Abstract
Eukaryotic precursor‐messenger RNAs (pre‐mRNAs) undergo splicing to remove intragenic regions (introns) and ligate expressed regions (exons) together. Unlike exons in the mature messenger RNAs (mRNAs) that are used for translation, introns that are spliced out of pre‐mRNAs were generally believed to lack function and to be degraded. However, recent studies have revealed that a large group of spliced introns can escape complete degradation and are processed to generate noncoding RNAs (ncRNAs), including different types of small RNAs, long‐noncoding RNAs, and circular RNAs. Strikingly, exonic sequences can be also back‐spliced from pre‐mRNAs to form stable circular RNAs. Together, the findings that ncRNAs can be spliced out of mRNA precursors not only expand the ever‐growing repertoire of ncRNAs that originate from different genomic regions, but also reveal the unexpected transcriptomic complexity and functional capacity of eukaryotic genomes. WIREs RNA 2015, 6:651–660. doi: 10.1002/wrna.1307
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
A fundamental feature of eukaryotic protein‐coding genes is that they are in pieces.1 It is crucial that intragenic regions (introns1) are spliced out of the precursor‐messenger RNA (pre‐mRNA) and expressed regions (exons1) are ligated together to form a final mature messenger RNA (mRNA) that encodes for a protein. The pre‐mRNA splicing offers flexibility in regulating gene expression,1 and in higher eukaryotes, alternative splicing of a single pre‐mRNA yields multiple mature mRNAs and therefore multiple protein products.2 Genome‐wide studies have suggested that nearly all human multiexonic protein‐coding genes undergo alternative splicing3, 4 to significantly increase the transcriptomic/proteomic complexity and hence their functional diversity.2, 5 Additionally, large/long‐intergenic/intervening noncoding RNAs (lincRNAs) were recently demonstrated to be alternatively spliced as well,6 although their splicing efficiency is relatively low.7 (For simplicity, this review will use pre‐RNA to refer to an unspliced precursor RNA irrespective of whether the primary transcript is used to generate an mRNA or lincRNA.)
Unlike the ligated exons in the mature RNAs, the intron lariats are generally debranched and ultimately degraded after splicing (Figure 1(a)). From a coding perspective, introns were generally regarded as ‘junk’ as they do not influence the sequence of the end product.8 However, it is now widely recognized that introns are not just passively removed during splicing, but play important roles in regulation of gene expression. Introns harbor diverse cis‐regulatory elements that affect pre‐RNA splicing,5 and have various fates that affect gene expression. For instance, introns can be retained in the final RNA product. A recent study reported that intron retention can tune mammalian transcriptomes by suppression of inappropriately expressed transcripts.9 Self‐splicing group I and group II introns are catalytically active as ribozyme to guide their own excision.10, 11, 12 These self‐splicing group I and group II intronic sequences, after spliced out, can be further processed to yield circular molecules10, 13; however, such circular transcripts are unstable and their functions are at best limited.14, 15
Figure 1.
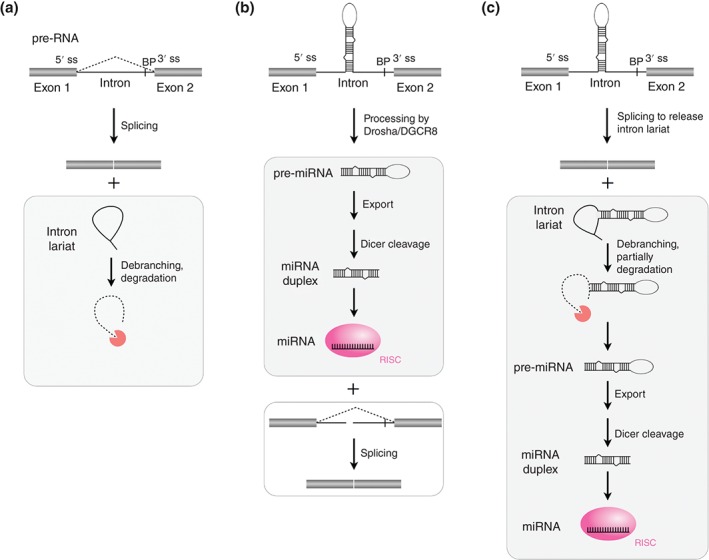
