Abstract
The molecular phylogeny of brown algae was examined using concatenated DNA sequences of seven chloroplast and mitochondrial genes (atpB, psaA, psaB, psbA, psbC, rbcL, and cox1). The study was carried out mostly from unialgal cultures; we included Phaeostrophion irregulare and Platysiphon glacialis because their ordinal taxonomic positions were unclear. Overall, the molecular phylogeny agreed with previously published studies, however, Platysiphon clustered with Halosiphon and Stschapovia and was paraphyletic with the Tilopteridales. Platysiphon resembled Stschapovia in showing remarkable morphological changes between young and mature thalli. Platysiphon, Halosiphon and Stschapovia also shared parenchymatous, terete, erect thalli with assimilatory filaments in whorls or on the distal end. Based on these results, we proposed a new order Stschapoviales and a new family Platysiphonaceae. We proposed to include Phaeostrophion in the Sphacelariales, and we emended the order to include this foliose member. Finally, using basal taxa not included in earlier studies, the origin and divergence times for brown algae were re‐investigated. Results showed that the Phaeophyceae branched from Schizocladiophyceae ~260 Ma during the Permian Period. The early diverging brown algae had isomorphic life histories, whereas the derived taxa with heteromorphic life histories evolved 155–110 Ma when they branched from the basal taxa. Based on these results, we propose that the development of heteromorphic life histories and their success in the temperate and cold‐water regions was induced by the development of the remarkable seasonality caused by the breakup of Pangaea. Most brown algal orders had diverged by roughly 60 Ma, around the last mass extinction event during the Cretaceous Period, and therefore a drastic climate change might have triggered the divergence of brown algae.
Keywords: divergence time, molecular phylogeny, Phaeophyceae, Platysiphon, Platysiphonaceae fam. nov., Stschapoviales ord. nov.
Platysiphon glacialis (Rosenvinge) H. Kawai et T. Hanyuda, a little‐studied brown alga endemic to the Arctic region, was first described by Wilce (1962) based on vegetative specimens collected at Inglefield Bay, Thule district, West Greenland. There have been few reports on this species since then (Pedersen 1976, Mathieson et al. 2010), and neither the reproductive structures nor the life history have been fully elucidated (Wilce and Bradley 2007). The species is distinctive owing to its laterally expanded blade terminated by a terete portion, both of which have numerous clumps of hair‐like assimilatory filaments arranged more or less in whorls on the distal end. On the basis of the gross morphology of juvenile thalli, Wilce (1962) placed the species in the Striariaceae of the Dictyosiphonales (Ectocarpales s.l.); however, the absence of pyrenoids suggested a more distant phylogenetic position from Ectocarpales s.l. (R.T. Wilce, unpublished data).
Phaeostrophion irregulare Setchell & N.L. Gardner is another enigmatic species also considered distant from Ectocarpales s.l. based on morphology and molecular phylogenetic analysis (Kawai et al. 2005). The alga was placed in its own family Phaeostrophiaceae, but the ordinal assignment was suspended because of insufficient resolution in the molecular analysis.
Historically, the divergence time of brown algal orders and families has been unclear because brown algal fossil evidence for evolutionary interpretations is scanty. Their soft tissues are composed of polysaccharides such as alginates, fucoidans and cellulose, and these tissues generally decompose before fossilization can occur. Furthermore, only a few calcified taxa exist (i.e., Padina spp. and Newhousia imbricata Kraft, G.W. Saunders, I.A. Abbott & Haroun) but they typically grow in exposed habitats where sedimentation is not common. The oldest generally accepted phaeophycean fossil is Julescranea grandicornis B.C. Parker & E.Y. Dawson from the Tertiary Period, described from the upper Miocene Epoch; this fossil is intermediate in appearance between the laminarialean genera Pelagophycus and Nereocystis (Parker and Dawson 1965).
Using RNA sequence data, the divergence time of brown algae from other heterokont lineages (e.g., diatoms) was first estimated to be 150–200 Ma (Lim et al. 1986). Silberfeld et al. (2010) published a time‐calibrated molecular phylogenetic tree based on 10 mitochondrial, chloroplast and nuclear gene DNA sequences that inferred a diversification of the brown algal crown groups in the Lower Cretaceous Period; however, their analyses did not include basal taxa such as the Discosporangiales and Ishigeales. Brown and Sorhannus (2010) published a chronogram of the Ochrophyta (Heterkontophyta) using 18S rDNA sequences; their analyses included Schizocladiophyceae, the sister group of Phaeophyceae, as a basal taxon, and their estimated divergence time was ~170–200 Ma.
In this paper, we re‐investigated Platysiphon glacialis and Phaeostrophion irregulare using a seven chloroplast and mitochondrial genes. In addition, we re‐examined the brown algal tree of life by including basal taxa, and we re‐estimated the divergence times for brown algal orders.
Materials and Methods
Culture strains used for DNA extractions were housed in the Kobe University Macroalgal Culture Collection. Strains were grown in polystyrene Petri dishes containing 50 mL PESI medium (Tatewaki 1966) at 10°C and 15°C long day (LD; 16:8 light:dark) conditions illuminated by daylight‐type white fluorescent lighting of ~50 μmol photons · m−2 · s−1. Specimens of Platysiphon glacialis were collected by SCUBA from a small bay at the eastern entrance of Ragged Channel, north Baffin Island, Nunavik, Canada, and quickly dried in silica gel. The culture strain codes and origins of the field‐collected specimens used for molecular phylogenetic studies are listed in Table S1 in the Supporting Information.
Genomic DNA was extracted from the silica gel‐dried algal tissue using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer's instructions. PCR amplifications of the chloroplast atpB, psaA, psaB, psbA, psbC, rbcL, and mitochondrial cox1 genes were carried out using the KOD FX (ToYoBo, Osaka, Japan) PCR enzyme and the TaKaRa PCR Thermal Cycler Dice (Takara Shuzo, Shiga, Japan). Primers used for PCR and/or sequencing are listed in Table 1. After PEG purification (Lis 1980), PCR products were sequenced using the CE DTCS Quick Start Kit (Beckman Coulter, Fullerton, CA, USA) and the CEQ8000 DNA analysis system (Beckman Coulter) according to the manufacturer's instructions. For the molecular phylogenetic analyses, published and newly determined sequence data of the Phaeophyceae and the Schizocladiaceae were used (Table S1). Schizocladia ischiensis E.C. Henry, K. Okuda & H. Kawai, which is sister to Phaeophyceae was chosen as the outgroup. Alignments were prepared using the program MAFFT v. 6 (Katoh and Toh 2008) and then manually adjusted prior to phylogenetic analyses.
Table 1.
