Abstract
The present study investigates the human leucocyte antigen (HLA) allele and haplotype frequencies in Japanese population. We carried out the frequency analysis in 5824 families living across Japanese archipelago. The studied population has mainly been typed for the purpose of transplant, especially the hematopoietic stem cell transplantation (HSCT). We determined HLA class I (A, B, and C) and HLA class II (DRB1) using Luminex technology. The haplotypes were directly counted by segregation. A total of 44 HLA‐A, 29 HLA‐C, 75 HLA‐B, and 42 HLA‐DRB1 alleles were identified. In the HLA haplotypes of A‐C‐B‐DRB1 and C‐B, the pattern of linkage disequilibrium peculiar to Japanese population has been confirmed. Moreover, the haplotype frequencies based on family study was compared with the frequencies estimated by maximum likelihood estimation (MLE), and the equivalent results were obtained. The allele and haplotype frequencies obtained in this study could be useful for anthropology, transplantation therapy, and disease association studies.
Keywords: allele frequency, family study, haplotype frequency, human leucocyte antigen, Japanese population
Introduction
The human leukocyte antigen (HLA) gene family is characterized by extreme degree of genetic polymorphism and linkage disequilibrium (LD). The varieties in polymorphism and LD patterns of HLA gene family show a tendency to be unique in each ethnic group 1, 2. HLA antigens have been known to play an important role in immune responses. In hematopoietic stem cell transplantation (HSCT), HLA matching between donors and recipients lowers the risk of graft rejection and graft‐versus‐host disease (GVHD) 3, 4. Morishima et al. suggested that the genetic difference derived from HLA haplotype is associated with acute GVHD in allogeneic HSCT 5. Therefore, HLA haplotype cannot be excluded from consideration during donor selection because of potential contribution from proteins encoded by non‐HLA genes inherited with HLA genes.
Several studies reported analysis of HLA allele and haplotype frequency data in the Japanese population 6, 7, 8, 9. However, these studies failed to present accurate and detailed information due to the haplotypes estimation using software or small sample size, or both. This necessitates developing a method which can produce accurate and detailed gene distribution. The present study aims to obtain a more exact and detailed HLA haplotype distribution from 18,604 members of 5824 Japanese families, whose HLA haplotypes were determined by descent. Our study also attempts to determine the frequency of specific haplotypes, C‐B, A‐B‐DRB1, and A‐C‐B‐DRB1, used in donor search. In addition, it was ascertained whether the haplotype frequencies estimated by maximum likelihood estimation (MLE) would be equivalent to the frequencies found in the present family study.
Materials and method
Subjects
A total of 18,604 members (including patients and normal subjects) from 5824 families, distributed in all parts of Japan, were enrolled for this study. Among these families, there were patients, considered for transplantation, especially HSCT. The families were divided into three groups (Table 1): (i) families with both parents with one or more children, (ii) families with one parent with one or more children, and (iii) families with no parents but having two or more children. The families with more than two generations were counted as separate families. Informed consent was obtained from all the participants of this study by the clinicians who ordered HLA typing.
Table 1.
Breakdown of the family structure
| Number of children | Number of familiesa | |
|---|---|---|
| Parents | 1–5 | 2077 |
| One parent | 1–8 | 1968 |
| No parent | 2–7 | 1779 |
| Total | 5824 |
The families with more than two generations are counted as separate families.
For comparing the haplotype frequencies obtained by family study and using mle software, unrelated 4500 people were chosen at random from the total subjects of present study (18,604 members). They were genetically unrelated, because one person was chosen from each of 4500 random chosen families. The overlaps of blood relationship with three or more generation were avoided in these families. The allele and haplotype frequencies calculated from these 4500 people were very similar to the frequencies from the total subjects [allele frequencies (AF) data not shown].
Samples
DNA samples were obtained from peripheral lymphocytes or buccal cells using a JetQuick® Blood & Cell Culture Kits (GENOMED, Löhne, Germany) or QuickGene DNA Tissue Kit (KURABO, Osaka, Japan) according to the manufacturer's protocols.
HLA allele typing
HLA (‐A, ‐C, ‐B, and ‐DRB1) four‐digit allele typing was performed using Luminex 200 system (Luminex, Austin, TX) and WAKFlow HLA Typing kit (Wakunaga, Hiroshima, Japan) 10, 11, 12. HLA alleles were assigned automatically using wakflow Typing software (Wakunaga, Hiroshima, Japan). The primer sequences of wakflow Typing kit are specifically designed to make allele determination easier in Japanese population, and by default the analysis with wakflow Typing software is based on the AF of the donors registered with Japan Marrow Donor Program (JMDP) which are available on the website, www.bmdc.jrc.or.jp 9. Therefore, this method can determine alleles with frequencies of 0.1% and greater in the Japanese population. A few alleles which could not be determined by this method as rare alleles were considered as secondary; these alleles were determined using Luminex 200 system and LAB Type SSO kit (One Lambda, Los Angeles, CA) assigned using the HLA Fusion software (One Lambda). In brief, exon 2 for HLA‐DRB1; exons 2 and 3 for HLA‐A, ‐B, and ‐C were amplified in these methods.
Haplotype determination
The haplotypes were determined by segregation. This study was designed with the aim to assess genetic linkage with high certainty. Nevertheless, the results also included some partial haplotypes because of the possibility of one or more recombination in a family. The haplotype frequencies were calculated by using haplotypes without taking into account the recombination. Thus, we counted the haplotypes of parents and not of children because fathers and mothers are genetically unrelated, while the haplotypes created by recombination were those of children only. Some haplotypes of parents whose children had recombinant haplotypes could not been determined because of two patterns of their combinations. These haplotypes were determined and counted as not less frequent but frequent haplotype phase. Specifically, assuming that four haplotypes are Hp1, Hp2, Hp3, and Hp4, those frequencies are HF1, HF2, HF3, and HF4, and two estimable phases are ‘Hp1, Hp2’ and ‘Hp3, Hp4’, HF1 was multiplied by HF2, HF3 was multiplied by HF4, and the haplotypes of the estimable phase with larger product were counted.
