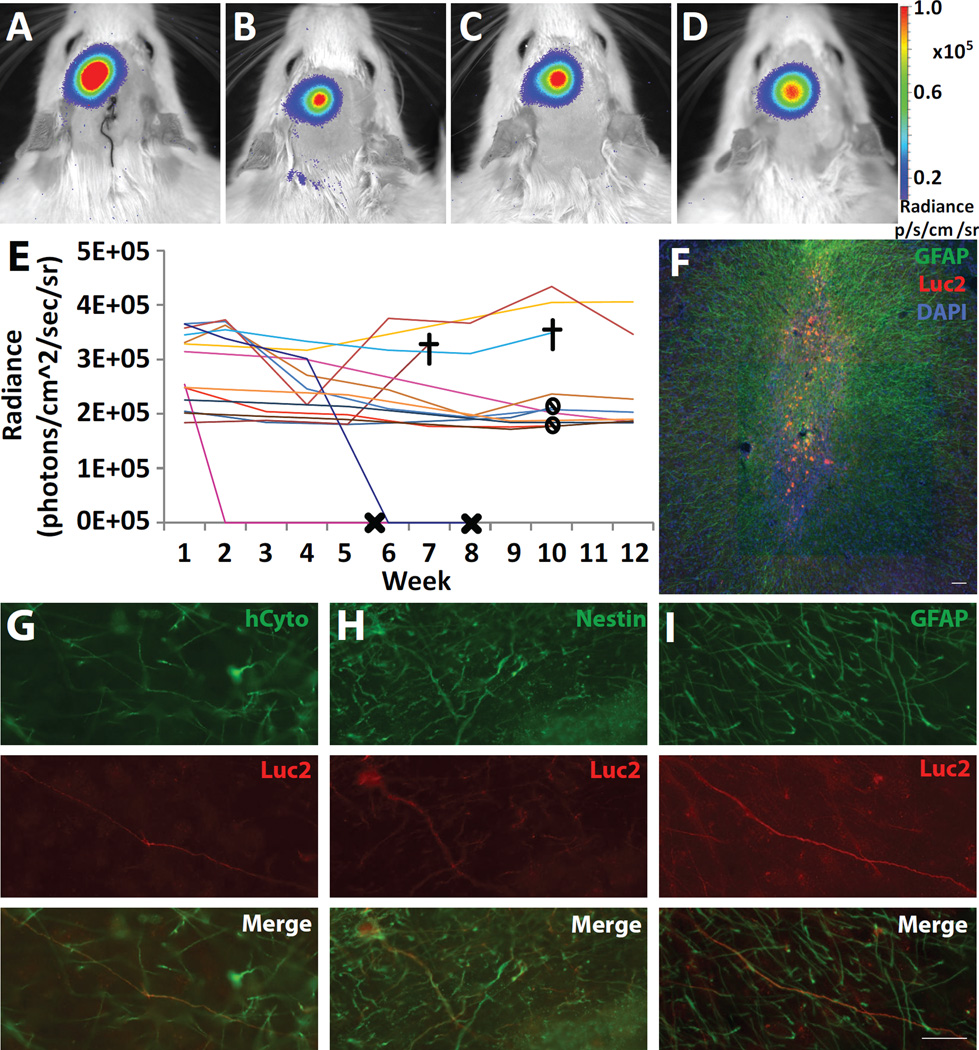
Next, we wanted to establish whether hNPCLuc2 could be tracked long term, for up to 10–12 weeks in the rat striatum. For this purpose, 9×105 hNPCLuc2 were transplanted into the striatum of 14 animals. Non-luciferase-expressing hNPC, as negative controls, were transplanted contralaterally into six of these. The animals were imaged seven days following transplantation and then incrementally until end point at 10–12 weeks. In 12 of the 14 animals, bioluminescence signal was detected using IVIS throughout this period of time using both 2-dimensional BLI (Fig. 3A–D) and 3D DLIT (Fig. S1). The average radiance over time was quantified throughout the study.
Fig. 3.
