Abstract
Vascular tissues are very important for providing both mechanical strength and long‐distance transport. The molecular mechanisms of regulation of vascular tissue development are still not fully understood. In this study we identified ANAC005 as a membrane‐associated NAC family transcription factor that regulates vascular tissue development. Reporter gene assays showed that ANAC005 was expressed mainly in the vascular tissues. Increased expression of ANAC005 protein in transgenic Arabidopsis caused dwarf phenotype, reduced xylem differentiation, decreased lignin content, repression of a lignin biosynthetic gene and genes related to cambium and primary wall, but activation of genes related to the secondary wall. Expression of a dominant repressor fusion of ANAC005 had overall the opposite effects on vascular tissue differentiation and lignin synthetic gene expression. The ANAC005‐GFP fusion protein was localized at the plasma membrane, whereas deletion of the last 20 amino acids, which are mostly basic, caused its nuclear localization. These results indicate that ANAC005 is a cell membrane‐associated transcription factor that inhibits xylem tissue development in Arabidopsis.
Keywords: ANAC005, Arabidopsis, NAC, vascular tissues development
Edited by: Jan Traas, University of Lyon, France
INTRODUCTION
About 2,500 transcription factors (TFs) have been identified in the Arabidopsis genome (Perez‐Rodriguez et al. 2010). These include about 105 members of the NAC family of plant‐specific transcription factors, which are named for NAM (No apical meristem), ATAF (Arabidopsis transcription activation factor) and CUC (Cup‐shaped cotyledon) (Ooka et al. 2003). NAC proteins contain a conserved N‐terminal NAC domain and a C‐terminal transcription regulatory region (Ernst et al. 2004; Olsen et al. 2005). A typical NAC domain can be divided into five subdomains: the C and D subdomains are highly conserved and responsible for DNA binding; the A subdomain may be involved in the formation of homodimer or heterodimer; the B and E subdomains may be responsible for the functional diversity of NAC genes (Ooka et al. 2003; Ernst et al. 2004; Jensen et al. 2010; Chen et al. 2011). However, several atypical NAC proteins have been identified, including proteins with the NAC domain only (Christiansen et al. 2011), some NAC proteins containing two tandemly repeated NAC domains (Jensen et al. 2010), and NAC proteins containing a zinc finger domain to the N‐terminal of NAC domain (Mitsuda et al. 2004; Jensen et al. 2010).
The activity of transcription factors can be regulated by several processes, including posttranscriptional modification, protein‐protein interactions, and nuclear transport (Poon and Jans 2005; Espenshade and Hughes 2007). Many transcription factors have been found physically tethered to the membrane and released by proteolytic cleavage (Hoppe et al. 2001; Liu et al. 2007a, 2007b; Iwata et al. 2009). A genome‐wide analysis predicted 85 membrane‐associated transcription factors (MTFs) in Arabidopsis (Kim et al. 2010). These include 18 NAC proteins predicated to contain α‐helical transmembrane motifs (TMs) in the C‐terminal region that may mediate association with the cell membrane, and they are named NAC with Transmembrane Motif 1 (NTM1), and NTM1‐like (NTL) factors (Kim et al. 2007b). It is notable that virtually all of the reported plant MTFs are closely related with plant responses to various abiotic stress conditions. NTM1/NTL12 and NTM2/NTL13 are released from membrane by proteolytic cleavage in response to salt treatment (Kim et al. 2006; Park et al. 2011). A truncated NTM1 that is unable to associate with membrane causes reduced cell numbers and defects in cell division. Membrane association of NTL8 is induced by salinity and GA deficiency, and NTL8 inhibits seed germination and flowering (Kim et al. 2007a, 2008). The processing of NTL9 is promoted by osmotic stress, and NTL9 in turn regulates some senescence‐associated genes (Lim et al. 2007; Yoon et al. 2008). Release of NTL6 from the membrane is activated by cold, drought, high salinity and abscisic acid (ABA) (Clarke et al. 2004; Kim et al. 2007b). Interestingly, pathogenesis‐related genes were significantly induced in these transgenic plants, suggesting that NTL6 might be an integrator of biotic and abiotic stress responses (Seo et al. 2010). These studies suggest that plant MTFs are closely related with plant responses to various abiotic stress conditions (Kim et al. 2010).
Several NAC family proteins have been shown to be involved in the regulation of vascular development. SND1 (secondary wall‐associated NAC domain protein 1) is a NAC family gene that is specially expressed in interfascicular fibers and xylary fibers. Ectopic overexpression of SND1 induces the expression of several secondary wall biosynthetic genes (such as CESA7 and CESA8) and the thickening of secondary wall. A dominant repressor form of SND1 reduces the thickness of secondary wall (Zhong et al. 2006, 2007). NST1 (NAC secondary wall thickening promoting factor 1), NST2 and NST3 are three homologous NAC proteins that activate secondary cell wall formation redundantly (Mitsuda et al. 2007, 2005; Zhong et al. 2007; Mitsuda and Ohme‐Takagi 2008). Similar to SND1, overexpression of NST1 and NST3 induces the thickening of a secondary cell wall in aboveground tissues. Expression of dominant negative forms of NST1 and NST3 repress the cell wall thickening in interfascicular fiber tissue independently. The cell wall thickening of presumptive interfascicular fiber tissue is also suppressed in double mutant of nst1‐1 nst3‐1 (Mitsuda et al. 2007; Zhong et al. 2007; Mitsuda and Ohme‐Takagi 2008). NST1 and NST2 redundantly activate secondary cell wall formation in endothecium of anther (Mitsuda et al. 2005). VASCULAR‐RELATED NAC‐DOMAIN proteins belong to a subfamily of NACs that includes seven members. VND6 and VND7 play an important role in the regulation of metaxylem vessel and protoxylem formation (Kubo et al. 2005; Yamaguchi et al. 2010, 2008, 2011). VNI2 is another NAC protein that interacts with VND7 in yeast two‐hybrid screen. VNI2 inhibits the formation of xylem vessel through reducing the expression of secondary wall related genes that are regulated by VND7 (Yamaguchi et al. 2010).
