Abstract
Interactions between enamel matrix proteins are important for enamel biomineralization. In recent in situ studies, we showed that the N-terminal proteolytic product of ameloblastin co-localized with amelogenin around the prism boundaries. However, the molecular mechanisms of such interactions are still unclear. Here, in order to determine the interacting domains between amelogenin and ameloblastin, we designed four ameloblastin peptides derived from different regions of the full-length protein (AB1, AB2 and AB3 at N-terminus, and AB6 at C-terminus) and studied their interactions with recombinant amelogenin (rP172), and the tyrosine-rich amelogenin polypeptide (TRAP). A series of amelogenin Trp variants (rP172(W25), rP172(W45) and rP172(W161)) were also used for intrinsic fluorescence spectroscopy. Fluorescence spectra of rP172 titrated with AB3, a peptide encoded by exon 5 of ameloblastin, showed a shift in λmax in a dose-dependent manner, indicating molecular interactions in the region encoded by exon 5 of ameloblastin. Circular dichroism (CD) spectra of amelogenin titrated with AB3 showed that amelogenin was responsible for forming α-helix in the presence of ameloblastin. Fluorescence spectra of amelogenin Trp variants as well as the spectra of TRAP titrated with AB3 showed that the N-terminus of amelogenin is involved in the interaction between ameloblastin and amelogenin. We suggest that macromolecular co-assembly between amelogenin and ameloblastin may play important roles in enamel biomineralization.
Abbreviations: Amel, Amelogenin; Ambn, Ameloblastin; TRAP, Tyrosine Rich Amelogenin Polypeptide; CD, Circular Dichroism; HPLC, High Performance Liquid Chromatography
Keywords: Ameloblastin, Amelogenin, Enamel, Matrix protein, Protein-protein interaction
Graphical abstract
Highlights
-
•
An in vitro approach to study ameloblastin-amelogenin interactions is presented.
-
•
Intrinsic fluorescence of tryptophan and Circular Dichroism were utilized.
-
•
We report that amelogenin has ameloblastin-binding ability via its N-terminal close to Trp 25.
-
•
We report that ameloblastin has amelogenin-binding ability via a peptide encoded by exon 5.
-
•
Macromolecular co-assembly between amelogenin and ameloblastin may play important roles in enamel biomineralization.
1. Introduction
The highly ordered structure of tooth enamel is regulated by the cells and extracellular matrix proteins that work together to control crystal initiation and organized growth [1]. These enamel matrix proteins are secreted by ameloblasts during the secretory stage of amelogenesis to create a mineralization front where the enamel crystals grow in length. The matrix proteins are degraded during the maturation stage by proteinases, and eventually removed as the enamel crystals grow in width and thickness [2], [3].
The major structural proteins in enamel are amelogenin (Amel), ameloblastin (Ambn), amelotin (Amtn) and enamelin (Enam) [1]. Amelogenin is the most abundant, accounting for approximately 90% of the enamel matrix protein. Mutations in the amelogenin gene result in X-linked amelogenesis imperfecta [4], [5]. In 16-week-old mice, amelogenin deficiency resulted in an enamel thickness of <10% of normal enamel, with disorganized prism structure [6]. Ameloblastin is the second most abundant protein, accounting for roughly 5% of enamel matrix protein [7]. Genomic deletion of AMBN exon 6 in humans results in hypoplastic amelogenesis imperfecta [8]. In the Ambn mutant mouse that expressed a truncated Ambn variant without the segment encoded by exons 5 and 6, ameloblasts detached from the tooth surface at the early secretory stage, and the mutation resulted in defective enamel formation[9], [10]. In vitro experiment further showed that the segment encoded by exon 5 may be involved in ameloblastin self-assembly [11].
Evidence for the notion that amelogenin and ameloblastin may have cooperative function was provided by recent double knock out animal model studies [10]. In 7-day-old Amel X-/-/Ambn-/- mice, the ameloblast layer was irregular and detached from the enamel surface as in Ambn-/- mice, and the enamel width was significantly thinner than that in Amel X-/- or Ambn-/- mice [12]. In a different report, dual-immunogold labeling showed the co-distribution of ameloblastin and amelogenin, suggesting a functional association between these two proteins [13]. It has been shown that the amelogenin trityrosyl-motif peptide localized at the N-terminal region of amelogenin binds to ameloblastin [14]. Immunochemical assays revealed that ameloblastin co-localized with amelogenin during the early stages of tooth development [15].
Recently, using an advanced quantitative co-localization assay, we reported that amelogenin co-localized with ameloblastin at the mineralization front during the secretory stage of enamel formation [16]. We further demonstrated in vitro that addition of recombinant full-length amelogenin to recombinant full-length ameloblastin induced conformational change in the latter, hinting at intermolecular interactions between the two proteins [16]. Our recent in vivo study further shows that the N-terminal proteolytic product of ameloblastin co-localizes with the N-terminus of amelogenin around the prism boundary [17]. However, it is not known how these proteins interact and whether any structural changes result from the interaction. In order to provide additional support for direct amelogenin-ameloblastin interactions and to determine the interacting domains between them, we designed four ameloblastin peptides derived from different regions of the full-length protein. We used intrinsic fluorescence spectroscopy and circular dichroism to study their interaction with amelogenin and the N-terminal tyrosine-rich amelogenin polypeptide (TRAP) [18].