MicroRNAs (MiRNAs) are processed from spliced introns. (a) Eukaryotic precursor RNAs (pre‐RNAs) undergo splicing (dash lines) to remove introns (lines) and ligate exons (bars) together to form either mature mRNAs that are subsequently translated or noncoding RNAs. After splicing, intron lariats are generally debranched and ultimately degraded. ss, splice site. BP, branchpoint. (b) Drosha/DGCR8‐dependent model of canonical mirtron processing. (c) Splicing‐dependent model of mirtron processing. Notably, some mirtrons that derive from small introns have the hairpin exactly ending at the splice sites to resemble pre‐miRNAs, thus do not need to be trimmed by exonucleases.40, 49
In addition to generating RNA circles from self‐splicing introns10, 13 and tRNA introns,16 a variety of noncoding RNAs (ncRNAs) are processed from nuclear pre‐RNA through the spliceosomal pathway. These intragenic ncRNAs include many, but not all, microRNAs (miRNAs),17, 18 small nucleolar RNAs (snoRNAs),19, 20 RNase P RNA subunit,21 new type of long‐noncoding RNAs (lncRNAs)22 and circular RNAs from either excised introns23 or excised exons.24, 25 Different from ncRNAs independently transcribed from intergenic loci by RNA polymerase II (RNA Pol II),6, 26 the expression of these intragenic ncRNAs is dependent on the transcription and splicing of their host pre‐RNAs. Importantly, such ncRNAs play important roles in altering gene expression both in cis 23 and in trans.22, 27, 28 This review focuses on the biogenesis of intragenic ncRNA species excised from the inside of nuclear pre‐RNAs by splicing.
A LARGE NUMBER OF miRNAs ARE PROCESSED FROM SPLICED INTRONS
The miRNAs are endogenous ncRNA species of ~22 nts that function as guide molecules in post‐transcriptional gene silencing.28, 29, 30 The miRNAs play a key role in both physiological and pathological processes, such as self‐renewal of embryonic stem cells (ESCs), development, and cancers.31 In the canonical mammalian miRNA biogenesis pathway, RNA Pol II transcribes a primary miRNA (pri‐miRNA) transcript, which is 5′ capped and 3′ polyadenylated.26, 32 This pri‐miRNA is processed by Drosha/DGCR8 microprocessor to produce a miRNA precursor (pre‐miRNA).33, 34 Through an association with Exportin‐5, the pre‐miRNA is subsequently exported from nucleus to cytoplasm,35 where this hairpin intermediate is cleaved by Dicer to yield a miRNA/miRNA* duplex.36, 37 Finally, the guide (but not the passenger) strand of the miRNA/miRNA* duplex is incorporated into the RNA‐induced silencing complex (RISC) to repress gene expression based on miRNA–mRNA sequence complementarity.38, 39, 40
In addition to the canonical biogenesis pathway, miRNAs can be also produced from introns of protein‐coding genes (termed as mirtrons) in both invertebrate41, 42 and mammals.43, 44 Indeed, a significant population of human and murine miRNAs originate from mirtrons.45, 46, 47 A Drosha/DGCR8‐dependent and splicing‐independent model has been proposed to yield mirtrons. In this model, the intronic pre‐miRNA hairpin is cleaved from the pre‐mRNA by Drosha/DGCR8 prior to the splicing catalysis,18 and then enters the miRNA biogenesis pathway (Figure 1(b)). Interestingly, the split intron by Drosha/DGCR8 showed little effect on the following exon linkage or mRNA maturation.18 As located within host genes, the expression of some mirtrons is coregulated by transcription and splicing of their host pre‐RNAs.48 In another splicing‐dependent pathway (Figure 1(c)), after cotranscribed with the host gene, the pre‐miRNA hairpin is excised out of host pre‐RNA with spliceosome, trimmed by exonucleases (for tailed mirtrons only) and exported to the cytoplasm where it can be further processed by Dicer.40, 49 Notably, some mirtrons that derive from small introns have the hairpin exactly ending at the splice sites to resemble pre‐miRNAs, thus do not need to be trimmed by exonucleases.40, 49