For the comparison to the result by MLE, the haplo.em program, which was evaluated by haplo.stats (version 1.6.0) software operated in the R language, was used 13, 14, 15, 16. For genetic markers measured on unrelated subjects with linkage phase unknown, this program computes the maximum likelihood estimates of haplotype probabilities using the progressive insertion algorithm that progressively inserts batches of loci into haplotypes of growing lengths.
Statistical analysis
The haplotypes were counted manually using Microsoft Excel® spreadsheets. The haplotypes that extended three or more generations were counted once (Figure 1). The allele and haplotype frequencies were calculated by using 19,183 haplotypes, counted as mentioned above. Relative LD values (RD) were computed for each haplotypes 17, 18. The exact test for deviation from Hardy–Weinberg Equilibrium were evaluated by genepop software, version 4.2 19, 20, which uses a Markov Chain (MC) algorithm (dememorization = 10,000, batches = 10,000, and iterations per batch = 10,000) to estimate the P‐value.
Figure 1.
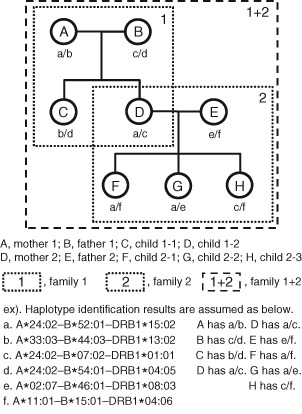
Avoiding haplotype duplication. The extended haplotypes of a and c are redundant in families 1 and 2. In this case, six haplotypes were counted as family 1 + 2.
The expected prevalence (P) of the allele or the haplotype under Hardy–Weinberg proportions were calculated from AF by using the following equation: P = 1 − (1 − AF)2.
Results
HLA‐A, ‐B, ‐C, and ‐DRB1 AF
Table 2 presents the list of the AF of HLA‐A, ‐B, ‐C, and ‐DRB1 loci 21. We identified 44 HLA‐A, 75 HLA‐B, 29 HLA‐C, and 42 HLA‐DRB1 alleles and found A*24:02 to be 36.48%, the highest in Japanese population; thus, it is distributed in approximately 60% of the population. The alleles underlined in Table 2 need specific attention for HLA allele matching in unrelated HSCT between the Japanese, as they are present at high frequencies within a serotype (allele family) match.
Table 2.
HLA‐A, ‐C, ‐B, ‐DRB1 allele frequencies in Japana
| HLA‐A serological specificity | HLA‐A allele | GF (%) | HLA‐C serological specificity | HLA‐C allele | GF (%) | HLA‐B serological specificity | HLA‐B allele | GF (%) | HLA‐DRB1 serological specificity | HLADRB1 allele | GF (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A*01:01 | 0.401 | Cw1 | C*01:02 | 17.604 | B7 | B*07:02 | 5.630 | DR1 | DRB1*01:01 | 5.823 |
| A2 | A*02:01 | 11.620 | C*01:03 | 0.318 | B*07:05 | 0.026 | DRB1*01:02 | 0.016 | |||
| A*02:06 | 9.081 | Cw2 | C*02:02 | 0.026 | B*07:31 | 0.005 | DR4 | DRB1*04:05 | 13.491 | ||
| A*02:07 | 3.461 | Cw4 | C*04:01 | 4.410 | B8 | B*08:01 | 0.016 | DRB1*04:06 | 3.388 | ||
| A*02:18 | 0.078 | Cw5 | C*05:01 | 0.381 | B13 | B*13:01 | 1.173 | DRB1*04:03 | 3.055 | ||
| A*02:28 | 0.010 | Cw6 | C*06:02 | 0.808 | B*13:02 | 0.287 | DRB1*04:10 | 2.153 | |||
| A*02:05 | 0.005 | Cw7 | C*07:02 | 12.714 | B18 | B*18:01 | 0.005 | DRB1*04:01 | 0.954 | ||
| A*02:13 | 0.005 | C*07:04 | 0.928 | B22 | B*56:03 | 0.177 | DRB1*04:07 | 0.573 | |||
| A203 | A*02:03 | 0.063 | C*07:01 | 0.073 | B*55:12 | 0.005 | DRB1*04:04 | 0.255 | |||
| A210 | A*02:10 | 0.386 | Cw8 | C*08:01 | 7.355 | B27 | B*27:04 | 0.188 | DRB1*04:02 | 0.005 | |
| A3 | A*03:01 | 0.396 | C*08:03 | 1.361 | B*27:05 | 0.063 | DRB1*04:11 | 0.005 | |||
| A*03:02 | 0.094 | C*08:02 | 0.026 | B*27:06 | 0.005 | DR6 | DRB1*14:45 | 0.005 | |||
| A11 | A*11:01 | 9.117 | Cw9 | C*03:03 | 13.053 | B35 | B*35:01 | 8.263 | DR7 | DRB1*07:01 | 0.339 |
| A*11:02 | 0.219 | Cw10 | C*03:04 | 12.391 | B*35:05 | 0.016 | DR8 | DRB1*08:03 | 8.320 | ||
| A*11:05 | 0.005 | C*03:02 | 0.568 | B*35:04 | 0.005 | DRB1*08:02 | 4.207 | ||||
