Summary
Single Nucleotide Polymorphisms in FTO intron 1 have been associated with obesity risk, leading to the hypothesis that FTO is the obesity‐related gene. However, other studies have shown that the FTO gene is part of the regulatory domain of the neighboring IRX3 gene and that enhancers in FTO intron 1 regulate IRX3. While Irx3 activity was shown to be necessary in the hypothalamus for the metabolic function of Irx3 in mouse, no enhancers with hypothalamic activity have been demonstrated in the risk‐associated region within FTO. In order to identify potential enhancers at the human FTO locus in vivo, we tested regulatory activity in FTO intron 1 using BAC transgenesis in zebrafish. A minimal gata2 promoter‐GFP cassette was inserted 1.3 kb upstream of the obesity associated SNP rs9939609 in a human FTO BAC plasmid. In addition to the previously identified expression domains in notochord and kidney, human FTO BAC:GFP transgenic zebrafish larvae expressed GFP in the ventral posterior tuberculum, the posterior hypothalamus and the anterior brainstem, which are also expression domains of zebrafish irx3a. In contrast, an in‐frame insertion of a GFP cassette at the FTO start codon resulted in weak ubiquitous GFP expression indicating that the promoter of FTO does likely not react to enhancers located in the obesity risk‐associated region. genesis 53:640–651, 2015. © 2015 The Authors. genesis Published by Wiley Periodicals, Inc.
Keywords: mammal, organism, genetics, genomics, GWAS, tissue
INTRODUCTION
Genome‐wide association studies have identified obesity‐associated single nucleotide polymorphisms (SNPs) in a 47kb block of linkage disequilibrium (LD block) at the end of FTO intron 1 (Dina et al., 2007; Frayling et al., 2007). As a result, FTO was thought to be the causal gene whose function was associated with weight gain (Fawcett and Barroso, 2010). Many studies since have attempted to establish a plausible biological mechanism underlying the function of FTO in obesity, but no pathway has been put forth (Tung et al., 2014; van Gestel et al., 2014). It was suggested early on that the association of the LD block to FTO was misleading, and that categorically assigning risk SNPs to the closest open reading frame [termed the “nearest neighboring gene hypothesis” (Flint, 2013)], may generate false leads in regions of long‐range cis‐regulation (Ragvin et al., 2010). Regions encompassing developmental regulatory genes with multiple cis‐regulatory inputs are termed genomic regulatory blocks (GRBs; Engstrom et al., 2007; Kikuta et al., 2007a, 2007b; Navratilova and Becker, 2009) and frequently span several genes, of which typically only one, identifiable by its transcriptional features, is the target of the regulatory elements in the region (Akalin et al., 2009). Computational analysis showed that FTO is part of a GRB targeting IRX3, encoding a developmental transcription factor, in all vertebrate genomes (Ragvin et al., 2010).
Testing of several conserved sequences from the risk LD block in transgenic zebrafish revealed that these drove expression patterns characteristic of IRX3 (Ragvin et al., 2010). In a comprehensive IRX3/Irx3 genomic and functional analysis, Smemo et al. (2014) have provided compelling proof that the IRX3 promoter interacts directly with sequences in the LD block. In addition, disrupted function of Irx3 in the mouse hypothalamus led to increased energy expenditure accompanied by activation of brown adipose tissue in the genetically modified mice. While Irx3 expression was detected in the arcuate nucleus of the hypothalamus, three enhancer sequences isolated from the LD block drove expression in lung and several brain regions, but not in hypothalamus (Smemo et al., 2014).
We asked whether FTO intron 1 relates to developmental IRX3 gene regulatory control in the hypothalamus, and aimed to reveal gene regulatory information in this genomic region (Fig. 1). We used an enhancer detection approach in a human bacterial artificial chromosome (BAC) followed by transgenesis in zebrafish. SNP rs9939609 is one of the largest effect obesity‐associated variants in the human genome and has been related to 3 kg weight gain on average in homozygous carriers (Frayling et al., 2007). SNP rs9939609 is located within the critical LD block and is about 84 Kb downstream of the FTO promoter toward the end of the 105 kb intron 1. A BAC encompassing 183 kb of human genomic sequence including FTO upstream sequence and intron 1 in its entirety was modified by galK mediated homologous recombination in bacterial cells to insert a gata2‐promoter‐GFP cassette 1.3 kb upstream of SNP rs9939609. Integration of the modified BAC into the zebrafish genome through microinjection of the DNA into one‐cell stage zebrafish embryos facilitated the generation of transgenic lines that revealed the available regulatory information through the expression of GFP. The galK recombination technique had been optimized previously for its use in zebrafish transgenesis by the insertion of inverted Tol2 sequences into the BAC that facilitate genomic integration (Kawakami et al., 2000; Suster et al., 2011, 2009). Transgenic embryos with the modified BAC inserted into their genomes expressed GFP specifically in the posterior hypothalamus and in the anterior brainstem in a pattern similar to zebrafish irx3a. To compare this expression pattern to developmental endogenous FTO expression, transgenic zebrafish carrying a BAC with a GFP cassette in frame with FTO exon 1 were created. These transgenic embryos had weak ubiquitous expression. We conclude that enhancers in the vicinity of rs9939609 in FTO intron 1 regulate IRX3 expression in the developing hypothalamus.
Figure 1.