2. Materials and methods
2.1. Wild type and variant Amelogenin expression and purification
Wild type (rP172) and variant recombinant porcine amelogenin proteins were expressed in E. coli strain BL21(DE3)pLysS (Stratagene, CA), and precipitated by 20% ammonium sulfate following the method described previously [19], [20]. The ammonium sulfate precipitate was dissolved in water containing 0.1% TFA and purified using a high performance liquid chromatography (HPLC) system (Varian, CA) equipped with a Phenomenex C4 column (10×250 mm, 5 µm). The purified proteins were lyophilized, kept at −20 °C, and dissolved in water before use. Our group designed three double-variant strains of amelogenin; rP172(W45Y, W161Y) will be referred to as rP172(W25), rP172(W25Y, W161Y) will be referred to as rP172(W45), and rP172(W25Y, W45Y) will be referred to as rP172(W161) [21]. The positions of tryptophan residues of rP172 and its variants are illustrated in Table 1.
Table 1.
The tryptophan residues of wild type amelogenin and its variants.
| Name | Location of Tryptophan residues | ||
|---|---|---|---|
| rP172 | W25 | W45 | W161 |
| rP172(W25) | W25 | ||
| rP172(W45) | W45 | ||
| rp172(W161) | W161 | ||
2.2. Peptide synthesis
Four peptides (AB1, AB2, AB3, and AB6) were designed based on the amino acid sequence of mouse ameloblastin (Ambn), and synthesized by Chempeptide Limited (Shanghai, China). AB1 includes 40 amino acid residues encoded by exon 3 and 4, AB2 is next to AB1 and includes 37 amino acid residues encoded by exon 5, AB3 is a variant of AB2 with two tryptophan residues substituted by two tyrosine residues, and AB6 includes 44 amino acid residues encoded by exon 13 and located at the C-terminus of Ambn. AB3 has no tryptophan residue, which is a fluorophore, and it is used instead of AB2 in the experiments where amelogenin-ameloblastin interactions are studied. The purity of peptides was determined by HPLC equipped with Kromasil-C18 column (4.6×250 mm, 5 µm), and the concentration was determined by Pierce BCA protein assay kit (Thermo Scientific, IL).
TRAP consisting of 45 amino acid residues at the N-terminus of full-length mouse amelogenin was synthesized at USC Microchemical Core Facility as previously described [22]. The amino acid sequences of AB1, AB2, AB3, AB6, and TRAP are shown in Table 2. The two substituted tyrosine residues are labeled in red.
Table 2.
The amino acid sequences and masses for ameloblastin (AB1, AB2, AB3, and AB6) and amelogenin (TRAP) peptides. AB3 is a silenced variant of AB2 in the fluorescence studies.
| Peptides | Sequence | Mass/Da |
|---|---|---|
| AB1 | 1VPAFPQQPGAQGMAPPGMASLSLETMRQLGSLQGLNALSQ40 | 4080.75 |
| AB2 |
41YSRLGFGKALNSL LHGLLPPHNSFP LHGLLPPHNSFP IGPREHETQQ77 IGPREHETQQ77
|
4284.89 |
| AB3 |
41YSRLGFGKALNSL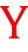 LHGLLPPHNSFP LHGLLPPHNSFP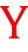 IGPREHETQQ77 IGPREHETQQ77
|
4238.82 |
| AB6 | 214YGTLFPRFGGFRQTLRRLNQNSPKGGDFTVEVDSPVSVTKGPEK257 | 4884.53 |
| TRAP | 1MPLPPHPGHPGYINFSYEVLTPLKWYQNMIRHPYTSYGYEPMGGW45 | 5324.1 |
2.3. Intrinsic fluorescence spectroscopy
Lyophilized amelogenin and peptides were first dissolved in water to make stock solutions with higher concentration of protein or peptide. All the samples were prepared by diluting the protein or the peptide into buffer (5 mM Tris-HCl) at pH of 7.3. Table 2 summarizes the molecular masses of the synthetic peptides.
To investigate whether amelogenin interacts with ameloblastin, samples containing 10 μM rP172 and 0, 5, 10, or 15 μM AB1, 10 μM rP172 and 0, 5, 10, or 15 μM AB3, and 10 μM rP172 and 0, 5, 10, or 15 μM AB6 were prepared separately. For collecting fluorescence spectra of rP172 titrated with AB3, 10 μM rP172(W25) was mixed with 0, 5, 10, or 15 μM AB3, 10 μM rP172(W45) was mixed with 0, 5, 10, or 15 μM AB3, and 10 rP172(W161) was mixed with 0, 5, 10, or 15 μM AB3. For collecting fluorescence spectra of TRAP titrated with AB3, 10 μM TRAP was mixed with 0, 1, 5, or 10 μM AB3. After preparation, all samples were kept at room temperature for 1 h before measurement.
The fluorescence emission spectra were collected using a QuantaMaster 4 spectrofluorometer (Photon Technology International, NJ) equipped with a 1-cm path-length cuvette. The excitation wavelength was 295 nm, the window sizes of the excitation and the emission shutters were set to 5 nm, and the final spectra were the average of three scans. All fluorescence experiments were carried out at room temperature.