List of primers used for PCR and sequencing
| Code | F/R | Sequence (5′–3′) | Annealing position |
|---|---|---|---|
| ycf3‐F1P* | F | CAAGCDYTAAATAATATWGCTG | ycf3 (319‐340) |
| atpB‐F1P* | F | GCWAAAGCNCATGGTGGTGT | atpB (535‐554) |
| atpB‐F1.2P* | F | AARGTMGTWGAYYTATTAGC | atpB (427‐446) |
| atpB‐F1.3P* | F | GTHMGHGCNATTGCNATGAGTGC | atpB (187‐209) |
| atpB‐F2P* | F | GCDGTRGAYCCHTTAGATTCA | atpB (1,057‐1,077) |
| atpB‐2R† | R | AGCTTGWACAAATCTAAAAATA | atpB (810‐789) |
| atpB‐R2P* | R | GCWATAATATCTTGTAATTC | atpB (1,184‐1,165) |
| atpB‐R2.2P* | R | TCKACHACTAADCGRTCTTC | atpB (1,232‐1,213) |
| atpB‐R3P* | R | CAYRTAYAAATCRTTTCCTTC | atpB (609‐589) |
| atpB‐R4P* | R | GCWGRKCKATGRATAGGTAA | atpB (374‐355) |
| atpB‐R1P* | R | TTTGCTTTAGMDATWGCTTC | atpB (1,430‐1,411) |
| psaA130F‡ | F | AACWACWACTTGGATTTGGAA | psaA (126‐146) |
| psaA‐P2§ | R | NCCWGACCAAGMWARACAAC | psaA (639‐620) |
| psaA‐P2.2* | R | TCDGCRTTYTGRAACCAYTC | psaA (578‐559) |
| psaA‐P3§ | R | GCTGGDTTRTARTCACGAACC | psaA (1,253‐1,233) |
| psaA‐P3.2* | R | GCWGGRTTATARTCRCGDACC | psaA (1,253‐1,233) |
| psaA970R‡ | R | GCYTCTARAATYTCTTTCA | psaA (977‐959) |
| psaA970R2* | R | TRCTATGDCCRATNCCCCAA | psaA (958‐939) |
| psaA1760R‡ | R | CCTCTWCCWGGWCCATCRCAWGG | psaA (1,736‐1,714) |
| psaA1760R2* | R | CCRTCACAHGGRAAWCGGAA | psaA (1,724‐1,705) |
| psaA‐F3P* | F | CWGCWGAYTTTATGGTTCAYC | psaA (1,583‐1,603) |
| psaA‐P4§ | R | NGATTCAATHARYTCTTGCC | psaA (2,094‐2,075) |
| psaA‐P5§ | F | CAYCATATHCATGCNTTYAC | psaA (1,600‐1,619) |
| psaA‐F1P* | F | ATGCAATCNGAYRTTTGGGG | psaA (1,834‐1,853) |
| psaB‐F1P* | F | ATGCAATCNGAYRTTTGGGG | psaB (21‐40) |
| psaB‐F3P* | F | WGATGCWCAYMGWCCMCCTG | psaB (915‐934) |
| psaB‐F3.2P* | F | GSTATHGGNCAYAAYATGAAAGA | psaB (886‐908) |
| psaB‐R2P* | R | KAWTGCATACATATGYTGAG | psaB (1,065‐1,046) |
| psaB‐R3P* | R | CAATGCCAATARAAHGTAACCC | psaB (1,787‐1,766) |
| psaB‐R3.2P* | R | CAATARAAHGTAACCCAWCC | psaB (1,781‐1,762) |
| psaB‐R1P* | R | TTCCAGCHGTTGAWGCWATA | psaB (2,194‐2,175) |
| psaB‐F4P* | F | CCWTATGCHTTYATGGCAAAAGAT | psaB (1,069‐1,092) |
| psbA‐F‡ | F | ATGACTGCTACTTTAGAAAGACG | psbA (1‐23) |
| psbA‐F1P* | F | ACCGTTTATACATYGGTTGG | psbA (77‐96) |
| psbA500F‡ | F | CTCTGATGGWATGCCWYTAGG | psbA (504‐524) |
| psbA600R‡ | R | CCAAATACACCAGCAACACC | psbA (620‐601) |
| psbA600R2* | R | AWACACCAGCAACACCAGCC | psbA (616‐597) |
| psbA‐R1P* | R | TACGYTCRTGCATTACTTCC | psbA (1,003‐984) |
| psbA‐R1‡ | R | GCTAAATCTARWGGGAAGTTGTG | psbA (1,031‐1,009) |
| psbC‐P1.2* | F | CCACGTGGAAACGCYCTTTA | psbC (33‐52) |
| psbC‐P1* | F | TAGCTCATGCAGGYWTAATGG | psbC (152‐172) |
| psbC‐P3* | R | CTTGCCAAGGTTGRATATCATT | psbC (1,168‐1,147) |
| psbC‐P3.2* | R | ATWCCTTTTTCRAARCCAGC | psbC (1,379‐1,360) |
| psbC‐P4* | F | GCGTGAYAAAAAYAAAATGAC | psbC (444‐464) |
| psbC‐P5* | R | AARAATGGRAAWGAYTCTTC | psbC (431‐412) |
| psbC‐P6* | F | GAAGCWTCTCAAKCDCARGC | psbC (925‐944) |
| rbcL‐P2¶ | F | GAWCGRACTCGAWTWAAAAGTG | rbcL (19‐40) |
| rbc‐R2.5** | R | CCTTCATAAACAACACG | rbcL (587–571) |
| Ral‐R952¶ | R | CATACGCATCCATTTACA | rbcL (969–952) |
| rbcL‐P5* | F | CWTAYYTAAAAACWTTCCAAG | rbcL (440‐460) |
| rbcL‐P4* | R | AGKTGRTGCATYTGRCCACA | rbcL (1,178–1,159) |
| rbcL‐Rh3†† | F | TTAAYTCTCARCCDTTYATGCG | rbcL (629–650) |
| rbcL‐P1¶ | F | GKGTWATTTGTAARTGGATGCG | rbcL (944–965) |
| rbcL‐P3** | F | CARTTYGGWGGWGGTACDATTGG | rbcL (1,210‐1,232) |
| rbcL‐P3.2* | F | GARGGTCCTGADATTYTACGT | rbcL (1,327‐1,347) |
| rbcL‐Pla1 | F | TATTCTCGTGCAGTTGGTAG | rbcL (778‐797) |
| rbcL‐Pla4 | F | ATTTACTAACRGCTTGTGAC | rbcL (227‐246) |
| rbcL‐Pla5 | R | AAAGTATTATAGAATCCTCG | rbcL (1,055‐1,036) |
| rbcS‐P1¶ | R | GGATCATCTGYCCATTCTACAC | rbcS (122–101) |
| GazF2‡‡ | F | CCAACCAYAAAGATATWGGTAC | cox1 (104‐125) |
| cox1‐P1.2* | F | GATHTTYTTTATGGTDATGCC | cox1 (276‐296) |
| cox1‐P2* | R | GGDATACGDCGHGGCATACC | cox1 (1,400‐1,381) |
| cox1‐P3* | R | CNGTAAACATRTGRTGVGCC | cox1 (961‐942) |
| cox1‐P4* | F | DGCRGCNTTTACNATGTTTG | cox1 (1,230‐1,249) |
| cox1‐P5* | R | TAATACCNCCRGCYAAAACWGG | cox1 (700‐679) |
| cox1‐P5.2* | R | GCHGTDATTAADACHGACCA | cox1 (656‐637) |
| trnI‐P1.2* | R | GCTTATCAGGCGTACACTCT | trnI (20‐39) |
| trnI‐P1* | R | TTGAACGAWCGVCTTTACGC | trnI (38‐57) |
Concatenated DNA sequences (data set 1: 44OTUs, seven genes, total 9,706 bp) were subjected to maximum parsimony (MP), maximum likelihood (ML), and Bayesian analyses. The MP tree was constructed in MEGA 5 (Tamura et al. 2011) using Tree‐Bisection‐Reconnection with a search level of 1 and initial trees by random addition (100,000 reps). To assess support for each clade, bootstrap analyses (Felsenstein 1985) were performed with 1,000 replicates. For ML analysis, we used RAxML GUI v. 1.31 (Silvestro and Michalak 2012) run to conduct 10,000 Rapid Bootstrap searches followed by an ML search, with the GTR + G + I model for each codon position of each gene. In addition, ML analyses were performed using two additional data sets for testing the differences in divergence time depending on the OTUs as follow. In data set 2 (39 OTUs), the basal taxa of Phaeophyceae (Discosporangium mesarthrocarpum (Meneghini) Hauck and Choristocarpus tenellus (Kützing) Zanardini (Discosporangiales), Ishige okamurae Yendo (Ishigeales), and Onslowia endophytica Searles (Onslowiales) and their sister taxon Schizocladia ischiensis (Schizocladiophyceae), which were not included in Silberfeld et al. (2010), were removed from data set 1, and a total of 9,706 bp was analyzed for the seven genes as mentioned above. In data set 3 (50 OTUs), the sister taxa of Phaeophyceae/Schizocladiophyceae such as Rhizosolenia, Corethron (Bacillariophyceae), Mallomonas, Synura (Synurophyceae), Tribonema (Xanthophyceae), and Phaeothamnion (Phaeothamniophyceae) were