In this study, we studied the function of ANAC005. We found that ANAC005 is expressed preferentially in the vascular tissue, and increasing the expression of ANAC005 proteins caused oval‐shaped leaves and short petioles, and reduced xylem tissue differentiation, whereas a dominant repressor form of ANAC005 enhanced xylem tissue differentiation, accompanied with altered expression of genes involved in vascular tissue development. While ANAC005 was not predicted to be an MTF, it is localized at the cell membrane through its C‐terminal domain, which is highly basic. Our results indicate that ANAC005 is a membrane‐associated NAC family transcription factor that plays an important role in vascular development in Arabidopsis. This study also identifies a novel motif that mediates membrane association of transcription factor.
RESULTS
ANAC005 is predominantly expressed in xylem
ANAC005 is a NAC gene preferentially expressed in xylem according to a tissue specific microarray analysis (Zhao et al. 2005). We first performed quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) to examine the expression of ANAC005 in different Arabidopsis tissues and organs. ANAC005 was ubiquitously expressed at a similar level in 1‐week‐old seedlings, roots and rosette leaves of 3‐week‐old plants, stem of 4‐week‐old plants and other organs of 6‐week‐old plants (Figure 1A). To further investigate the expression pattern of ANAC005, a 2.7 kb genomic sequence, including 1.1 kb promoter region and the 1.6 kb coding region, was fused to the Escherichia coli β‐glucuronidase (GUS) reporter gene and transformed into Arabidopsis. As shown in Figure 1B–H, the proANAC005::ANAC005‐GUS transgenic plants showed GUS activity in the vascular system of the cotyledons, rosette leaves, petioles, and inflorescence stem. A section of the inflorescence stem shows the highest GUS signal in the thin‐walled cells (presumably developing tracheary element) adjacent to the thick‐walled tracheary elements in xylem. No GUS signal was observed in the mature tracheary elements, suggesting that ANAC005 is highly expressed during tracheary element development. Weak GUS signals were also observed in the phloem cells of vascular tissue (Figure 1H). GUS expression was also detected preferentially in vascular tissues of cauline leaves, petals, and stigma. These results indicate that ANAC005 is mainly expressed in the vascular tissues of all organs in Arabidopsis. These results are consistent with the reported microarray result showing that ANAC005 is expressed at a higher level in xylem compared with phloem and non‐vascular system (Zhao et al 2005).
Figure 1.
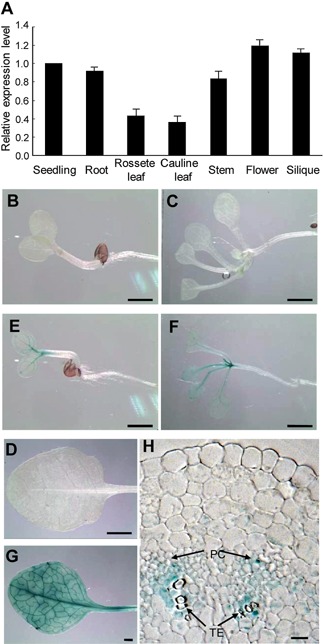
ANAC005 is expressed preferentially in vascular bundles (A) Analysis of ANAC005 expression level in different organs by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The expression level in the seedling is set to 1.0, and error bars represent SD of three biological replicates. (B–H) GUS staining of wild type and ProANAC005:ANAC005‐GUS plants. (B) 3‐day‐old wild type seedling. (C) 10‐day‐old wild type seedling. (D) 3‐day‐old ProANAC005:ANAC005‐GUS seedling. (E) 10‐day‐old ProANAC005:ANAC005‐GUS seedling. (F) rosette leaf of ProANAC005:ANAC005‐GUS plants. (G) Close‐up image of cotyledon from 5‐day‐old ProANAC005:ANAC005‐GUS seedling. (H) transverse section of inflorescence stem from 5‐week‐old ProANAC005:ANAC005‐GUS plants. PC, phloem cell; TE, tracheary element. Scale bars = 1 mm in (B–F), 20 µm in (G) and (H).
