Abstract
Aggregative multicellularity, resulting in formation of a spore-bearing fruiting body, evolved at least six times independently amongst both eukaryotes and prokaryotes. Amongst eukaryotes, this form of multicellularity is mainly studied in the social amoeba Dictyostelium discoideum. In this review, we summarise trends in the evolution of cell-type specialisation and behavioural complexity in the four major groups of Dictyostelia. We describe the cell–cell communication systems that control the developmental programme of D. discoideum, highlighting the central role of cAMP in the regulation of cell movement and cell differentiation. Comparative genomic studies showed that the proteins involved in cAMP signalling are deeply conserved across Dictyostelia and their unicellular amoebozoan ancestors. Comparative functional analysis revealed that cAMP signalling in D. discoideum originated from a second messenger role in amoebozoan encystation. We highlight some molecular changes in cAMP signalling genes that were responsible for the novel roles of cAMP in multicellular development.
Keywords: evolution of multicellularity, encystation, sporulation, cyclic nucleotide, dual component signalling
Graphical abstract
Highlights
-
•
Many eukaryotes and prokaryotes aggregate to form a community in which cells together construct a fruiting body.
-
•
Quorum sensing, two-component signalling and signalling mediated by the cyclic nucleotides cAMP and c-di-GMP play major roles in the social behaviour of prokaryotes.
-
•
Similar signalling mechanisms also regulate aggregation and cell-type specialisation during fruiting body formation in the eukaryote Dictyostelium discoideum.
-
•
Comparative genomic and gene functional analysis revealed that the cAMP and two-component signalling mechanisms that control Dictyostelium development originated from a stress response in the unicellular ancestors.
Aggregative Multicellularity in Eukaryotes
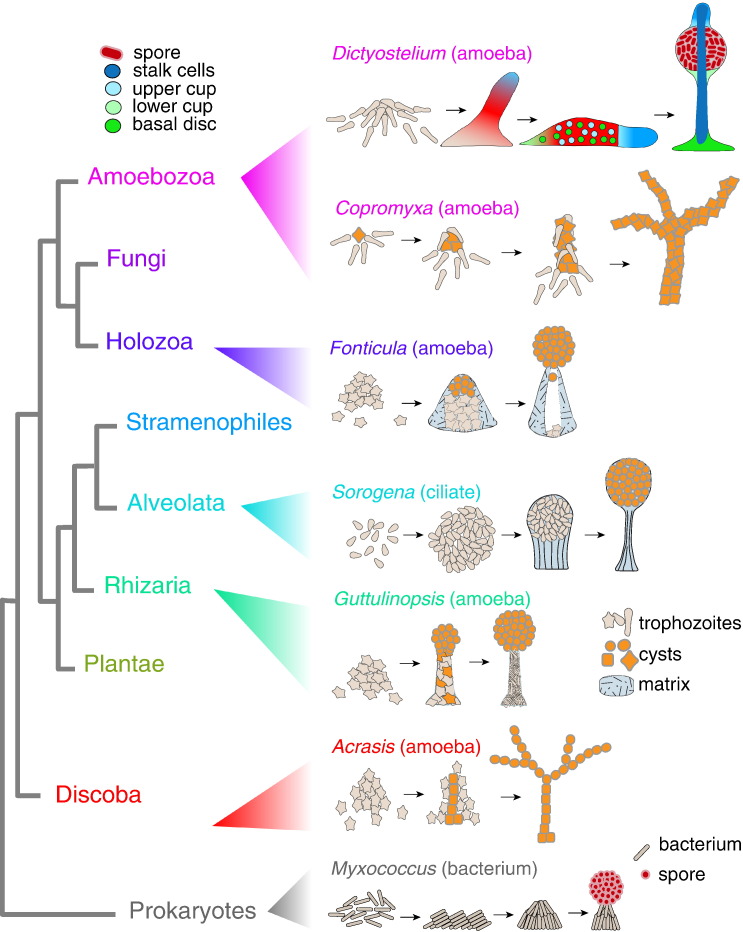
We are much more familiar with large multicellular organisms in the eukaryote domain, such as animals, plants and fungi, than with the unicellular organisms from which they evolved. The genetic diversity of eukaryotes is nevertheless much larger than the combined diversity of the multicellular forms [1], [2]. The eukaryotes comprise an immense range of morphologically distinct unicellular organisms and, in addition to animals, plants and fungi, at least six examples of organisms that independently made the transition from unicellularity to multicellularity (Fig. 1). Because these multicellular forms are rarely larger than a few centimetres, they are not commonly known.
Fig. 1.
Organisms with aggregative multicellularity.
Unicellular eukaryotes mostly have a simple life cycle consisting of a trophozoite feeding stage and a dormant cyst stage. In most eukaryote divisions and in prokaryotes, multicellular forms that aerially lift dormant spores or cysts in fruiting bodies evolved (sorocarps). Starving Myxococcus bacteria aggregate to form fruiting bodies, in which part of the rod-shaped bacteria differentiate into spherical spores. Acrasis amoebae aggregate and start encysting at the base of the structure. Other amoebae crawl to the top, rearrange themselves into chains and then encyst. Guttulinopsis amoebae construct a column surrounded by an elastic sheath. Some amoebae move to the top and differentiate as spores, whilst the remaining amoebae gradually disintegrate. The ciliate Sorogena aggregates by cell adhesion to form a mound encased in a mucous sheath. The sheath contracts to form a stalk that lift up the cells, which then encyst. Fonticula amoebae deposit a cone-shaped matrix around the aggregates. The amoebae then differentiate into spores and are expulsed through the apex. Copromyxa amoebae move towards a few encysted founder cells. Once aggregated, cells crawl on top of existing cysts and then encyst themselves. Dictyostelium aggregates first form migrating slugs. Inside the slug, the cells differentiate into precursors for the spore, stalk, basal disc and upper and lower cup cells. During fruiting formation, the precursor cell types move to their appropriate locations and complete the differentiation process.
Almost all multicellular organisms pass through a unicellular stage at least once in their life cycle. This single cell then divides repeatedly to generate the multicellular form. In animals, plants and fungi, the offspring of the first cell, a fertilised egg or a spore, remains attached to each other. However, in most other multicellular organisms, the cells disperse after cell division to maximise their access to food. They only come together again, when starved or otherwise stressed, to build a multicellular fruiting body or sorocarp with resilient cysts or spores.