Analysis of long term hNPCLuc2 tracking. IVIS images of an animal transplanted with 9×105 hNPCLuc2 in the left striatum and imaged at weeks (A) 1, (B) 5, (C) 9 and (D) 12. (E) Average radiance of luciferase signal over time plot for all fourteen animals included in long-term studies. Symbols are X for loss of signal (N=2) that was later confirmed histologically, stop sign for the end of the 10 week study and cross for animal death (N=2) that was unrelated to the study. (F) In the representative animal (same as A–D), histological analysis of GFAP (green) and luciferase (red) expression with DAPI (blue) confirms good survival in the striatum. (G) Human cytoplasmic marker (hCyto) and luciferase analysis confirms cell survival and continued expression over the 12 week time period. (H) Nestin and (I) GFAP analysis shows that luciferase-expressing cells remained as undifferentiated neural progenitors and that some differentiated towards a glial phenotype, respectively. Scale bar 20 µm (G–I) and 100 µm (F).
Detection of bioluminescence signal from two animals ceased at two and six weeks post-transplantation, respectively. These animals were re-imaged approximately 10 days later to ensure that lack of signal was not a result of improper luciferin delivery. The additional scan confirmed signal loss, and further histological evaluation showed that lack of bioluminescence signal (Fig. 3E) corresponded with an absence of hCyto expression (data not shown). The absence of hCyto-expressing cells suggested graft rejection. However, given that animals were sacrificed nearly 2 weeks following the loss of BLI signal, the cells had likely already been cleared, not allowing for confirmation of cell death using apoptosis or necrosis markers. Alternatively, lost bioluminescence signal could result from cell migration rather than a lack of cell survival. To address this, we carefully analyzed the histological slices in the vicinity of the graft from these animals to confirm that there was no significant cell migration. Other than the cells transplanted near the white matter tracks, which could be easily identified migrating along the corpus callosum, we did not observe any evidence of hNPC migrating in the striata of wild type animals, suggesting that the signal loss was likely a result of cell death rather than cell migration.
In all other animals, survival of luciferase-expressing cells was evident in the rat striatum (Fig. 3F). Additional histochemical analysis confirmed luciferase expression specific to hNPCLuc2 using human cytoplasmic marker (hCyto, Fig. 3G). Nestin and GFAP staining revealed that some cells remained as undifferentiated neural progenitors while others began differentiating into astrocytes, respectively (Fig. 3H and I). Overall, out of the fourteen animals transplanted with hNPCLuc2, approximately 86% (twelve) had surviving grafts at the end of the long term study which lasted between 10 and 12 weeks, with one animal dying of unrelated causes at 7 weeks post-transplantation with surviving hNPCLuc2 grafts. This experiment confirmed that hNPCLuc2 can be detected in vivo for at least 12 weeks following striatal injections, exhibiting the potential for long-term tracking. In addition, it showed that cell death can reliably be established from the loss of bioluminescence signal during the study.
3.5 Migration Detection of hNPCLuc2 in Optical Imaging System
After validating that viable hNPCLuc2 can be identified in vivo in the rat striatum long-term, we examined whether migration of these cells can be detected. QA, a neurotoxin that has been shown to induce lesions that model HD, was employed based on our previous studies demonstrating radial hNPC migration as a result of ipsilateral QA-induced lesions (Behrstock, Ebert et al. 2008). On the other hand, hNPC behavior following transplants contralateral to the lesion has not been investigated. One week after QA injection, 9×105 hNPCLuc2 were transplanted contralaterally or ipsilaterally to the toxin. The animals were imaged immediately after transplantation and during week one, two, and every other week following transplantation until end point, 12 weeks. Migration was assessed by defining an ROI based on anatomical landmarks and determining a ratio of signal intensity between the area to which the cells were intended to migrate and the area where the cells were transplanted (Fig. 4A and B). The brain tissues were also histologically analyzed for migration. Ipsilateral migration could not be detected histologically or by ROI-based analysis in any of the animals. Interestingly, it was found that contralateral migration along the corpus callosum occurred in one animal belonging to the ipsilateral migration group. For this reason, all animals transplanted with hNPCLuc2 and imaged for 12 weeks were also analyzed for migration both histologically and with the ROI-based method. Altogether, contralateral hNPCLuc2 migration was observed in four animals (Fig. 4C and 4D) while four animals had hNPCLuc2 transplants that did not migrate (Fig. 4E). The schematic representation of both group’s grafts and subsequent hNPCLuc2 migration is shown (Fig. F and G). The ROI analysis of the same animals confirmed that the ratio of signal in the opposite side of cell transplantation to that of original side of cell transplantation was increasing in animals with histologically observed contralateral migration, but not in the animals where migration was not histologically detected. The averages of these ratios were analyzed, and significant differences were found between the two groups with respect to time and cell migration overall. Post hoc analysis also determined a significant difference at weeks 9 and 10 between the two groups (Fig. 4H). This study demonstrated that although contralateral migration is not readily visible by eye on bioluminescence images, it can be detected with an ROI-based approach.