2.4. Circular dichroism (CD) spectroscopy
We mixed 2.5 μM of rP172 with 0, 1.25, 2.5 or 3.75 μM AB3 separately, and kept the solutions at room temperature for 1 h before measurement. The final solutions contained 5 mM Tris-HCl and their pH was 7.3. The CD spectra were collected using a J-815 spectrometer (Jasco, Japan) with a 1-mm path-length cuvette. The low concentration of Tris–HCl and short path-length reduced the effect of Tris–HCl at wavelength lower than 200 nm. The wavelength range was 190–250 nm. The final spectra were the average of three scans. The CD spectra of buffer with different concentrations of AB3 were collected first. These spectra were used as reference data, and were subtracted from the spectra of rP172 titrated with corresponding concentration of AB3. Finally, the CD spectra of rP172 in the presence of AB3 were collected and recorded. The experiments were carried out at room temperature.
3. Results
3.1. Identifying ameloblastin domains that interact with full-length amelogenin
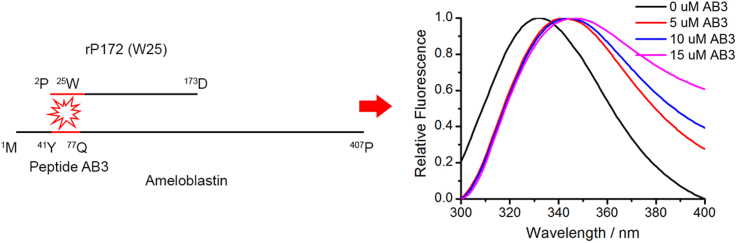
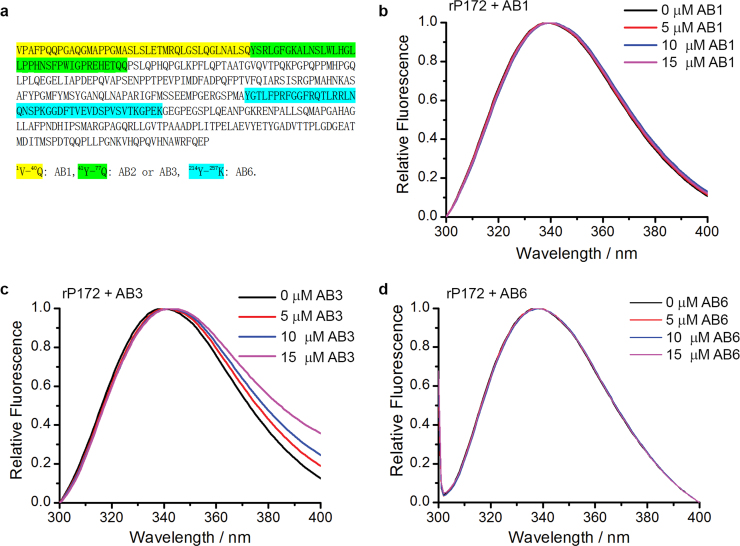
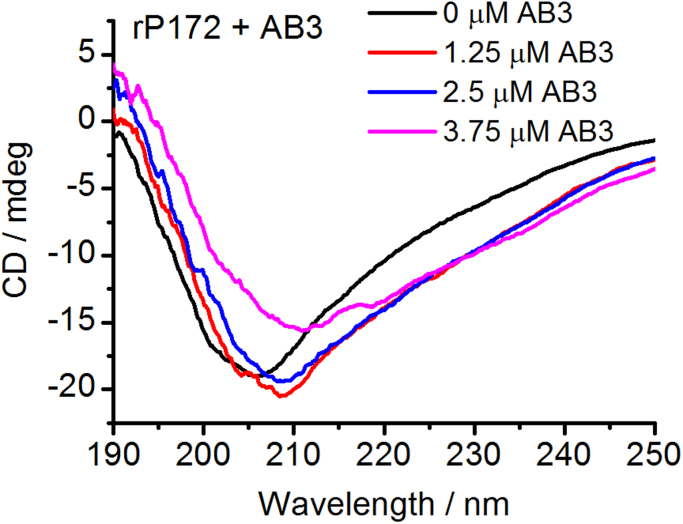
In order to identify the Amel-interacting domains on Ambn, recombinant porcine Amel rP172 (which is an 89% analogue to mouse Amel) was titrated with peptides AB1, AB3 and AB6 at pH 7.3 5 mM Tris–HCl buffer (Fig. 1(a)). CD analysis confirmed that the secondary structures of AB3 are similar to those of the wild type sequence (AB2), suggesting that substitution of the two tryptophan residues did not affect folding (Fig. S1). Thus, AB3 was used instead of the wild type sequence of Abmn (AB2) which preserves two tryptophan residues (Table 2). When rP172 was titrated with the AB1 comprising the N-terminal 40 amino acid residues of Ambn, or with the AB6 comprising the sequence at the C-terminus of Ambn, the emission maximum wavelength (λmax) of rP172 changed only slightly (Fig. 1b and d). However, as the concentration of AB3 increased from 0 µM to 15 µM, the λmax of amelogenin full-length spectra shifted from 339.7±0.58 to 342.3±0.58 nm (Fig. 1(c)). This redshift suggests that the microenvironments of the tryptophan residues of Amel become more hydrophilic in the presence of AB3[23], and that AB3, the segment within N-terminus encoded by exon 5, interacts with Amel. This is also supported by observations of CD spectra of Amel in the presence of Ambn peptides after subtraction of the peptides contribution. It was observed that AB3 induced a change in Amel secondary structure (Fig. 2). As the concentration of AB3 increased from 0 μM to 3.75 μM, θ222 decreased from −21,872 deg cm2 dmol−1 to −29,155 deg cm2 dmol−1, and λmax shifted from 205.9 nm to 211.7 nm (Fig. 2). Analysis of the percentage of secondary structures in amelogenin using the SELCON3 algorithm showed that the percentage of α-helix increased in the presence of AB3 (Table 3) [24]. Interestingly, the CD spectra of Amel in the presence of the 1–40 N-terminal peptide AB1 showed no significant changes in amelogenin secondary structures (Fig. S2).