This life cycle, termed alternatively aggregative or sorocarpic multicellularity, is not unique to eukaryotes and is also used by the myxobacteria in prokaryotes [3]. Eukaryotes that display aggregative multicellularity are Acrasis in the division Discoba [1], [4], Fonticula alba in Holozoa [5], Guttulinopsis spp. in Rhizaria [6], Sorogena stoianovitchae in Alveolata [7], [8] and Copromyxa and Dictyostelia spp. in Amoebozoa [9], [10].
For some genera, such as Acrasis and Copromyxa, the starving cells crawl on top of each other and differentiate into spores or cysts (Fig. 1). In others, such as Fonticula and Sorogena, the aggregated cells first deposit a structured extracellular matrix to support the spore mass. Guttulinopsis spp. show a primitive form of cell specialisation. Amoebas destined to become spores crawl to the top of the aggregate, whilst those that are left behind synthesise fibrous material to support the spores and then decay [11], [12]. Amongst aggregating eukaryotes, the Dictyostelia display the most sophisticated form of multicellularity, with a freely moving “slug” stage and up to five different cell types [13].
Evolution of Multicellular Complexity in Dictyostelia
The Dictyostelia are the largest group of eukaryotes with aggregative multicellularity, with around 150 known species. Molecular phylogenetic inference subdivides species into four major and two or three minor groups [14], [15]. Groups 1–3 consist mostly of species that form multiple small fruiting bodies from a single aggregate and/or fruiting bodies with multiple side branches. The fruiting bodies mainly form directly after aggregation, with no or little slug migration. The process of cell differentiation is relatively simple. After aggregation, the cells differentiate first into prespore cells and only cells that have reached the tip of the structure then transdifferentiate into stalk cells [16], [17]. The Acytostelids, which form clade 2A of group 2, do not form a cellular stalk. Here the prespore cells express both spore- and stalk-specific markers and collectively construct a central cellulose stalk tube. They next move up this tube and mature into spores [18].
In addition to forming multicellular fruiting bodies with spores, many species in groups 1–3 can still encapsulate individually as cysts, and have thus retained the survival strategy of their unicellular ancestors. The ability to encyst is lost in group 4, which additionally shows a pronounced increase in multicellular complexity. Species in group 4 generally form a large unbranched fruiting body from a single aggregate. Extensive migration of the sorogen or “slug” often precedes fruiting body formation, and cell differentiation is highly regulated. In the slugs, the amoebas differentiate into prespore and prestalk cells in proportions that reflect the ratio of spore to stalk cells in the fruiting body. The prestalk and prespore cells are at first intermixed, but they later sort out to form a well-defined anterior–posterior prestalk/prespore pattern. Additional cell types differentiate in the posterior, which will later form a basal disc to support the stalk and an upper cup and a lower cup to support the spore head. Group 4 is also unique in using cAMP as the chemoattractant for aggregation. In groups 1–3, the dipeptide glorin is most commonly used and more rarely folate, pterin or unknown compounds [16], [17].
In addition to encystation and fruiting body formation, Dictyostelia also have a sexual life cycle, where amoebas of opposite mating type fuse to form a zygote. The zygote then attracts and cannibalises cells of the same species and uses their contents to build a very resilient multilayered cell wall [19]. Species scattered over all four groups form these zygotic cysts or macrocysts, suggesting that this is an ancient survival strategy of Dictyostelia [15].
Cell–Cell Signalling during the Dictyostelium discoideum Life Cycle
Quorum sensing regulates the growth to development transition
The mechanisms that enable and regulate the multicellular life cycle of Dictyostelia were investigated almost exclusively in the model organism Dictyostelium discoideum, a member of group 4. Its popularity is due to the fact that procedures for genetic transformation were first developed for this species [20], soon to be followed by a wide range of molecular genetic and cell biological methodologies.
Starvation is the major trigger for entry into aggregative development, but the process is fine-tuned by the ability of amoebas to monitor their own cell density relative to that of their bacterial prey (Fig. 2). The growing cells secrete a glycoprotein, PSF (prestarvation factor) at a constant rate [21], which acts as a quorum sensing factor coordinating gene expression relative to cell density [22]. A combination of low bacterial density and high PSF induces expression of the protein kinase YakA [23], which inhibits binding of the translational repressor PufA to the 3′-end of the catalytic subunit of cAMP-dependent protein kinase (PkaC) [24]. PkaC is consequently translated and proceeds to induce expression of genes that are required for aggregation, such as the cAMP receptor carA, the adenylate cyclase acaA and the extracellular cAMP phosphodiesterase pdsA [25]. In addition to PSF, the starving cells secrete a polyketide MPBD (4-methyl-5-pentylbenzene-1,3-diol), which is also required for rapid expression of aggregation genes [26], and a protein, CMF (conditioned medium factor), which is essential for CarA-mediated signal transduction [27]. MPBD is synthesised by the polyketide synthase StlA and plays a second role in spore maturation in later development.
cAMP oscillations and cell–cell interactions prepare cells for post-aggregative development
CarA, AcaA and PdsA are key components of the network that autonomously generates pulses of cAMP [28]. These pulses are initially secreted by a few starving cells and propagate as waves through the cell population [29]. Cells move chemotactically towards a local cAMP source and collect into mounds. The utmost tip of the mound continues to emit cAMP pulses, which, by attracting cells from underneath, causes the cell mass to form the cylindrical sorogen or slug and later the fruiting body [30].
In addition to inducing chemotaxis, the cAMP pulses upregulate expression of genes that are required during and after aggregation by acting on the transcription factor GtaC [31], [32]. These genes are carA, acaA, pkaR and regA and the cell adhesion genes csaA, tgrB1 and tgrC1. TgrB1 and TgrC1 are members of a family of transmembrane proteins with highly polymorphic extracellular domains. Heterophilic interactions between compatible TgrB and TgrC proteins induce competence for post-aggregative cell differentiation (Fig. 2), and additionally serve the purpose of kin recognition, preventing non-related strains from participating in the same fruiting structure and forming an unfair share of spores compared to stalk cells [33], [34], [35].
cAMP, DIF-1 and c-di-GMP induce cell-type specialisation
After aggregation, a second adenylate cyclase, AcgA, is translationally upregulated in the posterior of the slug, where increased cAMP levels induce the differentiation of prespore cells [36], [37]. The prespore cells start to synthesise spore wall materials in Golgi-derived vesicles and additionally express the enzymes StlB, DmtA and ChlA that synthesise the chlorinated cyclohexanone DIF-1 [38], [39], [40]. DIF-1 synthesis causes differentiation of other posterior cells into prestalk O (pstO) cells, which later form the upper cup of the fruiting body, and into prestalk B (pstB) cells, which will form the lower cup and basal disc. A polyketide produced by either StlB or StlA, which is neither DIF-1 nor MPBD, is required for expression of genes at the anterior of the prestalk region. However, StlA and/or StlB are not required for the differentiation of stalk cells [39], [41].