Besides miRNAs, some of Piwi‐interacting RNAs (piRNAs) and endogenous small interfering RNAs (siRNAs) are also likely to be generated from introns and exons,46 while the detailed mechanisms require further investigation. Another type of house‐keeping ncRNA, the catalytic RNA subunit of RNase P (RPR), has been reported to be processed from the last intron of an RNA Pol II transcript of the gene ATPsynC in insects/crustaceans,21 while other animal RPR genes are independently transcribed from RNA polymerase III. The evolutionary driving force for this divergence over 500 million years ago is unknown.21
THE MAJORITY OF HUMAN snoRNAs ARE PROCESSED FROM SPLICED INTRONS
SnoRNAs are a family of conserved nuclear ncRNAs (~70–200 nts in length) that are usually located in nucleoli and participate in the modification of small nuclear RNAs (snRNAs)/ribosomal RNAs (rRNAs) or in the processing of rRNAs during ribosomal maturation.27, 50, 51 Two types of snoRNAs, box C/D and box H/ACA snoRNAs, are defined by their conserved sequence motifs.27 Hundreds of human cellular sno and scaRNAs (snoRNA variants that localize to Cajal bodies) have been annotated by snoRNA‐LBME‐db.52 In yeast, most snoRNAs are produced from independent transcripts by RNA Pol II.20 While in human, only a small portion of annotated snoRNAs is likely produced as independent RNA Pol II transcripts. Instead, the vast majority of human snoRNAs reside within introns of their host (coding or noncoding) genes.52, 53 During splicing and exonucleolytic trimming from debranched introns, the assembly with the snoRNA‐associated proteins (snoRNPs) protects the snoRNA sequences from further exonucleolytic degradation27, 54, 55 (Figure 2 (a)). The processing of intronic snoRNA is coupled to splicing; indeed, snoRNAs positioned about 70 nts upstream to the 3′ splice site is critical for efficient expression.56 In addition, the expression of individual snoRNAs from multi‐snoRNA host genes is coordinated with alternative splicing and nonsense‐mediated RNA decay (NMD), resulting in unbalanced expressions of intronic snoRNAs and their cognate spliced RNA from the same host gene locus.57
Figure 2.
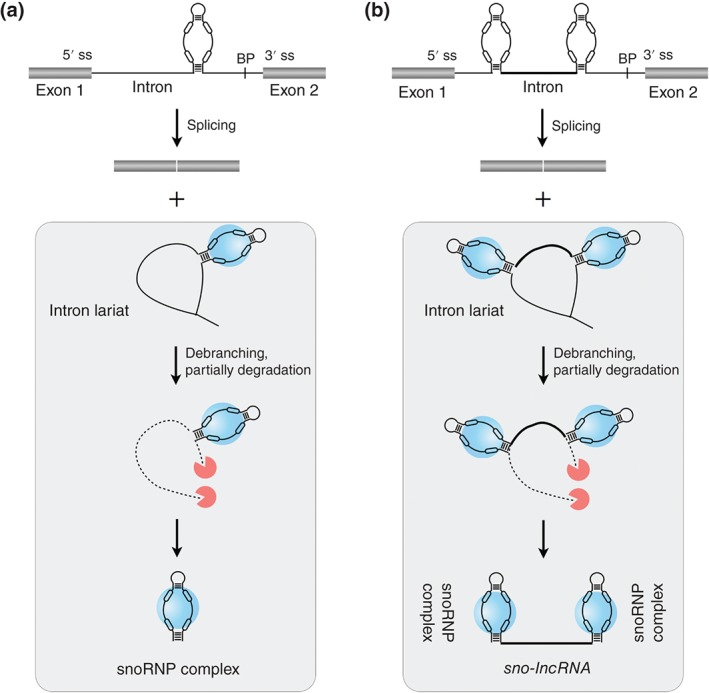
Small nucleolar RNAs (SnoRNAs) and snoRNA‐ended long‐noncoding RNAs (sno‐lncRNAs) are processed from spliced introns. (a) SnoRNAs are processed from spliced introns. During splicing and exonucleolytic trimming from debranched introns, the assembly of snoRNA with the snoRNA‐associated proteins (snoRNPs, blue spheres) protects it from further exonucleolytic degradation and leads to the formation of mature snoRNPs. (b) Sno‐lncRNAs are processed from spliced introns and flanked with snoRNAs at both ends. Introns containing two snoRNAs are processed from their ends by the snoRNP machinery (blue spheres) and the intronic sequences between these two snoRNAs are protected, thus leading to the formation of lncRNAs with snoRNA ends.