Fig. 1.
a. A cartoon showing locations of ameloblastin derived peptides AB1 , AB2 (or AB3) and AB6 in the full-length sequence. Fluorescence spectra of 10 μM of rP172 titrated with different concentrations of peptides b. AB1; c. AB3; d. AB6.
Fig. 2.
CD spectra of 2.5 μM rP172 titrated with 0, 5, 10, or 15 μM AB3.
Table 3.
The secondary structure content of amelogenin in the presence of AB3 as analyzed by SELCON3 algorithm and based on the CD results.
| AB3 [μM] | Helix 1 | Helix 2 | Total Helix | Strand 1 | Strand 2 | Total Sheet | Turns | Unordered | Total structure |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.071 | 0.14 | 0.211 | 0.172 | 0.071 | 0.243 | 0.194 | 0.375 | 1.023 |
| 5 | 0.084 | 0.095 | 0.179 | 0.07 | 0.033 | 0.103 | 0.11 | 0.655 | 1.046 |
| 10 | 0.09 | 0.143 | 0.233 | 0.088 | 0.039 | 0.127 | 0.15 | 0.512 | 1.021 |
| 15 | 0.209 | 0.136 | 0.345 | 0.043 | 0.069 | 0.112 | 0.236 | 0.318 | 1.011 |
These observations suggest that Amel interacts with the amino acid sequence encoded by exon 5 of Ambn (namely: AB3), and not the N- or C-terminus.
3.2. Identifying amelogenin domains that interact with ameloblastin peptide AB3
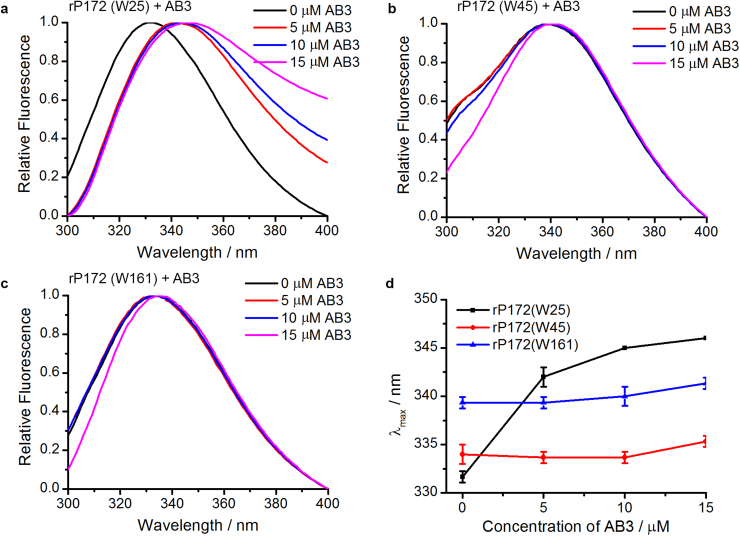
In order to identify the AB3 peptide interacting domains on Amel, three amelogenin variants were created, having only one Trp each, at positions 25, 45 and 161 (Table 1). These amelogenin variants, rP172(W25), rP172(W45) and rP172(W161), were each titrated by AB3. The λmax of each Trp variant was followed separately. In dynamic light scattering (DLS) studies, we have demonstrated that these Trp variants have similar self-assembly behavior to the wild type rP172 at both pH 5.5 (forming oligomers) and pH 8.0 (forming nanospheres) [21]. Further, circular dichroism studies of the variants in their monomeric forms have confirmed that their secondary structures are highly comparable to that of their wild-type counterpart [25]. Thus, these variants should be structurally and functionally similar to the wild type rP172. The Trp variants were titrated with AB3 and the λmax representing one Trp at a time were recorded. The results show that rP172(W25) (representing the N-terminal Trp at position 25) exhibited a significant redshift from 331±0.58 to 346±0 nm when the concentration of AB3 increased from 0 µM to 15 µM (Fig. 3(a)), while the λmax of rP172(W45) (representing Trp at position 45) and that of rP172(W161) (representing the C-terminal Trp) did not change significantly (Fig. 3b-c). The plot of λmax versus the contraction of AB3 peptide shows these trends more clearly (Fig. 3(d)). The redshift of rP172(W25) suggests that the microenvironment around Trp 25 became more hydrophilic in the presence of AB3, while that around the Trp 45 and Trp 161 did not change significantly. These observations suggest that Amel interacts with AB3 via its N-terminal domain around Trp 25.
Fig. 3.
a-c. Fluorescence spectra of rP172 variants titrated with 0, 5, 10, or 15 μM of variant peptide AB3: a. 10 μM rP172(W25). b. 10 μM rP172(W45). c.10 μM rP172(W161). d. Fluorescence shifts (λmax: the emission maximum wavelength) of rP172 variants (rP172(W25), rP172(W45), rP172(W161) as the function of AB3 concentration, titrated with 0, 5, 10, or 15 μM AB3 variant.