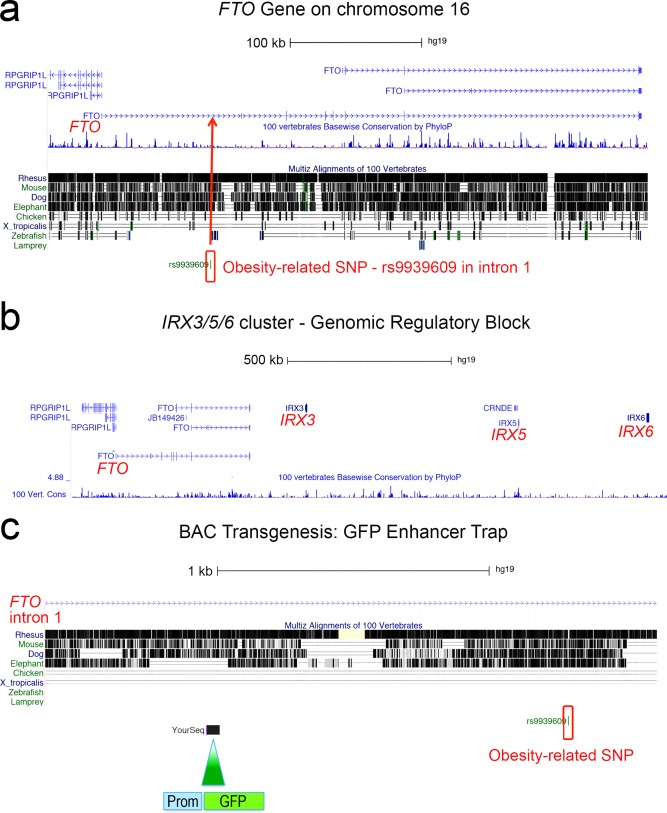
Modified UCSC Genome Browser windows. (a) Obesity‐associated SNP rs9939609 is localized in FTO intron 1 (Frayling et al., 2007). (b) FTO is in conserved synteny with IRX cluster containing IRX3, IRX5, and IRX6. (c) A human BAC containing FTO intron was used to insert a promoter‐GFP cassette upstream of rs9939609.
RESULTS
Obesity Associated SNP rs9939609‐Characterization and Modification of the BAC
The first intron of FTO spans 105910 Kb. Based on the SNP clusters associated with obesity, the approximate location of the 47 kb LD block is at chr16:53,799,315‐53,846,130 (hg38 assembly), which is ∼60 kb downstream of the FTO start codon. Obesity‐associated SNP rs9939609 (A/T) is about 86 kb downstream of the FTO ATG. BAC clone AC007909 contains 183149 bp of human genomic sequence in reverse orientation. In this BAC, the FTO ATG is at position 153,179, so the BAC contains ∼30 kb upstream of FTO and 150 kb downstream sequence in FTO including exon 4. The BAC was confirmed by Spe1 restriction analysis and gel bands were compared to a digest performed in silico. GFP reporter gene cassettes were inserted into this BAC by homologous recombination (Warming et al., 2005; see Methods). Constructs and representative transgenic embryos are shown in Figure 2.
Figure 2.
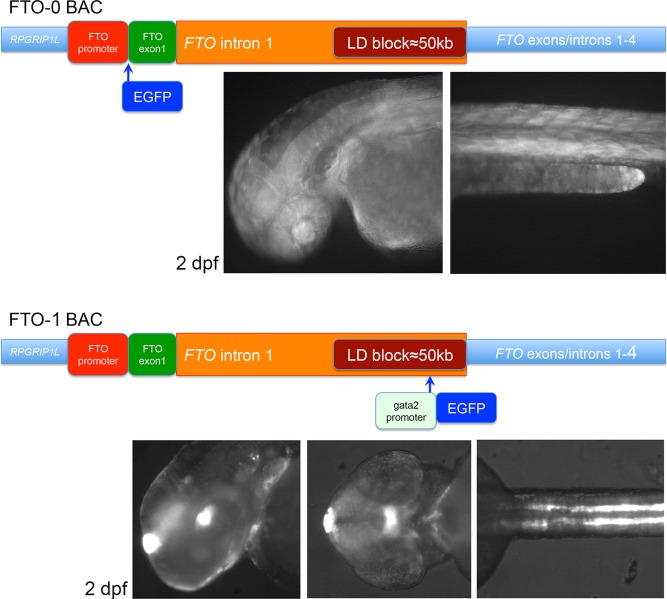
Schematics of the homologous recombinations into the FTO locus. FTO‐0: EGFP was inserted in frame with FTO ATG. The transgenic embryos and larvae had weak ubiquitous expression. FTO‐1 uses enhancer detection as shown in Figure 1c. The transgenic embryos and larvae had expression in the epiphysis, in the posterior tuberculum and in the hypothalamus, in the inner ear, notochord, and in the kidneys.
FTO Promoter Does Not React to Enhancers Located in FTO First Intron
A GFP sequence minus its ATG was inserted in frame with the ATG of FTO into the BAC (FTO‐0, Fig. 2). Transgenic larvae with this BAC inserted in their genomes had weak ubiquitous expression that appeared strongest in the eye and in muscle fibers. This is in line with in situ hybridization analyses showing that mouse Fto has low‐level ubiquitous expression (Gerken et al., 2007; Smemo et al., 2014). The BAC transgene also confirmed an earlier result with a very similar BAC construct, showing that replacing the first exon of Fto with a lacZ cassette recapitulated the endogenous Fto expression pattern (Smemo et al., 2014). In the mouse embryo, Fto displays near ubiquitous expression, while Irx3 is specifically expressed in several brain regions (Diez‐Roux et al., 2011).