added, and a total of 1,425 bp were analyzed for the rbcL gene. In data sets 4–10 (44 OTUs) the following genes were removed sequentially from the seven genes used in data set 1 and each data set analyzed using six genes: data set 4, –atpB, 8,535 bp; data set 5: –psaA, 7,778 bp; data set 6: –psaB, 8,013 bp; data set 7: –psbA, 8,735 bp; data set 8: –psbC, 8,400 bp; data set 9, –psaA, 8,287 bp; data set 10, –cox1, 8,488 bp. The comparisons of the ML tree topologies (AU test) were performed using RAxML v. 7.2.8 (Stamatakis 2006) and CONSEL (Shimodaira and Hasegawa 2001). Bayesian analysis was run using MrBayes v. 3.2.2 (Ronquist et al. 2012). With the aid of the Kakusan4 program (Tanabe 2011), the best‐fit evolutionary model for each codon position of each gene was determined by comparing different evolutionary models via the corrected Bayesian Information Criterion (Schwarz 1978). The Bayesian analysis was initiated with a random starting tree and four chains of Markov chain Monte Carlo (MCMC) iterations were run simultaneously for 10,000,000 generations, keeping one tree every 100 generations. The first 10,000 trees sampled were discarded as “burn‐in,” based on the stationarity of ln L as assessed using Tracer v. 1.5 (Rambaut and Drummond 2007). A consensus topology and posterior probability values were calculated from the remaining trees.
Molecular dating analyses for data sets 1–3 were performed using the MCMCTREE program in the PAML 4 package (Yang 2007). The best‐scoring ML trees based on the above‐mentioned three data sets in this study were used for divergence time estimation. The ML estimates of branch lengths were obtained using the BASEML (in PAML) programs under the GTR + G substitution models for the data set (concatenated DNA sequences separated into three codon partitions). Two priors, the overall substitution rate (rgene gamma) and rate‐drift parameter (sigma2 gamma), were set at G (1, 25) and G (1, 4.5) for data sets 1 and 2, and G (1, 20) and G (1, 4.5) for data set 3, using the strict molecular clock assumption with 195.5 Ma (data set 1 and 3) or 155 Ma (data set 2). The former (195.5 Ma) was the mean of the estimated divergence time of Shizocladiophyceae and Phaeophyceae in Brown and Sorhannus (2010) and the latter (155 Ma) was the estimated origin of the Phaeophyceae by Medlin et al. (1997). The independent rates model (Rannala and Yang 2007) was used to specify the prior of rates among internal nodes (clock = 2 in MCMCTREE). Three nodes in the tree were constrained in geological time based on knowledge from the fossil record and prior molecular clock analyses. For data set 1, the maximum (267 Ma) and the minimum (124 Ma) time constraints, which were the estimated divergence times of the Shizocladiaphyceae‐Phaeophyceae clade (Brown and Sorhannus 2010), were chosen for the most basal node on our tree. On the basis of the fossil record (Parker and Dawson 1965, Rajanikanth 1989), we defined the lower boundaries at 13 and 99.6 Ma for the stem nodes of Sargassaceae and Dictyotales, respectively. For the data set 2, the maximum (185 Ma) and the minimum (125 Ma) time constraints for the root of the tree, which were based on the estimated origin of the Phaeophyceae (155 Ma, Medlin et al. 1997) and a standard deviation of 30 Ma, were used in the analysis, as well as the lower boundaries at 13 and 99.6 Ma for Sargassaceae and Dictyotales, respectively. These setting for data set 2 were similar to that of Silberfeld et al. (2010). For data set 3, based on the fossil record and diatom biomarker, two calibration constraints (93–90 Ma: Rhizosolenia‐Corethron divergence based on the estimated time of the abrupt increase in the C25 HBI alkene (Sinninghe Damste et al. 2004), 49–40 Ma: first appearance of Synura uvella Ehrenberg and Mallomonas insignis Penard in the fossil record (Silver and Wolfe 2007) were used in the analysis, as well as the lower boundaries at 99.6 Ma for Dictyotales. MCMC approximation with a burn‐in period of 50,000 cycles was run, and sampled every 50 cycles to create a total of 100,000 samples. The effective sample sizes of all parameters were greater than 200 and judged to be sufficient by Tracer v.1.5.
Results
Molecular phylogenetic analyses
The molecular phylogenetic tree by ML (Fig. 1) and MP (Fig. S1 in the Supporting Information) analyses based on the concatenated sequences of seven genes (9,706 bp) resulted in similar tree topologies for most of the major nodes with high statistical support: i.e., Discosporangiales was the most basal order in the Phaeophyceae with the Ishigeales the next branching lineage. Phaeostrophion was a sister group to the Sphacelariales, and Platysiphon clustered with Halosiphon and Stschapovia. The phylogenetic positions of Onslowiales and Syringodermatales differed between the two analyses. Onslowiales branched next to Ishigeales in the ML tree but was sister to Dictyotales in the MP tree although the statistical support was low. In the ML tree, the large clade of more derived orders had Desmarestiales basal, and Laminariales and Ectocarpales forming a well‐supported clade. Tilopteriadales, composed of Cutleria, Saccorhiza and Tilopteris, were distinct from the monophyletic clade of Halosiphon, Platysiphon and Stschapovia in both ML and Baysian trees. The Baysian tree supported the monophyly of most orders by high posterior probabilities (Fig. 2).
Figure 1.

Maximum likelihood (ML) tree based on the concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis (left) and posterior probabilities from Baysian analysis (right). Asterisk (*) indicates 100 (ML) and 1.00 (Bayes). Only posterior probabilities >0.90 and bootstrap values >50% are shown.
Figure 2.
