Abstract
Although a few well-characterized polyketide synthases (PKSs) have been functionally reconstituted in vitro from purified protein components, the use of this strategy to decode “orphan” assembly line PKSs has not been described. To begin investigating a PKS found only in Nocardia strains associated with clinical cases of nocardiosis, we reconstituted in vitro its five terminal catalytic modules. In the presence of octanoyl-CoA, malonyl-CoA, NADPH, and S-adenosyl methionine, this pentamodular PKS system yielded unprecedented octaketide and heptaketide products whose structures were partially elucidated using mass spectrometry and NMR spectroscopy. The PKS has several notable features, including a “split, stuttering” module and a terminal reductive release mechanism. Our findings pave the way for further analysis of this unusual biosynthetic gene cluster whose natural product may enhance the infectivity of its producer strains in human hosts.
Graphical abstract
Virtually all known polyketide antibiotics were discovered through cultivation of poorly characterized soil microorganisms. The explosive pace of sequencing “orphan” polyketide synthase (PKS) gene clusters has motivated a fundamentally new approach to natural product discovery by activating the biosynthetic capacity of these cryptic gene clusters in native or heterologous hosts.1,2 An alternate strategy might involve bypassing altogether the constraints of microbial cultivation and gene cluster regulation by reconstituting the enzymatic machinery in vitro, but this has not yet been attempted to our knowledge for a reasonably complex orphan PKS.
Recently, we functionally reconstituted the hexamodular 6-deoxyerythronolide B synthase, a prototypical assembly line PKS.3 Encouraged by its catalytic efficiency in vitro, as well as other recent efforts to reconstitute natural product biosynthesis in vitro,4,5 we sought to apply the approach to decode a particularly remarkable group of closely related orphan synthases. Our target PKS family has only been observed in nine clinical strains of Nocardia isolated from patients diagnosed with nocardiosis6,7 (Table S1). Although the genomes of numerous Nocardia spp. (including some pathogenic species) have been sequenced to date,8 our target PKS (hereafter referred to as “NOCAP” for nocardiosis-associated polyketide synthase) represents a distinct clade in an evolutionary tree of ~1000 nonredundant assembly line synthases.9 Because these strains of Nocardia are potentially hazardous and lack well-established microbiological or genetic protocols, analysis of the NOCAP synthase in its natural hosts was impractical. The potential use of E. coli as a heterologous host was considered; however, several features of the NOCAP synthase (e.g., modules with trans-acting AT domains, a split PKS module, a reductive chain-release mechanism) have not been functionally reconstituted in E. coli to our knowledge. We therefore chose to investigate this PKS by directly reconstituting in vitro its constituent modules, given the considerably greater potential for troubleshooting an inactive complex system through direct biochemical analysis.
The NOCAP synthase has several unusual architectural features, including a split module,10 cis- as well as trans-acyltransferases (ATs),11–13 a terminal thioester reductase (TR),14 and a thioesterase (TE) domain fused to the trans-AT (Figure 1). It is encoded by three genes, nocap_PKS1–3, that are part of a larger gene cluster (Figures S1), including genes encoding deoxysugar biosynthesis and transfer (Scheme S1) and a “Module X” presumed to either synthesize a primer unit for the multimodular assembly line or a separate oligoketide that is ultimately incorporated into the final natural product via a convergent biosynthetic mechanism (Figure S2).
Figure 1.