3.3. The interaction between TRAP and AB3
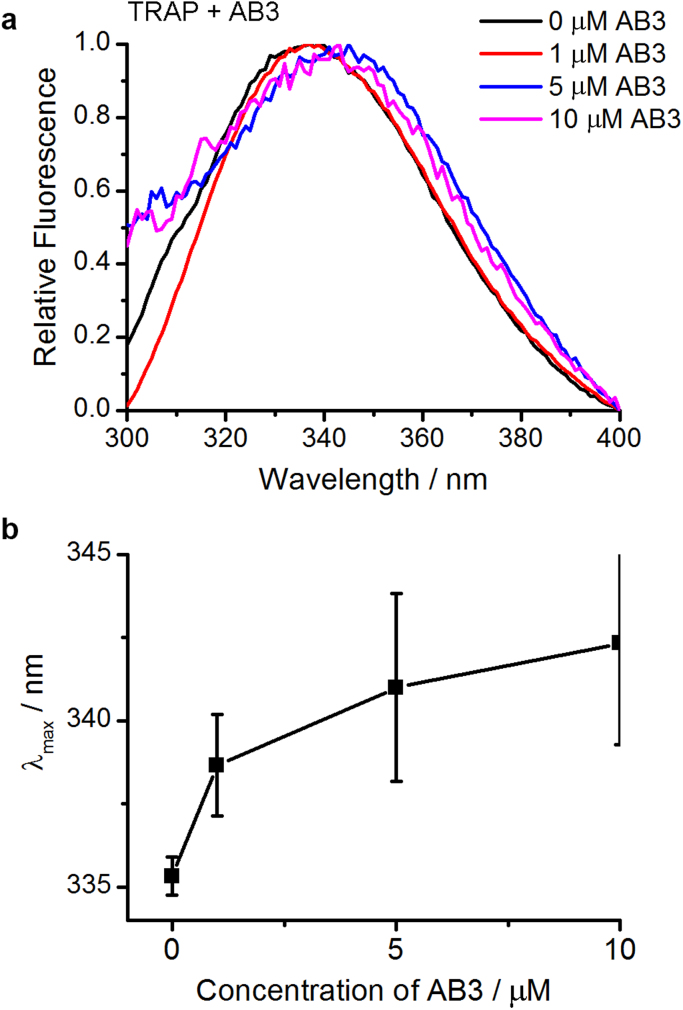
In a separate experiment, the Amel-derived peptide TRAP, containing Trp 25 and Trp 45, was titrated with AB3. When the concentration of AB3 increased from 0 µM to 10 µM, a redshift was observed from 335.3±0.58 to 342.3±3.1 nm (Fig. 4(a)). Note that the signal to noise drops low for the 5 and 10 μM additions of AB3 to TRAP. It is possible that TRAP-AB3 complexes start to aggregate at higher concentrations of AB3 decreasing the TRAP concentration in solution. The plot of λmax versus the concentration of TRAP makes the trend more clear, even though the error is large (Fig. 4(b)). Consistent with the results of double variant rP172(W25), the microenvironment of the tryptophan residues of TRAP became more hydrophilic.
Fig. 4.
a. Fluorescence spectra of 10 μM TRAP peptide titrated with 0, 5, 10, or 15 μM of variant peptide AB3; b. Fluorescence shifts (λmax: the emission maximum wavelength) of 10 μM TRAP titrated with 0, 1, 5, or 10 μM AB3 variant peptide.
4. Discussion
Recent in vivo and in vitro studies provide supporting evidence that extracellular matrix protein-protein interactions may play crucial roles in the process of crystal nucleation, growth and enamel tissue organization [12], [14], [16], [17], [26], [27], [28]. By in vivo co-localization and in vitro biophysical approaches, we have supported the idea that amelogenin forms a complex with enamelin or ameloblastin at the mineralization front during the secretory stage of enamel development, implying that such intermolecular complexes may function to initiate mineral formation [16], [27], [28], [29]. Recent in situ quantitative co-localization and FRET analysis in our laboratory revealed that the N-terminal proteolytic product of ameloblastin co-localized with amelogenin around the prism boundary [17]. Here, in order to support mechanisms of protein-protein interactions in enamel formation and to better identify the interaction domains on both amelogenin and ameloblastin, the in vitro interactions between ameloblastin peptides and full-length recombinant amelogenins were studied.
Fluorescence spectra of the wild type amelogenin titrated with the ameloblastin AB3 (Y41-Q77) showed a redshift, while no shifts were observed with AB1 (V1-Q40) and AB6 (Y214-K256). These data suggest that AB1, encoded by exon 3 and exon 4, does not interact with amelogenin, while AB3, encoded by exon 5 (close to the N-terminus of ameloblastin) does interact with amelogenin. AB6, encoded by exon 13 (at the C-terminus of ameloblastin), does not interact with amelogenin. The redshift (from 339.7±0.58 to 342.3±0.58 nm) in the case of AB3 suggests that the tryptophan amino residues of amelogenin are more exposed to water or the charged residues of AB3 in the presence of AB3. Because amelogenin self-assembles at pH 7.3, the redshift implies that AB3 interaction may dissociate the amelogenin nanospheres to oligomers leading to exposure of the Trp residues. We have reported similar effects of enamelin on rP172 and rP148 amelogenin leading to their dis-assembly into oligomers. These studies may suggest that oligomeric forms of amelogenin together with ameloblastin and enamelin proteolytic products may well be in vivo functional entities [21], [29].