| A24 | A*24:02 | 36.475 | Cw12 | C*12:02 | 11.182 | B*35:35 | 0.005 | DRB1*08:09 | 0.042 | ||
| A*24:20 | 0.709 | C*12:03 | 0.083 | B37 | B*37:01 | 0.495 | DR9 | DRB1*09:01 | 14.283 | ||
| A*24:08 | 0.042 | Cw14 | C*14:02 | 6.871 | B38 | B*38:02 | 0.281 | DR10 | DRB1*10:01 | 0.474 | |
| A*24:07 | 0.021 | C*14:03 | 6.688 | B*38:01 | 0.016 | DR11 | DRB1*11:01 | 2.518 | |||
| A*24:04 | 0.016 | Cw15 | C*15:02 | 3.081 | B39 | B*39:04 | 0.209 | DRB1*11:06 | 0.016 | ||
| A*24:25 | 0.010 | C*15:05 | 0.016 | B*39:23 | 0.031 | DRB1*11:08 | 0.005 | ||||
| A*24:05 | 0.005 | C*15:10 | 0.005 | B*39:05 | 0.005 | DR12 | DRB1*12:01 | 3.774 | |||
| A2403 | A*24:03 | 0.005 | Cw17 | C*17:01 | 0.010 | B3901 | B*39:01 | 3.321 | DRB1*12:02 | 1.814 | |
| A26 | A*26:01 | 7.350 | undefined | C*03:23 | 0.010 | B3902 | B*39:02 | 0.308 | DR13 | DRB1*13:02 | 5.948 |
| A*26:03 | 2.288 | C*03:43 | 0.010 | B41 | B*41:02 | 0.005 | DRB1*13:01 | 0.589 | |||
| A*26:02 | 1.809 | C*07:15 | 0.010 | B44 | B*44:03 | 6.751 | DRB1*13:07 | 0.005 | |||
| A*26:05 | 0.068 | C*03:64 | 0.005 | B*44:02 | 0.375 | DR14 | DRB1*14:54 | 3.274 | |||
| A*26:06 | 0.010 | C*04:15 | 0.005 | B45 | B*45:01 | 0.005 | DRB1*14:05 | 2.090 | |||
| A29 | A*29:01 | 0.010 | C*16:04 | 0.005 | B46 | B*46:01 | 4.765 | DRB1*14:06 | 1.303 | ||
| A30 | A*30:01 | 0.209 | B*46:02 | 0.005 | DRB1*14:07 | 0.109 | |||||
| A*30:04 | 0.021 | B48 | B*48:01 | 2.878 | DRB1*14:02 | 0.057 | |||||
| A31 | A*31:01 | 8.403 | B50 | B*50:01 | 0.016 | DRB1*14:12 | 0.026 | ||||
| A32 | A*32:01 | 0.021 | B51 | B*51:01 | 8.925 | DRB1*14:29 | 0.016 | ||||
| A33 | A*33:03 | 7.507 | B5102 | B*51:02 | 0.235 | DR1403 | DRB1*14:03 | 1.569 | |||
| A*33:01 | 0.010 | B5103 | B*51:03 | 0.005 | DR1404 | DRB1*14:04 | 0.010 | ||||
| A34 | A*34:01 | 0.010 | B52 | B*52:01 | 11.093 | DR15 | DRB1*15:02 | 10.650 | |||
| A66 | A*66:01 | 0.005 | B53 | B*53:01 | 0.005 | DRB1*15:01 | 7.778 | ||||
| A68 | A*68:01 | 0.010 | B54 | B*54:01 | 7.491 | DRB1*15:04 | 0.005 | ||||
| Null | A*02:53N | 0.010 | B55 | B*55:02 | 2.648 | DR16 | DRB1*16:02 | 0.897 | |||
| Undefined | A*24:46 | 0.010 | B*55:04 | 0.130 | DR17 | DRB1*03:01 | 0.146 | ||||
| A*11:43 | 0.005 | B56 | B*56:01 | 0.850 | undefined | DRB1*04:57 | 0.005 | ||||
| A*24:88 | 0.005 | B57 | B*57:01 | 0.005 | DRB1*08:23 | 0.005 | |||||
| A*26:18 | 0.005 | B58 | B*58:01 | 0.579 | |||||||
| A*31:11 | 0.005 | B59 | B*59:01 | 2.043 | |||||||
| B60 | B*40:01 | 5.348 | |||||||||
| B*40:07 | 0.010 | ||||||||||
| B*40:52 | 0.005 | ||||||||||
| B61 | B*40:02 | 7.945 | |||||||||
| B*40:06 | 4.791 | ||||||||||
| B*40:03 | 0.407 | ||||||||||
| B*40:50 | 0.016 | ||||||||||
| B*40:53 | 0.005 | ||||||||||
| B*40:11 | 0.005 | ||||||||||
| B62 | B*15:01 | 7.585 | |||||||||
| B*15:07 | 0.652 | ||||||||||
| B*15:27 | 0.109 | ||||||||||
| B*15:28 | 0.016 | ||||||||||
| B*15:35 | 0.016 | ||||||||||
| B*15:25 | 0.010 | ||||||||||
| B*15:38 | 0.005 | ||||||||||
| B64 | B*14:01 | 0.016 | |||||||||
| B65 | B*14:02 | 0.010 | |||||||||
| B67 | B*67:01 | 1.225 | |||||||||
| B71 | B*15:18 | 1.522 | |||||||||
| B75 | B*15:11 | 0.881 | |||||||||
| B*15:02 | 0.031 | ||||||||||
| B77 | B*15:13 | 0.005 | |||||||||
| B78 | B*78:02 | 0.005 | |||||||||
| B81 | B*81:01 | 0.005 | |||||||||
| Unknownb | B*54:21b | 0.005 | |||||||||
| Null | B*15:26N | 0.005 | |||||||||
| undefined | B*35:64 | 0.005 | |||||||||
| B*51:36 | 0.005 | ||||||||||
| B*52:05 | 0.005 | ||||||||||
| B*52:11 | 0.005 |
Haplotype segregation analysis based on family study
Haplotype frequencies of HLA‐A‐C‐B‐DRB1
Table 3 lists 60 haplotypes with frequencies higher than 0.2% in the population. Approximately, 38% of the entire Japanese population is expected to carry one or two of the five most common haplotypes.
Table 3.