4 Discussion
In the present study, we have shown for the first time that luciferase-expressing hNPC grafts can be detected in intact rat striatum using bioluminescence imaging. Furthermore, this imaging technique permits tracking of cell migration along the corpus callosum. Detecting transplanted cells is essential in evaluating and understanding stem cell therapy. Luciferase expression makes it possible to detect the cells of interest in the rat brain and reveals whether they remain alive. These new findings show that bioluminescence imaging can be efficiently utilized in preclinical studies of cell-based therapy for neurodegenerative diseases such as HD and PD.
4.1 Imaging hNPC Transiently Expressing Luciferase
Experiments with transiently luciferase-expressing hNPC demonstrated that these cells can be detected in the rat brain following transplantations into the cortex and striatum. Striatal cell transplantations are relevant for various disease models such as HD and PD (Behrstock, Ebert et al. 2006, Behrstock, Ebert et al. 2008). It was previously unclear whether the optical signal from luciferase-expressing hNPC could penetrate the brain, skull and surrounding tissue. To account for this possibility, luciferase-expressing hNPC were transplanted into the cortex as well as the striatum. Cell transplantations into the hind limb served as a positive control as it avoids the skull and the blood-brain barrier that can make signal acquisition challenging. This study revealed that cell signal could be detected in both the brain and hind limb muscle, with the hind limb signal being greater despite over an order of magnitude higher cell number in the brain (Fig. 1E). This was presumably due to reduced luciferin concentration in the brain compared to the muscle as a result of the blood-brain barrier (Aswendt, Adamczak et al. 2013). Furthermore, the skull acted as an additional, less penetrable barrier than the muscle tissue. The signal from cells in the brain was nevertheless easily detectable, further highlighting sensitivity as a major strength of bioluminescence imaging (Fig. 1F). Further exploration of hNPCLuc2 survival and long term detection in hind limb was not pursued extensively in this study as the hind limb was used primarily as a positive control for hNPCLuc2 visualization. This subject matter has been considered, however, using luciferase-expressing human mesenchymal stem cells (Krakora, Mulcrone et al. 2013).
4.2 Inducing Stable Expression of Luciferase
Studies using with hNPC transiently expressing luciferase indicated that BLI can be used to detect these cells in the brain. However, we determined that luciferase expression in hNPC following transfection significantly decreased by day 10, with less than 5% of cells still expressing the protein (Fig. 1B). For cell tracking over extended time periods which is used in cell therapy experiments, long-term expression of luciferase would be required. Lentiviral transduction is an established method to achieve stable protein expression in cells (Capowski, Schneider et al. 2007). Following lentiviral production, approximately 55% of hNPCLuc2 stably expressed luciferase for up to 15 weeks. It was vital to confirm that long-term protein expression did not alter the capacity of hNPC to proliferate and differentiate. Compared to retroviral techniques, which have been shown to activate proto-oncogenes by the inserted gene, lentiviral methods of stable transduction do not appear to have a negative effect on proliferation (Stein, Ott et al. 2010). Our data support this result, showing that hNPCLuc2 proliferation and differentiation capacities remained the same as that of hNPCWT (Fig. 2D). Overall, lentiviral infection successfully induced stable expression of luciferase in hNPC without changing their proliferation and differentiation potential.