The signal for stalk cell differentiation is c-di-GMP, which is synthesised by diguanylate cyclase A in both prestalk and stalk cells [42]. Diguanylate cyclases were previously only found in prokaryotes, where c-di-GMP is the intracellular intermediate for a range of stimuli that induce biofilm formation and other cellular responses [43].
Sensor histidine kinase and PKA-mediated signalling controls spore and stalk cell maturation
Fruiting bodies are formed by organised amoeboid movement, but the amoebas are meanwhilst becoming immobilised by cell walls as they are differentiating into spore and stalk cells. Several pathways acting in parallel therefore tightly control terminal differentiation. These pathways ultimately converge on the activation of PKA by cAMP. PKA activity is essential for both stalk and spore maturation and additionally prevents the germination of spores under conditions unfavourable for growth [44], [45], [46], [47]. AcgA and a third adenylate cyclase, AcrA, synthesise cAMP at this stage [48], but cAMP hydrolysis by the cytosolic phosphodiesterase RegA plays the most dominant role in regulation of PKA activity. The phosphodiesterase activity of RegA is activated by phosphorylation of its N-terminal response regulator domain by sensor histidine kinases (SHKs) [49], [50]. Most of the signals that control spore and stalk differentiation act either on SHKs to phosphorylate and activate RegA or on sensor histidine phosphatases (SHPs) to dephosphorylate and thereby inhibit RegA.
Stalk cell differentiation is under negative regulation of ammonia, which is produced in large quantities by protein degradation in the starving cells [51]. Ammonia activates the SHK DhkC, thereby activating RegA and inhibiting PKA [52]. Ammonia is lost from the aerially projecting tip of the early fruiting body, thus inactivating RegA and lifting PKA inhibition. Spore maturation requires release of the protein AcbA by prespore cells, which is cleaved by prestalk cells to yield the peptide SDF-2 [53]. SDF-2 in turn activates the SHP DhkA on prespore cells, which dephosphorylates RegA and thereby activates PKA [54].
Cells in fruiting bodies also secrete an adenine analogue, discadenine, which acts both to stimulate spore maturation and to inhibit spore germination. Genetic evidence indicates that the effects of discadenine are mediated by the SHK DhkB and AcrA [55], [56]. AcrA has two response regulator domains, but neither of these is required for AcrA activity [57]. It is therefore not yet clear how DhkB activates AcrA. A third factor contributing to spore maturation and preventing spore germination is the ambient high osmolarity of the spore head, which induces PKA activation by two different pathways. Firstly, high osmolarity is perceived by the extracellular osmosensor of AcgA, activating cAMP synthesis. Secondly, high osmolarity activates the SHP DokA, which in turn inactivates RegA [46], [58], [59]. A surprisingly large number of seemingly redundant pathways control the maturation and germination of spores (see also Ref. [60]). This likely reflects that the multicellular life cycle of Dictyostelia is a survival strategy that culminates into the differentiation of viable dispersible spores, which should only germinate when food is plentiful.
Prokaryote-type signalling is prevalent in Dictyostelium development
Several signal molecules with major functions in Dictyostelium development, such as cAMP and c-di-GMP, are also widely used in the prokaryote domain [61], with particularly c-di-GMP playing a major role in the association of bacteria in multicellular communities [43]. Two-component signalling systems, which consist minimally of a sensor histidine kinase/phosphatase and response regulator, represent the major mechanism for environmental sensing in prokaryotes [62] and are particularly important in controlling spore and stalk encapsulation in Dictyostelium. In addition, synchronisation of gene expression by quorum sensing is of crucial importance in early development of both Dictyostelium and myxobacteria [63]. One reason for the use of prokaryote-type signalling in Dictyostelia could be that these signalling mechanisms are particularly suited for the Dictyostelium life style, another that the Dictyostelium mechanisms directly evolved from prokaryote counterparts. More insight in the extent to which D. discoideum signalling is conserved within the Dictyostelia as a group and more deeply in their amoebozoan ancestors and other eukaryotes is required to resolve this question.
Evolutionary Reconstruction of Developmental Signalling in Dictyostelia
Comparative genomics
The genetic diversity within Dictyostelia indicates that they evolved from the last common ancestor about 0.6 billion years ago [64]. All life forms are the product of selection acting on random mutations to favour reproduction in a particular niche. This implies that there is no logic to a complex regulatory process other than the order in which its component parts evolved from an earlier state. To understand why a particular process is built up the way it is, it is essential to first reconstruct its ancestral state and next retrace how genetic change altered the process in derived lineages.
Comparative analysis of genomes that span the genetic diversity of the group of interest can yield information on the core set of genes that are present in all members and specific changes that occurred in different lineages [65]. By correlating genetic change with phenotypic innovations and testing putative causal relationship by gene replacement, it is possible to reconstruct how developmental control mechanisms evolved and generated increasing phenotypic complexity.
The genomes of species that represent all major groups of Dictyostelia have been sequenced and assembled to a high level of completion [64], [66] (G. Gloeckner and P. Schaap, unpublished results). Draft genome sequences of additional group 4 and group 2 species are also available [67], [68], as well as the genome sequence of the unicellular amoebozoan Acanthamoeba castellanii [69]. The Dictyostelid genomes are all 31–34 Mb in size, with the exception of the Dictyostelium lacteum genome in group 2 with a size of 22 Mb. The D. lacteum genome has the same number of genes (~ 12,000) as the others but contains less intergenic sequence and introns. The genome of Acanthamoeba is with 45 Mb and 15,400 genes considerably larger than that of the Dictyostelia, indicating that the evolution of multicellularity in Dictyostelia did not require more genes.
Global analysis of gene families involved in cell signalling shows that, amongst Dictyostelia, group 4 has about 30% more G-protein-coupled receptors than groups 1–3. However, the numbers of genes encoding heterotrimeric and monomeric G-proteins, sensor histidine kinases and transcription factors are about the same [64]. Polyketide synthase genes are 3 to 10-fold reduced in groups 1 and 2 compared to group 4, with each group showing considerable gene gain and loss [64], [68]. Acanthamoeba has 30% less G-protein-coupled receptors than D. discoideum but 30% more protein kinases and three times the number of sensor histidine kinases. There is a large family of 67 adenylate cyclases in Acanthamoeba, which is not present in Dictyostelia and a single ortholog of AcrA. Strikingly, Acanthamoeba has metazoan-type tyrosine kinases, which are not present in Dictyostelia, and three times the number of proteins with SH2 domains that interact with phospho-tyrosines [69]. In sheer number of cell signalling genes, the strictly unicellular Acanthamoeba therefore exceeds Dictyostelium. This suggests that innovation of gene function is probably more important for the evolution of multicellularity than a mere gain in gene numbers.