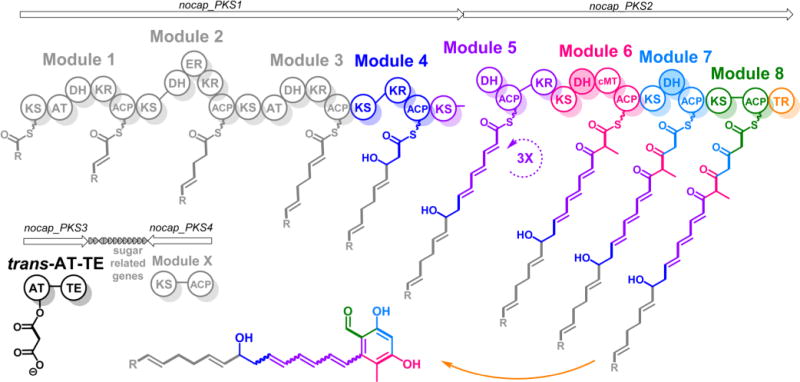
NOCAP synthase is comprised of eight PKS modules distributed across two large proteins. A third didomain protein has a trans-acyltransferase (AT) and thioesterase (TE). NOCAP_PKS1 has four modules and the ketosynthase (KS) of a fifth. NOCAP_PKS2 includes the dehydratase (DH), ketoreductase (KR), and acyl carrier protein (ACP) domains of module 5 along with three more trans-AT modules. The KR domain of module 5 is atypically located C-terminal to its ACP domain. Module 6 has a C-methyltransferase (cMT). The putative (shaded) DH domains of modules 6 and 7 are likely vestigial, as dehydratase active site residues were not observed. Chain release is by an NAD(P)H-dependent thioester reductase (TR). The proposed biosynthetic scheme is based on observed polyketide products of modules 4–8 (highlighted in color). Because the activity of modules 1–3 was not reconstituted in this study, the predicted structure of the tetraketide intermediate (shown in gray) is tentative. The R-group indicates that the primer unit of module 1 is unknown. All modules besides modules 1 and 3 require the trans-AT to supply malonyl extender units. The function of module X (NOCAP_PKS4) is also unknown.
RESULTS AND DISCUSSION
In Silico Analysis and Design
At the onset of our studies, nocap_PKS1–2 was assumed to encode a collinear, octamodular assembly line (Figure 1). To initiate experimental analysis of this PKS, the sequences of individual modules, starting from each KS domain and ending with the corresponding acyl carrier protein (ACP) domain, were aligned with those of canonical PKS modules. Putative catalytic domains were identified based on characteristic residues.15 This in silico analysis provided a basis for deconstructing large gene products into fragments that could be expressed in E. coli (Table S2; Figure S3). Docking domains from the 6-deoxyerythronolide B synthase were fused at the N- and C-termini of each KS and ACP domain, respectively, because these peptides play a critical role in intermodular chain translocation,16 and because the NOCAP synthase lacks interpolypeptide ACP-KS interfaces. Each protein was purified from E. coli (Figure S4) prior to use in vitro.
Extender Unit Analysis
Modules 1 and 3 of the NOCAP synthase harbor their own (putatively malonyl-specific) AT domains, whereas others belong to a family of “AT-less” modules originally observed in the leinamycin synthase.12 These AT-less modules are presumably transacylated by the trans-AT protein encoded by nocap_PKS3 (Figure 1). Using an α-ketoglutarate dehydrogenase-coupled kinetic assay,17 the trans-AT was found to be specific for malonyl-CoA, with comparable kinetics in the presence or absence of its downstream TE domain (Figure S5). Therefore, malonate was used as the source of extender units in subsequent studies.
Primer Unit Analysis
Because the NOCAP synthase lacks a dedicated active site for chain initiation, we screened modules 1–4 and module X individually for their ability to elongate a range of radiolabeled acyl-CoA substrates (C2–C16). Chain elongation was not observed for modules 1–3 or module X, although KS1 and KS2 showed acylation by fatty acyl chains C8–C16 (data not shown). When module 4+KS5 was coincubated with the DH5-ACP5 didomain protein in the presence of the trans-AT+TE,14 C-lauroyl-CoA, malonyl-CoA, and NADPH, the radiolabel was transferred to DH5-ACP5 (Figure S6). This unexpected finding provided convenient entry into the modules 4–8 segment of the NOCAP synthase. While this approach precludes identification of the natural aglycone, functional analysis of the partial assembly line provided considerable insight into the molecular logic of the NOCAP synthase.
In Vitro Reconstitution and LC-MS Product Analysis of NOCAP Synthase Modules 4–8
Full catalytic activity of modules 4–8 was reconstituted from four proteins (Figure 2A): module 4+KS5, the DH-ACP-KR tridomain of module 5 along with module 6 (pJK35; Table S2), modules 7 and 8 along with the terminal TR domain (pJK97; Table S2), and the trans-AT+TE. The four proteins were incubated with a range of alternative primer units (C2–C16), malonyl-CoA, NADPH, and S-adenosyl methionine (SAM). Malonyl-CoA was either added directly or generated in situ by adding malonate, ATP, and malonyl-CoA synthetase to the reaction mixture (Figure 2B).18 Control experiments were performed in which each substrate and protein component was omitted.