4.3 Imaging hNPC Stably Expressing Luciferase
Similar imaging results were observed in vitro and in vivo following transplantation of stable hNPCLuc2 compared to transiently transfected cells. Successful detection of hNPCLuc2 injected into the striatum alone showed that cortical injections were not necessary for signal detection. To our knowledge, this is the first time that non-tumor cells have been detected in the striatum of WT rats using BLI (Kondo, Goldman et al. 2009). This result is especially meaningful since hNPC proliferation has been shown to considerably slow post-transplantation, suggesting that the cell number most likely did not significantly increase (Ostenfeld, Caldwell et al. 2000). Furthermore, it was found that the minimum cell number that could be imaged in rat striatum was between 9×103 and 9×104 (Fig. 2K and L). This detection limit is a useful starting point for future experimental designs. It must be emphasized that only approximately 55% of the hNPCLuc2 were expressing luciferase, which indicates that the detection cut-off may be lower if a 100% luciferase-expressing population were selected and transplanted (i.e. it may be possible to detect fewer cells using the same transplantation and imaging parameters otherwise). Conversely, all of the cell numbers examined in vivo could also be detected in vitro, with a two-fold higher signal intensity compared to in vivo data (Fig. 2H). This finding is reflective of luminescence signal absorption and scatter that occurs in any tissue type, as well as the previously discussed decrease in signal due to specific obstacles of brain imaging. In this part of the study, we determined lower limits of detection in vitro and in vivo, although they may vary based on experimental settings and should be considered in future studies.
4.4 Visualizing hNPCLuc2 for up to Twelve Weeks in Rat Striatum
We further show that hNPCLuc2 can be tracked for up to twelve weeks in the WT rat striatum, while a loss of signal reflects cell graft rejection. Once bioluminescence signal could not be detected for at least two successive scan time points, the animals with signal loss were sacrificed for histological analysis. In those cases, no live hNPCLuc2 were found, confirming that hNPCLuc2 must be viable for luciferase-catalyzed optical light production and subsequent signal detection. Importantly, this shows that luciferase expression and successive bioluminescence scanning can be used for in vivo cell survival analysis. Analysis of mean signal intensities in rats spanning all long-term experiments revealed that bioluminescence signal varied between animals based on several factors. Animals receiving cortical transplants demonstrated higher signal intensity at the commencement of the study compared to animals receiving striatal transplants. One exception to this rule was that extremely large, dense striatal grafts of hNPCLuc2 produced high levels of bioluminescence signal. At the study completion, three animals had higher signal than the rest, two of which were the animals mentioned above with particularly large, dense striatal grafts and one with a large cortical graft (Fig. S2). The rest of the animals, with lower signal level at end point had either smaller cortical or average sized striatal grafts. This indicates that signal intensity over time may predict graft size, structure and survival, but other methods such as 3D DLIT may have to be used to separate the dual effects of graft size and structure.
4.5 Detecting hNPCLuc2 Migration to Contralateral Side of the Rat Brain
Finally, we demonstrate for the first time that contralateral migration of hNPCLuc2 in WT rat brain can be identified using bioluminescence imaging. QA was intended to initiate migration of hNPCLuc2 in both contralateral and ipsilateral directions. However, our findings show that QA alone did not induce obvious cell migration in either direction. While this may be a result of different transplantation coordinates compared to our previous studies, it could also be due to inability to assess hNPC migrating in the rostral-caudal direction through histological analysis of coronal brain sections (Behrstock, Ebert et al. 2008). Furthermore, radial migration that may have occurred could not be detected using our ROI-based analysis since the cells migrated equally in both rostral and caudal directions. However, while lesions contralateral to cell transplants did not appear to induce contralateral migration, transplantation near the corpus callosum white matter tract seemed to be an important factor in contralateral hNPCLuc2 migration. All of the animals undergoing scans for 12 weeks were analyzed both histologically and using ROI-based analysis to assess whether hNPCLuc2 migrated along the corpus callosum (Fig. 4A and C). The image analysis method described above indicated with statistical significance that in four animals, hNPCLuc2 migrated contralaterally. Histological analysis confirmed hNPCLuc2 migration along the corpus callosum to the contralateral side of the brain. No migration was observed histologically in other subjects. Past studies have shown that bioluminescence imaging is capable of tracking luciferase-expressing hNPC migration in nude mice (Tang, Shah et al. 2003, Waerzeggers, Klein et al. 2008). However, hNPCLuc2 migration in rats has not been established prior to this study. Moreover, most studies attempting to visualize hNPC in BLI employ nude animals ensuring that hair cannot interfere with optical imaging signal. In our report, we use WT animals treated with cyclosporine immunosuppressive therapy whose hair is simply removed prior to in vivo imaging. While further studies must be done to establish a method that can be used to detect ipsilateral migration, our technique appears to successfully detect contralateral migration.