The Expression Domains in FTO‐1 Transgenic Lines Resemble Expression Features of Vertebrate Irx3
A gata2 promoter‐GFP cassette was inserted 1.3 kb upstream of SNP rs9939609 in the BAC (FTO‐1, Fig. 2) and recombinant BAC DNA was integrated in the zebrafish genome. Transgenic lines were established and the distribution of the GFP (Figs. 2 and 3) was compared with published expression data and to in situ hybridization signals in sections of 3 dpf whole mount in situ hybridized zebrafish larvae (Fig. 4).
Figure 3.
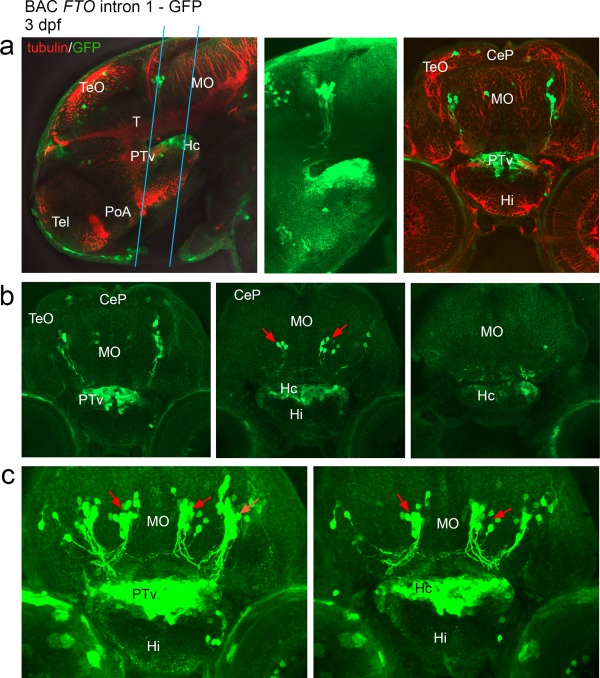
Confocal scan of transgenic larvae at 3 dpf having a promoter‐GFP cassette inserted upstream of SNP rs9939609 in FTO intron 1 (a–c). Expression is detected in the ventral part of posterior tuberculum, in the caudal hypothalamus and in the anterior brainstem. (a) Representative optical sections through 3 dpf larvae that were stained for GFP and tubulin, in lateral and transverse views. The blue lines indicate the range of sections shown in b,c. (b) A row of optical sections through the GFP expression domain as indicated in “a.” (c) Z‐stack images. The first z‐stack image includes hypothalamus and PTv, “b” is further posterior and includes the caudal hypothalamus only. Red arrows: Neurons in the hindbrain are strongly labeled and project ventrally as well as contralaterally. Abbreviations: (CeP) Cerebellar plate, (Hc) caudal hypothalamus, (Hi) intermediate hypothalamus, (MO) Medulla oblongata, (PoA) preoptic area, (PTv) ventral part of posterior tuberculum, (T) midbrain tegmentum, (Tel) Telencephalon, (TeO) Tectum opticum. From Mueller and Wullimann, Atlas of early Zebrafish brain development, 2005, Elsevier.
Figure 4.
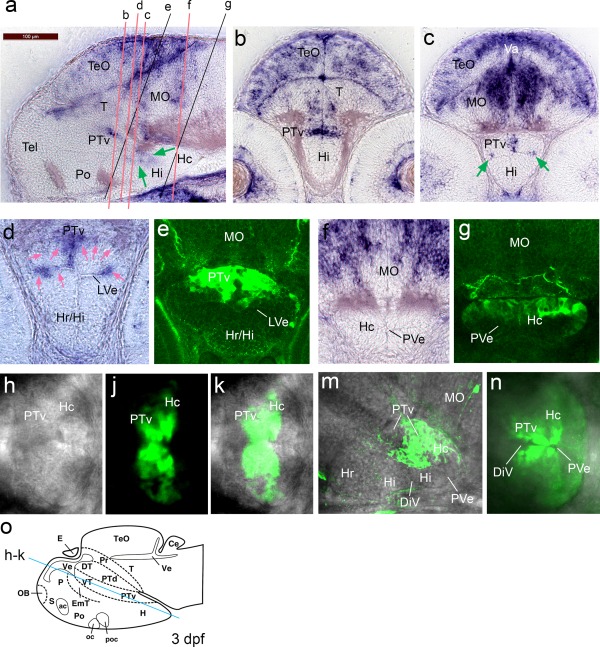
Irx3a in situ hybridization signals in 3 dpf embryos compared to GFP distribution in anti‐GFP stained FTO‐1 specimen that were analyzed by confocal microscopy. Emphasis is on the hypothalamus. (a–c) irx3a in situ hybridization in 3 dpf embryos, lateral (a) and transverse (b, c) sections. Expression of irx3a in the brain is within the optic tectum, tegmentum, cerebellum, hindbrain, posterior tuberculum, and hypothalamus. The signal in the hypothalamus is localized around the ventricular zone. The green arrows in “a” point to hybridization signal around the ventricular zone of the intermediate and caudal hypothalamus. Arrows in “c” point to irx3a activity in the lateral ventricular recess of the hypothalamus (LVe). (d and e) Sections with focus on the ventral posterior tuberculum and the hypothalamus in high magnification. The hybridization signal is strongest in the lateral ventricular recess of the hypothalamus and the medial region of the PTv, only very faint staining was detected dorsolaterally of the PTv (pink arrows). In the transgenic line the GFP appears more widely distributed. (f and g) The caudal hypothalamus appears free of irx3a transcripts, but GFP can be detected in this region. (h to k) Optical section of a scan performed horizontally through a live embryo (dorsal view) with focus on the posterior tuberculum and hypothalamus. The blue line in “o” indicates the approximate level of the section. (h) Bright field image. (j) The GFP is stronger around the ventricular zone. (k) Maximal intensity projection of the composite image shows that the posterior part of the caudal hypothalamus is free of GFP. (m) Lateral confocal scan. Maximal intensity projection of the medial expression domain. The GFP appears dorsal of the LVe. (n) Horizontally performed scan with ventral view. Maximal intensity projection of the GFP expression domain. The GFP is strongest around the ventricular zone. Abbreviations: DiV, diencephalic ventricle, LVe lateral recess ventricle of hypothalamus, (Hc) caudal hypothalamus, (Hi) intermediate hypothalamus, (Hr) rostral hypothalamus, (MO) medulla oblongata, (Po) preoptic region, (PTv) ventral part of posterior tuberculum, (PVe) posterior ventricular recess of H, (T) midbrain tegmentum, (Tel) telencephalon, (TeO) tectum opticum, (Va) valvula cerebelli. From Mueller and Wullimann, Atlas of early Zebrafish brain development, 2005, Elsevier.