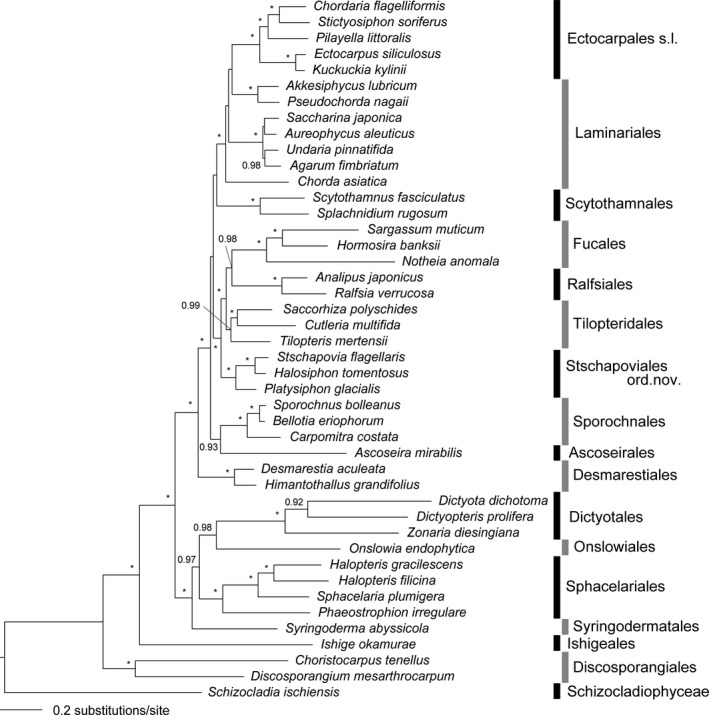
Bayesian consensus tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from Baysian analysis. Only posterior probabilities >0.90 are shown and asterisk (*) indicates 1.00.
The time trees inferred from three data sets are presented in Figure 3, Figures S2 and S3 in the Supporting Information. Estimated divergence times for the major nodes in Figure 3 and Figure S2 were summarized in Table 2. The branching of the Phaeophyceae and Schizocladiophyceae from their common ancestor (node 1) was estimated to have occurred in late Permian Period (mean age 258.0 Ma in Fig. 3 and 268.5 Ma in Fig. S3). Estimated divergence times of comparable nodes between Figure 3 and Figure S2 were similar, and the time differences were 0.3–8.5 Ma (Table 2).
Figure 3.
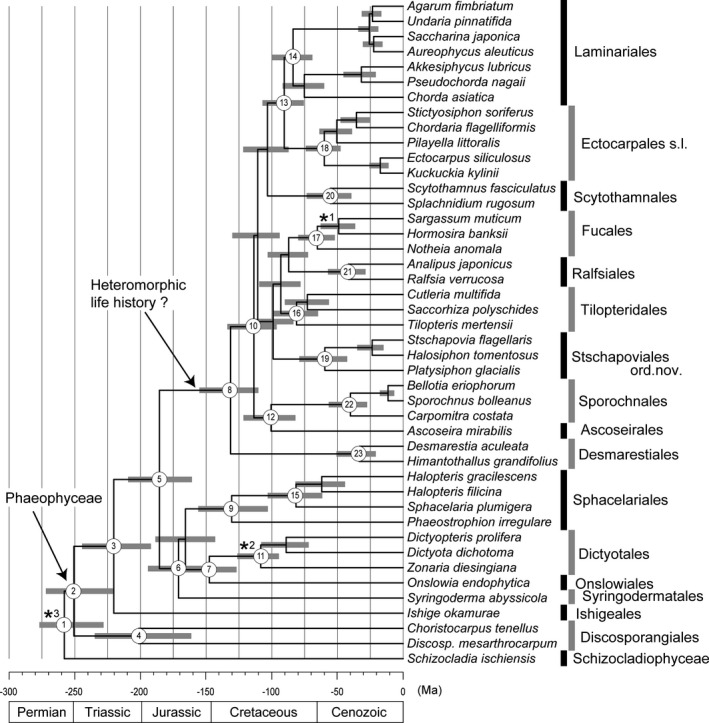
Time tree derived from relaxed molecular clock method implemented in MCMCTREE in PAML 4.7 (Yang 2007). Horizontal bars indicate 95% credible intervals of divergence time estimates. Asterisks on nodes correspond to calibration points. Asterisks 1 and 2 indicate calibration points with fossils (Parker and Dawson 1965 and Rajanikanth 1989, respec‐tively), and minimum time constraints used for nodes were 13 and 99.6 Ma, respectively. Asterisk 3 shows calibration point based on previous molecular clock study (Brown and Sorhannus 2010), and maximum (267 Ma) and minimum (124 Ma) time constraints used on node. Estimated ages and their 95% credible intervals are listed in Table 2 with node numbers.
Table 2.
Comparisons of estimated divergence time in this study (data set 1 and 2) and previous studies; estimated divergence time are expressed in Ma as posterior mean ± 95% credible interval. Node numbers in this table correspond to those shown in Figure 3
| Node | This study (data set 1) | This study (data set 2) | Silberfeld et al. (2010) | Brown and Sorhannus (2010)* |
|---|---|---|---|---|
| 1 | 258.0 (228.1–277.0) | – | – |
196 (131–267) 186 (124–253) |
| 2 | 250.7 (220.4–272.0) | – | – |
~160 (~100–225)†
~155 (~95–210)† |
| 3 | 220.3 (192.0–244.5) | – | – | – |
| 4 | 201.3 (161.3–234.8) | – | – | – |
| 5 | 185.5 (160.8–209.4) | 177.0 (156.6–187.9) | ~180 (~150–215)† | – |
| 6 | 171.0 (147.8–194.3) | 172.3 (151.6–185.3) | ~170 (~140–205)† | – |
| 7 | 147.6 (126.8–169.7) | – | – | – |
| 8 | 131.4 (110.2–155.2) | 128.2 (108.8–147.4) | 128.9 (98.7–162.0) | – |
| 9 | 130.6 (103.0–156.0) | 129.0 (104.1–149.9) | – | – |
| 10 | 113.7 (96.3–133.9) | 112.2 (96.0–129.4) | 118.7 (89.5–148.4) | – |
| 11 | 108.4 (94.6–126.3) | 108.8 (95.4–124.9) | ~110 (~100–130)† |
~55 (~20–90)†
~50 (~20–80)† |
| 12 | 100.6 (82.0–121.6) | 93.1 (89.3–121.5) | – | – |
| 13 | 90.5 (75.6–107.1) | 94.6 (79.0–110.4) | 98.0 (72.2–125.0) | – |
| 14 | 83.8 (69.1–100.3) | 87.1 (71.5–103.0) | 84.4 (59.9–110.4) | – |
| 15 | 81.6 (61.6–103.2) | 81.3 (61.9–101.4) | – | – |
| 16 | 81.2 (64.8–98.4) | 80.7 (65.0–96.4) | 85.3 (59.4–111.7) | – |
| 17 | 65.3 (52.0–79.9) | 64.6 (51.6–78.4) | 73.7 (54.6–96.0) | – |
| 18 | 60.1 (47.6–74.2) | 61.0 (48.2–75.0) | 69.7 (51.4–89.3) | – |
| 19 | 59.6 (42.6–79.1) | 57.7 (41.2–76.1) | – | – |
| 20 | 55.5 (39.4–73.5) | 55.3 (39.2–73.0) | 54.9 (33.1–79.4) | – |
| 21 | 41.8 (28.6–57.3) | 41.7 (28.3–57.2) | 62.0 (37.5–89.7) | – |
| 22 | 40.2 (27.4–56.7) | 38.7 (26.5–54.1) | 38.6 (22.0–55.5) | – |
| 23 | 33.3 (20.8–50.8) | 33.9 (20.9–52.2) | 52.7 (34.4–73.7) | – |
*Upper and lower divergence times were estimated by (broad) lognormal and (tight) exponential calibration sets, respectively.