Figure 2.
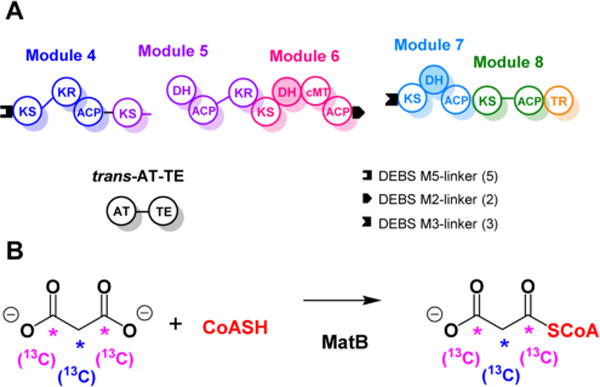
(A) Reconstituted NOCAP synthase modules 4–8 and the trans-AT-TE proteins. Docking domains from the 6-deoxyerythronolide B synthase (DEBS) were fused onto the C-terminus of ACP6 and the N-termini of KS4 and KS7. PKS abbreviations are as in Figure 1. (B) Stable isotope 13C-labeled malonyl-CoA was generated in situ enzymatically using MatB, S. coelicolor malonyl-CoA synthetase.
Reaction products were analyzed by LC-MS. Using XCMS software,19 putative products absent from control reactions were identified. First, their polyketide origins were confirmed by replacing malonate in the reaction mixture with [2-13C]-, [1,3-13C2]-, or [13C3]-malonate. On the basis of the observed mass shifts in the presence of alternative isotopically labeled substrates, the most abundant products were tentatively identified as heptaketides and octaketides. From reaction mixtures that either included or omitted SAM, pairs of heptaketides (1–2) and octaketides (3–4) differing by 14 a.m.u. were identified (Table 1; Figures 3 and S7). In the absence of SAM, neither methylated compound was observed, the desmethyl heptaketide being the dominant product (Figure S9). In contrast, in the presence of SAM, the methylated octaketide 4 appeared as the dominant product (Figure 3). The UV spectra of these compounds are shown in Figure S8. An observed triketide 5 (Table 1, Figure 3) was also in reactions containing only the module 7–8-TR and trans-AT+TE proteins, so it was predicted to be an α-pyrone from decarboxylative priming of module 7 (Figure S10).20 Replacing the trans-AT+TE didomain protein with its truncated trans-AT derivative resulted in 2–10× reduction in product formation (data not shown), suggesting a nonessential role for the TE domain in natural product biosynthesis.21 Analogous products were observed in the presence of other primer unit sources, albeit in reduced amounts (Figure S9 and Table S3).
Table 1.
Polyketide Products of Modules 4–8 of the NOCAP Synthase in the Presence of Octanoyl-CoA
| calcd [M + H]+ | observed m/z | formula | compound |
|---|---|---|---|
| 333.2061 | 333.2060 | C20H28O4 | 1 |
| 347.2217 | 347.2219 | C21H30O4 | 2 |
| 359.2217 | 359.2212 | C22H30O4 | 3 |
| 373.2373 | 373.2373 | C23H32O4 | 4 |
| 211.1330 | 211.1330 | C12H18O3 | 5 |
Figure 3.
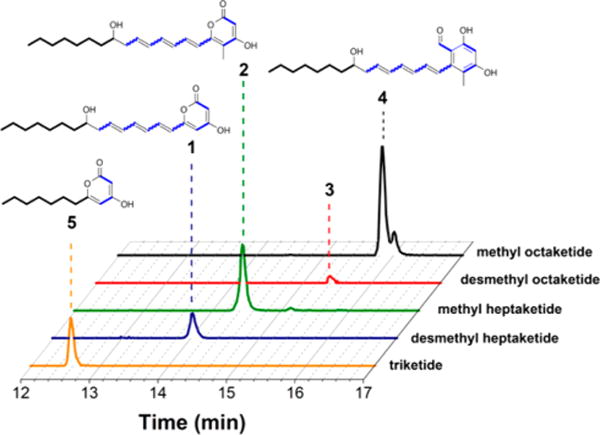
LC-MS traces of octanoyl-CoA derived products of NOCAP synthase modules 4–8. Extracted ion chromatograms of products 1–5 (Table 1) are shown, all from the same reaction analyzed on a phenyl-hexyl reverse-phase HPLC column attached to an Agilent 6520 Q-TOF mass spectrometer.