Comparative functional analysis—Genes involved in intracellular cAMP signalling
Genome comparisons provide very broad information on gene gain and loss. However, deeper and more targeted analysis of conservation and change in genes with known functions has thus far yielded the greatest insight into the evolution of developmental signalling in Dictyostelia. The most striking aspect of Dictyostelium development is the prominent role of cAMP (Fig. 2). As a secreted signal, it coordinates cell movement during aggregation and fruiting body formation and induces expression of aggregation genes and prespore genes. In a classical second messenger role, it mediates effects of many different stimuli that control the differentiation of spore and stalk cells and the germination of the spores. The proteins involved in the second messenger role of cAMP are the adenylate cyclases AcgA and AcrA; the cAMP phosphodiesterase RegA; the sensor histidine kinases DhkA, DhkB, DhkC and DokA; and the catalytic (C) and regulatory (R) subunits of PKA. AcgA and acrA are conserved in all Dictyostelid genomes, and acrA, regA, pkaC and pkaR are also present in Acanthamoeba. DhkB, dhkC and dokA are conserved throughout Dictyostelia, but there is no dhkA ortholog in group 1. None of Dictyostelium enzymes have clear orthologs amongst the 48 Acanthamoeba histidine kinases [64], [68], [69].
To assess whether conserved genes also have similar functions across Dictyostelia or even Amoebozoa, we analysed their roles by gene knockout in the group 2 species Polysphondylium pallidum or by pharmacological intervention with protein function in Acanthamoeba castellanii. As is the case in D. discoideum, disruption of pkaC in P. pallidum prevents entry into multicellular development, but P. pallidum amoebae also lose the ability to encyst [70], [71]. As described above, group 4 species, such as D. discoideum, have lost this ancestral survival strategy. In D. discoideum, the combined deletion of AcrA and AcgA prevents spore differentiation [37]. However, in P. pallidum, acra−acga− double mutants lose encystation but not spore differentiation. This is probably due to the presence of two additional acaA genes in P. pallidum of which one is expressed in prespore cells. Loss of RegA accelerates multicellular development in D. discoideum [49], and this is also the case in P. pallidum. However, P. pallidum regA− amoebae also encyst precociously, when sufficient food is still available [72]. A specific inhibitor of A. castellanii RegA also causes precocious encystation, and this is accompanied by elevated intracellular cAMP [72].
When combined, these studies show that cAMP acting on PKA has a core function in triggering encystation of single-celled amoebas in response to nutrient stress. In Dictyostelia, cAMP levels are negatively regulated by RegA and positively regulated by AcrA and AcgA (Fig. 3). In Acanthamoeba, RegA and probably AcrA have similar functions. In the course of Dictyostelid evolution, the roles of PKA, AcrA, AcgA and RegA were co-opted to additionally regulate the differentiation of spores and stalk cells. RegA, pkaC, pkaR, adenylate cyclases and a large family of sensor histidine kinases are also present in Naegleria gruberi [73], an unrelated amoeboflagellate from the division Discoba, the closest eukaryote relatives to prokaryotes [1]. Like most protozoa, Naegleria also encyst in response to stress. A role for cAMP in Naegleria encystation has yet to be demonstrated, but the conservation of the relevant cAMP signalling genes in this organism suggests that this is likely.
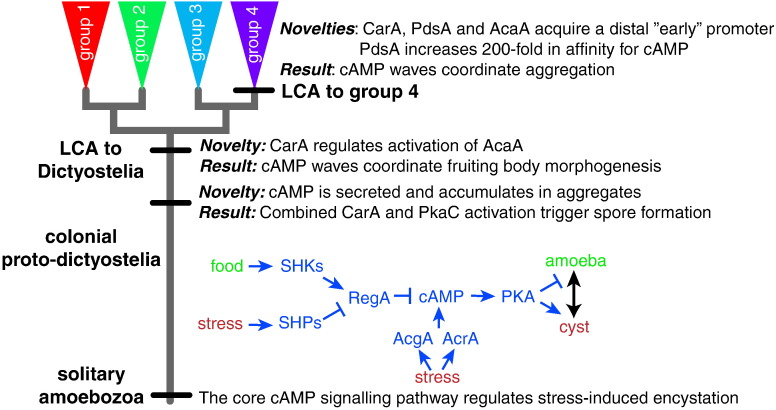
Fig. 3.
Hypothetical scenario for the evolution of developmental cAMP signalling in Dictyostelia.
The cAMP signalling mechanisms that coordinate Dictyostelium development likely evolved from a core function of cAMP as intermediate for stress-induced encystation in the unicellular ancestor. In this role, stress acts on sensor histidine phosphatases to inhibit the cAMP phosphodiesterase RegA, allowing cAMP levels, produced by AcgA or AcrA, to increase and activate PKA, which subsequently induces encystation. The roles of secreted cAMP in induction of spore formation and in coordination of fruiting body morphogenesis evolved later, with the chemoattractant role of cAMP during aggregation only emerging in the last common ancestor (LCA) to group 4.
cAMP-mediated encystation and its regulation by sensor histidine kinases/phosphatases may therefore be very deeply conserved in eukaryotes. During regulation of encystation and cyst germination, sensor histidine kinases would typically sense conditions favourable for growth and reduce cAMP levels by activating RegA, whilst the sensor histidine phosphatases would act as stress sensors and inhibit RegA (Fig. 3). In the Dictyostelid lineage, the sensor histidine kinases acquired novel functions in cell–cell communication (Fig. 2). These novel functions subjected spore and stalk cell differentiation to strict spatiotemporal control, a defining feature of multicellular development.