Most prominently, 48 hpf FTO‐1 transgenic embryos had GFP expression in an area of the ventral posterior tuberculum and dorso‐posterior hypothalamus, in the epiphysis and in the pronephric ducts indicating that enhancers surrounding the insertion have regulatory activity in these structures (Fig. 2). GFP expression was also observed in the notochord, which is in line with previous analyses that identified an enhancer with activity in this structure 20 kb downstream of rs9939609, whereas a kidney enhancer could be located to 13 kb upstream of the SNP (Ragvin et al., 2010). The BAC transgenic lines did not have reporter expression in the pancreas, which is contrary to our previous data. With the exception of the GFP expression in the epiphysis, which cannot be explained at this point, the other expression domains relate to published Irx3 features. Irx3 expression and function in the developing kidneys is well established (Alarcon et al., 2008; Houweling et al., 2001). Expression of irx3a in the posterior tuberculum/dorso‐posterior hypothalamus in 48 hpf zebrafish embryos is documented in zfin.org through data created in a large scale in situ hybridization screen (Thisse et al., 2004). As shown in the publicly available database Eurexpress (www.eurexpress.org) in the mouse fetal brain (E14.5) Irx3 mRNA is distributed in a very similar area, here determined as prethalamus and hypothalamus (dorso‐posterior part).
The GFP expression in the developing brain was analyzed in detail (Fig. 3) and compared to irx3a transcript distribution (Fig. 4). Irx3a mRNA was detected in optic tectum, tegmentum, cerebellum, ventral posterior tuberculum, the intermediate and caudal hypothalamus, and the anterior brainstem (Fig. 4a–d). The GFP was expressed within the irx3a expression domain in the ventral posterior tuberculum, in the caudal hypothalamus and in two bilateral clusters of neurons in the anterior brainstem (medulla oblongata). One of these clusters was more anterior and appeared laterally while the second cluster was further medial, and both neuronal clusters had axons that projected ventrally. Scattered expression in the tectum appeared at 3 dpf and became stronger beyond that stage (data not shown).
The posterior tuberculum as well as the caudal hypothalamus appeared more widely GFP‐labeled than was indicated by irx3a mRNA distribution. While the mRNA signal in the ventral posterior tuberculum appeared only medial (Fig. 4c,d), in the transgenes the GFP extended further laterally (Fig. 4e). With the GFP expression extending posteriorly from the ventral posterior tuberculum into the caudal hypothalamus, the same phenomenon was observed – while transcripts were detected medially and laterally in the caudal hypothalamus, the GFP was expressed more widely in this region and also further posterior (compare Fig. 4f,g), perhaps reflecting endurance of GFP protein in cells that have since migrated. Another possibility is that the mRNA detection may not have been sufficiently sensitive to detect areas of weaker transcription. Alternatively, it has to be considered that it is human DNA that regulated the activity of the reporter and that human regulatory sequence might be differently used in the zebrafish. However, we think that the BAC transgenic GFP expression indicates genuine Irx3 enhancer activity, because the mRNA and GFP signals are in comparable areas and the in situ hybridization signals detected in the mouse (Eurexpress) appear more broad than the signals we have detected in the zebrafish. Since the Irx3 locus is highly conserved in vertebrate genomes, it is not surprising that enhancers retain functions during evolution, but it cannot be excluded that with evolution of brain morphology exact anatomical functions would vary.
Adult FTO‐1 transgenic fish were analyzed for GFP expression in the brain. Immunostained sections revealed GFP in neurons in the anterior brain stem (Fig. 5). These neurons projected ventrally, and appeared similar to the ones observed in developmental stages.
Figure 5.
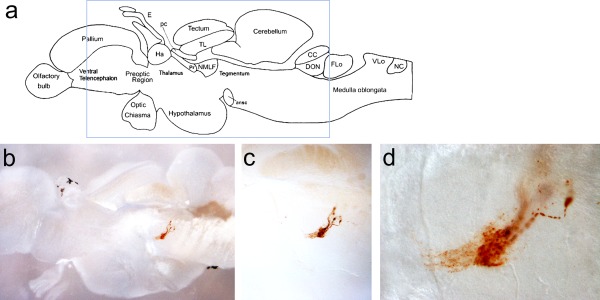
DAB anti‐GFP immunostain of FTO‐1 BAC adult brain sagittal sections. (a) Schematics of the adult zebrafish telencephalon (From Wullimann et al., Neuroanatomy of the Zebrafish Brain, 2004, Birkhaeuser Verlag). The blue rectangle indicates the area shown in the section in “b.” (b–d) Light microscopic images of FTO‐1 BAC transgenic brain with magnifications focusing on DAB‐stained neurons in the brainstem projecting ventrally.