ANAC005 localizes to the cell membrane through its C‐terminus
To determine the subcellular localization of ANAC005, we made an ANAC005‐GFP (green fluorescent protein) fusion protein construct, driven by the Cauliflower mosaic virus (CaMV) 35S promoter, and transformed it into Nicotiana benthamiana. In the epidermal cells of Nicotiana benthamiana transformed with 35S::GFP, GFP fluorescence was visible in both nucleus and cytoplasm (Figure 2B). By contrast, the ANAC005‐GFP fluorescence was only detected at the plasma membrane (Figure 2C, D). Protein structure analysis showed that the N‐terminal of ANAC005 has a NAM domain including A, B, C, D and E motifs; its C‐terminus is a putative transcription regulation domain (Figure S1). ANAC005 does not belong to the 18 NAC proteins previously identified as putatively MTFs. We further examined the ANAC005 sequence using the TopPred‐v2 program, and identified a putative transmembrane motif (TM motif) in the middle region (aa268‐288). In order to test whether the putative TM motif determines the localization of ANAC005, we made several truncated ANAC005 proteins fused with GFP protein (Figure 2E–I). When expressed in Nicotiana benthamiana driven by the CaMV 35S promoter, full‐length ANAC005‐GFP was localized exclusively at the plasma membrane (Figure 2C). In contrast, ANAC005(1‐272)‐GFP (with deletion of C‐terminal 90 amino acids), ANAC005(1‐302)‐GFP (with deletion of C‐terminal 60 amino acids), ANAC005(1‐332)‐GFP (with deletion of C‐terminal 30 amino acids), and ANAC005(1‐342)‐GFP (with deletion of C‐terminal 20 amino acids) were all localized in the nucleus (Figure 2E–H). However, deletion of the C‐terminal 10 amino acids resulted in localization in both nucleus and plasma membrane (Figure 2I). These results show that C‐terminal 20 amino acids of ANAC005 play an important role in its localization of to the plasma membrane, whereas the N‐terminal region specifies nuclear localization.
Figure 2.
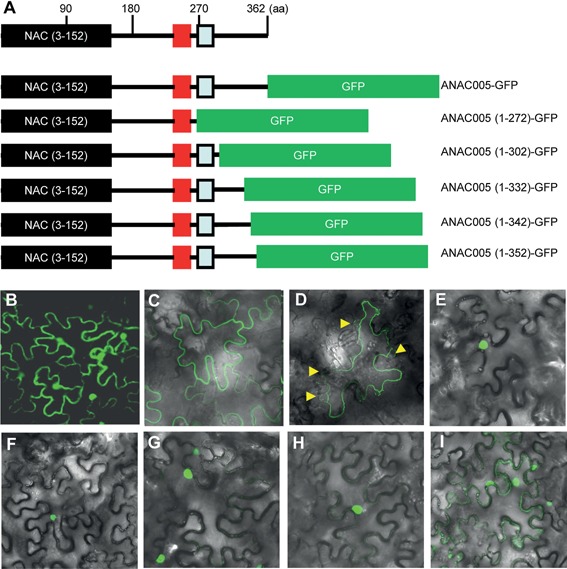
Subcellular localization of ANAC005‐green fluorescent protein (GFP) protein (A) Diagram of ANAC005 protein and GFP fusion proteins. Red box shows transcription activation domain; blue box shows putative transmembrane domain. (B, C, E−I) Fluorescence microscopic image of Nicotiana benthamiana leaf epidermal cells transformed with 35S:GFP (B), 35S:ANAC005‐GFP (C), 35S:ANAC005(1‐272)‐GFP (E), 35S:ANAC005(1‐302)‐GFP (F), 35S:ANAC005(1‐332)‐GFP (G), 35S:ANAC005(1‐342)‐GFP (H) and 35S:ANAC005(1‐352)‐GFP (I). (D) Fluorescence microscopic image of plasmolysis of Nicotiana benthamiana leaf epidermal cells transformed with 35S:ANAC005‐GFP. Yellow arrows marker the places where plasmolysis happen.
ANAC005 represses the growth of Arabidopsis and influences its morphogenesis
The proANAC005::ANAC005‐GUS transgenic plants showed dwarf phenotypes with round rosette leaves, and the severity of phenotypes correlated with the intensities of GUS staining, indicating that expression of ANAC005‐GUS caused the dwarf phenotypes (Figure 3A, B). The tissue‐specific pattern of GUS staining, however, remained the same in lines that showed different levels of GUS and phenotype severity (Figures 3B, S3B). Similar dwarf phenotypes were observed in transgenic plants expressing the non‐fusion ANAC005 or ANAC005‐GFP fusion proteins, but the non‐fusion ANAC005 appeared to cause milder phenotypes compared to the GUS and GFP fusions (Figure 3C). Compared with wild type plants, the transgenic plants expressing high levels of ANAC005 have shorter petiole and oval leaves (Figure 3C, D). Scanning electron microscopy (SEM) analysis showed the cell elongation is reduced in ANAC005‐GFP transgenic plants (Figure 3E). Furthermore, the ANAC005‐GFP transgenic plants show a bigger angle between inflorescence stem and pedicle (Figure 4A, C). When expressed using the CaMV 35S promoter, ANAC005 causes similar dwarf phenotypes as driven by ANAC005 promoter (Figure S2).
Figure 3.
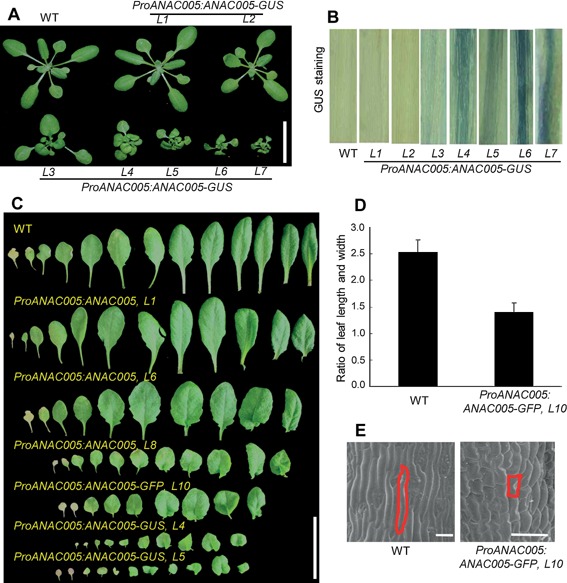
Increasing ANAC005 expression caused dwarf phenotypes (A) Three‐week‐old plants grown under long‐day conditions. Scale bar = 2cm. (B) GUS‐stained leaf petiole of 3‐week‐old plants grown in soil. (C) Comparison of rosette leaves. Leaves are arranged from the first leaf at the left to the latest leaf at the right. Scale bar = 2cm. (D) Ratio of leaf length and leaf width of wild type and ProNAC005:NAC005‐GFP transgenic plant grown under long‐day condition. (E) Cell length of leaf petiole in wild type and ProNAC005:NAC005‐GFP transgenic plants. Scale bar = 100 µm.