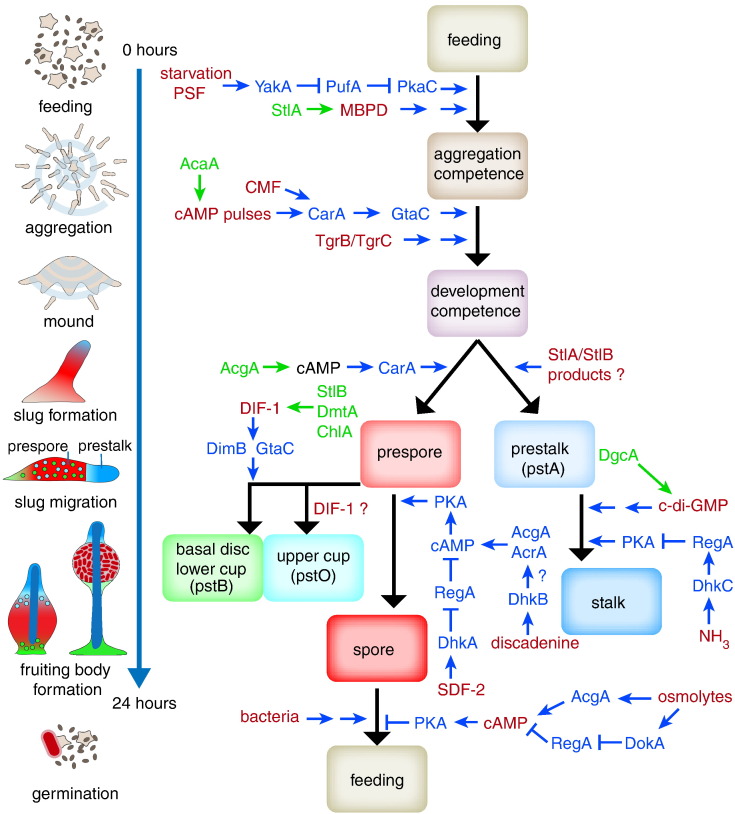
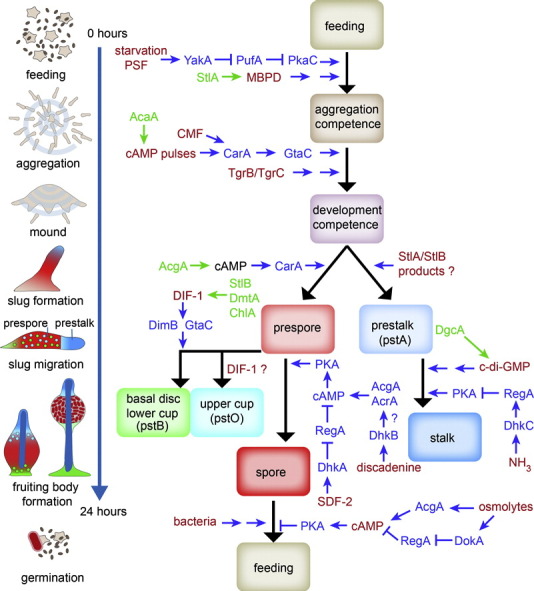
Fig. 2.
Cell signalling during Dictyostelium development.
(a) The asexual life cycle of D. discoideum. (b) Cell signalling mechanisms. The schematic shows the signals (in red) that control the life cycle transitions and the differentiation of amoebae in spores and somatic cell types. The enzymes that synthesise secreted signal molecules are shown in green text, and proteins and small molecules involved in the intracellular signal transduction pathway are in blue text. Blue arrows and t-crosses denote stimulatory and inhibitory effects, respectively. Double blue arrows signify that no components of the signal transduction pathway are known. All pathways are described in detail in the main text. Abbreviations: PSF: prestarvation factor; MPBD: 4-methyl-5-pentylbenzene-1,3-diol; cAMP: 3′-5′-cyclic adenosine monophosphate; CMF: conditioned medium factor; Tgr: transmembrane, IPT, IG, E-set, repeat protein; DIF-1: differentiation inducing factor 1; c-di-GMP: 3′,5′-cyclic diguanylic acid; NH3: ammonia; SDF-2: spore differentiation factor 2; StlA: Steely A; AcaA: adenylate cyclase A; StlB: Steely B; AcgA: adenylate cyclase G; DmtA: des-methyl-DIF-1 methyltransferase; ChlA: chlorination A; DgcA: diguanylate cyclase A; YakA: DYRK family protein kinase; PufA: pumilio RNA-binding protein; PkaC: cAMP-dependent protein kinase, catalytic subunit; CarA: cAMP receptor 1; GtaC: GATA-binding transcription factor C; DimB: transcription factor DIF-insensitive mutant B; RegA: cAMP phosphodiesterase RegA; AcrA: adenylate cyclase R DhkA: histidine phosphatase A; DhkB: histidine kinase B; DhkC: histidine kinase C; DokA: osmosensing histidine phosphatase.
Comparative functional analysis—Genes involved in extracellular cAMP signalling
The use of cAMP as extracellular signal is thus far unique for Dictyostelia, and its role as chemoattractant in aggregation is unique for group 4 [16]. The cell surface receptor CarA, the extracellular cAMP phosphodiesterase PdsA and the adenylate cyclase AcaA are specific hallmarks of extracellular signalling, although cAMP produced by AcaA can also have second messenger roles [74]. Orthologs of carA were only detected in Dictyostelia, with independent gene duplications occurring in groups 1, 3 and 4 [75], [76]. In group 4, carA is expressed from two separate promoters. A promoter, proximal to the start codon, directs carA expression after aggregation, and a more distal promoter directs expression during aggregation [77]. In groups 1–3, carA orthologs are mainly expressed after aggregation [75], suggesting that Dictyostelia initially used secreted cAMP only after aggregation.
This was confirmed by studies showing that deletion of both copies of a duplicated carA gene in P. pallidum (group 2) left aggregation intact but prevented normal fruiting body morphogenesis. Deletion of pdsA in P. pallidum also did not affect aggregation, whilst disrupting fruiting body morphogenesis [78]. This indicates that, in contrast to aggregation, which is in P. pallidum coordinated by the chemoattractant glorin, post-aggregative cell movement is coordinated by cAMP [16], [76]. The non-hydrolysable cAMP analogue Sp-cAMPS desensitises CarA, thereby disrupting pulsatile cAMP signalling [79]. In group 4 species, Sp-cAMPS therefore effectively blocks aggregation. However, in groups 1, 2 and 3 species, Sp-cAMPS only disrupts fruiting body morphogenesis [16], [75]. Combined, these studies indicate that Dictyostelia first used pulsatile cAMP signalling to coordinate fruiting body morphogenesis. Only group 4 additionally started to use secrete cAMP to coordinate aggregation. The pdsA and acaA genes show a similar complex promoter structure as the carA gene with proximal promoters directing post-aggregative expression and distal promoters directing expression during aggregation [80], [81]. One evolutionary change that contributed to the use of cAMP as attractant in group 4 was therefore the addition of a distal promoter to existing cAMP signalling genes (Fig. 3).