The aberrant expression of snoRNAs and their associated proteins is linked to human diseases. An extreme example of noncoding genes with snoRNAs is located at the human imprinted 15q11‐q13 locus, which has been implicated in Prader‐Willi/Angelman syndrome (PWS).19, 20 Within this imprinted region, two clusters of tandemly repeated snoRNAs (29 SNORD116s and 42 SNORD115s) and several single snoRNA genes (such as SNORD109A) are processed from downstream introns of a gigantic, 470‐knt long‐paternal transcript.19, 58 The minimal paternal deletion region associated with PWS (108 kb) removes SNORD109A, the SNORD116 cluster of 29 similar snoRNAs and Imprinted in Prader‐Willi syndrome ncRNA (IPW), and the most current published model suggests that the deficiency of SNORD116s is associated with PWS.59, 60, 61 However, although most snoRNAs guide rRNA or snRNA modifications by a base pairing mechanism, SNORD116s show minimal complementarity to rRNAs or snRNAs,58 and thus are unlikely to function in guiding rRNA/snRNA modification. In this case, the molecular mechanism of how SNORD116 snoRNAs are possibly involved in the PWS remains elusive.
PAIRED snoRNAs STABILIZE A NEW TYPE OF lncRNAs FROM SPLICED INTRONS
While most introns are unstable after being spliced out of pre‐RNAs, a large number of lncRNA candidates have been predicted by computational analysis to originate from postspliced introns.46 In addition, profiling of the nonpolyadenylated (poly(A)−) RNAs have revealed mature RNA transcripts from excised introns,62 such as sno‐lncRNAs.22 Unlike the majority of lncRNAs that contain 5′ cap structures and 3′ poly(A) tails, sno‐lncRNA is a new type of lncRNAs that are derived from spliced introns and are flanked by snoRNAs at both termini.22 As they do not contain poly(A) tails at their 3′ ends, sno‐lncRNAs have been missed by most polyadenylated (poly(A)+) RNA‐seq.62 Mechanically, after splicing, introns containing two snoRNAs are processed from their ends by the snoRNP machinery and the internal intronic sequences between the two snoRNAs are protected, leading to the formation of lncRNAs with snoRNA ends22 (Figure 2 (b)).
Sno‐lncRNAs are widely expressed in cells and tissues and can be produced by either box C/D or box H/ACA snoRNAs in human genome.22, 53 Strikingly, the most abundant sno‐lncRNAs in human embryonic stem cells (hESCs) reside in the PWS deletion region.22 There are five sno‐lncRNAs that are produced from the SNORD116 cluster in hESCs. Rather than localizing to nucleoli or Cajal bodies, PWS‐region sno‐lncRNAs strongly accumulate near to their sites of synthesis, suggesting that they are functionally different from snoRNAs. Importantly, these PWS‐region sno‐lncRNAs regulate alternative splicing by interacting with splicing factor Fox2.22 For example, knocking down these sno‐lncRNAs resulted in the aberrant splicing regulation of known Fox2‐targeted cassette exons, many of which are from genes with a clear connection to neuronal function.22 Likely, in PWS patients where the PWS region sno‐lncRNAs are not expressed due to the paternal deletion, altered patterns of Fox2‐regulated splicing may happen along development, possibly causing neurogenetic disorder in PWS patients. Thus, the finding of PWS region sno‐lncRNAs and their potential role in altering Fox2‐regulated alternative splicing lead to a possible association between a new class of lncRNAs and PWS pathogenesis.
Although the primary sequences are highly conserved from mouse to human, mouse SNORD116s are scattered in individual introns.53 As one intron containing two snoRNAs is a prerequisite for the biogenesis of a sno‐lncRNA,53, 63 the lack of PWS region snoRNA pairs within single introns in the mouse genome may result in undetectable PWS region sno‐lncRNAs in mouse transcriptomes.53 Finally, genome‐wide analysis suggests that only a small portion of paired human snoRNAs are identified in single introns based on the current splicing annotations. Considering the widespread tissue‐/cell‐specific alternative splicing,64, 65 it is reasonable to expect identification of more sno‐lncRNAs when additional RNA‐seq datasets become available.
CIRCULAR RNAs FROM SPLICED INTRONS
In addition to sno‐lncRNAs, another type of intron‐derived ncRNAs, circular intronic RNAs (ciRNAs), has been also identified from poly(A)− RNA‐seq datasets.23 The ciRNAs are produced from excised intron lariats that fail to be debranched after splicing, leading to a covalent circle with 2′,5′‐phosphodiester bond between 5′ splice donor site and the branchpoint site (Figure 3(a)). Different from lariat RNAs containing a variety length of 3′‐tails,66 The ciRNAs are derived from partially processed lariats that do not likely contain 3′ linear appendage,23 as only one sharp band on the native high resolution PAGE (polyacrylamide gel electrophoresis) could be detected with or without RNase R (an enzyme that can degrade linear and Y‐structure RNAs, while preserving the loop portion of a lariat RNA67) treatment.23 Moreover, evidence at both bioinformatic and experimental levels has suggested that the formation of ciRNAs depends on a consensus RNA motif containing a 7‐nt GU rich element near 5′ splice site and an 11‐nt C‐rich element near the branchpoint (Figure 3(a)). However, it is still unclear how these cis‐elements function to resist debranching and what other trans‐factors are involved in this process.