In order to evaluate the effect of the discrepancies between the phylogenetic signals of each gene, we assessed the effects of removing one gene from the seven genes used in data set 1 (data sets 4–10). The topologies of two ML trees (i.e., Figs. S4 and S5 in the Supporting Information) were the same as in Figure 1. In contrast, in the other five ML trees (Figs. S6–S10 in the Supporting Information), the positions of two to eight different taxa were not congruent with those in Figure 1. The AU tests indicated that the differences in the tree topologies between these five ML trees and Figure 1 were not significant.
Discussion
Molecular phylogeny of Phaeophyceae
The overall phylogenetic relationships of representative brown algal orders revealed by the concatenated sequence data of the seven chloroplast and mitochondrial genes generally agreed with previous studies using multigene sequences (Phillips et al. 2008, Silberfeld et al. 2010). In general those taxa with isomorphic life histories (i.e., Discosporangiales, Ishigeales, Onslowiales, Phaeostrophionaceae, Sphacelariales, and Dictyotales) were basal, except the heteromorphic Syringodermatales (Henry and Müller 1983, Henry 1984, Kawai and Yamada 1990), and those principally having heteromorphic life histories or secondarily lacking an alternation of generations in their life histories (i.e., Desmarestiales, Scytothamnales, Ecotcarpales, Laminariales, Ascoseirales, Sporochnales, Tilopteridales, and Fucales) formed a more derived clade, with mostly isomorphic Ralfsiales being the exception. However, Heteroralfsia saxicola (Okamura & Yamada) H. Kawai has a heteromorphic life history (Kawai 1989).
In the present analyses, Platysiphon glacialis was sister to the clade of Halosiphon tomentosus and Stschapovia flagellaris in all of the ML, MP, and Baysian trees. In the MP analysis, the Halosiphon/Platysiphon/Stschapovia clade clustered with the Tilopteridales clade composed of Cutleria, Saccorhiza and Tilopteris, but the bootstrap support was low (<50%). Conversely, in the ML and Baysian trees the Halosiphon/Platysiphon/Stschapovia clade was not monophyletic with Tilopteridales, but was sister to the clade including Fucales, Ralfsiales, and Tilopteridales supported by a moderate bootstrap values.
Phaeostrophion irregulare was thought to be distant from Ectocarpales s.l. both in the morphology and molecular phylogeny based on rbcL and partial 18S and 26S rDNA sequences (Kawai et al. 2005). Thereby, the authors proposed to treat Phaeostrophiaceae in its own family, but an ordinal assignment was not made. In the present analyses, Phaeostrophion formed a clade with the Sphacelariales. Although they have different thallus constructions (thin terete or filamentous vs. foliose), they share the occurrence of an apical meristematic cell(s), chloroplast morphology, and isomorphic life history (Kawai et al. 2005). Bodanella lauterborni W. Zimmermann, which made a clade with Phaeostrophion in the rbcL analyses of Phillips et al. (2008), was not included in the present analyses. At present, we find no distinctive phenotypic characters except for the foliose thallus constructions to exclude Phaeostrophion from Sphaceraliales and, we tentatively place the Phaeostrophiaceae in the Sphacelariales.
Platysiphonaceae H. Kawai, T. Hanyuda et R. T. Wilce fam. nov.
Typus: Platysiphon glacialis (Rosenvinge) H. Kawai et T. Hanyuda comb. nov. Basionym: Punctaria glacialis Rosenvinge 1910. In: Rosenvinge, L. K. 1910. On the marine algae from North‐East Greenland (N. of 76. N. lat.) collected by ‘Danmark‐Expedition’ Medd. Gr.nl. 43: 118, figs 6,7.
Description: Erect thalli simple, terete, parenchymatous with multicellular hair‐like filaments at the tip and on surfaces of distal portions, later becoming flattened and blade‐like basally; cells containing many discoid chloroplasts without pyrenoids.
Stschapoviales H. Kawai ord. nov.
Typus: Stschapovia flagellaris A. D. Zinova 1954: 242, figs 1–6. In: Zinova, A. D. (1954). Novye smejstvo, rod i vid u burykh vodoroslej. Trudy Botanicheskogo Instituta Akademii Nauk SSSR, Ser. 2, Sporovye Rasteniya 9:223–44.
Description: Erect thalli simple, terete, parenchymatous with multicellular hair‐like filaments at the tip and on surfaces of distal portions. Erect thalli may become thicker or flattened and blade‐like basally and form reproductive structures on the surface. Cells containing many discoid chloroplasts without pyrenoids. Life history heteromorphic or without alternation of generations.
All members of the Halosiphon/Platysiphon/Stschapovia clade have rather narrow geographic distributions in the cold‐water regions of the Northern Hemisphere that are normally or occasionally influenced by frozen surface seawater. They also share several morphological features, including parenchymatous, terete, erect thalli at least in early stages of development, no obvious growth zones or apical cells, and multicellular hair‐like filaments (assimilatory filaments) that are arranged in whorls (Wilce 1962, Kawai and Sasaki 2004). It is noteworthy that Stschapovia shows developmental processes in erect thalli similar to Platysiphon. Thalli of Stschapovia are first thin throughout, but then the proximal portion of the thallus becomes thickened (flattened in Platysiphon); the reproductive structures form only on thickened portions (Wilce 1962, Kawai and Sasaki 2004, Kawai et al. 2015). Multicellular assimilatory filaments are abundant on juvenile terete thalli, but they are absent on proximal thickened portions (Kawai and Sasaki 2004, Kawai et al. 2015).
The life history and reproductive structures of the Stschapoviales are diverse. Halosiphon is heteromorphic, alternating between a macroscopic sporophyte that forms unilocular zoidangia and a microscopic, oogamous gametophyte that is similar to the Laminariales and Phyllariaceae (Tilopteridales) (Maier 1984, Kawai 2014). Stschapovia is not yet fully characterized, but uniquely it lacks an alternation of generations. Stschapovia reproduces by monospore‐like cells although sperm‐like flagellated cells are formed, resembling Tilopteris and Phaeosiphoniella (Tilopteridales) (Kuhlenkamp and Müller 1985, Hooper et al. 1988, Kawai and Sasaki 2004). Platysiphon life stages are also incompletely known, but flagellated cells are provably not released, and sexual reproductive cells resemble Stschapovia and tilopteridacean species (Kawai et al. 2015). The similarity of Stschapoviales and Tilopteridales reproductive features are likely an adaptation to ice coverage in winter, and hence they share these features by convergent evolution.
Among the three monotypic families comprising the clade including Platysiphon, Halosiphonaceae (type: Halosiphon tomentosus (Lyngbye) Jaasund 1957), Platysiphonaceae (type: Platysiphon glacialis (Rosenvinge) H. Kawai et T. Hanyuda 2015), and Stschapoviaceae (type: Stschapovia flagellaris Zinova 1954), Stschapovia has nomenclatural priority. Therefore, we proposed establishing a new order, Stschapoviales, to accommodate the three families.