For carA, the addition of the distal promoter was probably the only change needed for use of CarA during aggregation, since defective aggregation of a D. discoideum cara− mutant was fully restored by expression of a group 3 carA [75]. However, for a D. discoideum pdsa− mutant, expression of a group 3 pdsA only partially restored its aggregation-defective phenotype. Both groups 2 and 3 PdsAs have a 200-fold lower affinity for cAMP than D. discoideum PdsA. It is likely that D. discoideum PdsA requires its higher affinity to hydrolyse the lower cAMP concentrations in the aggregation field. In short, both changes in gene regulation and in gene function accompanied the novel role of cAMP in group 4 aggregation.
Apart from defective morphogenesis and disorganised stalk cell differentiation, the P. pallidum car-null fruiting bodies contained cysts instead of spores in their “spore” heads. Similar to D. discoideum, P. pallidum expresses spore coat genes in response to stimulation with extracellular cAMP, but this response was lost in the car null mutant [76]. As shown above, PKA activation by intracellular cAMP is sufficient for encystation, whereas spores additionally require extracellular cAMP. By deleting cAMP receptors, the P. pallidum cells reverted to the ancestral pathway of encystation.
Dictyostelids secrete most of the cAMP that they synthesise. When cells starve in isolation, secreted cAMP levels remain low and only PKA is activated, yielding cysts. However, when cells collect in aggregates, secreted cAMP accumulates to sufficient levels to activate cAMP receptors and to induce spores instead. High extracellular cAMP is therefore a signal for the aggregated state, causing cells to differentiate into spores instead of cysts. Induction of spore formation is probably the most ancestral role of secreted cAMP. The more complex mechanisms needed to produce cAMP pulses are likely to have evolved later to form the architecturally sophisticated fruiting bodies that are characteristic for the Dictyostelia (Fig. 3).
Concluding Remarks
Aggregative multicellularity is the most common evolutionary transition from a unicellular to a multicellular life style. Many taxonomically diverse prokaryotes respond to environmental change by forming communities known as biofilms, whilst one taxon, the Myxobacteria, aggregates to form fruiting structures with spores. The latter form, also called sorocarpic multicellularity, evolved at least six times independently across most divisions of eukaryotes.
The molecular mechanisms that regulate sorocarpic development have mainly been studied in two organisms: the social amoeba D. discoideum and the myxobacterium Myxococcus xanthus. Despite the vast evolutionary distance between these organisms, these mechanisms have a number of features in common.
In both organisms, the formation of aggregates is initiated by starvation at high cell density, the latter being assessed by quorum sensing. Both secreted factors and direct cell–cell interactions play essential roles in coordinating the developmental programme, which for both species culminates in the differentiation of resilient spores. In both organisms, the secretion of a polysaccharide-rich matrix, otherwise known as slime, is essential for providing structural coherence, traction for cell movement and adhesion to substrata. Two-component signalling critically regulates sporulation in Dictyostelium, and this also appears to be the case in Myxococcus [82]. Dictyostelium uses c-di-GMP as a secreted signal to induce stalk formation. c-di-GMP induces biofilm formation in prokaryotes and roles for this molecule in extracellular matrix deposition in Myxococcus development are just emerging [83].
In D. discoideum, the regulation of sporulation by two-component signalling converges on controlling the levels of cAMP and thereby the activity of PKA, which is essential for spore formation. Comparative genomic analysis shows that the components of these pathways are not only conserved in all Dictyostelia but also in the amoebozoan ancestors and at least one other division of eukaryotes. Comparative functional analysis indicated that the original function for cAMP activation of PKA was to induce encystation of unicellular protozoa in response to environmental stress. Comparative studies also indicated that the manifold roles of both intracellular and secreted cAMP in regulating the developmental programme of Dictyostelium gradually emerged from this original role [84].
At this moment, the mechanisms controlling aggregative multicellularity in other eukaryote divisions are unknown. The similarities between Dictyostelium and Myxococcus may simply result from convergent evolution, rather than deep evolutionary conservation. However, the rapid increase in sequenced genomes for a wide variety of protists, combined with novel methods for gene manipulation, such as CRISPR-Cas9 [85] and RNA interference [86], may generate further insight in the universality of the mechanisms that control aggregative multicellularity.
Acknowledgements
Q.D., C.S. and Z.C. are supported by grant 100293/Z/12/Z from the Wellcome Trust. Y.K. is supported by grant BB/K000799/1 from the Biotechnology and Biological Sciences Research Council.
Edited by I. B. Holland
Contributor Information
Qingyou Du, Email: q.du@dundee.ac.uk.
Yoshinori Kawabe, Email: y.kawabe@dundee.ac.uk.
Christina Schilde, Email: c.schilde@dundee.ac.uk.
Zhi-hui Chen, Email: z.y.chen@dundee.ac.uk.
Pauline Schaap, Email: p.schaap@dundee.ac.uk.