Figure 3.
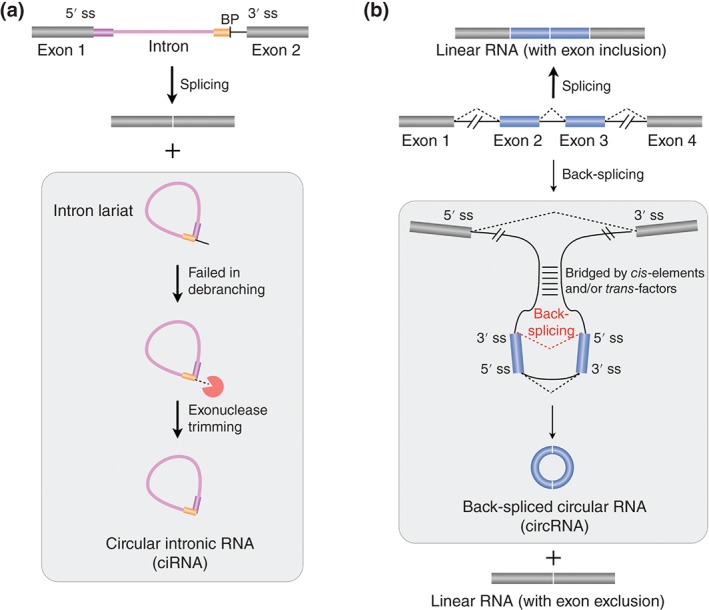
Two types of circular RNAs are processed from excised introns or exons. (a) Circular intronic RNAs (CiRNAs) are processed from excised introns. CiRNAs fail to be debranched after splicing, leading to a covalent circle with 2′,5′‐phosphodiester bond between 5′ splice donor site and the branchpoint site. The formation of ciRNAs depends on a consensus RNA motif containing a 7‐nt GU rich element near 5′ splice site (magenta bar) and an 11‐nt C‐rich element near branchpoint (yellow bar). (b) Back‐spliced circular RNAs (CircRNAs) are processed from excised exons. Different from canonical splicing (dashed lines in black), which ligates an upstream 5′ splice site (5′ ss) with a downstream 3′ splice site (3′ ss) to form a linear RNA (top), back‐splicing (dashed line in red) connects downstream a 5′ ss reversely with an upstream 3′ ss to yield a circular RNA with normal 3′,5′‐phosphodiester bond and an alternatively spliced linear RNA with exon exclusion (bottom). Both complementary sequences and protein factors can facilitate back‐splicing by bridging downstream 5′ ss close to upstream 3′ ss. See text for details.
Intron‐lariat‐derived human ciRNAs are abundantly localized in the nucleus and are largely associated with the nuclear insoluble fractionation.23 Some abundant ciRNAs play a cis‐regulatory role in promoting the transcription of their host genes by associating with the elongation RNA Pol II machinery.23 Additionally, a recent study on some back‐spliced circular RNAs that contain both exons and introns has suggested a similar function on transcription regulation.68 Finally, stable intronic sequence RNAs (sisRNAs) were also revealed from both oocyte nucleus69 and cytoplasm70 of Xenopus tropicalis. However, whether these sisRNAs can form similar circle structures as ciRNAs remains to be further investigated.