HLA–A–C–B–DRB1 haplotype frequencies in Japanesea
| A–C–B–DRB1 | HF (%) | RD | A–C–B–DRB1 | HF (%) | RD |
|---|---|---|---|---|---|
| A*24:02–C*12:02–B*52:01–DRB1*15:02 | 8.377 | 0.79 | A*02:07–C*01:02–B*46:01–DRB1*09:01 | 0.308 | 0.09 |
| A*33:03–C*14:03–B*44:03–DRB1*13:02 | 4.473 | 0.75 | A*33:03–C*03:02–B*58:01–DRB1*13:02 | 0.308 | 0.54 |
| A*24:02–C*07:02–B*07:02–DRB1*01:01 | 3.722 | 0.66 | A*26:01–C*03:03–B*35:01–DRB1*04:10 | 0.302 | 0.14 |
| A*24:02–C*01:02–B*54:01–DRB1*04:05 | 2.539 | 0.33 | A*02:01–C*03:03–B*15:11–DRB1*09:01 | 0.292 | 0.33 |
| A*02:07–C*01:02–B*46:01–DRB1*08:03 | 1.866 | 0.54 | A*24:02–C*04:01–B*15:01–DRB1*04:06 | 0.292 | 0.09 |
| A*11:01–C*04:01–B*15:01–DRB1*04:06 | 1.345 | 0.40 | A*24:02–C*14:02–B*51:01–DRB1*14:03 | 0.276 | 0.17 |
| A*24:02–C*01:02–B*59:01–DRB1*04:05 | 1.058 | 0.51 | A*24:02–C*03:04–B*40:01–DRB1*04:05 | 0.266 | 0.04 |
| A*11:01–C*01:02–B*54:01–DRB1*04:05 | 1.001 | 0.13 | A*26:02–C*03:03–B*15:01–DRB1*14:06 | 0.261 | 0.20 |
| A*26:01–C*03:04–B*40:02–DRB1*09:01 | 0.745 | 0.10 | A*24:02–C*03:04–B*40:02–DRB1*04:05 | 0.255 | 0.03 |
| A*24:02–C*08:01–B*40:06–DRB1*09:01 | 0.709 | 0.14 | A*02:01–C*07:02–B*07:02–DRB1*01:01 | 0.250 | 0.04 |
| A*24:02–C*14:02–B*51:01–DRB1*09:01 | 0.652 | 0.09 | A*02:06–C*07:02–B*07:02–DRB1*01:01 | 0.250 | 0.04 |
| A*31:01–C*14:02–B*51:01–DRB1*08:02 | 0.579 | 0.14 | A*11:01–C*01:02–B*54:01–DRB1*08:03 | 0.240 | 0.03 |
| A*33:03–C*14:03–B*44:03–DRB1*08:03 | 0.547 | 0.08 | A*11:01–C*01:02–B*55:02–DRB1*04:05 | 0.240 | 0.09 |
| A*26:02–C*08:01–B*40:06–DRB1*09:01 | 0.542 | 0.30 | A*31:01–C*14:02–B*51:01–DRB1*04:05 | 0.240 | 0.03 |
| A*02:01–C*03:04–B*13:01–DRB1*12:02 | 0.532 | 0.45 | A*02:01–C*15:02–B*51:01–DRB1*15:01 | 0.235 | 0.08 |
| A*24:02–C*01:02–B*46:01–DRB1*08:03 | 0.532 | 0.11 | A*03:01–C*05:01–B*44:02–DRB1*13:01 | 0.235 | 0.63 |
| A*02:06–C*08:01–B*40:06–DRB1*09:01 | 0.464 | 0.10 | A*11:01–C*07:02–B*67:01–DRB1*15:01 | 0.235 | 0.19 |
| A*11:01–C*07:02–B*39:01–DRB1*08:03 | 0.433 | 0.13 | A*24:02–C*14:03–B*44:03–DRB1*13:02 | 0.235 | 0.04 |
| A*26:01–C*03:04–B*40:02–DRB1*08:02 | 0.427 | 0.10 | A*01:01–C*06:02–B*37:01–DRB1*10:01 | 0.229 | 0.57 |
| A*02:06–C*03:03–B*35:01–DRB1*15:01 | 0.422 | 0.05 | A*24:02–C*03:03–B*35:01–DRB1*15:01 | 0.229 | 0.03 |
| A*24:02–C*12:02–B*52:01–DRB1*09:01 | 0.391 | 0.03 | A*24:02–C*03:04–B*40:01–DRB1*11:01 | 0.229 | 0.09 |
| A*31:01–C*14:02–B*51:01–DRB1*14:03 | 0.391 | 0.25 | A*31:01–C*14:02–B*51:01–DRB1*09:01 | 0.229 | 0.03 |
| A*02:06–C*07:02–B*39:01–DRB1*15:01 | 0.386 | 0.12 | A*26:03–C*03:03–B*15:01–DRB1*09:01 | 0.224 | 0.10 |
| A*24:02–C*03:04–B*40:02–DRB1*09:01 | 0.370 | 0.04 | A*02:06–C*01:02–B*54:01–DRB1*04:05 | 0.219 | 0.03 |
| A*02:01–C*01:02–B*54:01–DRB1*04:05 | 0.360 | 0.05 | A*02:06–C*03:03–B*35:01–DRB1*09:01 | 0.219 | 0.02 |
| A*26:03–C*03:03–B*15:01–DRB1*15:01 | 0.344 | 0.15 | A*02:01–C*01:02–B*46:01–DRB1*08:03 | 0.214 | 0.04 |
| A*11:01–C*07:02–B*67:01–DRB1*16:02 | 0.339 | 0.38 | A*02:01–C*08:01–B*40:06–DRB1*09:01 | 0.214 | 0.04 |
| A*02:06–C*01:02–B*59:01–DRB1*04:05 | 0.323 | 0.16 | A*02:06–C*08:01–B*48:01–DRB1*04:07 | 0.214 | 0.37 |
| A*24:02–C*03:03–B*15:07–DRB1*04:03 | 0.323 | 0.50 | A*31:01–C*04:01–B*56:01–DRB1*09:01 | 0.214 | 0.25 |
| A*24:02–C*07:04–B*15:18–DRB1*04:01 | 0.318 | 0.34 | A*31:01–C*07:02–B*07:02–DRB1*01:01 | 0.209 | 0.04 |
HF, haplotype frequencies; HLA, human leucocyte antigen; RD, relative linkage disequilibrium value.