References
- 1.He D., Fiz-Palacios O., Fu C.J., Fehling J., Tsai C.C., Baldauf S.L. An alternative root for the eukaryote tree of life. Curr. Biol. 2014;24:465–470. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 2.Pawlowski J., Audic S., Adl S., Bass D., Belbahri L., Berney C., Bowser S.S., Cepicka I., Decelle J., Dunthorn M., Fiore-Donno A.M., Gile G.H., Holzmann M., Jahn R., Jirku M., Keeling P.J., Kostka M., Kudryavtsev A., Lara E., Lukes J., Mann D.G., Mitchell E.A., Nitsche F., Romeralo M., Saunders G.W., Simpson A.G., Smirnov A.V., Spouge J.L., Stern R.F., Stoeck T., Zimmermann J., Schindel D., de Vargas C. CBOL protist working group: Barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 2012;10:e1001419. doi: 10.1371/journal.pbio.1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velicer G.J., Vos M. Sociobiology of the myxobacteria. Annu. Rev. Microbiol. 2009;63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 4.Brown M.W., Silberman J.D., Spiegel F.W. A contemporary evaluation of the acrasids (Acrasidae, Heterolobosea, Excavata) Eur. J. Protistol. 2012;48:103–123. doi: 10.1016/j.ejop.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Brown M.W., Spiegel F.W., Silberman J.D. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol. Biol. Evol. 2009;26:2699–2709. doi: 10.1093/molbev/msp185. [DOI] [PubMed] [Google Scholar]
- 6.Brown Matthew W., Kolisko M., Silberman Jeffrey D., Roger Andrew J. Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Curr. Biol. 2012;22:1123–1127. doi: 10.1016/j.cub.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Lasek-Nesselquist E., Katz L.A. Phylogenetic position of Sorogena stoianovitchae and relationships within the class colpodea (Ciliophora) based on SSU rDNA sequences. J. Eukaryot. Microbiol. 2001;48:604–607. doi: 10.1111/j.1550-7408.2001.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 8.Sugimoto H., Endoh H. Analysis of fruiting body development in the aggregative ciliate Sorogena stoianovitchae (Ciliophora, Colpodea) J. Eukaryot. Microbiol. 2006;53:96–102. doi: 10.1111/j.1550-7408.2005.00077.x. [DOI] [PubMed] [Google Scholar]
- 9.Baldauf S.L., Roger A.J., Wenk-Siefert I., Doolittle W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 10.Brown M.W., Silberman J.D., Spiegel F.W. “Slime molds” among the Tubulinea (Amoebozoa): Molecular systematics and taxonomy of Copromyxa. Protist. 2011;162:277–287. doi: 10.1016/j.protis.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Raper K.B., Worley A.C., Kessler D. Observations on Guttulinopsis vulgaris and Guttulinopsis nivea. Mycologia. 1977;69:1016–1030. [Google Scholar]
- 12.Erdos G.W., Raper K.B. Ultrastructural aspects of two species of Guttulinopsis. Am. J. Bot. 1978;65:552–561. [Google Scholar]
- 13.Schilde C., Schaap P. The Amoebozoa. Methods Mol. Biol. 2013;983:1–15. doi: 10.1007/978-1-62703-302-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romeralo M., Cavender J.C., Landolt J.C., Stephenson S.L., Baldauf S.L. An expanded phylogeny of social amoebas (Dictyostelia) shows increasing diversity and new morphological patterns. BMC Evol. Biol. 2011;11:84. doi: 10.1186/1471-2148-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaap P., Winckler T., Nelson M., Alvarez-Curto E., Elgie B., Hagiwara H., Cavender J., Milano-Curto A., Rozen D.E., Dingermann T., Mutzel R., Baldauf S.L. Molecular phylogeny and evolution of morphology in the social amoebas. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romeralo M., Skiba A., Gonzalez-Voyer A., Schilde C., Lawal H., Kedziora S., Cavender J.C., Glockner G., Urushihara H., Schaap P. Analysis of phenotypic evolution in Dictyostelia highlights developmental plasticity as a likely consequence of colonial multicellularity. Proc. Biol. Sci./Proc. R. Soc. 2013;280:20130976. doi: 10.1098/rspb.2013.0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilde C., Skiba A., Schaap P. Evolutionary reconstruction of pattern formation in 98 Dictyostelium species reveals that cell-type specialization by lateral inhibition is a derived trait. EvoDevo. 2014;5:34. doi: 10.1186/2041-9139-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohri K., Kiyota Y., Kuwayama H., Urushihara H. Temporal and non-permanent division of labor during sorocarp formation in the social amoeba Acytostelium subglobosum. Dev. Biol. 2013;375:202–209. doi: 10.1016/j.ydbio.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 19.O'Day D.H., Keszei A. Signalling and sex in the social amoebozoans. Biol. Rev. Camb. Philos. Soc. 2012;87:313–329. doi: 10.1111/j.1469-185X.2011.00200.x. [DOI] [PubMed] [Google Scholar]
- 20.Nellen W., Silan C., Firtel R.A. DNA-mediated transformation in Dictyostelium discoideum: Regulated expression of an actin gene fusion. Mol. Cell. Biol. 1984;4:2890–2898. doi: 10.1128/mcb.4.12.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke M., Kayman S.C., Riley K. Density-dependent induction of discoidin-I synthesis in exponentially growing cells of Dictyostelium discoideum. Differentiation. 1987;34:79–87. doi: 10.1111/j.1432-0436.1987.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 22.Kleerebezem M., Quadri L.E., Kuipers O.P., de Vos W.M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 23.Souza G.M., Lu S.J., Kuspa A. Yaka, a protein kinase required for the transition from growth to development in Dictyostelium. Development. 1998;125:2291–2302. doi: 10.1242/dev.125.12.2291. [DOI] [PubMed] [Google Scholar]
- 24.Souza G.M., daSilva A.M., Kuspa A. Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development. 1999;126:3263–3274. doi: 10.1242/dev.126.14.3263. [DOI] [PubMed] [Google Scholar]
- 25.Schulkes C., Schaap P. cAMP-dependent protein kinase activity is essential for preaggegative gene expression in Dictyostelium. FEBS Lett. 1995;368:381–384. doi: 10.1016/0014-5793(95)00676-z. [DOI] [PubMed] [Google Scholar]
- 26.Narita T.B., Chen Z.H., Schaap P., Saito T. The hybrid type polyketide synthase SteelyA is required for cAMP signalling in early Dictyostelium development. PLoS One. 2014;9:e106634. doi: 10.1371/journal.pone.0106634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuen I.S., Jain R., Bishop J.D., Lindsey D.F., Deery W.J., Van Haastert P.J.M., Gomer R.H. A density-sensing factor regulates signal transduction in Dictyostelium. J. Cell Biol. 1995;129:1251–1262. doi: 10.1083/jcb.129.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda M., Lu S., Shaulsky G., Miyazaki Y., Kuwayama H., Tanaka Y., Kuspa A., Loomis W.F. Periodic signaling controlled by an oscillatory circuit that includes protein kinases ERK2 and PKA. Science. 2004;304:875–878. doi: 10.1126/science.1094647. [DOI] [PubMed] [Google Scholar]