Structure Elucidation by NMR
Further structural analysis was performed by NMR spectroscopy. Enzymatic reactions performed on a 12 mL scale using octanoyl-CoA and alternative malonate isotopomers as substrates were extracted with ethyl acetate, with polyketide yields on the order of 1–3 μg/mL. A 2D NMR analysis of such small samples was possible only because of this enriched 13C-labeling strategy. The desmethyl heptaketide (1), methyl heptaketide (2), and methyl octaketide (4) were purified by preparative HPLC. Each compound showed significant absorption at 362 nm (Figure S8), suggestive of a conjugated polyene skeleton.22 NMR spectra are shown in Figures S12–S38. The uniformly 13C-labeled sample of 1 enabled identification of eight carbon atoms (C-6 to C-12) by HSQC/HMQC.
HCCH–COSY analysis of the same sample established the connectivity of C-6 to C-11, thereby revealing a conjugated olefin moiety (Figure 4; Table S4). 1H–1H COSY provided further support for this polyene functionality, while also connecting the aliphatic octanoyl tail (0.8–1.6 ppm) to a hydroxyl substituent at C-13 (4.45 ppm), a geminal H-13 (3.47 ppm), and the C-12 methylene group (2.27 ppm). HMBC analysis of the uniformly 13C-labeled sample showed low signal-to-noise due to shorter relaxation times (or pulse sequence suppression due to one-bond 1H–13C couplings) but still established a correlation between OH-13 and C-12 (40.9 ppm). Meanwhile, two-bond correlation H2BC,23 complementary to HMBC, correlated H-13 to C-12. To compensate for the lack of observed HMBC/H2BC correlations from the uniformly 13C-labeled sample, a sample of 1 labeled with [1,3-13C2]-malonate, yielding 13C-labels at only odd-numbered positions (Figure 4), was also prepared. The HMQC and 1-D 1H results for this sample were consistent for the C-6 to C-13 stretch. New HMBC and H2BC correlations connected H-12 to C-11 and H-6 (6.14 ppm) to C-5 (155.3 ppm), and further verified the polyene connectivity between C-6 and C-11. The assignment of OH-13 was confirmed by observation of its signal attenuation in a 1-D 1H experiment with presaturation applied on the residual water peak in the sample.
Figure 4.
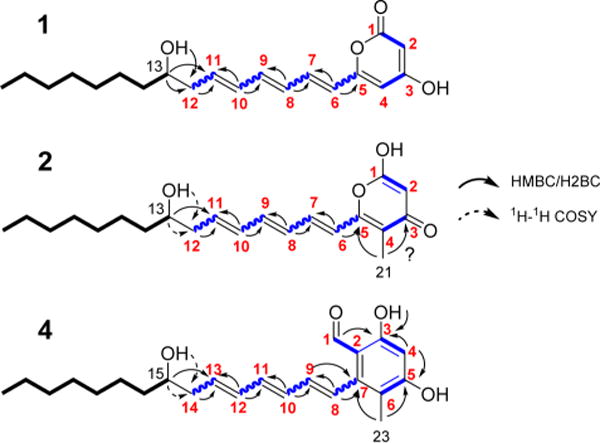
Summary of NMR analysis of polyketides 1, 2, and 4. HMBC and H2BC (solid arrows) and highlighted 1H–1H COSY (dashed arrows) correlations are shown for desmethyl heptaketide 1, methyl heptaketide 2, and methyl octaketide 4. 1H and 13C chemical shifts are listed in Table S4. Carbon positions 1–12 (bold, red) could be 13C-labeled.