In summary, we show that the GFP expression pattern in the developing brain regulated by a human BAC containing the FTO risk LD block resembles that of zebrafish irx3a, but not that of fto. We can further state that FTO intronic sequence surrounding the SNP rs9939609 contains enhancers that regulate developmental gene activity in the posterior hypothalamus.
DISCUSSION
The insertion of an enhancer detection cassette close to rs9939609 in a human BAC and the generation of transgenic zebrafish lines carrying this modified construct in the genome has provided evidence that enhancers driving the activity of IRX3 in the developing hypothalamus are in the neighborhood of the obesity associated SNP. This was of special interest since increased IRX3 expression was related to the rs9939609 variant and Irx3 hypothalamic function was identified to regulate fat composition through control of energy homeostasis (Smemo et al., 2014). Even though this was an extensive study, hypothalamic enhancers could not be identified in FTO intron 1. Also another previous study that tested individual enhancers in this region has come up with expression domains that are compatible with Irx3/irx3a expression domains, but no enhancer was shown to be active in the hypothalamus (Ragvin et al., 2010). Several individual sequences might need to interact to generate expression in the hypothalamus. Since the critical SNP is located in the middle of the LD block at the end of FTO intron 1, but the BAC contains also introns 2 and 3, our approach raises the question if sequences further downstream could harbour the critical enhancer(s). This cannot be excluded, however the hypothalamic reporter expression is similar in strength to the GFP in the pronephric ducts (Fig. 2), which is known to be regulated by an enhancer located 20 kb upstream of the SNP (Ragvin et al., 2010). This is no proof, but from our enhancer detection screen and another transposon‐based enhancer detection approach we find that enhancer activity decreases with increasing distance from the gene (Kikuta et al., 2007b; Kokubu et al., 2009). Other support for this notion comes from p300 and H3K27ac ChIP sequencing data produced in embryonic and fetal mouse forebrain (Visel et al., 2013). These data do not reveal active enhancers in intron 2 and 3, but in intron 1. Ultimate proof could come from the testing of further single sequences or a BAC sequence deletion analysis.
The reporter expression in the epiphysis cannot be related to Irx3 activity, with our own and published data. We have used a human BAC in zebrafish, which are distantly related organisms and this could hamper analysis, however the fact that irx3a in situ hybridization and BAC transgene expression are largely overlapping would argue against this possibility. Nevertheless, it is possible that the BAC contains human sequences that have evolved since the split of a common ancestor, and thus could lead to expression patterns that are not detected with zebrafish irx3a.
Even though our emphasis here was on hypothalamic enhancer function, IRX3 enhancer activity in the anterior brain stem could also be of interest as the bilateral clusters of neurons projecting ventrally might be related to metabolic functions as well. The FTO‐1 BAC transgenic line created here thus might present a tool to further explore IRX3/irx3a neuronal/metabolic functions.
In conclusion, we have shown here that human BAC plasmids can be used to reveal regulatory information in transgenic zebrafish, and that the human FTO gene contains IRX3 enhancers that are active in the developing hypothalamus. With Irx3 function in the hypothalamus being related to obesity, it could be speculated that developmental hypothalamic enhancer function might be compromised in humans carrying the rs9939609 variant. However, the ontogeny related to later physiological functions will be subject of future studies.
METHODS
Work with Zebrafish
Zebrafish were maintained and used for experiments in accordance with terms and conditions of University of Sydney Animal Ethics committee. The fish were kept at a 14 h light/8 h dark cycle at 28.5°C. The larvae were staged according to established zebrafish protocols (Kimmel et al., 1995).
BAC Analysis
A BAC (pBACe3.6 vector) clone with ID AC007909 contains human genomic DNA including FTO upstream sequences and exons 1 to 4. The sequence was extracted from UCSC genome browser to be analyzed with APE software. This clone was ordered through BACPAC Resources Center (https://bacpac.chori.org/, clone ID: RP11‐515G11) and was received as DH10 bacterial agar stab. For its use in galK selection recombination, the BAC DNA had to be transformed into SW102 cells.
Cells containing the BAC were streaked on an agar plate to receive a single colony and a 5 ml liquid culture containing 12.5 µg chloramphenicol was grown to yield sufficient cells for DNA preparation. The DNA was purified using a BAC Mini Prep Protocol and Qiagen Plasmid preparation Kit solutions (P1, P2 and P3; 250 µl each). BAC DNA solutions were mixed by inversion only and the incubation times were always 5 min. After neutralization, the supernatant was cleared by 10 min full speed centrifugation, followed by another centrifugation step after the supernatant had been transferred to a new tube. DNA was precipitated with 750 µl Isopropanol and 10 min incubation on ice. The pellet was washed twice with 70% ethanol and dissolved in 50 µl TE. To confirm the correct clone 40 µl (∼1 µg) was used for Spe1 restriction analysis, while 1 µl was used for PCR. 1 µl was used for transformation into electrocompetent SW102 cells in which homologous recombination reactions are performed.
BAC Recombineering
Overview
For directed homologous recombination of reporter gene cassettes into the human FTO gene contained in a BAC, we used a galactokinase (galK) positive and negative selection technique (Warming et al., 2005). SW102 cells lack the galK gene, essential for utilizing galactose. Since galK function can be added in trans, recombination of the gene into a BAC restores the ability of SW102 cells to grow on a galactose minimal medium and is used to identify successfully recombineered clones. A second recombineering step replacing the galK cassette with a double stranded DNA of interest uses negative selection. Successfully recombineered clones were identified by growth on a 2‐deoxy‐galactose (DOG) minimal medium. Cells that still contain BACs containing the galK gene metabolize the DOG into a toxic product (2‐deoxy‐galactose‐1‐phosphate) and are unable to grow. This protocol was adapted for zebrafish transgenesis by adding a step that inserts inverted Tol2 sequences into the BAC plasmid backbone to facilitate efficient genomic integration after the DNA is injected into one cell stage embryos along with Tol2 transposase (Suster et al., 2011, 2009).