Figure 4.
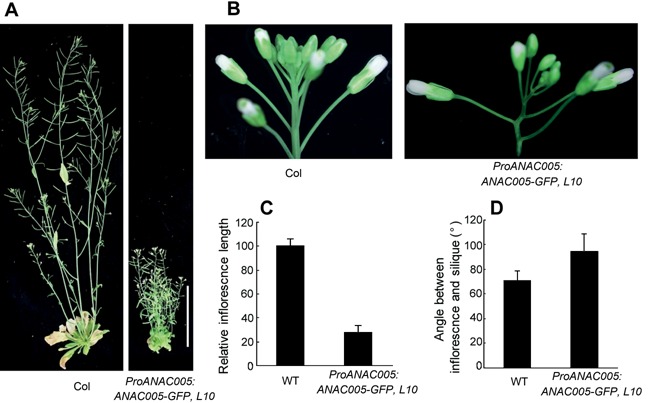
Phenotypes of plants expressing ANAC005‐GFP (A) 8‐week‐old plants grown under longer‐day conditions. Scale bar = 4 cm. (B) Inflorescence of plants grown under longer‐day conditions for 6 weeks. (C) Final height of the wild type and ProANAC005:ANAC005‐GFP transgenic plants. (D) Angle between inflorescence and silique stems.
We generated ANAC005 knockdown plants through artificial microRNA and RNA interference. But no phenotype was detected in the transgenic plant. With eight homologous genes in the subfamily (Ooka et al. 2003), ANAC005 is likely to have redundant functions with its homologs. We thus adopted the SRDX chimeric repressor method to study ANAC005 function (Hiratsu et al. 2003, 2004, 2002). Full‐length coding sequence of ANAC005 was fused to the SRDX transcription repression domain (ANAC005‐SRDX) and expressed in transgenic Arabidopsis using the CaMV 35S promoter. To our surprise, the 35S:ANAC005‐SRDX transgenic plants show no obvious morphological phenotype.
ANAC005 inhibits xylem differentiation
Since ANAC005 is preferentially expressed in the vascular bundle, we investigated whether ANAC005 regulates vascular development. We performed sectioning and microscopic analysis of the influorescence stems of both ProANAC005:ANAC005‐GUS and 35S:ANAC005‐SRDX plants. As shown in Figure 5A–D, the ProANAC005: ANAC005‐GUS plants showed reduced xylem differentiation, whereas the 35S:ANAC005‐SRDX plants showed increased xylem development. Consistent with the altered xylem development, the lignin content was increased in the 35S:ANAC005‐SRDX plants but reduced in the ProANAC005:ANAC005‐GUS plants (Figure 5E).
Figure 5.
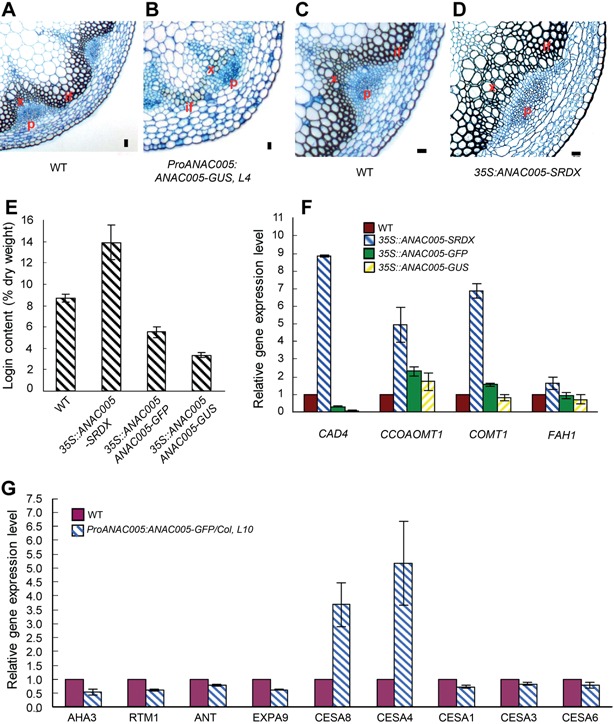
ANAC005 inhibits xylem differentiation (A‐D) Resin‐embedded transverse sections of the basal portion of the inflorescence stems of wild type (A) and ProANAC005::ANAC005‐GUS plants (B), wild type (C) and 35S:ANAC005‐SRDX plants (D). X, xylem; P, phleom; C, cambium; IF, interfascicular fiber. Scale bars = 20µm. (E) The lignin content of wild type (WT) and different transgenic plants. (F, G) Relative expression level of genes involved in lignin synthesis from 7‐day‐old plants (F) and genes related to vascular development from stem of 5‐week‐old plants (G) of wild type and transgenic plants.