Four‐locus haplotypes with HF >0.2% are listed.
These data have also been submitted to Allele Frequency Net Database (AFND) 22. The four‐loci haplotypes with frequencies equal to or more than 0.01% and the AF can be found at the AFND website, www.allelefrequencies.net 22.
HLA‐A‐B‐DRB1 haplotype sets with the same serotypes
Table 4 lists the sets of three‐loci haplotypes at frequencies of 0.2% or greater, which would have the same serotype.
Table 4.
Haplotype sets assigned same serotype of HLA–A–B–DRB1a
| Set no. | Haplotype | HF (%) |
|---|---|---|
| 1 | A*02:01‐B*07:02‐DRB1*01:01 | 0.26 |
| A*02:06‐B*07:02‐DRB1*01:01 | 0.25 | |
| 2 | A*02:01‐B*46:01‐DRB1*08:03 | 0.23 |
| A*02:07‐B*46:01‐DRB1*08:03 | 1.87 | |
| 3 | A*02:01‐B*54:01‐DRB1*04:05 | 0.38 |
| A*02:06‐B*54:01‐DRB1*04:05 | 0.22 | |
| 4 | A*02:01‐B*40:06‐DRB1*09:01 | 0.29 |
| A*02:06‐B*40:06‐DRB1*09:01 | 0.48 | |
| 5 | A*24:02‐B*40:02‐DRB1*09:01 | 0.53 |
| A*24:02‐B*40:06‐DRB1*09:01 | 0.95 | |
| 6 | A*24:02‐B*15:01‐DRB1*04:06 | 0.31 |
| A*24:02‐B*15:07‐DRB1*04:03 | 0.34 | |
| 7 | A*24:02‐B*35:01‐DRB1*04:03 | 0.23 |
| A*24:02‐B*35:01‐DRB1*04:05 | 0.22 | |
| 8 | A*26:01‐B*40:02‐DRB1*09:01 | 0.81 |
| A*26:01‐B*40:06‐DRB1*09:01 | 0.22 | |
| A*26:02‐B*40:06‐DRB1*09:01 | 0.54 |
HF, haplotype frequency; HLA, human leucocyte antigen.
The analysis objects are HF with more than 0.2%.
Haplotype frequencies of HLA‐C‐B
Table 5 lists 64 haplotypes with frequencies higher than 0.1 % in the population. Half of these haplotypes have RD values more than 0.7, suggesting conservation of HLA‐C‐B linkage. B*40:02 and B*40:06 which correspond to the same serotype (B61) have high frequencies alleles of B61 and need attention for matching in HSCT. Focusing on these, Table 5 shows the frequency of C*03:04‐B*40:02 as 6.26%, while the frequency of B*40:02 is 7.95% (Table 2); therefore, the frequency of C*03:04‐B*40:02 linkage would account for 79% of B*40:02 alleles. Similarly, linkage of B*40:06 with C*08:01 would account for 81% of the presence of B*40:06. Thus, it is important to differentiate serotypes such as B61 when the constituting alleles are in linkage with different HLA‐C alleles.
Table 5.
HLA–C–B haplotype frequencies in Japanesea
| C–B | HF (%) | RD | C–B | HF (%) | RD |
|---|---|---|---|---|---|
| C*12:02–B*52:01 | 10.963 | 0.99 | C*08:01–B*15:18 | 0.495 | 0.27 |
| C*01:02–B*54:01 | 7.147 | 0.94 | C*03:04–B*51:01 | 0.469 | −0.58 |
| C*14:02–B*51:01 | 6.761 | 0.98 | C*04:01–B*35:01 | 0.464 | 0.02 |
| C*14:03–B*44:03 | 6.641 | 0.99 | C*04:01–B*40:01 | 0.464 | 0.05 |
| C*03:04–B*40:02 | 6.256 | 0.76 | C*03:04–B*40:03 | 0.396 | 0.97 |
| C*03:03–B*35:01 | 6.115 | 0.70 | C*05:01–B*44:02 | 0.370 | 0.99 |
| C*07:02–B*07:02 | 5.573 | 0.99 | C*03:04–B*35:01 | 0.339 | −0.67 |
| C*01:02–B*46:01 | 4.316 | 0.89 | C*03:04–B*40:06 | 0.339 | −0.43 |
| C*08:01–B*40:06 | 3.904 | 0.80 | C*15:02–B*40:06 | 0.334 | 0.06 |
| C*07:02–B*39:01 | 3.196 | 0.96 | C*01:03–B*46:01 | 0.318 | 1.00 |
| C*03:03–B*15:01 | 3.190 | 0.33 | C*07:02–B*39:02 | 0.308 | 1.00 |
| C*03:04–B*40:01 | 2.867 | 0.47 | C*15:02–B*40:02 | 0.292 | 0.02 |
| C*04:01–B*15:01 | 2.606 | 0.56 | C*06:02–B*13:02 | 0.287 | 1.00 |
| C*01:02–B*55:02 | 2.252 | 0.82 | C*07:02–B*38:02 | 0.276 | 0.98 |
| C*01:02–B*59:01 | 2.012 | 0.98 | C*03:04–B*15:01 | 0.266 | −0.72 |
| C*08:01–B*48:01 | 1.553 | 0.50 | C*03:03–B*55:02 | 0.261 | −0.25 |
| C*15:02–B*51:01 | 1.345 | 0.38 | C*03:03–B*48:01 | 0.240 | −0.36 |
| C*07:02–B*67:01 | 1.220 | 1.00 | C*01:02–B*56:01 | 0.235 | 0.12 |
| C*03:04–B*13:01 | 1.069 | 0.90 | C*03:03–B*40:01 | 0.235 | −0.66 |
| C*08:01–B*35:01 | 1.069 | 0.07 | C*08:03–B*54:01 | 0.229 | 0.10 |
| C*03:03–B*40:02 | 1.048 | 0.00 | C*01:02–B*51:01 | 0.224 | −0.86 |
| C*07:02–B*40:01 | 0.923 | 0.05 | C*07:02–B*39:04 | 0.209 | 1.00 |