Clones and plasmids
The galK plasmids and SW102 cells are intellectual property of the National Cancer Institute (Frederick) and were ordered through the Biological Resources Branch of the NCI. SW102 cells were received in LB medium containing tetracycline. A few microliters of these were streaked on an agar plate containing 12.5 µl/ml tetracycline and the plate was incubated at 32°C to grow a single colony to proceed with. The galK plasmid came as DNA solution to be transformed in DH1alpha cells for amplification.
The gata2 promoter and GFP cassette were amplified using an enhancer‐testing plasmid as template [described in (Ishibashi et al., 2013)].
The iTol2 cassettes contain either ampicillin or kanamycin resistance genes to select positive clones after homologous recombination (iTol2AMP, iTol2KAN) and are inserted into a pCR8GW‐TOPO vector. For DNA amplification, transformed cells containing the plasmid were grown in LB with 75 µg/ml Spectinomycin.
SW102 electrocompetent cells
SW102 cells were grown at 32°C in LB medium containing 20 µg/ml tetracycline. A single colony, grown for 16–18 h on an agar plate, was used to innoculate a 5 ml overnight culture. The next day 2.5 ml of this culture were used to inoculate a 500 ml culture that was grown to optic density (OD600) of 0.6. The culture was split and chilled on ice for 20 min. Centrifuging for pelleting and resuspension of the pellet in 200 ml ice‐cold sterile water was performed twice. Finally the pellet was resuspended in 5 ml ice‐cold sterile 10% glycerol. Cells were pooled into 10 ml aliquots, centrifuged and the pellet resuspended in final 1 ml of cold sterile 10% glycerol. OD was measured for a 1:100 dilution. Aliquots of 50 µl were stored at −80°C.
Electroporation procedure
An Eppendorf electroporator 2510 was used. Five µl of the cassette were added to 25 µl electrocompetent cells and the mix was put into a cuvette. Electroporations were performed with 1.8 kV voltage; 1 ml LB containing 20 mM l‐glucose was added to the cuvette and the content was transferred to a fresh culture tube to be cultured at 37°C for 1 h.
BAC modification steps
BAC DNA was electroporated into electrocompetent SW102 cells and cells were streaked on LB agar plates containing chloramphenicol for selection.
SW102 cells containing the BAC were made electrocompetent and 25 µl were used for electroporation of the galK cassette. The electroporated cells were recovered in 1 ml LB medium and then washed and diluted in M9 medium. The dilutions were plated on M63 minimal media plates containing galactose. After 3 days incubation a few colonies were streaked onto MacConcey‐galactose‐chloramphenicol indicator plates. A bright red colony was expected positive and was used to inoculate 5 ml LB/chloramphenicol medium. SW102 cells were always grown at 32°C.
SW102 cells containing the galK cassette in the correct position were made electrocompetent and 25 µl were used for electroporation of the GFP/gata2‐GFP cassette. Recovery and washing of the cells was performed as described in ‘2’. The dilutions were streaked on M63 minimal media plates containing 2‐deoxy‐galactose and chloramphenicol. After 3 days incubation 10 colonies were picked to check if the cassette was successfully integrated (Spe1 enzymatic digest and PCR, the PCR product was confirmed by sequencing).
SW102 cells containing the GFP/gata2‐GFP cassette in the correct position were made electrocompetent and 25 µl were used for electroporation of the iTol2 cassette. The cells were grown on LB medium with Amp selection.
Cassette amplification and primer design with overhangs for homologous recombination into the FTO locus (overhangs in small letters)
The cassettes for homologous recombination were PCR amplified from plasmids with primers that had 50 bp overhangs of a sequence homologous to the sites of integration. All PCR reactions were Dpn1 digested to remove plasmid DNA and the PCR products were subsequently gel‐purified and sequenced for confirmation; 10–30 ng (∼2.5 µl) were used for electroporation.
Primers for amplification of galK and GFP (gata2‐GFP, respectively) cassettes had the same 50 bp 5′‐overhangs.
galK cassette in FTO‐0 (Insertion of GFP in frame)
Forward: 5′‐cgagggatctacgcagcttgcggtggcgaaggc ggctttagtggcagcatg‐CCTGTTGACAATTAATCATCGGCA
Reverse: 5′‐ccgacataccttagcttcgcgctctcgttcctcggcagtc ggggtgcgctTCAGCACTGTCCTGCTCCTT‐3′
galK cassette in FTO‐1
Forward: 5′‐tatgtgtgac cagcctcaatagattttattcattagtaacttggttgattCCTGTTGACAAT TAATCATCGGCA
Reverse: 5′‐actttcattcagtaattattttatgtctacttcatgtcaggca ttgttccTCAGCACTGTCCTGCTCCTT
GFP cassette (without ATG) in FTO‐0
Forward: 5′‐cgagggatctacgcagcttgcggtggcgaaggcggctttagtggcagcat g‐GTGAGCAAGGGCGAGGAGCTGT‐3′
Reverse: 5′‐ccgacataccttagcttcgcgctctcgttcctcggcagtc ggggtgcgct‐GACTTAATTAACCATGGCGTACGC‐3′
gata2‐GFP cassette in FTO‐1
Forward: 5′‐tatgtgt gaccagcctcaatagattttattcattagtaacttggttgattAAGCTTGCG GCGATATTCAT‐3′
Reverse: 5′‐actttcattcagtaattattttatgtctacttcatgtcaggcat tgttccGACTTAATTAACCATGGCGTACGC‐3′
Homologous recombination of the iTol2 cassette into pBACe3.6 vector
After successful integration of the GFP/gata2‐GFP/gata2 promoter had been confirmed by PCR and sequencing, inverted Tol2 sequences with an Ampicillin resistance cassette were recombined into the BAC backbone. The primers had 50 bp 5′‐overhangs homologous to the sequences of the pBACe3.6 vector.