Evolutionary divergence times
Regarding the evolutionary divergence time for the Phaeophyceae, Silberfeld et al. (2010) proposed a chronogram based on the Bayesian molecular phylogenetic tree of the concatenated DNA sequences of 10 genes (i.e., mitochondrial cox1, cox3, nad1, nad4, atp9, chloroplast rbcL, psaA, psbA, atpB, and nuclear 26S rDNA) analyzed using the BEAST program. They used three calibration points based on brown algal fossil records (i.e., Padina of Dicyotales, Sargassaceae of Fucales, and Nereocystis/Pelagophycus‐like species of Laminariales). Furthermore, they followed the estimation of Medlin et al. (1997) for the divergence time of Phaeophyceae to be 155 Ma, although this was solely based on a molecular clock model. The analyses by Silberfeld et al. (2010), which focused on the crown taxa, did not include the basal taxa of the Phaeophyceae or the sister taxon. On the other hand, Brown and Sorhannus (2010) reported a chronogram of the Ochrophyta (Heterokontophyta) based on 18S rDNA sequences, analyzed using the BEAST program, and calibrated principally by diverse microalgal fossils (e.g., diatoms, Haptophyta, etc.) and red algae. When discussing their analyses, they suggested a divergence time for the Schizocladiophyceae and Phaeophyceae as ~172–186 Ma in the exponential age priors and as 180–198 Ma in the lognormal age priors (Brown and Sorhannus 2010).
In our analyses, which included the basal groups Discosporangiales and Ishigeales as well as the sister taxon (Schizocladiophyceae) of Phaeophyceae (Fig. 3), the divergence time of Phaeophyceae from Schizocladiophyceae was estimated to be ~260 Ma in the Permian Period, which was somewhat earlier than the estimation by Brown and Sorhannus (2010), perhaps due to the influence of the time constraints for the fossil Padina. The evolutionary success of Phaeophyceae may be attributed to the development of elaborate multicellular thalli with complex life histories. For achieving the multicellar thalli with tissue differentiation, one of the key elements was the development of cytoplasmic connections (plasmodesmata) allowing the transport of photosynthetic products and regulation signals. Although some heterokonts phylogenetically close to Phaeophyceae, such as Xanthophyceae and Schizocladiophyceae, also form multicellular thalli, they lack plasmodesmata. By evolving multicellularity with tissue differentiation, Phaeophyceae could obtain higher tolerance to harsh environmental conditions (i.e., desiccation, osmotic shock, and temperature stresses). These adaptations allowed the brown algae to survive the mass extinction during the Permian‐Triassic extinction event, and they diversified subsequently.
Within Phaeophyceae, compared to the chronogram by Silberfeld et al. (2010), the divergence time in our analyses generally shifted to more recent times (~7.5 Ma for the clades that diverged later than Discosporangiales and Ishigeales; Table 2). Basal taxa of brown algae generally share an isomorphic life history and apical growth (i.e., Discosporangiales, Ishigeales, Onslowiales, Dicytotales, and Sphacelariales). Taxa with diffuse and intercalary growth as well as a heteromorphic life history diverged later from the basal lineage, and the divergence time of such taxa (e.g., Desmarestiales, Sporochnales, Laminariales, Ectocarpales, etc.) was estimated to be 155–110 Ma. The environmental factors that resulted in the evolution of heteromorphic life histories is still unclear, but global climate changes and considerable tectonic activities, such as the breakup of the Pangaea during the period resulting in colder seas and seasonality, may have played a role. The fact that taxa with isomorphic life histories, such as Dictyotales, are more common in the tropical and subtropical regions, whereas taxa with heteromorphic life histories, such as Desmarestiales and Laminariales, are more common in temperate and cold‐water regions suggest that heteromorphic life histories are adapted for the survival in the areas where considerable seasonal changes occur in water temperature and transparency. Therefore, considering the estimated divergence time of the derived phaeophycean taxa and the tectonic events, we may hypothesize that the development of heteromorphic life histories in Phaeophyceae and their success in the temperate and cold‐water regions was induced by the development of the remarkable seasonality caused by the breakup of Pangaea. Of course such seasonality could have been present before the formation of Pangaea, but Phaeophyceae did not exist prior to Pangea.
Most brown algal orders had diverged by roughly 60 Ma, around the last mass extinction event during the Cretaceous (70–65 Ma; Schulte et al. 2010, Renne et al. 2013). Therefore, a drastic climate change might have triggered the divergence of brown algae, although the mechanisms causing mass extinctions of marine biodiversity is controversial (Alegret et al. 2012).
We are grateful to Dr. Eric C. Henry for helpful advice in writing the manuscript. A part of this work was supported by the NSF grant DEB‐0212138 (PI: R.A. Andersen), and JSPS Grants‐in‐Aid for Scientific Research (no. 22370034) to H. K.
Supporting information
Figure S1. Maximum parsimony (MP) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S2. Time tree derived from relaxed molecular clock method implemented in MCMCTREE in PAML 4.7 (Yang 2007) for data set 2. Horizontal bars indicate 95% credible intervals of divergence time estimates. Asterisks on nodes correspond to the calibration points. Asterisks 1 and 2 indicate calibration points with fossils (Parker and Dawson 1965 and Rajanikanth 1989, respectively), and minimum time constraints used for nodes were 13 and 99.6 Ma, respectively. Asterisk 3 shows calibration point based on previous molecular clock study (Medlin et al. 1997), and maximum (185 Ma) and minimum (125 Ma) time constraints used on node. Estimated ages and their 95% credible intervals are listed in Table 3 with node numbers.
Figure S3. Timetree derived from the relaxed molecular clock method implemented in MCMCTREE in PAML 4.7 (Yang 2007) for the data set 3. Horizontal bars indicate 95% credible intervals of the divergence time estimates. Asterisks on the nodes correspond to the calibration points. Asterisk 1 shows the calibration point with fossils (Rajanikanth 1989), and the minimum time constraints on the node was 99.6 Ma. Asterisks 2 and 3 indicate the calibration points based on the fossil and diatom biomarker (Sinninghe Damste et al. 2004 and Silver and Wolfe 2007, respectively), and the time constraints used for the nodes were 49–40 Ma and 93–90 Ma, respectively.
Figure S4. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast psaA, psaB, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S5. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S6. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaB, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S7. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S8. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S9. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, and psbC genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S10. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, psbC, and rbcL genes). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Table S1. Origin of samples and sequence data used for molecular analyses, including their DDBJ/GenBank accession numbers.