| C*08:03–B*48:01 | 0.923 | 0.67 | C*01:02–B*56:03 | 0.177 | 1.00 |
| C*07:04–B*15:18 | 0.907 | 0.98 | C*15:02–B*51:02 | 0.151 | 0.63 |
| C*03:03–B*15:11 | 0.850 | 0.96 | C*01:02–B*40:02 | 0.141 | −0.90 |
| C*15:02–B*40:01 | 0.704 | 0.18 | C*12:02–B*27:04 | 0.141 | 0.72 |
| C*01:02–B*15:01 | 0.626 | −0.53 | C*15:02–B*15:01 | 0.141 | −0.40 |
| C*03:03–B*15:07 | 0.610 | 0.93 | C*08:03–B*15:01 | 0.136 | 0.03 |
| C*03:02–B*58:01 | 0.568 | 1.00 | C*03:03–B*55:04 | 0.120 | 0.91 |
| C*04:01–B*56:01 | 0.537 | 0.61 | C*07:02–B*40:02 | 0.115 | −0.89 |
| C*07:02–B*15:01 | 0.500 | −0.48 | C*04:01–B*15:27 | 0.109 | 1.00 |
| C*06:02–B*37:01 | 0.495 | 1.00 | C*04:01–B*48:01 | 0.109 | −0.14 |
HF, haplotype frequencies; HLA, human leucocyte antigen; RD, relative linkage disequilibrium value.
C‐B haplotypes with HF >0.1% are listed.
Observed recombination
A total observation number of recombination events were 136 in 134 families. These were divided into two groups: (i) the haplotypes of 103 parents (75.7%) could be determined, (ii) the haplotypes of 33 parents (24.3%) could not be determined and thus were inferred. Table 6 summarizes the observation number of recombination events in informative families which contain the parents and three or more children. The genotypes of these parents are heterozygote at all loci of HLA‐A, ‐B, ‐C, and ‐DRB1. Group (ii) is not included in these informative families. The transmission of recombination (%R/T) shows the HLA‐A‐DRB1 recombination probability as 1.08% per child. Furthermore, Table 6 also indicates the recombination probabilities of HLA‐A‐C and B‐DRB1 are 0.54%.
Table 6.
Number of HLA–A/C, C/B, B/DRB1 recombination evens by each family structure
| Number of children | Number of families | A/Ca n (%R/T) | B/DRB1a n (%R/T) | A/DRB1a n (%R/T) | |
|---|---|---|---|---|---|
| Parents | 5 | 5 | b | b | b |
| 4 | 28 | 2 (0.89) | 1 (0.45) | 3 (1.34) | |
| 3 | 233 | 7 (0.50) | 8 (0.57) | 15 (1.07) | |
| Total | 836 | 266 | 9 (0.54) | 9 (0.54) | 18 (1.08) |
HLA, human leucocyte antigen.
X/Y, number of recombination between X and Y; %R/T, % of recombination per transmission.
No observation of recombination.
Comparison to the result by MLE
Table 7 shows the haplotype frequencies of HLA‐A‐C‐B‐DRB1 based on family study (result‐FS) and based on MLE (result‐MLE) with frequencies more than 0.5%. In the frequent haplotypes with frequencies not less than 0.12%, result‐MLE tends to be higher than result‐FS. In the low‐frequent haplotypes with less than 0.12%, result‐MLE tends to be lower than result‐FS. In addition, result‐MLE could not be detected in 585 haplotypes of the 2099 haplotypes with frequencies less than 0.12% in result‐FS.
Table 7.
Comparison to maximum likelihood estimation (n = 4500)a
| A–C–B–DRB1 | FS (%) | MLE (%) |
|---|---|---|
| A*24:02‐C*12:02‐B*52:01‐DRB1*15:02 | 8.144% | 8.298% |
| A*33:03‐C*14:03‐B*44:03‐DRB1*13:02 | 4.444% | 4.478% |
| A*24:02‐C*07:02‐B*07:02‐DRB1*01:01 | 3.689% | 3.857% |
| A*24:02‐C*01:02‐B*54:01‐DRB1*04:05 | 2.344% | 2.490% |
| A*02:07‐C*01:02‐B*46:01‐DRB1*08:03 | 2.044% | 2.053% |
| A*11:01‐C*04:01‐B*15:01‐DRB1*04:06 | 1.322% | 1.326% |
| A*24:02‐C*01:02‐B*59:01‐DRB1*04:05 | 1.100% | 1.167% |
| A*11:01‐C*01:02‐B*54:01‐DRB1*04:05 | 0.833% | 0.815% |
| A*26:01‐C*03:04‐B*40:02‐DRB1*09:01 | 0.778% | 0.892% |
| A*24:02‐C*08:01‐B*40:06‐DRB1*09:01 | 0.733% | 0.830% |
| A*24:02‐C*14:02‐B*51:01‐DRB1*09:01 | 0.722% | 0.803% |
| A*31:01‐C*14:02‐B*51:01‐DRB1*08:02 | 0.589% | 0.619% |
| A*33:03‐C*14:03‐B*44:03‐DRB1*08:03 | 0.578% | 0.589% |
| A*26:02‐C*08:01‐B*40:06‐DRB1*09:01 | 0.556% | 0.526% |
| A*24:02‐C*01:02‐B*46:01‐DRB1*08:03 | 0.522% | 0.504% |
| A*02:01‐C*03:04‐B*13:01‐DRB1*12:02 | 0.511% | 0.502% |
FS, haplotype frequencies based on family study; MLE, haplotype frequencies estimated by maximum likelihood estimation.