Forward: 5′‐gcgtaagcggggcacatttcattacctctttctccgcacc cgacatagatCCCTGCTCGAGCCGGGCCCAAGTG
Reverse: 5′‐cgcggggcatgactattggcgcgccggatcgatcctta attaagtctactaATTATGATCCTCTAGATCAGATCT
Zebrafish Transgenesis
Two microliter of purified BAC DNA (250 ng/µl) in a mix with 2 µl of Transposase mRNA (125 ng/µl) were injected into fertilized zebrafish eggs. The embryos were screened the next day with a fluorescent stereomicroscope. One hundred and fifty positive embryos were sorted and raised to adulthood. For positive founder screening the fish were outcrossed with wild type fish. Two to three positive founder fish were identified for each modified BAC and the F1 offspring was analyzed at 1‐3 dpf. GFP‐positive embryos were mounted in 2% Methylcellulose in water and imaged with an inverted fluorescent microscope.
Confocal Microscopy
A Zeiss LSM 710 confocal microscope using Zen software was used for high resolution imaging. The embryos were mounted in 1–1.5% low melting point agarose (Sigma A9414) in a glass‐bottom petri dish. Confocal scanning files were processed with ImageJ software.
In Situ Hybridization Analysis
The plasmid for creating the irx3a in situ hybridization probe (kindly provided by Dr. José Luis Gómez‐Skarmeta) was linearized with BamHI and transcribed with T7 polymerase. Wild type embryos were collected at 3 and 4 dpf and fixed for 3 h in 4% PFA at room temperature. Embryos were then rinsed in PBT (PBS including 0.1% Tween 20; 3 × 5 min washes) and dehydrated through a series of methanol washes (25% methanol/PBT, 50% methanol/PBT, 75% methanol/PBT, 3 × 100% methanol) and stored at −20°C. For hybridization, the embryos were rehydrated through reverse methanol washes and rinsed 3× in PBT, permeabilized with 10 mg/ml Proteinase K for 25 minutes and then post fixed in 4%PFA for 20 min. Embryos were then incubated in hybridization solution for 2 h at 65°C before adding the irx3a probe at ∼1 ng/µl for hybridizing overnight. The hybridized probe was detected using anti DIG‐AP Fab fragments (1:5000; Roche) at 4°C overnight and colorimetric detection was performed the following day with a staining solution containing NBT and BCIP. Once the color had reached the desired intensity, the embryos were washed and fixed in 4%PFA.
Immunohistochemistry
Anti GFP and anti tubulin immunohistochemical stainings were performed as described previously (Punnamoottil et al., 2008; Wilson et al., 1990). An incross of the BAC‐FTO‐1‐GFP line was set up and the embryos were collected and screened for GFP expression at 3 dpf. The embryos were then fixed in 4%PFA and dehydrated through a series of methanol washes. If not stored at −20°C, the embryos were immediately rehydrated by reverse washes. Following permeabilization in 2 mg/ml collagenase for 20 min and refixation, the embryos were incubated overnight at 4°C in blocking solution containing Rabbit polyclonal anti GFP antibody (1:1000; AMSBio TP401) and mouse monoclonal anti acetylated tubulin antibody (1:500; Sigma T7451). Primary antibodies were detected through incubation with secondary antibodies (Life Technologies: Alexa Fluor® 488 goat anti‐rabbit, A‐11034; Alexa Fluor® 594 Chicken Anti‐Mouse, A‐21201) overnight at 4°C.
Sectioning
Immunostained and in situ hybridized embryos were sectioned using a vibratome set for 25 or 30 µm sections. The embedding medium was made of 0.5% gelatine/30% albumin solution (frozen aliquots) and a 10% volume of glutaraldehyde to be mixed in a silicon well. Embryos were immediately positioned (either for saggital or transverse sectioning), because the mixture set quickly. The embryos were then mounted on the vibratome platform and sections were collected and mounted on slides.
Adult Brain Analysis
The fish were killed on ice and brains were dissected from the skull to be subsequently fixed in 4% paraformaldehyde. The brains were then washed in PBT (PBS/0.8% Triton‐X) and dehydrated and rehydrated in a series of Methanol/PBT before embedding in gelatin/albumin mixture as described above. Sections of 75 µm were cut with a vibratome and collected and processed for immunostaining in serial wells with nets. The immunostaining followed the regular protocol with collagenase treatment applied for 25 min and 10% goat serum/1%DMSO in PBT used as blocking solution. Anti‐GFP antibody detection used the Vectastain ABC System (Punnamoottil et al., 2008). For the staining reaction DAB aliquots of 25 mg/500 µl were diluted in 30 ml PBS and 1 µl 0.3% H2O2 per ml was added for activation.
ACKNOWLEDGMENTS
The authors thank Dr. Nai Yang in the Breast Cancer Laboratory at the Walter and Eliza Hall Institute for sending galK plasmids and SW102 cells and José‐Luis Gomez Skarmeta for the irx3a probe. The authors declare that there are no competing financial interests in relation to the described work.
Conflict of Interest: The authors declare that there are no competing financial interests in relation to the described work.