CIRCULAR RNAs FROM BACK‐SPLICED EXONS
Profiling of poly(A)− RNAs has surprisingly revealed signals from not only excised introns but also excised exons,62 which were further proven as circular RNAs.24 Genome‐wide analyses with specific computational approaches, which identify junction reads with reversed genomic orientation, successfully identified thousands of circRNAs from back‐spliced exons (circRNAs) in various cell lines and from different species.25, 71, 72, 73, 74, 75 Most circRNA exons are located in the middle of annotated genes25 and excised from pre‐RNA by back‐splicing. Different from canonical splicing that ligates an upstream 5′ splice site (5′ ss) with a downstream 3′ ss to form a linear RNA, back‐splicing connects a downstream 5′ ss with an upstream 3′ ss to yield a circular RNA with 3′,5′‐phosphodiester bond24, 25, 71, 72, 76 (Figure 3(b)). Although catalyzed by the canonical spliceosomal machinery,77 the efficiency for circRNAs formation is often very low, possibly due to the unfavorable spliceosome assembly for back‐splicing.25, 73, 76
Back‐splicing competes with canonical splicing for circRNA biogenesis,76 leading to the ‘lariat intermediate’ or ‘direct back‐splicing’ models.24, 71, 73, 78 The main difference between these two models relates to the question of timing: which takes place first: canonical splicing or back‐splicing?76 In the ‘lariat intermediate’ model, the transcribed pre‐RNA first undergo canonical splicing to generate a linear RNA with skipped exon(s) and a long intron‐lariat intermediate containing these skipped exon(s). This long intron‐lariat intermediate is further processed by back‐splicing to generate a circRNA. In contrast, pre‐RNA might be ‘directly back‐spliced’ to first generate a circRNA and an unusual exon‐intron(s)‐exon intermediate, which can be further processed to linear RNAs with skipped exon(s) or degraded. In fact, both mechanistic possibilities might be used in a context (organism)‐dependent fashion. In lower eukaryotes, such as Schizosaccharomyces pombe, circRNA are suggested to favorably generate through the ‘lariat intermediate’ mechanism with short flanking introns.79 While, in human and mouse, complementary sequences25, 80 (mostly repetitive Alu elements in human) across long flanking introns can facilitate ‘direct back‐splicing’ by bridging downstream 5′ ss close to upstream 3′ ss to generate circRNAs76 (Figure 3(b)). In addition to cis‐elements, RNA‐binding proteins were also reported to regulate circRNA biogenesis.75, 81, 82, 83 It is possible that cis‐elements and trans‐factors might work together to synergistically alter back‐spliced circularization, which requires further investigation.
Despite lowly expressed in general, some circRNAs are more abundant than their linear counterparts.73 It has been recently reported that circRNAs are highly enriched in brain (from fly to mammals) with a potential to regulate synaptic function and to be used as biomarkers74, 83, 84; however, the underlying mechanism for enhanced expression in the brain is largely undetermined. Such a differential expression might reflect an array of possible functions for this new class of RNAs. First, some circRNAs can function as miRNA72, 85 or protein sponges,81 but a large scale analysis revealed that only a limited number of circRNAs can potentially act as sponges for miRNAs.86 Second, with the competition between splicing and back‐splicing,76, 81, 87, 88 The circRNA biogenesis might also regulate the alternative splicing of linear RNAs.15 Third, the potential of circRNAs on translation might further expand the diversity of proteome. Artificial circRNAs with internal ribosome entry sites (IRESs) generated from expression vectors are translatable.89, 90 However, endogenous circRNAs have not yet been reported to associate with ribosomes for translation.71, 86 Finally, similar to the intron‐derived ciRNAs,23 some circRNAs with retained introns can promote transcription of their host genes by interacting with U1 snRNP and RNA Pol II.68 Despite recent studies have revealed some biological roles of certain circRNAs, further investigation is required to gain a comprehensive understanding of what most other circRNAs really do in cells.
CONCLUSION
Although generally believed that intragenic sequences (usually introns) are degraded after splicing and therefore functionally inconsequential, accumulated lines of evidence have shown that some spliced introns can be further processed to produce a variety of ncRNAs, including new types of lncRNAs. In addition, recent studies have shown that intragenic exons can be back‐spliced from inside of pre‐RNAs to form RNAs in circle.25, 71, 72, 73, 74, 81 Apparently, the production of these intragenic ncRNAs (short or long) are largely dependent on splicing to occur, but many questions remain to be addressed. How is the processing of intragenic ncRNAs linked with other RNA processing pathways, including transcription,91 NMD,57 and canonical splicing?25, 77, 81 How are these different pathways coregulated and crosstalked? How are different protein cofactors involved in the entire life cycle of these intragenic ncRNAs?92 Moreover, it appears that the expression of many intragenic ncRNAs is not conserved across species; it will be of particular significance to study how and when such sequences were embedded into or removed from their host genes in evolution. Finally, as many of these intragenic ncRNAs were identified from a combination of high‐throughput sequencing and newly developed computational methods, it will be not surprising to find other types of ncRNAs by applying novel genome‐wide approaches. Collectively, the finding that a ncRNA gene is embedded inside of another gene and can be activated to function by splicing sheds new light on the unanticipated complexity of transcriptome and the multifaceted regulation by splicing.