A cross-peak observed by HSQC and HMQC in the uniformly 13C-labeled sample of 1—and missing in the sample labeled at odd-numbered carbons—was tentatively assigned as between H-4 and C-4, although coupling was not observed from C-4 to C-5 or to the polyene moiety. The lack of peaks associated with carbon atoms 1–4 presumably reflected peak broadening due to enolization, proton exchange with water in the sample, and/or the acidity of H-2. As such, the structure of the pyrone moiety in 1 was principally established from mass spectrometric analysis (Figures S39–S40, S45, S46) as well as the logic of polyketide biosynthesis inferred from the absence of KR domains in modules 6–8 of the NOCAP synthase. We note, however, the resemblance between 1 and the pyrones generated by the fungal lovastatin synthase in the absence of a requisite enoylreductase.24 Indeed, our observed 13C and 1H chemical shifts for the pyrone moiety are consistent with those reported in that study.
1H–1H COSY, HMQC, and HMBC analysis also revealed a conjugated triene in methyl heptaketide 2. The methyl substituent was placed at C-4 based on correlations observed by HMBC between H-21 (1.77 ppm) and C-5 (150.8 ppm) as well as a weaker correlation between H-21 and C-3 (177.8 ppm; Figure 4, Table S4). Observation of a correlation between H-6 (6.50 ppm) and C-5 (150.8) ruled out methylation at C-6, thus lending further support to C-4 as the site of the methyl substituent. As in the case of 1, peaks corresponding to C-1 to C-3 were not clearly observed.
In the presence of SAM, the methyl octaketide (4) was the most abundant product. A sample of 4 with 13C labels at the odd-numbered positions was prepared. Again, an unsubstituted triene moiety was detected, along with correlations observed by HMBC between the methyl substituent (H-23 = 1.94 ppm) and C-7 (142.2 ppm) of the triene moiety (Figure 4, Table S4). Thus, the polyene tail of 4 appeared identical to its counterparts in 1 and 2. However, the C-7 chemical shift (142.2 ppm) was more upfield than the analogous C-5 (150–156 ppm) of 1 and 2, suggesting the absence of bonded oxygen. Notably, a free aldehyde was identified in 4 from the very downfield shifts (δH = 9.54, δC = 191.6 ppm) in the HMQC spectrum. This is consistent with the presence of a thioester reductase (TR) domain as the chain release mechanism (Figure 1). Further evidence in support of the proposed structure of 4 was obtained by NMR analysis in chloroform (CDCl3), which led to the observation of sharper peaks and key new correlations, presumably due to a slower exchange rate. Specifically, H-4 (6.30 ppm) was assigned and correlated to C-3 (158.2) as well as C-5 (155.5) via HMBC, with additional correlations from the methyl H-23 (2.12 ppm) to C-5 and from a very downfield OH-3 (12.28 ppm) to C-3 (Figure 4, Table S4). Additionally, a weaker HMBC correlation was seen from the aldehyde H-1 (9.92 ppm) to C-3 (158.2).
The polyketide structures proposed were also supported by MS/MS fragmentation analysis of 1 (Figures S39, S40), 2 (Figures S41, S42), and 4 (Figures S43, S44). Notably, the fragmentation pattern of methyl octaketide 4 was markedly different from that of heptaketides 1 or 2. Compound 4 showed strong UV absorption at 280 nm, not observed for 1 and 2 (Figure S8); this is consistent with a phenolic moiety.
Biosynthetic Insights
Notwithstanding the lack of clarity regarding the stereochemistry of 1–4, two intriguing biosynthetic features of the NOCAP synthase can be inferred from the deduced structures of its products. First, its ability to synthesize heptaketides and octaketides suggests that at least one of its modules must be iteratively used in the overall catalytic cycle. Such systems are known as “stuttering” modules.25–28 By mapping polyketide structures onto the assembly line (Figure 1), it is evident that module 5 catalyzes three rounds of chain elongation, ketoreduction, and dehydration before eventually translocating the intermediate onto module 6. If so, then it would be most interesting to establish whether the capacity of module 5 to stutter is mechanistically related to its disconnected architecture. A split module is occasionally found in trans-AT assembly line PKSs,13 but stuttering behavior for such split modules had not been observed previously (Figure 5). Such a naturally occurring variant could provide unique insights into the origins of vectorial biosynthesis on a PKS assembly line.29
Figure 5.