4.6 Novel Aspects of Current Study
Even though a number of preclinical and clinical studies using human neural progenitor/stem cells find promising results in treatment of neurodegenerative diseases, the inability to predict and understand the dynamics of cell homing and migration poses a serious challenge to the field. This study develops a method that can be used to answer some of these vital questions. We find that hNPCLuc2 can be visualized in vivo in the WT rat striatum. Multiple past studies have focused on detecting luciferase-expressing cells in nude mice (Caceres, Zhu et al. 2003, Ragel, Elam et al. 2008, Hafeez, Zhong et al. 2013). In this work, we have shown that this depth-dependent imaging method can be used to track hNPCLuc2 in a larger, WT rodent model, making BLI even more applicable for preclinical studies. This report also encourages the exploration of luciferase-based BLI in larger animals, such as pigs and monkeys. While this technology’s utility in the larger animal CNS must be tested, our results suggest that cells can be detected with BLI in larger animals that previously thought. Therefore, studies using pigs and monkeys may also benefit from BLI to further provide an understanding of cell behavior in vivo. Importantly, this method is valuable as a tool to discriminate between live and dead cells in vivo. Graft rejection is a major concern in therapeutic applications of hNPC (Riley, Federici et al. 2009). Having the capacity to distinguish between live and dead hNPCLuc2 in vivo without histological analysis is an incredibly useful tool as the field moves forward. Finally, detection of cell migration in the contralateral direction is the first step in being able to recognize cell movement within in the host tissue. These studies must be continued in order to develop more flexible and robust methods of migration detection. BLI still lacks resolution when compared with some other imaging modalities, as only approximate cell locations can be determined. Therefore, BLI would benefit considerably from multi-modality imaging, where a higher resolution modality such as MRI is employed concurrently to provide more accurate cell location information. Still, these encouraging results show that bioluminescence of luciferase-expressing cells is helpful in understanding cell location, viability and migration over a long period of time in living animals.
5 Conclusions
In this study, we show for the first time that viable hNPCLuc2 can be imaged in the rat striatum using bioluminescence imaging for up to twelve weeks. We show that this method can be used to ascertain whether the transplanted cells are alive or dead and to detect contralateral cell migration along the corpus callosum. In conclusion, our study consists of several novel discoveries that may serve to further develop the field of in vivo stem cell tracking in a therapeutic setting.
Supplementary Material
Three-dimensional diffuse luminescence tomography (3D DLIT) images of an animal tracked long term at (A) week 1 and (B) week 12.
Comparison of graft location, size and the corresponding signal intensity in all long-term animals. Graft size is depicted as + being the smallest and ++++ being the largest.
Highlights.
We track Luc2 human neural progenitor cells (hNPCLuc2) via bioluminescence imaging.
hNPCLuc2 can be visualized in rat striatum up to twelve weeks.
This method distinguishes dead versus live hNPCLuc2 in vivo in rat striatum.
Region of interest-based image analysis reveals hNPCLuc2 contralateral migration.
Acknowledgments
We gratefully acknowledge Dr. Bernard Schneider (École Polytechnique Fédérale de Lausanne) for packaging the lentiviral construct. We also thank Drs. Soshana Svendsen and Genevieve Gowing (Regenerative Medicine Institute, Cedars-Sinai Medical Center, Los Angeles, CA) for the help in preparing this manuscript. This work was supported by Promega Corp. and grants from University of Wisconsin-Madison Institute for Clinical & Translational Research and Stem Cell and Regenerative Medicine Center (M.S.) and National Institutes of Health (T32GM08349, C.L.).