ACKNOWLEDGMENT
I am grateful to L.‐L. Chen for discussion and V. Gopalan for critical editing of the article. L.Y. is supported by 31271390 and 31471241 from NSFC and 2014CB910600 from MOST.
Conflict of interest: The author has declared no conflicts of interest for this article.
REFERENCES
- 1. Gilbert W. Why genes in pieces? Nature 1978, 271:501. [DOI] [PubMed] [Google Scholar]
- 2. Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high‐throughput sequencing. Nat Genet 2008, 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- 4. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Black DL. Mechanisms of alternative pre‐messenger RNA splicing. Annu Rev Biochem 2003, 72:291–336. [DOI] [PubMed] [Google Scholar]
- 6. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon‐Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011, 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co‐transcriptional in the human genome but inefficient for lncRNAs. Genome Res 2012, 22:1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature 1980, 284:604–607. [DOI] [PubMed] [Google Scholar]
- 9. Braunschweig U, Barbosa‐Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos‐Pournatzis T, Frey B, Irimia M, Blencowe BJ. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 2014, 24:1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self‐splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena . Cell 1982, 31:147–157. [DOI] [PubMed] [Google Scholar]
- 11. Cech TR. Self‐splicing of group I introns. Annu Rev Biochem 1990, 59:543–568. [DOI] [PubMed] [Google Scholar]
- 12. Michel F, Ferat JL. Structure and activities of group II introns. Annu Rev Biochem 1995, 64:435–461. [DOI] [PubMed] [Google Scholar]
- 13. Grabowski PJ, Zaug AJ, Cech TR. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena . Cell 1981, 23:467–476. [DOI] [PubMed] [Google Scholar]
- 14. Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome‐wide discovery of circular RNAs in Archaea . Nucleic Acids Res 2012, 40:3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014, 20:1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo . RNA 2015, 21:1554–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez A, Griffiths‐Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004, 14:1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J 2007, 26:775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC‐SNURF‐SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet 2001, 10:2687–2700. [DOI] [PubMed] [Google Scholar]
- 20. Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol 2002, 14:319–327. [DOI] [PubMed] [Google Scholar]
- 21. Manivannan SN, Lai LB, Gopalan V, Simcox A. Transcriptional control of an essential ribozyme in Drosophila reveals an ancient evolutionary divide in animals. PLoS Genet 2015, 11:e1004893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol Cell 2012, 48:219–230. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013, 51:792–806. [DOI] [PubMed] [Google Scholar]
- 24. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012, 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence‐mediated exon circularization. Cell 2014, 159:134–147. [DOI] [PubMed] [Google Scholar]
- 26. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004, 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kiss T. Small nucleolar RNA‐guided post‐transcriptional modification of cellular RNAs. EMBO J 2001, 20:3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009, 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116:281–297. [DOI] [PubMed] [Google Scholar]
- 30. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005, 6:376–385. [DOI] [PubMed] [Google Scholar]
- 31. Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol 2013, 425:3582–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10:1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature 2004, 432:231–235. [DOI] [PubMed] [Google Scholar]
- 34. Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432:235–240. [DOI] [PubMed] [Google Scholar]
- 35. Yi R, Qin Y, Macara IG, Cullen BR. Exportin‐5 mediates the nuclear export of pre‐microRNAs and short hairpin RNAs. Genes Dev 2003, 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409:363–366. [DOI] [PubMed] [Google Scholar]
- 37. Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA‐interference enzyme Dicer in the maturation of the let‐7 small temporal RNA. Science 2001, 293:834–838. [DOI] [PubMed] [Google Scholar]
- 38. Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115:209–216. [DOI] [PubMed] [Google Scholar]
- 39. Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115:199–208. [DOI] [PubMed] [Google Scholar]
- 40. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009, 10:126–139. [DOI] [PubMed] [Google Scholar]
- 41. Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA‐class regulatory RNAs in Drosophila . Cell 2007, 130:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell 2007, 28:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJ. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res 2012, 40:438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 2010, 24:992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A. Critical association of ncRNA with introns. Nucleic Acids Res 2011, 39:2357–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Godnic I, Zorc M, Jevsinek Skok D, Calin GA, Horvat S, Dovc P, Kovac M, Kunej T. Genome‐wide and species‐wide in silico screening for intragenic MicroRNAs in human, mouse and chicken. PLoS One 2013, 8:e65165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11:241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Curtis HJ, Sibley CR, Wood MJ. Mirtrons, an emerging class of atypical miRNA. WIREs RNA 2012, 3:617–632. [DOI] [PubMed] [Google Scholar]