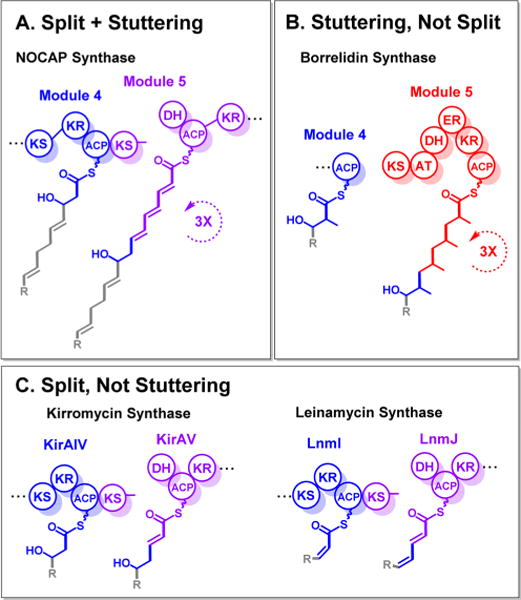
(A) Split and stuttering NOCAP synthase module 5. Though there are other examples of (B) stuttering26 and (C) split11 trans-AT assembly line PKS modules, NOCAP synthase module 5 is unique in being both split and stuttering.
Second, chain length flexibility of the NOCAP synthase appears to result from the optional activity of module 8, suggesting that the KS domain of module 8 can discriminate between a γ-methylated heptaketide and its desmethyl analog. In the absence of the methyl substituent, spontaneous chain release from ACP7 is the favored outcome, whereas the methylated chain undergoes facile chain translocation between modules 7 and 8. While a reductive release domain is occasionally observed in iterative PKSs and in nonribosomal peptide synthases (NRPSs),14,30 the only other example of reductive release in an assembly line PKS is in the case of coelimycin biosynthesis,14 where an aldehyde is also released from the assembly line before being processed further. Thus, the TR domain of the NOCAP synthase could be a versatile addition to engineering efforts involving assembly line PKSs.
Conclusion
This report represents the first example of an attempt to decode an orphan assembly line PKS through direct in vitro reconstitution of its constituent modules. While this approach has thus far been limited to biochemical analysis of biosynthetically characterized systems, our findings reinforce its utility in the context of natural product discovery. For example, the octaketide aglycone 4 resembles other resorcylaldehyde natural products of uncharacterized function,31–34 suggesting an evolutionary role for this chemical “warhead.” Octaketide 4 also harbors multiple nucleophilic sites (3-, 5-, and 15-OH), two of which may be glycosylated while the third may be acylated by the polyketide product of module X (Figure S11). While further effort will be required to identify the natural product of the complete gene cluster, the UV–vis features of 4 could aid in its isolation. More generally, as DNA sequencing reveals an ever-increasing number of fascinating biosynthetic pathways from biologically inaccessible life-forms, the power of in vitro reconstitution is especially useful for studying complex linear pathways where piecemeal characterization in heterologous hosts is precluded due to the absence of suitable substrates in vivo.
METHODS
For detailed descriptions, see the Supporting Information.
Cloning, Protein Expression, and Purification
Genes were PCR amplified from Nocardia genomic DNA and cloned into pET vectors (Novagen) using Gibson assembly or restriction enzymes and T4 ligase and E. coli DH5α. Proteins were overexpressed in E. coli BL21(DE3) strains and purified by Ni-NTA affinity chromatography followed by anion exchange chromatography.
In Vitro Assays and LC-MS
Purified proteins (3–12 μM) were incubated with relevant cofactors. Reactions (25–50 μL) were extracted with ethyl acetate, with the organic phase dried and reconstituted in 88 μL of methanol for analysis by LC-MS (Agilent 1260 HPLC with Agilent 6520 Accurate-Mass Q-TOF ESI mass spectrometer).
2D-NMR
All NMR spectra were acquired on an 800 MHz Agilent VNMRS spectrometer (Stanford Magnetic Resonance Laboratory). Samples were prepared in 250 μL of deuterated solvent (d6-DMSO or CDCl3) in a matched symmetrical Shigemi tube (5 mm) and acquired at 25 °C.