Abbreviations
- BLI
bioluminescence imaging
- IVIS
In Vivo Imaging System
- hNPC
human neural progenitor cells
- hNPC-Luc2
transient luciferase-expressing human neural progenitor cells
- hNPCLuc2
stable luciferase-expressing human neural progenitor cells
- hNPCWT
wild type human neural progenitor cells
- hNSC
human neural stem cells
- HD
Huntington’s disease
- PD
Parkinson’s disease
- QA
quinolinic acid
- CNS
central nervous system
- DMEM
Dulbecco’s modified Eagle medium
- PSA
penicillin/streptomycin/amphotericin
- EGF
epidermal growth factor
- FGF-2
fibroblast growth factor-2
- LIF
leukemia inhibitory factor
- p-HEMA
polyhydroxyethylmethacrylate
- CMV
cytomegalovirus
- SIN-W-PGK
self-inactivating lentiviral vector with posttranscriptional cis-acting regulatory elements of woodchuck hepatitis virus and mouse phosphoglycerate kinase 1 promoter
- PGK
phosphoglycerate kinase
- PFA
paraformaldehyde
- PBS
phosphate buffered saline
- NDS
normal donkey serum
- BSA
bovine albumin serum
- GFAP
glial fibrillary acidic protein
- hCyto
human cytoplasmic marker
- BrdU
bromodeoxyuridine
- TBST
Tris-buffered saline with 0.5% Tween 20
- TA
tibialis anterior
- BVC
bupivacaine
- IP
intraperitoneal
- ROI
region of interest
- R
right
- L
left
- RLU
relative light units
- ANOVA
analysis of variance
- SEM
standard error of the mean
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aswendt M, Adamczak J, Couillard-Despres S, Hoehn M. Boosting bioluminescence neuroimaging: an optimized protocol for brain studies. PLoS One. 2013;8(2):e55662. doi: 10.1371/journal.pone.0055662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrstock S, Ebert A, McHugh J, Vosberg S, Moore J, Schneider B, Capowski E, Hei D, Kordower J, Aebischer P, Svendsen CN. Human neural progenitors deliver glial cell line-derived neurotrophic factor to parkinsonian rodents and aged primates. Gene Ther. 2006;13(5):379–388. doi: 10.1038/sj.gt.3302679. [DOI] [PubMed] [Google Scholar]
- Behrstock S, Ebert AD, Klein S, Schmitt M, Moore JM, Svendsen CN. Lesion-induced increase in survival and migration of human neural progenitor cells releasing GDNF. Cell Transplant. 2008;17(7):753–762. doi: 10.3727/096368908786516819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury MS, Panagiotakos G, Chan BK, Tomishima M, Zanzonico P, Vider J, Ponomarev V, Studer L, Tabar V. Optical bioluminescence imaging of human ES cell progeny in the rodent CNS. J Neurochem. 2007;102(6):2029–2039. doi: 10.1111/j.1471-4159.2007.04681.x. [DOI] [PubMed] [Google Scholar]
- Caceres G, Zhu XY, Jiao JA, Zankina R, Aller A, Andreotti P. Imaging of luciferase and GFP-transfected human tumours in nude mice. Luminescence. 2003;18(4):218–223. doi: 10.1002/bio.729. [DOI] [PubMed] [Google Scholar]
- Capowski EE, Schneider BL, Ebert AD, Seehus CR, Szulc J, Zufferey R, Aebischer P, Svendsen CN. Lentiviral vector-mediated genetic modification of human neural progenitor cells for ex vivo gene therapy. J Neurosci Methods. 2007;163(2):338–349. doi: 10.1016/j.jneumeth.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Daadi MM, Li Z, Arac A, Grueter BA, Sofilos M, Malenka RC, Wu JC, Steinberg GK. Molecular and magnetic resonance imaging of human embryonic stem cell-derived neural stem cell grafts in ischemic rat brain. Mol Ther. 2009;17(7):1282–1291. doi: 10.1038/mt.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gera A, Steinberg GK, Guzman R. In vivo neural stem cell imaging: current modalities and future directions. Regen Med. 2010;5(1):73–86. doi: 10.2217/rme.09.79. [DOI] [PubMed] [Google Scholar]