We further analyzed vascular‐related genes in the inflorescence stem of the transgenic plants by qRT‐PCR. These genes include lignin synthesis genes (CAD4, CCOAOMT1, COMT1, FAH1), cambium marker (EXPA9 and ANT) (Gray‐Mitsumune et al. 2004; Schrader et al. 2004), phloem marker (AHA3 and RTM1) (DeWitt and Sussman 1995; Chisholm et al. 2001), primary cell wall cellulose synthesis genes (CESA1, CESA3 and CESA6) (Desprez et al. 2007), and secondary cell wall cellulose synthesis genes (CESA4 and CESA8) (Taylor et al. 2003). As shown in Figure 5F, CAD4 expression was highly increased in the 35S:ANAC005‐SRDX plants, but decreased in two ProANAC005: ANAC005‐GFP lines, consistent with the alteration of lignin content and xylem development in these plants. Expression levels of CCOAOMT1, COMT1, FAH1 were also increased in the 35S:ANAC005‐SRDX plants, but not obviously altered in the ProANAC005:ANAC005‐GFP plants. All these results support that ANAC005 regulates lignin synthesis and xylem differentiation. Interestingly, genes related to cambium, phloem and primary cell wall were downregulated, while genes related to the secondary cell wall were upregulated in the ProANAC005:ANAC005‐GFP plants compared to wild type (Figure 5G).
DISCUSSION
NAC proteins constitute a big family of plant‐specific transcription factors and function in diverse plant developmental processes (Ooka et al. 2003). In this article, we identified ANAC005 as a membrane‐bound NAC family transcription factor, and demonstrated its function in vascular tissue development.
Our results provide strong evidence that ANAC005 plays a role in vascular tissue development, specifically in controlling xylem development. First, previous microarray analysis (Zhao et al. 2005) and our promoter‐GUS reporter gene experiments showed preferential ANAC005 expression in xylem. Our promoter‐GUS reporter gene also showed a low level expression in phloem cells. Second, expression of ANAC005‐GUS and the ANAC005‐SRDX dominant negative form reduced and increased, respectively, xylem development, lignin content, and expression of lignin biosynthetic gene CAD4. The opposite effects of ANAC005 and ANAC005‐SRDX suggest that ANAC005 regulates xylem development as a transcription activator and it reduces CAD4 expression likely by activating a transcription repressor that inhibits CAD4 expression. Overexpression of ANAC005‐GFP also increased the expression of CESA4 and CESA8, which are also activated by the NAC factor SND1, a major regulator of secondary wall synthesis (Zhong et al. 2008; Taylor‐Teeples et al. 2015). Recent studies showed that SND1 directly activates genes involved in secondary wall synthesis, but also activates KNAT7, which represses lignin synthetic genes (Taylor‐Teeples et al. 2015). ANAC005 may play a similar or overlapping role as SND1, in activating CESA4 and CESA8, but represses lignin synthesis through other transcription factors such as KNAT7. It is noteworthy that the dominant negative ANAC005‐SRDS and the knat7 mutation promoted xylem development, whereas overexpression of ANAC005 had similar effects as overexpression of KNAT7 in repressing xylem development (Figure 5; Li et al. 2012). ANAC005‐SRDS and the knat7 also had similar effects on lignin synthetic gene expression (Figure 5; Li et al. 2012). It will be interesting to test in future studies whether ANAC005 regulates lignin synthesis and xylem differentiation through KNAT7.
ANAC005 is a membrane‐associated NAC factor. The ANAC005‐GFP protein showed exclusive plasma membrane localization, which is mediated by the C‐terminal 20 amino acids of ANAC005. Deletion of the C‐terminal 10 amino acids partially disrupted ANAC005 localization at the membrane, whereas deletion of 20 amino acids completely abolished membrane localization, leading to accumulation in the nucleus. This C‐terminal sequence is highly conserved in several close homologs, such as ANAC003, ANAC004, and ANAC0048, but not in any of the known MTFs. Among the known NAC MTFs, NTM2/NTL13 showed the highest overall sequence homology to ANAC005, with 47% sequence identity in the amino acids 5‐288 region of ANAC005. Unlike the NTM/NTL factors, which have C‐terminal hydrophobic TM domain, the C‐terminal 20‐aa motif of ANAC005 contains eight basic amino acid residues and shares no homology with the C‐terminal TM motifs of NTM/NTL factors. We therefore conclude that ANAC005 is associated with membrane through a novel mechanism.
The distinct C‐terminal motifs suggest that the regulation of ANAC005 membrane disassociation is likely also different from the NTM/NTL factors. Indeed, salt treatment had no effect on ANAC005 localization (data not shown). The vascular phenotypes of ANAC005‐overexpressors suggest that ANAC005 membrane disassociation might be regulated developmentally by a cell type‐specific mechanism. Expression of ANAC005‐GFP and ANAC005‐GUS fusion proteins from the native ANAC005 promoter or overexpression of non‐fusion ANAC005, inhibited cell elongation and caused dwarf phenotypes (Figure 4). However, overexpression of ANAC005‐SRDX fusion affected xylem patterning, but did not cause severe dwarf phenotypes, suggesting that different transcriptional activities (activation vs repression) may be involved in ANAC005's effects on cell elongation and vascular development. Future study will be required to understand the mechanism that regulates ANAC005 membrane disassociation and activation, and to elucidate ANAC005's function in patterning vascular tissue differentiation.