Supplementary Material
Acknowledgments
We thank C.T. Walsh for feedback on this project and E.S. Sattely for use of the LC-MS. This research was supported by a grant from the National Institutes of Health (R01 GM087934).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.6b00489.
Detailed methods, tables, figures (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laureti L, Song LJ, Huang S, Corre C, Leblond P, Challis GL, Aigle B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci U S A. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowry B, Robbins T, Weng CH, O’Brien RV, Cane DE, Khosla C. In vitro reconstitution and analysis of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2013;135:16809–16812. doi: 10.1021/ja409048k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 5.Ghssein G, Brutesco C, Ouerdane L, Fojcik C, Izaute A, Wang S, Hajjar C, Lobinski R, Lemaire D, Richaud P, Voulhoux R, Espaillat A, Cava F, Pignol D, Borezee-Durant E, Arnoux P. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science. 2016;352:1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 6.Iida S, Kageyama A, Yazawa K, Uchiyama N, Toyohara T, Chohnabayashi N, Suzuki S, Nomura F, Kroppenstedt RM, Mikami Y. Nocardia exalbida sp. nov., isolated from Japanese patients with nocardiosis. Int J Syst Evol Microbiol. 2006;56:1193–1196. doi: 10.1099/ijs.0.63850-0. [DOI] [PubMed] [Google Scholar]
- 7.Kageyama A, Yazawa K, Mukai A, Kohara T, Nishimura K, Kroppenstedt RM, Mikami Y. Nocardia araoensis sp. nov. and Nocardia pneumoniae sp. nov., isolated from patients in Japan. Int J Syst Evol Microbiol. 2004;54:2025–2029. doi: 10.1099/ijs.0.63074-0. [DOI] [PubMed] [Google Scholar]
- 8.Komaki H, Ichikawa N, Hosoyama A, Takahashi-Nakaguchi A, Matsuzawa T, Suzuki K, Fujita N, Gonoi T. Genome based analysis of type-I polyketide synthase and non-ribosomal peptide synthetase gene clusters in seven strains of five representative Nocardia species. BMC Genomics. 2014;15:323. doi: 10.1186/1471-2164-15-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien RV, Davis RW, Khosla C, Hillenmeyer ME. Computational identification and analysis of orphan assembly-line polyketide synthases. J Antibiot. 2014;67:89–97. doi: 10.1038/ja.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber T, Laiple KJ, Pross EK, Textor A, Grond S, Welzel K, Pelzer S, Vente A, Wohlleben W. Molecular analysis of the kirromycin biosynthetic gene cluster revealed beta-alanine as precursor of the pyridone moiety. Chem Biol. 2008;15:175–188. doi: 10.1016/j.chembiol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 12.Cheng YQ, Tang GL, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Natl Acad Sci U S A. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helfrich EJ, Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2016;33:231–316. doi: 10.1039/c5np00125k. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Escribano JP, Song LJ, Fox DJ, Yeo V, Bibb MJ, Challis GL. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem Sci. 2012;3:2716–2720. [Google Scholar]
- 15.Keatinge-Clay AT. The structures of type I polyketide synthases. Nat Prod Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 16.Wu N, Cane DE, Khosla C. Quantitative analysis of the relative contributions of donor acyl carrier proteins, acceptor ketosynthases, and linker regions to intermodular transfer of intermediates in hybrid polyketide synthases. Biochemistry. 2002;41:5056–5066. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- 17.Dunn BJ, Cane DE, Khosla C. Mechanism and specificity of an acyltransferase domain from a modular polyketide synthase. Biochemistry. 2013;52:1839–1841. doi: 10.1021/bi400185v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes AJ, Keatinge-Clay A. Enzymatic extender unit generation for in vitro polyketide synthase reactions: structural and functional showcasing of Streptomyces coelicolor MatB. Chem Biol. 2011;18:165–176. doi: 10.1016/j.chembiol.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Tautenhahn R, Bottcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinf. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobsen JR, Cane DE, Khosla C. Spontaneous priming of a downstream module in 6-deoxyerythronolide B synthase leads to polyketide biosynthesis. Biochemistry. 1998;37:4928–4934. doi: 10.1021/bi9729920. [DOI] [PubMed] [Google Scholar]
