Abstract
The use of appearance and shape priors in image segmentation is known to improve accuracy; however, existing techniques have several drawbacks. Active shape and appearance models require landmark points and assume unimodal shape and appearance distributions. Level set based shape priors are limited to global shape similarity. In this paper, we present a novel shape and appearance priors for image segmentation based on an implicit parametric shape representation called disjunctive normal shape model (DNSM). DNSM is formed by disjunction of conjunctions of half-spaces defined by discriminants. We learn shape and appearance statistics at varying spatial scales using nonparametric density estimation. Our method can generate a rich set of shape variations by locally combining training shapes. Additionally, by studying the intensity and texture statistics around each discriminant of our shape model, we construct a local appearance probability map. Experiments carried out on both medical and natural image datasets show the potential of the proposed method.
1 Introduction
The use of prior information about shape and appearance is critical in many biomedical image segmentation problems. These include scenarios where the object of interest is poorly differentiated from surrounding structures in terms of its intensity, the object and the background have complex, variable appearances and where a significant amount of noise is present. Active shape models (ASM) and its extension active appearance models (AAM) [1] are powerful techniques for segmentation using priors. However, the explicit shape representation used in these models has some drawbacks. Annotating landmark points with correct correspondences across all example shapes can be difficult and time consuming. The extensions of the technique to handle topological changes and segment multiply-connected objects are not straightforward. Moreover, ASM and AAM use linear analysis tools such as principal component analysis (PCA), which limits the domain of applicability of these techniques to unimodal densities. To overcome the limitations of ASMs, level set based shape priors were proposed [2,3]. Because of their implicit nature, level set methods can easily handle topological changes. However, due to their non-parametric nature the use of shape priors in level-set segmentation framework has limited capability. For example, during segmentation using level set based shape priors, the candidate shapes are forced to move towards the globally similar training shapes without any consideration for any local shape similarity [2,3]. In addition, the region based shape similarity metrics used in shape prior computations do not always correspond to the true shape similarity observed by humans [2,3]. Finally, appearance statistics in level sets framework is usually limited to a simple use of global histograms [4], and its extension to full appearance models is not straightforward.
We use an implicit and parametric shape model called Disjunctive Normal Shape Models (DNSM) [5], which were previously used for interactive segmentation, to construct novel shape and appearance priors. DNSM’s parametric nature allows the use of a powerful local prior statistics, while its implicit nature removes the need to use landmark points. The major contributions of this paper include new global and semi-local shape priors for segmentation using DNSM (Section 3), and a new local appearance model for image segmentation that includes both texture and intensity (Section 4). We describe the overall segmentation algorithm that uses the proposed priors in Section 5. Section 6 uses ISBI 2013 prostate central gland segmentation and MICCAI 2012 prostate segmentation datasets, and reports state-of-the-art results on both challenges.
2 Disjunctive Normal Shape Model
DNSMs approximate the characteristic function of a shape as a union of convex polytopes which themselves are represented as intersections of half-spaces. Consider the characteristic function of a D-diinensional shape f : RD → B where B = {0,1}. Let = Ω+ = {x ∈ RD : f(x) = 1} represent the foreground region. Ω+ can be approximated as the union of N convex polytopes . The ith polytope is defined as the intersection of M half-spaces. The half-spaces are defined as Hij = {x ∈ RD : hij(x)}, where hij(x) = 1 if , and hij(x) = 0 otherwise. Therefore, Ω+ is approximated by and equivalently f(x) is approximated by the disjunctive normal form [6]. Converting the disjunctive normal form to a differentiable shape representation requires the following steps. First, De Morgan’s rules are used to replace the disjunction with negations and conjunctions which yields . Since conjunctions of binary functions are equivalent to their product and negation is equivalent to subtraction from 1, f(x) can also be approximated as . The final step for obtaining a differentiable representation is to relax the discriminants hij to sigmoid functions (Σij), which gives
| (1) |
where x = {x, y, 1} for 2-dimensional (2D) shapes and x = {x, y, z, 1} for 3-dimensional (3D) shapes. The only free parameters are wijk which determine the orientation and location of the sigmoid functions(discriminants) that define the half-spaces. The level set f(x) = 0.5 is taken to represent the interface between the foreground (f(x) > 0.5) and background (f(x) < 0.5) regions. DNSMs can be used for segmentation by minimizing edge-based and region-based energy terms when no training data are available [5]. The contributions of this paper are the construction of shape and appearance priors for the DNSM from training data and their use in segmentation.