Note that all Trp residues (W25, W45, W161) contribute to the intrinsic fluorescence spectra of wild-type amelogenin in Fig. 1. The use of variants with two Tyr-for-Trp substitutions allowed us to distinguish the contribution of each Trp residue to the spectra as the result of their interactions with ameloblastin peptide AB3. Comparing the fluorescence spectra of amelogenin variants rP172(W25), rP172(W45) and rP172(W161) titrated with the AB3 confirms that the sequence around W25 is the domain for the interaction between amelogenin and ameloblastin AB3 peptide.
The fluorescence spectra of TRAP, containing both W25 and W45, titrated with AB3 confirmed that the ameloblastin domain encoded by exon 5 interacts with the N-terminus of amelogenin. It is therefore reasonable to assume that the contribution of intrinsic fluorescence in TRAP-AB3 interactions results from W25 as W45 is at the C-terminus of TRAP and is known to be a photolytic cleavage site [18]. TRAP is a proteolytic product of full-length amelogenin that accumulates in the extracellular matrix during the transition and early maturation stages [18]. The interactions demonstrated here between TRAP and AB3 may have physiological significance, as suggested by others, in causing amelogenesis imperfecta [30].
As both amelogenin and ameloblastin proteins are degraded in the progress of enamel maturation, we then hypothesized that the interactions between proteolytic products of amelogenin (mainly TRAP) and ameloblastin may provide the matrix that functions to stabilize the prism organization during the early stage of enamel maturation [17], [31], [32]. These results are consistent with our very recent report that amelogenin co-localized with the N-terminal proteolytic product of ameloblastin; FRET analysis showed that these two peptides are in close proximity to each other [17]. The N-terminus of amelogenin is highly conserved among species indicating that domains within this segment are critical for amelogenin function [33]. Further evidence to support this claim is based on the amelogenesis imperfecta cases in which mutations on the N-terminal sequence were reported [5], [34], [35], [36].
Not only amelogenin, but also ameloblastin degrades rapidly after secretion in such a manner that the C-terminal fragments leave the enamel, while the N-terminal fragments containing the exon 5 coded peptide stay in the sheath space between enamel rods [31], [32], [37]. Biochemical and biophysical studies suggest that this N-terminal segment, particularly the sequence encoded by exon 5, is involved in the self-assembly of ameloblastin [11]. Our results further suggest that the same segment may also be responsible for the interaction between ameloblastin and amelogenin.
The propensity of these extracellular matrix proteins to interact with each other and with other targets is not surprising as both amelogenin and ameloblastin are intrinsically disordered proteins [38], [39], [40]. Peptide AB3 has a relatively well defined structure in the middle, while both its N- and C-termini remain unstructured [11], [41]. It is interesting that the percentage of disordered structure in amelogenin decreases and that of α-helix increases in the presence of AB3, implying that these two intrinsically disordered proteins may interact with each other, making them more structured in order to function in vivo. Our recent studies also showed that both SDS and phospholipids induced amelogenin to form more secondary structure [42], [43]. The disorder-order transition was suggested to be vital for the interactions between amelogenin and its targets.
In summary, four peptides were designed according to the amino acid sequence of ameloblastin, and the interaction between these peptides and amelogenin were studied. The results suggest that ameloblastin interacts with the N-terminus of amelogenin via the sequence encoded by exon 5. Amelogenin becomes more structured when interacting with this domain of ameloblastin. We suggest that macromolecular co-assembly between amelogenin and ameloblastin may play important roles in enamel biomineralization.
Conflict of interest
The authors declare no competing financial interests.
Acknowledgements
We acknowledge Nanobiophysics Core Facility at the University of Southern California for access to circular dichroism and fluorescence spectroscopy. This research was supported by NIH-NIDCR R01 grants; DE-13414 and DE-020099 to JMO.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.bbrep.2016.05.007.
Appendix A. Supplementary material
Supplementary material Fig. S1. CD spectra of peptides AB1, AB2 and AB3 at pH7.3 5 mM Tris–HCl buffer showing the similarity between AB2 and AB3 and their differences with AB1.
.
Supplementary material Fig. S2. CD spectra of amelogenin rP172 in the presence of AB1 N-terminus ameloblastin peptide.
.
Supplementary material
.
Supplementary material
.