- Gowing G, Shelley B, Staggenborg K, Hurley A, Avalos P, Victoroff J, Latter J, Garcia L, Svendsen CN. Glial cell line-derived neurotrophic factor-secreting human neural progenitors show long-term survival, maturation into astrocytes, and no tumor formation following transplantation into the spinal cord of immunocompromised rats. Neuroreport. 2013 doi: 10.1097/WNR.0000000000000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafeez BB, Zhong W, Fischer JW, Mustafa A, Shi X, Meske L, Hong H, Cai W, Havighurst T, Kim K, Verma AK. Plumbagin, a medicinal plant (Plumbago zeylanica)-derived 1,4-naphthoquinone, inhibits growth and metastasis of human prostate cancer PC-3M–luciferase cells in an orthotopic xenograft mouse model. Mol Oncol. 2013;7(3):428–439. doi: 10.1016/j.molonc.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Tsuji K, Kim YR, Mueller FJ, Eom HS, Snyder EY, Lo EH, Weissleder R, Schellingerhout D. Neural stem cell transplant survival in brains of mice: Assessing the effect of immunity and ischemia by using real-time bioluminescent imaging. Radiology. 2006;241(3):822–830. doi: 10.1148/radiol.2413050466. [DOI] [PubMed] [Google Scholar]
- Kondo A, Goldman S, Lulla RR, Mania-Farnell B, Vanin EF, Sredni ST, Rajaram V, Soares MB, Tomita T. Longitudinal assessment of regional directed delivery in a rodent malignant glioma model. J Neurosurg Pediatr. 2009;4(6):592–598. doi: 10.3171/2009.7.PEDS09186. [DOI] [PubMed] [Google Scholar]
- Krakora D, Mulcrone P, Meyer M, Lewis C, Bernau K, Gowing G, Zimprich C, Aebischer P, Svendsen CN, Suzuki M. Synergistic effects of GDNF and VEGF on lifespan and disease progression in a familial ALS rat model. Mol Ther. 2013;21(8):1602–1610. doi: 10.1038/mt.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindvall O, Barker RA, Brustle O, Isacson O, Svendsen CN. Clinical translation of stem cells in neurodegenerative disorders. Cell Stem Cell. 2012;10(2):151–155. doi: 10.1016/j.stem.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz GS, Flebbe-Rehwaldt L, Gaensler KM. Reexpression following readministration of an adenoviral vector in adult mice after initial in utero adenoviral administration. Mol Ther. 2000;2(4):374–380. doi: 10.1006/mthe.2000.0136. [DOI] [PubMed] [Google Scholar]
- McBride JL, Behrstock SP, Chen EY, Jakel RJ, Siegel I, Svendsen CN, Kordower JH. “Human neural stem cell transplants improve motor function in a rat model of Huntington’s disease. J Comp Neurol. 2004;475(2):211–219. doi: 10.1002/cne.20176. [DOI] [PubMed] [Google Scholar]
- Ostenfeld T, Caldwell MA, Prowse KR, Linskens MH, Jauniaux E, Svendsen CN. Human neural precursor cells express low levels of telomerase in vitro and show diminishing cell proliferation with extensive axonal outgrowth following transplantation. Exp Neurol. 2000;164(1):215–226. doi: 10.1006/exnr.2000.7427. [DOI] [PubMed] [Google Scholar]
- Ragel BT, Elam IL, Gillespie DL, Flynn JR, Kelly DA, Mabey D, Feng H, Couldwell WT, Jensen RL. A novel model of intracranial meningioma in mice using luciferase-expressing meningioma cells. Laboratory investigation. J Neurosurg. 2008;108(2):304–310. doi: 10.3171/JNS/2008/108/2/0304. [DOI] [PubMed] [Google Scholar]