LITERATURE CITED
- Akalin A, Fredman D, Arner E, Dong X, Bryne JC, Suzuki H, Daub CO, Hayashizaki Y, Lenhard B. 2009. Transcriptional features of genomic regulatory blocks. Genome Biol 10:R38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon P, Rodriguez‐Seguel E, Fernandez‐Gonzalez A, Rubio R, Gomez‐Skarmeta JL. 2008. A dual requirement for Iroquois genes during Xenopus kidney development. Development 135:3197–3207. [DOI] [PubMed] [Google Scholar]
- Diez‐Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin‐Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nurnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia‐Lopez R, Echevarria D, Puelles E, Garcia‐Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dolle P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. 2011. A high‐resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9:e1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy‐Marchal C, Horber F, Potoczna N, Hercberg S, Le Stunff C, Bougneres P, Kovacs P, Marre M, Balkau B, Cauchi S, Chevre JC, Froguel P. 2007. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 39:724–726. [DOI] [PubMed] [Google Scholar]
- Engstrom PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. 2007. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res 17:1898–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett KA, Barroso I. 2010. The genetics of obesity: FTO leads the way. Trends Genet 26:266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J. 2013. Gwas. Curr Biol 23:R265–R266. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben‐Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. 2007. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316:889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso I, Lindahl T, Ponting CP, Ashcroft FM, O'Rahilly S, Schofield CJ. 2007. The obesity‐associated FTO gene encodes a 2‐oxoglutarate‐dependent nucleic acid demethylase. Science 318:1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houweling AC, Dildrop R, Peters T, Mummenhoff J, Moorman AF, Ruther U, Christoffels VM. 2001. Gene and cluster‐specific expression of the Iroquois family members during mouse development. Mech Dev 107:169–174. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Mechaly AS, Becker TS, Rinkwitz S. 2013. Using zebrafish transgenesis to test human genomic sequences for specific enhancer activity. Methods 62:216–225. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. 2000. Identification of a functional transposase of the Tol2 element, an Ac‐like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A 97:11403–11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Fredman D, Rinkwitz S, Lenhard B, Becker TS. 2007a. Retroviral enhancer detection insertions in zebrafish combined with comparative genomics reveal genomic regulatory blocks ‐ a fundamental feature of vertebrate genomes. Genome Biol 8 Suppl 1:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, Ghislain J, Pezeron G, Mourrain P, Ellingsen S, Oates AC, Thisse C, Thisse B, Foucher I, Adolf B, Geling A, Lenhard B, Becker TS. 2007b. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res 17:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubu C, Horie K, Abe K, Ikeda R, Mizuno S, Uno Y, Ogiwara S, Ohtsuka M, Isotani A, Okabe M, Imai K, Takeda J. 2009. A transposon‐based chromosomal engineering method to survey a large cis‐regulatory landscape in mice. Nat Genet 41:946–952. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF. 2005. Atlas of Early Zebrafish Brain Development: A Tool for Molecular Neurogenetics. Amsterdam: Elsevier; p 183. [Google Scholar]
- Navratilova P, Becker TS. 2009. Genomic regulatory blocks in vertebrates and implications in human disease. Brief Funct Genomic Proteomic 8:333–342. [DOI] [PubMed] [Google Scholar]
- Punnamoottil B, Kikuta H, Pezeron G, Erceg J, Becker TS, Rinkwitz S. 2008. Enhancer detection in zebrafish permits the identification of neuronal subtypes that express Hox4 paralogs. Dev Dyn 237:2195–2208. [DOI] [PubMed] [Google Scholar]
- Ragvin A, Moro E, Fredman D, Navratilova P, Drivenes O, Engstrom PG, Alonso ME, de la Calle Mustienes E, Gomez Skarmeta JL, Tavares MJ, Casares F, Manzanares M, van Heyningen V, Molven A, Njolstad PR, Argenton F, Lenhard B, Becker TS. 2010. Long‐range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci U S A 107:775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez‐Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez‐Skarmeta JL, Nobrega MA. 2014. Obesity‐associated variants within FTO form long‐range functional connections with IRX3. Nature 507:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster ML, Abe G, Schouw A, Kawakami K. 2011. Transposon‐mediated BAC transgenesis in zebrafish. Nat Protoc 6:1998–2021. [DOI] [PubMed] [Google Scholar]
- Suster ML, Sumiyama K, Kawakami K. 2009. Transposon‐mediated BAC transgenesis in zebrafish and mice. BMC Genomics 10:477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C. 2004. Spatial and temporal expression of the zebrafish genome by large‐scale in situ hybridization screening. Methods Cell Biol 77:505–519. [DOI] [PubMed] [Google Scholar]
- Tung YC, Yeo GS, O'Rahilly S, Coll AP. 2014. Obesity and FTO: Changing Focus at a Complex Locus. Cell Metab 20:710–718. [DOI] [PubMed] [Google Scholar]
- van Gestel MA, Sanders LE, de Jong JW, Luijendijk MC, Adan RA. 2014. FTO knockdown in rat ventromedial hypothalamus does not affect energy balance. Physiol Rep 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, Hansen DV, Nord AS, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer‐Frick I, Shoukry M, Afzal V, Kaplan T, Kriegstein AR, Rubin EM, Ovcharenko I, Pennacchio LA, Rubenstein JLR. 2013. A high‐resolution enhancer atlas of the developing telencephalon. Cell 152:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SW, Ross LS, Parrett T, Easter SS Jr. 1990. The development of a simple scaffold of axon tracts in the brain of the embryonic zebrafish, Brachydanio rerio. Development 108:121–145. [DOI] [PubMed] [Google Scholar]