MATERIALS AND METHODS
Plant material and growth
Arabidopsis thaliana ecotype Columbia‐0, and transgenic plants obtained in this study were grown at 22°C under white light (16‐h light/8‐h dark cycles) either on half‐strength MS medium or in the soil. Arabidopsis seeds were sterilized with 75% ethanol plus 0.01% Triton X‐100 for 15 min, then rinsed with 95% ethanol, and dried in the hood. The surface‐sterilized seeds were sown on 0.7% phytoagar plates containing half‐strength MS medium and 1% sucrose. The plates were kept at 4°C for 3 d and exposed to white light for 2 h before being transferred into the dark. Leaves, inflorescence stem, and silique were photographed and petioles, inflorescence stem, and angle between inflorescence and pedicle were measured using Image J software (http://rsb.info.nih.gov/ij/).
Vector construction and transformation
To obtain the overexpression vector of ANAC005, a 1,609‐bp genomic fragment containing full‐length ANAC005 open reading frame was amplified by PCR and then cloned into the BamHI and KpnI sites of the pSN1301 binary vector to place ANAC005 under the control of the CaMV 35S promoter. The primer sequence used was 5′‐CGGGATCCATGGCGAATCCGGTGGGTTT‐3′ and 5′‐GGGGTACCTCATGTTCTTAGGTGAATTTTCTTGAC‐3′.
To get localized expression of ANAC005, 2,676‐bp/2,679‐bp (containing terminator codon) genomic segment containing promoter and gene sequence was amplified by PCR and ligated into pENTRTM/SD/D‐TOPO vector, and then ligated into pMDC163 and C3 by recombinant clone. The primer sequence used was 5′‐CACCAGTATCACAACTATGGGTCTGACT‐3′ and 5′‐TGTTCTTAGGTGAATTTTCTTGAC‐3′; 5′‐CACCAGTATCACAACTATGGGTCTGACT‐3′ and 5′‐CTATGTTCTTAGGTGAATTTTCTTGAC‐3′.
The 35S:ANAC005‐GFP, 35S:ANAC005(1‐272)‐GFP, 35S:ANAC005(1‐302)‐GFP, 35S:ANAC005(1‐332)‐GFP, 35S:ANAC005(1‐342)‐GFP, and 35S:ANAC005(1‐352)‐GFP fusion construct was generated by inserting a full‐length ANAC005 cDNA without stop codon, a 816 bp cDNA segment without 270 bp segment in the 3′ terminal, a 906 bp cDNA segment without 180 bp cDNA segment in the 3′ terminal, a 996 bp cDNA segment without 90 bp segment in the 3′ terminal, a 1,026 bp cDNA segment without 60 bp in the 3′ terminal, or a 1,056 bp cDNA segment without 30 bp in the 3′ terminal into pENTRTM/SD/D‐TOPO vector, and then ligated into pMDC83 by recombinant clone. The forward primer was 5′‐CACCATGGCGAATCCGGTGGGTTT‐3′ and the reverse primers were 5′‐TGTTCTTAGGTGAATTTTCTTGAC‐3′, 5′‐TGTATCATCCTGTGAAAGAAAC‐3′, 5′‐TTTGTTCTTGATCGTCTCTTGAC‐3′, 5′‐ATGCTCACCAATCTCAGTCCCTTG‐3′, 5′‐TGAGTTAGGAGATTCCTGCAAG‐3′, and 5′‐AGTTGTTGCAGAAATCGGATCAG‐3′.
To knock down ANAC005 in Arabidopsis using RNAi, a specific sequence was amplified from wild type Arabidopsis genomic DNA with primers 5′‐GGGGTACCACTAGTAACACGAGCCATGTCGATG‐3′ and 5′‐CGGGATCCGAGCTCATGTTCTTAGGTGAATTTTCTTG‐3′. This fragment was first digested by BamHI and KpnI for the reverse insert to vector pTCK309. The forward insert was generated by SpeI and SacI digestion.
To get the dominant repressor of ANAC005, the full‐length cDNA sequence was amplified with primers 5′‐CGGGATCCATGGCGAATCCGGTGGGTTT‐3′ and 5′‐GGACTAGTTGTTCTTAGGTGAATTTTCTTGAC‐3′. Then the segment is ligated into p35SSRDX vector between BamHI and SpeI sites.
The constructs were transformed into the Agrobacterium tumefactions strain GV3101 and then introduced into Arabidopsis thaliana plants via floral dip method.
Total RNA extraction and quantitative RT‐PCR analysis
Total RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, California, USA). About 500 ng RNA was reverse‐transcribed by AMV reverse transcriptase (Takara Biotechnology, Dalian, China) following the manufacture's instruction. Quantitative RT‐PCR analyses were carried out on ABI7500 (Applied Biosystems, Foster City, California, USA) by using SYBR Green reagent (Toyobo, Osaka, Japan). Three biological repeats and three technical repeats were performed in each treatment. The UBC30 gene was used as internal reference for all the qRT‐PCR analysis.
Assays of GUS activity
The histochemical GUS assays were performed in a staining solution containing 0.5 mg/mL 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) in 0.1 M Na2HPO4, pH 7.0, 10 mM Na2EDTA, 0.5 mM potassium ferricyanide/ ferrocyanide, and 0.06% Triton X‐100 (Jefferson et al. 1987). Samples were infiltrated under vacuum for 10 min and then incubated at 37°C overnight. The staining buffer was removed, and the samples were cleared in 70% ethanol. All observations by light microscopy were made with the Olympus BX51 microscope system.