References
- Alegret, L. , Thomas, E. & Lohmann, K. 2012. End‐Cretaceous marine mass extinction not caused by productivity collapse. Proc. Natl. Acad. Sci. USA 109:728–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J. C. , Bidigare, R. R. , Christensen, S. J. & Andersen, R. A. 1998. Phaeothamniophyceae Classis Nova: a new lineage of chromophytes based upon photosynthetic pigments, rbcL sequence analysis and ultrastructure. Protist 149:245–63. [DOI] [PubMed] [Google Scholar]
- Brown, J. W. & Sorhannus, U. 2010. A molecular genetic timescale for the diversification of autotrophic stramenopiles (Ochrophyta): substantive underestimation of putative fossil age. PLoS ONE 5(9):e12759. doi:10.1371/journal.pone.0012759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugbjerg, N. & Andersen, R. A. 1997. A molecular phylogeny of the heterokont algae based on analyses of chloroplast‐encoded rbcL sequence data. J. Phycol. 33:1031–41. [Google Scholar]
- Draisma, S. G. A. , Prud'homme van Reine, W. F. & Kawai, H. 2010. A revised classification of the Sphacelariales (Phaeophyceae) inferred from a psbC and rbcL based phylogeny. Eur. J. Phycol. 45:308–26. [Google Scholar]
- Draisma, S. G. A. , Prud'homme van Reine, W. F. , Stam, W. T. & Olsen, J. L. 2001. A reassessment of phylogenetic relationships within the Phaeophyceae based on Rubisco large subunit and ribosomal DNA sequences. J. Phycol. 37:586–603. [Google Scholar]
- Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–91. [DOI] [PubMed] [Google Scholar]
- Hanyuda, T. , Suzawa, Y. , Suzawa, T. , Arai, S. , Sato, H. , Ueda, K. & Kumano, S. 2004. Biogeography and taxonomy of Batrachospermum helminthosum Bory (Batrachospermales, Rhodophyta) in Japan inferred from rbcL gene sequences. J. Phycol. 40:581–8. [Google Scholar]
- Henry, E. C. 1984. Syringodermatales ord. nov. and Syringoderma floridana sp. nov. (Phaeophyceae). Phycologia 23:419–26. [Google Scholar]
- Henry, E. C. & Müller, D. G. 1983. Studies of the life history of Syringoderma phinneyi sp. nov. (Phaeophyceae). Phycologia 22:387–93. [Google Scholar]
- Hooper, R. G. , Henry, E. C. & Kuhlenkamp, R. 1988. Phaeosiphoniella cryophila gen. et sp. nov., a third member of the Tilopteridales (Phaeophyceae). Phycologia 27:395–404. [Google Scholar]
- Jaasund, E. 1957. Marine algae from northern Norway. Bot. Not. 110:205–31. [Google Scholar]
- Jo, B. Y. , Shin, W. , Kim, H. S. , Silver, P. A. & Andersen, R. A. 2013. Phylogeny of the genus Mallomonas (Synurophyceae) and descriptions of five new species on the basis of morphological evidence. Phycologia 52:266–78. [Google Scholar]
- Katoh, K. & Toh, H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–98. [DOI] [PubMed] [Google Scholar]
- Kawai, H. 1989. Life history and systematic position of Heteroralfsia saxicola gen. et comb. nov. (Ralfsiaceae, Phaeophyceae). Phycologia 28:243–51. [Google Scholar]
- Kawai, H. 2014. Recent advances in the phylogeny and taxonomy of Laminariales. Perspect Phycol. ISSN 2198‐011X (online) 1:27–40. [Google Scholar]
- Kawai, H. , Hanyuda, T. , Draisma, S. G. A. & Müller, D. G. 2007. Molecular phylogeny of Discosporangium mesarthrocarpum (Phaeophyceae) with a reassessment of the Discosporangiales. J. Phycol. 43:186–94. [Google Scholar]
- Kawai, H. , Hanyuda, T. , Lindeberg, M. & Lindstrom, S. C. 2008. Morphology and molecular phylogeny of Aureophycus aleuticus gen. et sp. nov. (Laminariales, Phaeophyceae) from the Aleutian Islands. J. Phycol. 44:1013–21. [DOI] [PubMed] [Google Scholar]
- Kawai, H. , Hanyuda, T. , Ridgway, L. M. & Holser, K. 2013. Ancestral reproductive structure in basal kelp Aureophycus aleuticus . Sci. Rep. 3:2491. doi:10.1038/srep02491. ISSN (online):2045–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, H. , Hanyuda, T. , Yamagishi, T. , Kai, A. , Lane, C. , McDevit, D. , Küpper, F. & Saunders, G. W. 2015. Reproductive morphology and DNA sequences of the brown alga Platysiphon verticillatus support the new combination P. glacialis . J. Phycol 51:DOI: 10.1111/jpy.12331. [DOI] [PubMed] [Google Scholar]
- Kawai, H. , Kogishi, K. , Hanyuda, T. & Kitayama, T. 2012. Taxonomic revision of the genus Cutleria proposing a new genus Mutimo to accommodate M. cylindrica (Cutleriaceae, Phaeophyceae). Phycol. Res. 60:241–8. [Google Scholar]
- Kawai, H. , Maeba, S. , Sasaki, H. , Okuda, K. & Henry, E. 2003. Schizocladia ischiensis: a new filamentous marine chromophyte belonging to a new class, Schizocladiophyceae. Protist 154:211–28. [DOI] [PubMed] [Google Scholar]
- Kawai, H. & Sasaki, H. 2004. Morphology, life history, and molecular phylogeny of Stschapovia flagellaris (Tilopteridales, Phaeophyceae), and the erection of the Stschapoviaceae fam. nov. J. Phycol. 40:1156–69. [Google Scholar]
- Kawai, H. , Sasaki, H. , Maeba, S. & Henry, E. C. 2005. Morphology and molecular phylogeny of Phaeostrophion irregulare (Phaeophyceae) with proposal of Phaeostrophiaceae fam. nov., and review of Ishigeaceae. Phycologia 44:169–82. [Google Scholar]
- Kawai, H. & Yamada, I. 1990. The specific identity and life history of Japanese Syringoderma (Syringodermatales, Phaeophyceae). Bot. Mag. Tokyo 103:325–34. [Google Scholar]
- Kuhlenkamp, R. & Müller, D. G. 1985. Culture studies on the life history of Haplospora globosa and Tilopteris mertensii (Tilopteridales, Phaeophyceae). Br. Phycol. J. 20:301–12. [Google Scholar]
- Lane, C. E. , Lindstrom, S. C. & Saunders, G. W. 2007. A molecular assessment of northeast Pacific Alaria species (Laminariales, Phaeophyceae) with reference to the utility of DNA barcoding. Mol. Phylogenet. Evol. 44:634–48. [DOI] [PubMed] [Google Scholar]
- Le Corguillé, G. , Pearson, G. , Valente, M. , Viegas, C. , Gschloessl, B. , Corre, E. , Bailly, X. et al. 2009. Plastid genomes of two brown algae, Ectocarpus siliculosus and Fucus vesiculosus: further insights on the evolution of red‐algal derived plastids. BMC Evol. Biol. 9:253–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, B. L. , Kawai, H. , Hori, H. & Osawa, S. 1986. Molecular evolution of 5S ribosomal RNA from red and brown algae. Jpn. J. Genet. 61:169–76. [Google Scholar]
- Lim, P. E. , Sakaguchi, M. , Hanyuda, T. , Kogame, K. , Phang, S. M. & Kawai, H. 2007. Molecular phylogeny of crustose brown algae (Ralfsiales, Phaeophyceae) inferred from rbcL sequences resulting in proposal for Neoralfsiaceae fam. nov. Phycologia 46:456–66. [Google Scholar]
- Lis, J. T. 1980. Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods in Enzymology 65:347–53. [DOI] [PubMed] [Google Scholar]
- Maier, I. 1984. Culture studies of Chorda tomentosa (Phaeophyta, Laminariales). Br. Phycol. J. 19:95–106. [Google Scholar]
- Mathieson, A. C. , Moore, G. E. & Short, F. T. 2010. A floristic comparison of seaweeds from James Bay and Three contiguous Northeastern Canadian Arctic sites. Rhodora 112:396–434. [Google Scholar]
- Medlin, L. K. , Kooistra, W. H. C. F. , Potter, D. , Saunders, G. W. & Andersen, R. A. 1997. Phylogenetic relationships of the ‘golden algae’ (haptophytes, heterokont chromophytes) and their plastids. Plant Syst. Evol. 11(Suppl.):187–219. [Google Scholar]
- Parker, B. C. & Dawson, E. Y. 1965. Non‐calcareous marine algae from California Miocene deposits. Nova Hedwigia 10:273–95. [Google Scholar]
- Pedersen, P. M. 1976. Marine benthic algae from southernmost Greenland. Meddelelser om Grønland 199:1–80, 7 pls. [Google Scholar]
- Phillips, N. , Burrowes, R. , Rousseau, F. , de Reviers, B. & Saunders, G. W. 2008. Resolving evolutionary relationships among the brown algae using chloroplast and nuclear genes. J. Phycol. 44:394–405. [DOI] [PubMed] [Google Scholar]
- Rajanikanth, A. 1989. A fossil marine brown alga from the Gangapur formation, Pranhita‐Godavari graben. Curr. Sci. India 58:78–80. [Google Scholar]
- Rambaut, A. & Drummond, A. 2007. Tracer v1.5. Available at: http://tree.bio.ed.ac.uk/software/tracer/. Accessed March 4, 2014.