- 50. Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007, 8:574–585. [DOI] [PubMed] [Google Scholar]
- 51. Matera AG, Terns RM, Terns MP. Non‐coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 2007, 8:209–220. [DOI] [PubMed] [Google Scholar]
- 52. Lestrade L, Weber MJ. snoRNA‐LBME‐db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 2006, 34:D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang XO, Yin QF, Wang HB, Zhang Y, Chen T, Zheng P, Lu X, Chen LL, Yang L. Species‐specific alternative splicing leads to unique expression of sno‐lncRNAs. BMC Genomics 2014, 15:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tycowski KT, Shu MD, Steitz JA. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev 1993, 7:1176–1190. [DOI] [PubMed] [Google Scholar]
- 55. Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre‐mRNA introns. Genes Dev 1995, 9:1411–1424. [DOI] [PubMed] [Google Scholar]
- 56. Hirose T, Shu MD, Steitz JA. Splicing‐dependent and ‐independent modes of assembly for intron‐encoded box C/D snoRNPs in mammalian cells. Mol Cell 2003, 12:113–123. [DOI] [PubMed] [Google Scholar]
- 57. Lykke‐Andersen S, Chen Y, Ardal BR, Lilje B, Waage J, Sandelin A, Jensen TH. Human nonsense‐mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev 2014, 28:2498–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain‐specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA 2000, 97:14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. de Smith AJ, Purmann C, Walters RG, Ellis RJ, Holder SE, Van Haelst MM, Brady AF, Fairbrother UL, Dattani M, Keogh JM, et al. A deletion of the HBII‐85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum Mol Genet 2009, 18:3257–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader‐Willi syndrome. Eur J Hum Genet 2010, 18:1196–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader‐Willi phenotype caused by paternal deficiency for the HBII‐85 C/D box small nucleolar RNA cluster. Nat Genet 2008, 40:719–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genome‐wide characterization of non‐polyadenylated RNAs. Genome Biol 2011, 12:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yin QF, Hu SB, Xu YF, Yang L, Carmichael GG, Chen LL. SnoVectors for nuclear expression of RNA. Nucleic Acids Res 2015, 43:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barbosa‐Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338:1587–1593. [DOI] [PubMed] [Google Scholar]
- 65. Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 2012, 338:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Qian L, Vu MN, Carter M, Wilkinson MF. A spliced intron accumulates as a lariat in the nucleus of T cells. Nucleic Acids Res 1992, 20:5345–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R‐digested cellular RNA source that consists of lariat and circular RNAs from pre‐mRNA splicing. Nucleic Acids Res 2006, 34:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015, 22:256–264. [DOI] [PubMed] [Google Scholar]
- 69. Gardner EJ, Nizami ZF, Talbot CC Jr, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis . Genes Dev 2012, 26:2550–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Talhouarne GJ, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 2014, 20:1476–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495:333–338. [DOI] [PubMed] [Google Scholar]
- 73. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell‐type specific features of circular RNA expression. PLoS Genet 2013, 9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome‐wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age‐dependent neural accumulation. Cell Rep 2014, 9:1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015, 10:170–177. [DOI] [PubMed] [Google Scholar]
- 76. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015, 12:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep 2015, 10:103–111. [DOI] [PubMed] [Google Scholar]
- 78. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014, 32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon‐containing lariat precursor. elife 2015, 4:e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014, 28:2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ashwal‐Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell 2014, 56:55–66. [DOI] [PubMed] [Google Scholar]
- 82. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160:1125–1134. [DOI] [PubMed] [Google Scholar]
- 83. Rybak‐Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal‐Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015, 58:870–885. [DOI] [PubMed] [Google Scholar]
- 84. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015, 18:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495:384–388. [DOI] [PubMed] [Google Scholar]
- 86. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014, 15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yang L, Chen LL. Competition of RNA splicing: line in or circle up. Sci China Life Sci 2014, 57:1232–1233. [DOI] [PubMed] [Google Scholar]
- 88. Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol 2015, 427:2414–2417. [DOI] [PubMed] [Google Scholar]
- 89. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995, 268:415–417. [DOI] [PubMed] [Google Scholar]
- 90. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015, 21:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yin S, Yu Y, Reed R. Primary microRNA processing is functionally coupled to RNAP II transcription in vitro. Sci Rep 2015, 5:11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wilusz JE. Long noncoding RNAs: re‐writing dogmas of RNA processing and stability. Biochim Biophys Acta 2015. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]