3 DNSM Shape Priors
In this section, we describe how a DNSM shape prior can be constructed from a set of training shapes and used in the segmentation of new images. The set of parameters W = {wijk} of the DNSM are used to represent shapes; therefore, shape statistics will be constructed in this parameter space. In order to obtain pure shape statistics it is important to first remove the effects of pose variations (scale, rotation, and translation) in the training samples using image registration [7]. Then, the DNSM can be fit to the registered training shapes by choosing the weights that minimize the energy
| (2) |
where represents the individual polytopes of f(x), qt(x) is the ground truth (1 for object and 0 for background) of the tth training sample and η is a constant. The first term fits the model to the training shape, while the second term minimizes the overlap between the different polytopes. An η value of 0.1 is experimentally found to be suffieient to avoid the overlapping of the polytopes. We have found that a common initialization for all training shapes together with the second term is sufficient to keep the correspondence between the discriminants and polytopes across the training shapes. Figure 1(d-f) shows the correspondence achieved between the polytopes across the shapes in (a-c). This is an advantage over ASMs which can require manually placed landmark points to ensure correspondence. Another reason for minimizing the overlap between polytopes will be further discussed in Section 4. We minimize (2) using gradient descent to obtain Wt.
Fig. 1.

(a)-(c) are shapes from walking silhouettes dataset [3]. (d)-(f) show the non-overlapping polytopes(N=15) for shapes in (a)-(c) respectively, using DNSM. Each color corresponds to 1 polytope.
One of the major limitations of level set based shape priors is that the similarity between the candidate and training shapes are computed only globally [2,3]. Since no local shape similarity is considered, these approaches can not generate shape variations by locally combining training shapes. For instance, in Fig. 1, let shapes (a) and (b) be the training samples, and we want to segment the shape in (c). Since shape (c) is not in the training set, segmentation using global shape prior can only move the candidate shape towards the globally similar training sample. However, if shape similarities are considered locally at smaller spatial scales, then the hand positions of shape (c) are similar to shape (a), and the leg positions are similar to shape (b). Therefore, evaluating the similarity between shapes at semi-local scale, as will be defined in the next paragraph, helps to segment shape (c) in our example, by combining training shapes (a and b) at locations that are locally more similar to the candidate shape.
Let a given shape be represented by N polytopes and M discriminants per polytope using DNSM. Let us also assume that semi-local regions are represented by a single polytope (see Fig. 1(d-f)). We make this assumption for explanation purposes. It can be relaxed so that the semi-local region can be of any size. We will study the shape priors of each polytope independently by decoupling the entire shape in to N semi-local regions (polytopes). We can write the probability density function of the candidate’s ith polytope shape, represented by the weight Wi, given the discriminant parameters of the training shapes for the corresponding polytope Wit as
| (3) |
where T is the total number of training shapes, is a Gaussian kernel of standard deviation σi, d(Wi, Wif) is the ith polytope shape similarity distance between the candidate shape and tth training sample. We define the distance between two polytopes as
| (4) |
where Wijk is kth weight of the jth discriminant of the ith polytope, and WikA is the average of the kth weight across all discriminants in ith polytope. This normalization is necessary because the bias weights are typically much larger than the other weights. The shape energy for the ith polytope is defined as the negative logarithm of (3). During segmentation, the update to the discriminant weights, wijk of the ith polytope, is obtained by minimizing the polytope shape energy using gradient descent
| (5) |
Equation (5) shows that at local maxima, the candidate polytope shape is a weighted average of the corresponding polytope training shapes, where the weight depends on the similarity between the polytope of the candidate shape and that of the given training sample. Therefore, in the semi-local region represented by a given polytope, the shape prior term forces that part of the segmented image to move towards the semi-locally closest plausible shapes. The global shape prior used in level set based techniques [2,3] can be seen as a special case, where all polytopes are used together in (3). To use the global prior in our model, we let (4) be the distance between full parameter vectors d(W, Wt).