Semi‐thin section and microscopy
Arabidopsis stems were fixed overnight in FAA buffer (3.7% formalin, 5% acetic acid, 50% alcohol). Samples were embedded in Spurr's resin (SPI‐CHEM) and sectioned to about 0.5ìm. The microsections were stained with 0.05% toluidine blue‐O (Sigma‐Aldrich) before imaging under a microscope (Zeiss Imager M2).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ANAC005 (At1g02250), APL (At1g79430), RTM1 (At1g05760), AHA3 (At5g57350), ANT (At4g37750), EXPA9 (At5g02260), CESA8 (At4g18780), CESA4 (At5g44030), CESA1 (At4g32410), CESA3 (At5g05170) and CESA6 (At5G64740).
AUTHOR CONTRIBUTIONS
J.Z., Z.Y.W. and S.W.Z. designed the study, analyzed the data, and wrote the article. J.S.L and F.N.M. performed gene, protein expression analysis and physiological analysis. Z.Z.Z. and H.L. performed transgenic experiment and subcellular localization of proteins. W.H.L. and X.M.L. assisted in physiological analysis. J.Z. performed all the other studies.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Figure S1. ANAC005 is a classic NAC family protein (A) Structure analysis of predicted amino acid sequence of ANAC005. The yellow, light green, light blue, red, and dark green box indicate the motif of A to E of NAC domain. The gray box indicates the TAR (Transcription activation region) motif. The underline marker sequence is a predicated transmembrane motif (TM) according to the TopPred‐v2 program. (B) Sequence alignment of the predicted amino acids of ANAC005 and closely related members of NAC proteins. Shade residues indicate positions where amino acid are highly conserved. The yellow, light green, light blue, red, and dark green overlines indicate the motif of A to E of NAC domain. The gray line indicates the TAR motif. The black line indicates C‐terminal 20 amino acids of ANAC005, ANAC003, ANAC004 and ANAC048.
Figure S2. Overexpression of ANAC005 causes dwarf phenotypes Phenotypes (upper panel) and semiquantitative RT‐PCR analysis of ANAC005 expression (lower panel) of wild type (WT) and different lines of 35S::ANAC005. UBC30 was used as a control. Scale bar = 5 cm.
Figure S3. ANAC005 is expressed preferentially in vascular bundles (A–C) GUS‐stained 3‐day‐old seedlings of wild type (A), ProANAC005::ANAC005‐GUS plants without phenotype (B) and weak phenotype (C). Scale bar = 1 mm.
ACKNOWLEDGEMENTS
We thank Feng‐Qin Dong (Institute of Botany, Chinese Academy of Sciences) for kind assistance in historesin‐embedded sectioning and Li‐jia Qu (Peking University, China) for kindly providing us 35SSRDX vector. This work was supported by the National High Technology Research and Development Program of China (2012AA101108), National Natural Science Foundation of China (31171614) and Ministry of Agriculture of China (2014ZX08001).
Zhao J, Liu JS, Meng FN, Zhang ZZ, Long H, Lin WH, Luo XM, Wang ZY, Zhu SW ( 2015) ANAC005 is a membrane‐associated transcription factor and regulates vascular development in Arabidopsis . J Integr Plant Biol 58: 442–451
Available online on Jul. 14, 2015 at www.wileyonlinelibrary.com/journal/jipb
REFERENCES
- Chen Q, Wang Q, Xiong L, Lou Z ( 2011) A structural view of the conserved domain of rice stress‐responsive NAC1. Protein Cell 2: 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Parra MA, Anderberg RJ, Carrington JC ( 2001) Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long‐distance movement of tobacco etch virus. Plant Physiol 127: 1667–1675 [PMC free article] [PubMed] [Google Scholar]
- Christiansen MW, Holm PB, Gregersen PL ( 2011) Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res Notes 4: 302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SM, Mur LA, Wood JE, Scott IM ( 2004) Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana . Plant J 38: 432–447 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Hofte H, Gonneau M, Vernhettes S ( 2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana . Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, Sussman MR ( 1995) Immunocytological localization of an epitope‐tagged plasma membrane proton pump (H+‐ATPase) in phloem companion cells. Plant Cell 7: 2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst HA, Olsen AN, Larsen S, Lo Leggio L ( 2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade PJ, Hughes AL ( 2007) Regulation of sterol synthesis in eukaryotes. Annu Rev Genet 41: 401–427 [DOI] [PubMed] [Google Scholar]
- Gray‐Mitsumune M, Mellerowicz EJ, Abe H, Schrader J, Winzell A, Sterky F, Blomgvist K, McQueen‐Mason S, Teeri TT, Sundberg B ( 2004) Expansins abundant in secondary xylem belong to subgroup A of the alpha‐expansin gene family. Plant Physiol 135: 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme‐Takagi M ( 2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis . Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Mitsuda N, Matsui K, Ohme‐Takagi M ( 2004) Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis . Biochem Biophys Res Commun 321: 172–178 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme‐Takagi M ( 2002) The SUPERMAN protein is an active repressor whose carboxy‐terminal repression domain is required for the development of normal flowers. FEBS Lett 514: 351–354 [DOI] [PubMed] [Google Scholar]
- Hoppe T, Rape M, Jentsch S ( 2001) Membrane‐bound transcription factors: Regulated release by RIP or RUP. Curr Opin Cell Biol 13: 344–348 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N ( 2009) The Arabidopsis membrane‐bound transcription factor AtbZIP60 is a novel plant‐specific endoplasmic reticulum stress transducer. Plant Signal Behav 4: 514–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O'Shea C, Skriver K ( 2010) The Arabidopsis thaliana NAC transcription factor family: Structure‐function relationships and determinants of ANAC019 stress signalling. Biochem J 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Kim SG, Kim SY, Park CM ( 2007a) A membrane‐associated NAC transcription factor regulates salt‐responsive flowering via FLOWERING LOCUS T in Arabidopsis . Planta 226: 647–654 [DOI] [PubMed] [Google Scholar]
- Kim SG, Lee AK, Yoon HK, Park CM ( 2008) A membrane‐bound NAC transcription factor NTL8 regulates gibberellic acid‐mediated salt signaling in Arabidopsis seed germination. Plant J 55: 77–88 [DOI] [PubMed] [Google Scholar]