Four‐locus haplotypes with frequencies >0.5% in result‐FS are listed.
On the allele data used for this comparison, four loci showed Hardy–Weinberg Equilibrium: the P‐values of the exact test at HLA‐A, ‐B, ‐C, and DRB1 loci were 0.3184, 0.2557, 0.1449, and 0.4998, respectively.
Discussion
The AF presented in Table 2 show the high frequency of A*24:02 at 36.48%; therefore, around 60% of the population could be administered peptide vaccines such as WT1 peptide vaccine presented by HLA molecules coded A*24:02 23, 24. Moreover, around 80% of the population could be administered the vaccines 25, 26 presented by HLA molecules coded by one of three alleles as A*24:02 (36.5%), A*02:01 (11.6%), and A*02:06 (9.1%).
The haplotype frequencies are also characteristic of Japanese population. The haplotypes of high frequencies are well conserved. Approximately one third (38%) of Japanese population carry the five most common haplotypes. We believe that this is strongly influenced by the founder effect: the ancestors having the common haplotypes migrated to Japan, thus, their haplotypes have been concentrated. While searching in the AFND 22, we found that the common haplotypes are homologous to those residing in the neighboring countries, especially in South Korea 11, 12, 27, 28, 29, 30, 31. In Korean population, the frequent haplotypes are similar to the ones in Japanese population. The three major Japanese haplotypes A*24:02‐C*12:02‐B*52:01‐DRB1*15:02, A*33:03‐C*14:03‐B*44:03‐DRB1*13:02, and A*24:02‐C*07:02‐B*07:02‐DRB1*01:01 have also high frequencies in Korea (1.9%, 4.2%, 2.9%, respectively) 27, suggesting the migration of some ancestors though the Korean Peninsula.
The haplotype analysis not only helps in understanding the history of human migration but also in matching for unrelated donor searches for HSCT. In JMDP, the HLA compatibility with a donor is evaluated by both serotype and genotype. Furthermore, allele matching is a better evaluation of compatibility compared to the serotype matching; the rejection and GVHD risk of bone marrow transplant (BMT) have been found to be lower with allele‐level matching compared to the serotype‐level matching 3, 32. Accordingly, focusing on the alleles in every serotype, A2, A26, etc. increases the possibility of allele mismatch in spite of serotype match (Table 2). As shown in Table 4, HLA‐A2, B61, B15, and DR4 especially increase the allele mismatch risk. However, compared with South Korean haplotype analysis, South Korea has 15 sets of haplotypes to pay attention to in HSCT matching 27 but Japan has eight sets (Table 4). It also shows that for most HLA haplotypes at the serotype‐level, HLA haplotype matching is almost allele‐level matching in HSCT between the Japanese.
Although HLA‐C locus is not indispensable for registering information in HSCT, HLA‐C allele matching is important 32, 33. Even without information of donor HLA‐C allele, the C‐B haplotypes can predict HLA‐C allele, although not always, because of well‐conserved C‐B linkage (Table 5); the conservation is possible due to short genetic linkage distance. In other words, HLA‐B allele matching increases the possibility of HLA‐C allele matching. Accordingly, analysis in the HLA distribution in Japanese population may contribute in planning the strategies of HLA matching for HSCT.
Result‐MLE was similar to result‐FS (Table 7). Table 7 indicates the haplotype frequencies estimated by the software are very similar to real family‐derived haplotypes. If the detection of the low frequency haplotypes is needed, determination of haplotypes by descent or a large sample size appears to be necessary.
In conclusion, this study of determination of HLA allele and haplotype frequencies using family samples not only serves as a tool for elucidating linkage of each HLA locus but also acts as a tool in detecting HLA gene mutations in human germ cells such as recombination. The data obtained in this study will be useful in various fields such as anthropology, transplantation therapy, and disease association studies.
Conflicts of interest
The authors have no conflicts of interest.
Acknowledgments
We would like to thank Jun Ohashi from Department of Biological Sciences, Graduate School of Science, The University of Tokyo, for valuable comments and suggestions about choosing unrelated samples and the exact test for deviation from Hardy–Weinberg equilibrium.
The copyright line for this article was changed on 23 September 2016 after original online publication.