4 DNSM Appearance Priors
Histograms of the global appearances of the object and background are commonly used in image segmentation. However, medical objects usually have spatially varying intensity distributions. In this section, we construct a local appearance prior using DNSM. The first step in appearance training is representing the training shapes with DNSM using (2). Then, each pixel in the region of interest is assigned to its closest discriminant plane using point-to-plane distance. In order to have a proper local appearance prior, the different polytopes should cover non overlapping regions, which is achieved by the second term in (2), as can also be seen from Fig. 1(d-f). During training, two separate histograms are built for each discriminant: one for the foreground pixels and the other for background pixels. That is, for a shape represented by M × N DNSM, there will be 2 × M × N different intensity histograms. In addition to intensity, eight features (energy, entropy, correlation, difference moment, inertia, cluster shade, cluster prominence, and Haralick’s correlation) that summarize the texture of a given image are obtained using grey-level co-occurrence matrix texture measurements [8].
During segmentation, our goal is to compute the probability that the current pixel, with intensity I and texture vector T, belongs to the foreground region, based on the local appearance statistics obtained during the training. For each pixel, we first find its nearest discriminant ij, and then use the appearance statistics of that particular discriminant to compute the probability that the pixel belongs to the foreground
| (6) |
where Hij refer to the normalized intensity and texture histograms for foreground and background regions for discriminant ij. β is a constant value between 0 and 1, and it controls how much the texture term contributes to the appearance prior. See Fig. 2(b) for an example of appearance probability map obtained from both intensity and texture for central gland. The energy from appearance term, EAppr(W), for the segmentation is then given as
| (7) |
where f(x) is the level set value given as in (1), and S(x) is given in (6). During segmentation, the update to the discriminant weights, wijk from the appearance prior is obtained by minimizing (7) using gradient descent, which is given as
| (8) |
Fig. 2.
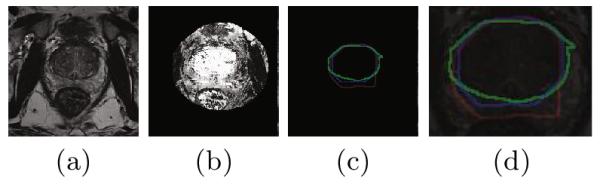
Central gland segmentation: a) MRI section; b) Local appearance probability map (the brightness corresponds to the probability of the point to be a central gland); c) Segmentation results with and without shape prior in blue and red respectively; green is the ground truth, d) Result in (c) overlaid on the MRI and zoomed in.
5 Segmentation Algorithm
The segmentation is achieved by minimizing the weighted average of the shape and appearance prior energy terms. By applying gradient descent on the combined energy, the update to the discriminants wijk is then given as
| (9) |
where and are as given in (5) and (8) respectively. α and γ are constants that determine the level of contributions from the shape and appearance priors. The steps involved in the segmentation algorithm can be summarized as follows:
Preprocessing: Intensity normalizations by histogram matching(for MRI).
Pose Estimation: The image of the appearance term (7), see Fig. 2(b), is used to find the approximate pose (location and size) of the object. This step improves segmentation accuracy, while also decreasing the number of iterations required to reach the final result.
Gradient Descent: Starting from the initial pose obtained in step 2 above, one gradient descent iteration involves: a) update the weights using the appearance prior term (8); b) register the current shape to the aligned training shapes [7]; c) update the weights using the shape prior term (5); d) register the current shape to its original pose. Note that registration of the current shape to the training shapes and back to its original pose are required for computation of the shape and appearance priors, respectively.