- Kim SG, Lee S, Seo PJ, Kim SK, Kim JK, Park CM ( 2010) Genome‐scale screening and molecular characterization of membrane‐bound transcription factors in Arabidopsis and rice. Genomics 95: 56–65 [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, Park CM ( 2007b) Exploring membrane‐associated NAC transcription factors in Arabidopsis: Implications for membrane biology in genome regulation. Nucleic Acids Res 35: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM ( 2006) A membrane‐bound NAC transcription factor regulates cell division in Arabidopsis . Plant Cell 18: 3132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T ( 2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bhargava A, Qiang W, Michael C. Friedmann MG, Natascha Forneris N, Rodney A, Savidge RA, Lee A, Johnson LA, Shawn D, Mansfield SD, Brian E, Ellis BE, Douglas CJ ( 2012) The Class II NOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus . New Phytol 194: 102–115 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG ( 2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH ( 2007a) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane‐associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH ( 2007b) Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 51: 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Hisabori T, Takeyasu K, Sato MH ( 2004) VOZ; isolation and characterization of novel vascular plant transcription factors with a one‐zinc finger from Arabidopsis thaliana . Plant Cell Physiol 45: 845–854 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme‐Takagi M ( 2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis . Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme‐Takagi M ( 2008) NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 56: 768–778 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme‐Takagi M ( 2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K ( 2005) NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S ( 2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana . DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Park J, Kim YS, Kim SG, Jung JH, Woo JC, Park CM ( 2011) Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis . Plant Physiol 156: 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Rodriguez P, Riano‐Pachon DM, Correa LG, Rensing SA, Kersten B, Mueller‐Roeber B ( 2010) PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res 38: D822–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IK, Jans DA ( 2005) Regulation of nuclear transport: Central role in development and transformation? Traffic 6: 173–186 [DOI] [PubMed] [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G ( 2004) A high‐resolution transcript profile across the wood‐forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Park CM ( 2010) A membrane‐bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal Behav 5: 481–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, Patil S, Kim TW, Ji H, Wong WH, Rhee SY, Wang ZY ( 2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis . Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR ( 2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor‐Teeples M, ML, Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, Handakumbura PP, Xiong G, Wang C, Corwin J, Tsoukalas A, Zhang L, Ware D, Pauly M, Kliebenstein DJ, Dehesh K, Tagkopoulos I, Breton G, Pruneda‐Paz JL, Ahnert SE, Kay SA, Hazen SP, Brady SM ( 2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Goue N, Igarashi H, Ohtani M, Nakano Y, Mortimer JC, Nishikubo N, Kubo M, Katayama Y, Kakegawa K, Dupree P, Demura T ( 2010) VASCULAR‐RELATED NAC‐DOMAIN6 and VASCULAR‐RELATED NAC‐DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol 153: 906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Kubo M, Fukuda H, Demura T ( 2008) Vascular‐related NAC‐DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55: 652–664 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme‐Takagi M, Kato K, Demura T ( 2011) VASCULAR‐RELATED NAC‐DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J 66: 579–590 [DOI] [PubMed] [Google Scholar]
- Yoon HK, Kim SG, Kim SY, Park CM ( 2008) Regulation of leaf senescence by NTL9‐mediated osmotic stress signaling in Arabidopsis . Mol Cells 25: 438–445 [PubMed] [Google Scholar]
- Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP ( 2005) The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root‐hypocotyl. Plant Physiol 138: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH ( 2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis . Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH ( 2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis . Planta 225: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH ( 2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis . Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Figure S1. ANAC005 is a classic NAC family protein (A) Structure analysis of predicted amino acid sequence of ANAC005. The yellow, light green, light blue, red, and dark green box indicate the motif of A to E of NAC domain. The gray box indicates the TAR (Transcription activation region) motif. The underline marker sequence is a predicated transmembrane motif (TM) according to the TopPred‐v2 program. (B) Sequence alignment of the predicted amino acids of ANAC005 and closely related members of NAC proteins. Shade residues indicate positions where amino acid are highly conserved. The yellow, light green, light blue, red, and dark green overlines indicate the motif of A to E of NAC domain. The gray line indicates the TAR motif. The black line indicates C‐terminal 20 amino acids of ANAC005, ANAC003, ANAC004 and ANAC048.
Figure S2. Overexpression of ANAC005 causes dwarf phenotypes Phenotypes (upper panel) and semiquantitative RT‐PCR analysis of ANAC005 expression (lower panel) of wild type (WT) and different lines of 35S::ANAC005. UBC30 was used as a control. Scale bar = 5 cm.
Figure S3. ANAC005 is expressed preferentially in vascular bundles (A–C) GUS‐stained 3‐day‐old seedlings of wild type (A), ProANAC005::ANAC005‐GUS plants without phenotype (B) and weak phenotype (C). Scale bar = 1 mm.