- Redmond DE, Bjugstad KB, Teng YD, Ourednik V, Ourednik J, Wakeman DR, Parsons XH, Gonzalez R, Blanchards BC, Kim SU, Gu Z, Lipton SA, Markakis EA, Roth RH, Elsworth JD, Sladek JR, Sidman RL, Snyder EY. Behavioral improvement in a primate Parkinson’s model is associated with multiple homeostatic effects of human neural stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(29):12175–12180. doi: 10.1073/pnas.0704091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J, Federici T, Park J, Suzuki M, Franz CK, Tork C, McHugh J, Teng Q, Svendsen C, Boulis NM. Cervical spinal cord therapeutics delivery: preclinical safety validation of a stabilized microinjection platform. Neurosurgery. 2009;65(4):754–761. doi: 10.1227/01.NEU.0000343524.45387.9E. discussion 761–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, Schmidt M, Kramer A, Schwable J, Glimm H, Koehl U, Preiss C, Ball C, Martin H, Gohring G, Schwarzwaelder K, Hofmann WK, Karakaya K, Tchatchou S, Yang R, Reinecke P, Kuhlcke K, Schlegelberger B, Thrasher AJ, Hoelzer D, Seger R, von Kalle C, Grez M. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16(2):198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Suzuki M, McHugh J, Tork C, Shelley B, Hayes A, Bellantuono I, Aebischer P, Svendsen CN. “Direct muscle delivery of GDNF with human mesenchymal stem cells improves motor neuron survival and function in a rat model of familial ALS. Mol Ther. 2008;16(12):2002–2010. doi: 10.1038/mt.2008.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, McHugh J, Tork C, Shelley B, Klein SM, Aebischer P, Svendsen CN. GDNF secreting human neural progenitor cells protect dying motor neurons, but not their projection to muscle, in a rat model of familial ALS. PLoS One. 2007;2(8):e689. doi: 10.1371/journal.pone.0000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Wright LS, Marwah P, Lardy HA, Svendsen CN. Mitotic and neurogenic effects of dehydroepiandrosterone (DHEA) on human neural stem cell cultures derived from the fetal cortex. Proc Natl Acad Sci U S A. 2004;101(9):3202–3207. doi: 10.1073/pnas.0307325101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen CN, Caldwell MA, Shen J, terBorg MG, Rosser AE, Tyers P, Karmiol S, Dunnett SB. Long-term survival of human central nervous system progenitor cells transplanted into a rat model of Parkinson’s disease. Experimental Neurology. 1997;148(1):135–146. doi: 10.1006/exnr.1997.6634. [DOI] [PubMed] [Google Scholar]
- Svendsen CN, ter Borg MG, Armstrong RJ, Rosser AE, Chandran S, Ostenfeld T, Caldwell MA. A new method for the rapid and long term growth of human neural precursor cells. J Neurosci Methods. 1998;85(2):141–152. doi: 10.1016/s0165-0270(98)00126-5. [DOI] [PubMed] [Google Scholar]
- Tang Y, Shah K, Messerli SM, Snyder E, Breakefield X, Weissleder R. In vivo tracking of neural progenitor cell migration to glioblastomas. Hum Gene Ther. 2003;14(13):1247–1254. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- Waerzeggers Y, Klein M, Miletic H, Himmelreich U, Li H, Monfared P, Herrlinger U, Hoehn M, Coenen HH, Weller M, Winkeler A, Jacobs AH. Multimodal imaging of neural progenitor cell fate in rodents. Mol Imaging. 2008;7(2):77–91. [PubMed] [Google Scholar]
- Wright LS, Li J, Caldwell MA, Wallace K, Johnson JA, Svendsen CN. Gene expression in human neural stem cells: effects of leukemia inhibitory factor. J Neurochem. 2003;86(1):179–195. doi: 10.1046/j.1471-4159.2003.01826.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three-dimensional diffuse luminescence tomography (3D DLIT) images of an animal tracked long term at (A) week 1 and (B) week 12.
Comparison of graft location, size and the corresponding signal intensity in all long-term animals. Graft size is depicted as + being the smallest and ++++ being the largest.