- Rannala, B. & Yang, Z. 2007. Inferring speciation times under an episodic molecular clock. Syst. Biol. 56:453–66. [DOI] [PubMed] [Google Scholar]
- Renne, P. R. , Deino, A. L. , Hilgen, F. J. , Kuiper, K. F. , Mark, D. F. , Mitchell, W. S. III , Morgan, L. E. , Mundil, R. & Smit, J. 2013. Time scales of critical events around the Cretaceou‐Paleogene boundary. Science 339:684–7. [DOI] [PubMed] [Google Scholar]
- Ronquist, F. , Teslenko, M. , van der Mark, P. , Ayres, D. L. , Darling, A. , Höhna, S. , Larget, B. , Liu, L. , Suchard, M. A. & Huelsenbeci, J. P. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte, P. , Alegret, L. , Arenillas, I. , Arz, J. A. , Barton, P. J. , Bown, P. R. , Bralower, T. J. et al. 2010. The Chicxulub asteroid impact and mass extinction at the Cretaceous‐Paleogene boundary. Science 327:1214–8. [DOI] [PubMed] [Google Scholar]
- Schwarz, G. 1978. Estimating the dimension of a model. Ann. Stat. 6:461–4. [Google Scholar]
- Shimodaira, H. & Hasegawa, M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–7. [DOI] [PubMed] [Google Scholar]
- Silberfeld, T. , Leigh, J. W. , Verbruggen, H. , Cruaud, C. , de Revier, B. & Rousseau, F. 2010. A multi‐locus time‐calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): investigating the evolutionary nature of the “brown algal crown radiation”. Mol. Phyl. Evol. 56:659–74. [DOI] [PubMed] [Google Scholar]
- Silver, P. A. & Wolfe, A. P. 2007. Eunotia spp. (Bacillariophyceae) from Middle Eocene lake sediments and comments on the origin of the diatom raphe. Can. J. Bot. 85:83–90. [Google Scholar]
- Silvestro, D. & Michalak, I. 2012. raxmlGUI: a graphical front‐end for RAxML. Org. Divers. Evol. 12:335–7. [Google Scholar]
- Sinninghe Damste, J. S. , Muyzer, G. , Abbas, B. , Rampen, S. W. , Masse, G. , Allard, W. G. , Belt, S. T. et al. 2004. The rise of the rhizosolenid diatoms. Science 304:584–7. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. 2006. RAxML‐VI‐HPC: maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–90. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. & Kumer, S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, A. S. 2011. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Res. 11:914–21. [DOI] [PubMed] [Google Scholar]
- Tatewaki, M. 1966. Formation of crustaceous sporophyte with unilocular sporangia in Scytosiphon lomentaria . Phycologia 6:62–6. [Google Scholar]
- Theriot, E. C. , Ashworth, M. , Ruck, E. , Nakov, T. & Jansen, R. K. 2010. A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research. Pl. Ecol. Evol. 143:278–96. [Google Scholar]
- Wilce, R. T. 1962. A new member of the Punctariaceae: Platysiphon verticillatus gen. nov., sp. nov. Bot. Tidsskr. 58:35–42. [Google Scholar]
- Wilce, R. T. & Bradley, P. M. 2007. Enigmatic reproductive structures in Platysiphon verticillatus Wilce (1962): an arctic endemic. In Abstracts, Joint Meeting of Phycological Society of America and International Society of Protistologists, Warwick, RI, pp. 67 Phycol. Soc. Am., Lawrence, KS. [Google Scholar]
- Yang, Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–91. [DOI] [PubMed] [Google Scholar]
- Yoon, H. S. , Hackett, J. D. & Bhattacharya, D. 2002. A single origin of the peridinin‐ and fucoxanthin‐containing plastids in dinoflagellates through tertiary endosymbiosis. Proc. Natl. Acad. Sci. USA 99:11724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinova, A. D. 1954. A new family, a new genus and a new species of brown algae. Contr. Bot. Inst. Komarov Acad. Nauk. USSR Ser. II. 9:223–44 (In Russian). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Maximum parsimony (MP) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S2. Time tree derived from relaxed molecular clock method implemented in MCMCTREE in PAML 4.7 (Yang 2007) for data set 2. Horizontal bars indicate 95% credible intervals of divergence time estimates. Asterisks on nodes correspond to the calibration points. Asterisks 1 and 2 indicate calibration points with fossils (Parker and Dawson 1965 and Rajanikanth 1989, respectively), and minimum time constraints used for nodes were 13 and 99.6 Ma, respectively. Asterisk 3 shows calibration point based on previous molecular clock study (Medlin et al. 1997), and maximum (185 Ma) and minimum (125 Ma) time constraints used on node. Estimated ages and their 95% credible intervals are listed in Table 3 with node numbers.
Figure S3. Timetree derived from the relaxed molecular clock method implemented in MCMCTREE in PAML 4.7 (Yang 2007) for the data set 3. Horizontal bars indicate 95% credible intervals of the divergence time estimates. Asterisks on the nodes correspond to the calibration points. Asterisk 1 shows the calibration point with fossils (Rajanikanth 1989), and the minimum time constraints on the node was 99.6 Ma. Asterisks 2 and 3 indicate the calibration points based on the fossil and diatom biomarker (Sinninghe Damste et al. 2004 and Silver and Wolfe 2007, respectively), and the time constraints used for the nodes were 49–40 Ma and 93–90 Ma, respectively.
Figure S4. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast psaA, psaB, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S5. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S6. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaB, psbA, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S7. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbC, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S8. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, and rbcL genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S9. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, and psbC genes, and mitochondrial cox1 gene). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Figure S10. Maximum likelihood (ML) tree based on concatenated DNA sequences (chloroplast atpB, psaA, psaB, psbA, psbC, and rbcL genes). Numbers on branches indicate bootstrap values from ML analysis. Only bootstrap values >50% are shown.
Table S1. Origin of samples and sequence data used for molecular analyses, including their DDBJ/GenBank accession numbers.