References
- 1. Cao K, Hollenbach J, Shi X et al. Analysis of the frequencies of HLA‐A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol 2001: 62: 1009–30. [DOI] [PubMed] [Google Scholar]
- 2. Imanishi T, Wakisaka A, Gojobori T. Genetic relationships among various human populations indicated by MHC polymorphisms In: Tsuji K, Aizawa M, Sasazuki T, eds. HLA 1991. Oxford: Oxford University Press, 1992, 627–32. [Google Scholar]
- 3. Lee SJ, Klein J, Haagenson M et al. High‐resolution donor‐recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood 2007a: 110: 4576–83. [DOI] [PubMed] [Google Scholar]
- 4. Horan J, Wang T, Haagenson M et al. Evaluation of HLA matching in unrelated hematopoietic stem cell transplantation for nonmalignant disorders. Blood 2012: 120: 2918–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morishima S, Ogawa S, Matsubara A et al. Impact of highly conserved HLA haplotype on acute graft‐versus‐host disease. Blood 2010: 115: 4664–70. [DOI] [PubMed] [Google Scholar]
- 6. Nakaoka H, Mitsunaga S, Hosomichi K et al. Detection of ancestry informative HLA alleles confirms the admixed origins of Japanese population. PLoS One 2013: 8: e60793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tokunaga K, Ishikawa Y, Ogawa A et al. Sequence‐based association analysis of HLA class I and II alleles in Japanese supports conservation of common haplotypes. Immunogenetics 1997: 46: 199–205. [DOI] [PubMed] [Google Scholar]
- 8. Saito S, Ota S, Yamada E, Inoko H, Ota M. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens 2000: 56: 522–9. [DOI] [PubMed] [Google Scholar]
- 9. Moriyama Y, Kato K, Mura T, Juji T. Analysis of HLA gene frequencies and HLA haplotype frequencies for bone marrow donors in Japan. MHC 2006: 12: 183–201. [Google Scholar]
- 10. Ogata S, Shi L, Matsushita M et al. Polymorphisms of human leucocyte antigen genes in Maonan people in China. Tissue Antigens 2007: 69: 154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao Y, Shi L, Shi L et al. Distribution of HLA‐A, ‐B, ‐Cw, and ‐DRB1 alleles and haplotypes in an isolated Han population in Southwest China. Tissue Antigens 2009: 73: 561–8. [DOI] [PubMed] [Google Scholar]
- 12. Shi L, Yao YF, Shi L et al. HLA alleles and haplotypes distribution in Dai population in Yunnan province, Southwest China. Tissue Antigens 2010: 75: 159–65. [DOI] [PubMed] [Google Scholar]
- 13. Shichi D, Kikkawa EF, Ota M et al. The haplotype block, NFKBIL1‐ATP6V1G2‐BAT1‐MICB‐MICA, within the class III ‐ class I boundary region of the human major histocompatibility complex may control susceptibility to hepatitis C virus‐associated dilated cardiomyopathy. Tissue Antigens 2005: 66: 200–8. [DOI] [PubMed] [Google Scholar]
- 14. Kamatani Y, Wattanapokayakit S, Ochi H et al. A genome‐wide association study identifies variants in the HLA‐DP locus associated with chronic hepatitis B in Asians. Nat Genet 2009: 41: 591–5. [DOI] [PubMed] [Google Scholar]
- 15. Zhao H, Qi Y, Wang Y et al. Interactive contribution of serine/threonine kinase 39 gene multiple polymorphisms to hypertension among northeastern Han Chinese. Sci Rep 2014: 4: 5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang ZT, Hu JJ, Fan R, Zhou J, Zhong J. RAGE gene three polymorphisms with Crohn's disease susceptibility in Chinese Han population. World J Gastroenterol 2014: 20: 2397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lewontin RC. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics 1964: 49: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics 1987: 117: 331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Heredity 1995: 86: 248–9. [Google Scholar]
- 20. Rousset F. GENEPOP'007: a complete re‐implementation of the GENEPOP software for Windows and Linux. Mol Ecol 2007: 8: 103–6. [DOI] [PubMed] [Google Scholar]
- 21. Marsh SGE, Albert ED, Bodmer WF et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 2010: 75: 291–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonzalez‐Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: A database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res 2011: 39: D913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsuboi A, Oka Y, Udaka K et al. Enhanced induction of human WT1‐specific cytotoxic T lymphocytes with a 9‐mer WT1 peptide modified at HLA‐A*2402‐binding residues. Cancer Immunol Immunother 2002: 51: 614–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oka Y, Tsuboi A, Taguchi T et al. Induction of WT1 (Wilms' tumor gene)‐specific cytotoxic T lymphocytes by WT1 peptide vaccine and the resultant cancer regression. Proc Natl Acad Sci U S A 2004: 101: 13885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaida M, Morita‐Hoshi Y, Soeda A et al. Phase 1 trial of Wilms tumor 1 (WT1) peptide vaccine and gemcitabine combination therapy in patients with advanced pancreatic or biliary tract cancer. J Immunother 2011: 34: 92–9. [DOI] [PubMed] [Google Scholar]
- 26. Ohno S, Okuyama R, Aruga A, Sugiyama H, Yamamoto M. Phase I trial of Wilms' Tumor 1 (WT1) peptide vaccine with GM‐CSF or CpG in patients with solid malignancy. Anticancer Res 2012: 32: 2263–9. [PubMed] [Google Scholar]
- 27. Lee KW, Oh DH, Lee C, Yang SY. Allelic and haplotypic diversity of HLA‐A, ‐B, ‐C, ‐DRB1, and ‐DQB1 genes in the Korean population. Tissue Antigens 2005: 65: 437. [DOI] [PubMed] [Google Scholar]
- 28. Yang G, Deng YJ, Hu SN et al. HLA‐A, ‐B, and ‐DRB1 polymorphism defined by sequence based typing of the Han population in Northern China. Tissue Antigens 2006: 67: 146–52. [DOI] [PubMed] [Google Scholar]
- 29. Chen S, Hu Q, Xie Y et al. Origin of Tibeto‐Burman speakers: evidence from HLA allele distribution in Lisu and Nu inhabiting Yunnan of China. Hum Immunol 2007: 68: 550–9. [DOI] [PubMed] [Google Scholar]
- 30. Lai MJ, Wen SH, Lin YH et al. Distributions of human leukocyte antigen‐A, ‐B, and ‐DRB1 alleles and haplotypes based on 46,915 Taiwanese donors. Hum Immunol 2010: 71: 777–82. [DOI] [PubMed] [Google Scholar]
- 31. Wen SH, Lai MJ, Yang KL. Human leukocyte antigen‐A, ‐B, and ‐DRB1 haplotypes of cord blood units in the Tzu Chi Taiwan Cord Blood Bank. Hum Immunol 2008: 69: 430–6. [DOI] [PubMed] [Google Scholar]
- 32. Flomenberg N, Baxter‐Lowe LA, Confer D et al. Impact of HLA class I and class II high‐resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA‐C mismatching is associated with a strong adverse effect on transplantation outcome. Blood 2004: 104: 1923–30. [DOI] [PubMed] [Google Scholar]
- 33. Kanda Y, Kanda J, Atsuta Y et al. Impact of a single human leucocyte antigen (HLA) allele mismatch on the outcome of unrelated bone marrow transplantation over two time periods. A retrospective analysis of 3003 patients from the HLA Working Group of the Japan Society for Blood and Marrow Transplantation. Br J Haematol 2013: 161: 566–77. [DOI] [PubMed] [Google Scholar]