6 Experiments
Prostate Central Gland Segmentation
We use the NCI-ISBI 2013 Challenge - Automated Segmentation of Prostate Structures [9] MRI dataset to evaluate the effect of our shape priors. Automated segmentation of the central gland in MRI is challenging due to its variability in size, shape, location, and its similarity in appearance with the surrounding structures. Figure 2 shows one slice of the original MR image, the local appearance probability map, and the segmentation results with and without shape priors. Local appearance prior is used in the experiments of this section. Table 1 compares our segmentation algorithm using global and semi-local shape priors, with the top performing results from the NCI-ISBI challenge. Our algorithm shows a larger improvement over the 1st ranked result, compared to the improvement of the 1st rank over the 2nd rank [9] result, on both mean distance and DICE measurements. The table also shows that the semi-local shape prior outperforms the global shape prior.
Table 1.
Central gland segmentation quantitative results
Full Prostate Segmentation
We use the MICCAI PROMISE2CII2 challenge dataset to compare the local and global appearance priors. The semi-local shape prior is used here since it was shown to provide better accuracy in the previous experiment. Since the prostate has two distinct regions, the central gland and the peripheral region, a single global histogram is suboptimal. Learning local appearance at different parts of the prostate during training improves accuracy, as shown in Table 2. Our approach performs comparably to or better than the best results from the challenge participants. Figure 3 shows sample segmentation result for one slice using local and global appearance priors.
Table 2.
Prostate segmentation quantitative results
Fig. 3.

Prostate segmentation: a) MRI section; b) Section of global appearance probability map; c) Local appearance probability map, d) Segmentation results with local and global appearance priors in blue and red respectively; green is the ground truth.
7 Conclusion
In this paper we presented shape and appearance priors based image segmentation using DNSM shape representation. Because of the implicit parametric nature of DNSM, we are able to learn the shape priors at semi-local and global scales. From the experiments we have seen that semi-local shape priors give better segmentation results. DNSMs also allow us to model appearance locally or globally. Our experimental results show that learning appearance statistics at small local neighborhoods give better results. Finally, our method is able to outperform state-of-the-art techniques in central gland and full prostate segmentations. Possible extensions of our work include, coupled segmentation of multiple objects and the joint modeling of shape and appearance.
Acknowledgments
This work is supported by NSF IIS-1149299, NIH 1R01-GM098151-01, TUBITAK-113E603 and TUBITAK-2221.
References
- 1.Cootes TF, Edwards GJ, Taylor CJ. Active appearance models. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1998:484–498. [Google Scholar]
- 2.Kim J, Cetin M, Willsky AS. Nonparametric shape priors for active contour-based image segmentation. Signal Processing. 2007;87(12):3021–3044. [Google Scholar]
- 3.Cremers D, Osher SJ, Soatto S. Kernel density estimation and intrinsic alignment for shape priors in level set segmentation. International Journal of Computer Vision. 2006;69(3):335–351. [Google Scholar]
- 4.Toth R, et al. SPIE Medical Imaging. Vol. 7962. SPIE; 2011. Integrating an adaptive region-based appearance model with a landmark-free statistical shape model: application to prostate mri segmentation. [DOI] [PubMed] [Google Scholar]
- 5.Ramesh M, Mesadi F, Cetin M, Tasdizen T. Disjunctive normal shape model. ISBI, IEEE International Symposium on Biomedical Imaging; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazewinkel M. Encyclopaedia of Mathematics. Vol. 1. Springer; 1997. Encyclopaedia of Mathematics: An Updated and Annotated Translation of the Soviet Mathematical Encyclopaedia. [Google Scholar]
- 7.Zitova B, Flusser J. Image registration methods: a survey. Image and Vision Computing. 2003;21(11):977–1000. [Google Scholar]
- 8.Haralick RM. Statistical and structural approaches to texture. Proceedings of the IEEE. 1979;67(5):786–804. [Google Scholar]
- 9.NCI-ISBI Challenge. Automated segmentation of prostate structures. 2013.
- 10.MICCAI Grand Challenge. Prostate mr image segmentation. 2012.