References
- 1.Moradian-Oldak J. Protein-mediated enamel mineralization. Front. Biosci.: J. Virtual Libr. 2012;17:1996. doi: 10.2741/4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett J., Simmer J. Proteinases in developing dental enamel. Crit. Rev. Oral. Biol. Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 3.Smith C. Cellular and chemical events during enamel maturation. Crit. Rev. Oral. Biol. Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 4.Hu J.-C., Chun Y.-H., Al Hazzazzi T., Simmer J.P. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 2007;186:78–85. doi: 10.1159/000102683. [DOI] [PubMed] [Google Scholar]
- 5.Collier P., Sauk J., Rosenbloom J., Yuan Z., Gibson C. An amelogenin gene defect associated with human X-linked amelogenesis imperfecta. Arch. Oral. Biol. 1997;42:235–242. doi: 10.1016/s0003-9969(96)00099-4. [DOI] [PubMed] [Google Scholar]
- 6.Gibson C.W., Yuan Z.-A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J.T., Decker S., Piddington R. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 7.Krebsbach P.H., Lee S.K., Matsuki Y., Kozak C.A., Yamada K.M., Yamada Y. Full-length sequence, localization, and chromosomal mapping of ameloblastin a novel tooth-specific gene. J. Biol. Chem. 1996;271:4431–4435. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- 8.Poulter J.A., Murillo G., Brookes S.J., Smith C.E., Parry D.A., Silva S., Kirkham J., Inglehearn C.F., Mighell A.J. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum. Mol. Genet. 2014;23:5317–5324. doi: 10.1093/hmg/ddu247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukumoto S., Kiba T., Hall B., Iehara N., Nakamura T., Longenecker G., Krebsbach P.H., Nanci A., Kulkarni A.B., Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wazen R.M., Moffatt P., Zalzal S.F., Yamada Y., Nanci A. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 2009;28:292–303. doi: 10.1016/j.matbio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wald T., Osickova A., Sulc M., Benada O., Semeradtova A., Rezabkova L., Veverka V., Bednarova L., Maly J., Macek P. Intrinsically disordered enamel matrix protein ameloblastin forms ribbon-like supramolecular structures via an N-terminal segment encoded by exon 5. J. Biol. Chem. 2013;288:22333–22345. doi: 10.1074/jbc.M113.456012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatakeyama J., Fukumoto S., Nakamura T., Haruyama N., Suzuki S., Hatakeyama Y., Shum L., Gibson C., Yamada Y., Kulkarni A. Synergistic roles of amelogenin and ameloblastin. J. Dent. Res. 2009;88:318–322. doi: 10.1177/0022034509334749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zalzal S.F., Smith C.E., Nanci A. Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 2008;27:352–359. doi: 10.1016/j.matbio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Ravindranath H.H., Chen L.-S., Zeichner-David M., Ishima R., Ravindranath R.M. Interaction between the enamel matrix proteins amelogenin and ameloblastin. Biochem. Biophys. Res. Commun. 2004;323:1075–1083. doi: 10.1016/j.bbrc.2004.08.207. [DOI] [PubMed] [Google Scholar]
- 15.Ravindranath R.M., Devarajan A., Uchida T. Spatiotemporal expression of ameloblastin isoforms during murine tooth development. J. Biol. Chem. 2007;282:36370–36376. doi: 10.1074/jbc.M704731200. [DOI] [PubMed] [Google Scholar]
- 16.Mazumder P., Prajapati S., Lokappa S.B., Gallon V., Moradian-Oldak J. Analysis of co-assembly and co-localization of ameloblastin and amelogenin. Front. Physiol. 2014;5:274. doi: 10.3389/fphys.2014.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.P. Mazumder, S. Prajapati, R. Bapat, J. Moradian-Oldak, Amelogenin-ameloblastin spatial interactions around maturing enamel rods, J. Dent. Res., 2016, 10.1177/0022034516645389 (In press). [DOI] [PMC free article] [PubMed]
- 18.Fincham A.G., Moradian-Oldak J. Amelogenin post-translational modifications: carboxy-terminal processing and the phosphorylation of bovine and porcine” TRAP” and” LRAP” amelogenins. Biochem. Biophys. Res. Commun. 1993;197:248–255. doi: 10.1006/bbrc.1993.2468. [DOI] [PubMed] [Google Scholar]
- 19.Ruan Q., Moradian-Oldak J. Development of amelogenin-chitosan hydrogel for in vitro enamel regrowth with a dense interface. J. Vis. Experiments. 2014:89. doi: 10.3791/51606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakshminarayanan R., Yoon I., Hegde B.G., Fan D., Du C., Moradian-Oldak J. Analysis of secondary structure and self-assembly of amelogenin by variable temperature circular dichroism and isothermal titration calorimetry. Proteins: Struct., Funct., Bioinform. 2009;76:560–569. doi: 10.1002/prot.22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bromley K.M., Kiss A.S., Lokappa S.B., Lakshminarayanan R., Fan D., Ndao M., Evans J.S., Moradian-Oldak J. Dissecting amelogenin protein nanospheres characterization of metastable oligomers. J. Biol. Chem. 2011;286:34643–34653. doi: 10.1074/jbc.M111.250928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan J., Leung W., Moradian-Oldak J., Zeichner-David M., Fincham A. Quantitative analysis of amelogenin solubility. J. Dent. Res. 1998;77:1388–1396. doi: 10.1177/00220345980770060301. [DOI] [PubMed] [Google Scholar]
- 23.Burstein E.A., Vedenkina N.S., Ivkova M.N. Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 1973;18:263–279. doi: 10.1111/j.1751-1097.1973.tb06422.x. [DOI] [PubMed] [Google Scholar]