- 21.Heathcote ML, Staunton J, Leadlay PF. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem Biol. 2001;8:207–220. doi: 10.1016/s1074-5521(01)00002-3. [DOI] [PubMed] [Google Scholar]
- 22.Skellam EJ, Stewart AK, Strangman WK, Wright JL. Identification of micromonolactam, a new polyene macrocyclic lactam from two marine Micromonospora strains using chemical and molecular methods: clarification of the biosynthetic pathway from a glutamate starter unit. J Antibiot. 2013;66:431–441. doi: 10.1038/ja.2013.34. [DOI] [PubMed] [Google Scholar]
- 23.Nyberg NT, Duus JO, Sorensen OW. Heteronuclear two-bond correlation: suppressing heteronuclear three-bond or higher NMR correlations while enhancing two-bond correlations even for vanishing 2J(CH) J Am Chem Soc. 2005;127:6154–6155. doi: 10.1021/ja050878w. [DOI] [PubMed] [Google Scholar]
- 24.Ma SM, Li JWH, Choi JW, Zhou H, Lee KKM, Moorthie VA, Xie XK, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Complete Reconstitution of a Highly Reducing Iterative Polyketide Synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, Hertweck C. Iteration as programmed event during polyketide assembly; molecular analysis of the aureothin biosynthesis gene cluster. Chem Biol. 2003;10:1225–1232. doi: 10.1016/j.chembiol.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Olano C, Wilkinson B, Sanchez C, Moss SJ, Sheridan R, Math V, Weston AJ, Brana AF, Martin CJ, Oliynyk M, Mendez C, Leadlay PF, Salas JA. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tu4055: cluster analysis and assignment of functions. Chem Biol. 2004;11:87–97. doi: 10.1016/j.chembiol.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Hardt IH, Steinmetz H, Gerth K, Sasse F, Reichenbach H, Hofle G. New natural epothilones from Sorangium cellulosum, strains So ce90/B2 and So ce90/D13: isolation, structure elucidation, and SAR studies. J Nat Prod. 2001;64:847–856. doi: 10.1021/np000629f. [DOI] [PubMed] [Google Scholar]
- 28.Tang L, Qiu RG, Li Y, Katz L. Generation of novel epothilone analogs with cytotoxic activity by biotransformation. J Antibiot. 2003;56:16–23. doi: 10.7164/antibiotics.56.16. [DOI] [PubMed] [Google Scholar]
- 29.Lowry B, Li X, Robbins T, Cane DE, Khosla C. A Turnstile Mechanism for the Controlled Growth of Biosynthetic Intermediates on Assembly Line Polyketide Synthases. ACS Cent Sci. 2016;2:14–20. doi: 10.1021/acscentsci.5b00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisch KM, Skellam E, Ivison D, Cox RJ, Bailey AM, Lazarus CM, Simpson TJ. Catalytic role of the C-terminal domains of a fungal non-reducing polyketide synthase. Chem Commun (Cambridge, U K) 2010;46:5331–5333. doi: 10.1039/c0cc01162b. [DOI] [PubMed] [Google Scholar]
- 31.Ahuja M, Chiang YM, Chang SL, Praseuth MB, Entwistle R, Sanchez JF, Lo HC, Yeh HH, Oakley BR, Wang CC. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Cox DG, Ding W, Huang G, Lin Y, Li C. Three new resveratrol derivatives from the mangrove endophytic fungus Alternaria sp. Mar Drugs. 2014;12:2840–2850. doi: 10.3390/md12052840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teles HL, Sordi R, Silva GH, Castro-Gamboa I, Bolzani VdS, Pfenning LH, Magalhães de Abreu L, Costa-Neto CM, Young MC, Araujo AR. Aromatic compounds produced by Periconia atropurpurea, an endophytic fungus associated with Xylopia aromatica. Phytochemistry. 2006;67:2686–2690. doi: 10.1016/j.phytochem.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Yang X, Lin Y, Yuan J, Lu Y, Zhu X, Li J, Li M, Lin Y, He J, Liu L. Meroterpenes and azaphilones from marine mangrove endophytic fungus Penicillium 303#. Fitoterapia. 2014;97:241–246. doi: 10.1016/j.fitote.2014.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