- 24.Sreerama N., Woody R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 25.Lokappa S.B., Chandrababu K.B., Moradian-Oldak J. Tooth enamel protein amelogenin binds to ameloblast cell membrane-mimicking vesicles via its N-terminus. Biochem. Biophys. Res. Commun. 2015;103(2):96–108. doi: 10.1016/j.bbrc.2015.07.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith C.E., Wazen R., Hu Y., Zalzal S.F., Nanci A., Simmer J.P., Hu J.C.C. Consequences for enamel development and mineralization resulting from loss of function of ameloblastin or enamelin. Eur. J. Oral. Sci. 2009;117:485–497. doi: 10.1111/j.1600-0722.2009.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan D., Iijima M., Bromley K.M., Yang X., Mathew S., Moradian-Oldak J. The cooperation of enamelin and amelogenin in controlling octacalcium phosphate crystal morphology. Cells Tissues Organs. 2011;194:194–198. doi: 10.1159/000324208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallon V., Chen L., Yang X., Moradian-Oldak J. Localization and quantitative co-localization of enamelin with amelogenin. J. Struct. Biol. 2013;183:239–249. doi: 10.1016/j.jsb.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan D., Du C., Sun Z., Lakshminarayanan R., Moradian-Oldak J. In vitro study on the interaction between the 32 kDa enamelin and amelogenin. J. Struct. Biol. 2009;166:88–94. doi: 10.1016/j.jsb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W., Gao C., Yan Y., DenBesten P. X-linked amelogenesis imperfecta may result from decreased formation of tyrosine rich amelogenin peptide (TRAP) Arch. Oral. Biol. 2003;48:177–183. doi: 10.1016/s0003-9969(02)00170-x. [DOI] [PubMed] [Google Scholar]
- 31.Uchida T., Murakami C., Wakida K., Satoda T., Dohi N., Takahashi O. Synthesis, secretion, degradation, and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J. Histochem. Cytochem. 1997;45:1329–1340. doi: 10.1177/002215549704501002. [DOI] [PubMed] [Google Scholar]
- 32.Nanci A., Zalzal S., Lavoie P., Kunikata M., Chen W.-Y., Krebsbach P., Yamada Y., Hammarström L., Simmer J., Fincham A. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J. Histochem. Cytochem. 1998;46:911–934. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- 33.Sire J.-Y., Delgado S., Fromentin D., Girondot M. Amelogenin: lessons from evolution. Arch. Oral. Biol. 2005;50:205–212. doi: 10.1016/j.archoralbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Kim J.-W., Simmer J., Hu Y., Lin B.-L., Boyd C., Wright J., Yamada C., Rayes S., Feigal R., Hu J.-C. Amelogenin p. M1T and p. W4S mutations underlying hypoplastic X-linked amelogenesis imperfecta. J. Dent. Res. 2004;83:378–383. doi: 10.1177/154405910408300505. [DOI] [PubMed] [Google Scholar]
- 35.Barron M.J., Brookes S.J., Kirkham J., Shore R.C., Hunt C., Mironov A., Kingswell N.J., Maycock J., Shuttleworth C.A., Dixon M.J. A mutation in the mouse Amelx tri-tyrosyl domain results in impaired secretion of amelogenin and phenocopies human X-linked amelogenesis imperfecta. Hum. Mol. Genet. 2010;19(7):1213–1247. doi: 10.1093/hmg/ddq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lench N.J., Winter G.B. Characterisation of molecular defects in X-linked amelogenesis imperfecta (AIH1) Hum. Mutat. 1995;5:251–259. doi: 10.1002/humu.1380050310. [DOI] [PubMed] [Google Scholar]
- 37.Murakami C., Dohi N., Fukae M., Tanabe T., Yamakoshi Y., Wakida K., Satoda T., Takahashi O., Shimizu M., Ryu O. Immunochemical and immunohistochemical study of the 27-and 29-kDa calcium-binding proteins and related proteins in the porcine tooth germ. Histochem. Cell Biol. 1997;107:485–494. doi: 10.1007/s004180050136. [DOI] [PubMed] [Google Scholar]
- 38.Delak K., Harcup C., Lakshminarayanan R., Sun Z., Fan Y., Moradian-Oldak J., Evans J.S. The Tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48:2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vymětal J., Slabý I., Spahr A., Vondrášek J., Lyngstadaas S.P. Bioinformatic analysis and molecular modelling of human ameloblastin suggest a two-domain intrinsically unstructured calcium-binding protein. Eur. J. Oral. Sci. 2008;116:124–134. doi: 10.1111/j.1600-0722.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 40.Wald T., Bednárová L., Osička R., Pachl P., Šulc M., Lyngstadaas S.P., Slaby I., Vondrášek J. Biophysical characterization of recombinant human ameloblastin. Eur. J. Oral. Sci. 2011;119:261–269. doi: 10.1111/j.1600-0722.2011.00913.x. [DOI] [PubMed] [Google Scholar]
- 41.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 42.Chandrababu K.B., Dutta K., Lokappa S.B., Ndao M., Evans J.S., Moradian-Oldak J. Structural adaptation of tooth enamel protein amelogenin in the presence of SDS micelles. Biopolymers. 2014;101:525–535. doi: 10.1002/bip.22415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bekshe Lokappa S., Balakrishna Chandrababu K., Dutta K., Perovic I., Spencer Evans J., Moradian-Oldak J. Interactions of amelogenin with phospholipids. Biopolymers. 2015;103:96–108. doi: 10.1002/bip.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Fig. S1. CD spectra of peptides AB1, AB2 and AB3 at pH7.3 5 mM Tris–HCl buffer showing the similarity between AB2 and AB3 and their differences with AB1.
Supplementary material Fig. S2. CD spectra of amelogenin rP172 in the presence of AB1 N-terminus ameloblastin peptide.
Supplementary material
Supplementary material