- 29.Tomchik K.J., Devreotes P.N. Adenosine 3′,5′-monophosphate waves in Dictyostelium discoideum: A demonstration by isotope dilution-fluorography. Science. 1981;212:443–446. doi: 10.1126/science.6259734. [DOI] [PubMed] [Google Scholar]
- 30.Dormann D., Weijer C.J. Propagating chemoattractant waves coordinate periodic cell movement in Dictyostelium slugs. Development. 2001;128:4535–4543. doi: 10.1242/dev.128.22.4535. [DOI] [PubMed] [Google Scholar]
- 31.Gerisch G., Fromm H., Huesgen A., Wick U. Control of cell-contact sites by cyclic AMP pulses in differentiating Dictyostelium cells. Nature. 1975;255:547–549. doi: 10.1038/255547a0. [DOI] [PubMed] [Google Scholar]
- 32.Cai H., Katoh-Kurasawa M., Muramoto T., Santhanam B., Long Y., Li L., Ueda M., Iglesias P.A., Shaulsky G., Devreotes P.N. Nucleocytoplasmic shuttling of a GATA transcription factor functions as a development timer. Science. 2014;343:1249531. doi: 10.1126/science.1249531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dynes J.L., Clark A.M., Shaulsky G., Kuspa A., Loomis W.F., Firtel R.A. LagC is required for cell–cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 1994;8:948–958. doi: 10.1101/gad.8.8.948. [DOI] [PubMed] [Google Scholar]
- 34.Benabentos R., Hirose S., Sucgang R., Curk T., Katoh M., Ostrowski E.A., Strassmann J.E., Queller D.C., Zupan B., Shaulsky G., Kuspa A. Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr. Biol. 2009;19:567–572. doi: 10.1016/j.cub.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirose S., Benabentos R., Ho H.-I., Kuspa A., Shaulsky G. Self-recognition in social Amoebae is mediated by allelic pairs of tiger genes. Science. 2011;333:467–470. doi: 10.1126/science.1203903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M., Van Driel R., Schaap P. Cyclic AMP-phosphodiesterase induces dedifferentiation of prespore cells in Dictyostelium discoideum slugs: Evidence that cyclic AMP is the morphogenetic signal for prespore differentiation. Development. 1988;103:611–618. [Google Scholar]
- 37.Alvarez-Curto E., Saran S., Meima M., Zobel J., Scott C., Schaap P. cAMP production by adenylyl cyclase G induces prespore differentiation in Dictyostelium slugs. Development. 2007;134:959–966. doi: 10.1242/dev.02775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson C.R., Kay R.R. The role of DIF-1 signaling in Dictyostelium development. Mol. Cell. 2000;6:1509–1514. doi: 10.1016/s1097-2765(00)00147-7. [DOI] [PubMed] [Google Scholar]
- 39.Saito T., Kato A., Kay R.R. DIF-1 induces the basal disc of the Dictyostelium fruiting body. Dev. Biol. 2008;317:444–453. doi: 10.1016/j.ydbio.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neumann C.S., Walsh C.T., Kay R.R. A flavin-dependent halogenase catalyzes the chlorination step in the biosynthesis of Dictyostelium differentiation-inducing factor 1. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5798–5803. doi: 10.1073/pnas.1001681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato Y.G., Kagami H.N., Narita T.B., Fukuzawa M., Saito T. Steely enzymes are involved in prestalk and prespore pattern formation. Biosci. Biotechnol. Biochem. 2013;77:2008–2012. doi: 10.1271/bbb.130294. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z.H., Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romling U., Galperin M.Y., Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harwood A.J., Hopper N.A., Simon M.-N., Driscoll D.M., Veron M., Williams J.G. Culmination in Dictyostelium is regulated by the cAMP-dependent protein kinase. Cell. 1992;69:615–624. doi: 10.1016/0092-8674(92)90225-2. [DOI] [PubMed] [Google Scholar]
- 45.Hopper N.A., Harwood A.J., Bouzid S., Véron M., Williams J.G. Activation of the prespore and spore cell pathway of Dictyostelium differentiation by cAMP-dependent protein kinase and evidence for its upstream regulation by ammonia. EMBO J. 1993;12:2459–2466. doi: 10.1002/j.1460-2075.1993.tb05900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Es S., Virdy K.J., Pitt G.S., Meima M., Sands T.W., Devreotes P.N., Cotter D.A., Schaap P. Adenylyl cyclase G, an osmosensor controlling germination of Dictyostelium spores. J. Biol. Chem. 1996;271:23623–23625. doi: 10.1074/jbc.271.39.23623. [DOI] [PubMed] [Google Scholar]
- 47.Virdy K.J., Sands T.W., Kopko S.H., van Es S., Meima M., Schaap P., Cotter D.A. High cAMP in spores of Dictyostelium discoideum: Association with spore dormancy and inhibition of germination. Microbiology. 1999;145:1883–1890. doi: 10.1099/13500872-145-8-1883. [DOI] [PubMed] [Google Scholar]
- 48.Soderbom F., Anjard C., Iranfar N., Fuller D., Loomis W.F. An adenylyl cyclase that functions during late development of Dictyostelium. Development. 1999;126:5463–5471. doi: 10.1242/dev.126.23.5463. [DOI] [PubMed] [Google Scholar]
- 49.Shaulsky G., Fuller D., Loomis W.F. A cAMP-phosphodiesterase controls PKA-dependent differentiation. Development. 1998;125:691–699. doi: 10.1242/dev.125.4.691. [DOI] [PubMed] [Google Scholar]
- 50.Thomason P.A., Traynor D., Cavet G., Chang W.-T., Harwood A.J., Kay R.R. An intersection of the cAMP/PKA and two-component signal transduction systems in Dictyostelium. EMBO J. 1998;17:2838–2845. doi: 10.1093/emboj/17.10.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang M., Schaap P. Ammonia depletion and DIF trigger stalk cell differentiation in intact Dictyostelium discoideum slugs. Development. 1989;105:569–574. [Google Scholar]
- 52.Singleton C.K., Zinda M.J., Mykytka B., Yang P. The histidine kinase dhkC regulates the choice between migrating slugs and terminal differentiation in Dictyostelium discoideum. Dev. Biol. 1998;203:345–357. doi: 10.1006/dbio.1998.9049. [DOI] [PubMed] [Google Scholar]
- 53.Anjard C., Loomis W.F. Peptide signaling during terminal differentiation of Dictyostelium. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7607–7611. doi: 10.1073/pnas.0501820102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang N., Soderbom F., Anjard C., Shaulsky G., Loomis W.F. SDF-2 induction of terminal differentiation in Dictyostelium discoideum is mediated by the membrane-spanning sensor kinase DhkA. Mol. Cell. Biol. 1999;19:4750–4756. doi: 10.1128/mcb.19.7.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abe H., Hashimoto K., Uchiyama M. Discadenine distribution in cellular slime molds and its inhibitory activity on spore germination. Agric. Biol. Chem. 1981;45:1295–1296. [Google Scholar]
- 56.Anjard C., Loomis W.F. Cytokinins induce sporulation in Dictyostelium. Development. 2008;135:819–827. doi: 10.1242/dev.018051. [DOI] [PubMed] [Google Scholar]
- 57.Chen Z.H., Schilde C., Schaap P. Functional dissection of adenylate cyclase R, an inducer of spore encapsulation. J. Biol. Chem. 2010;285:41724–41731. doi: 10.1074/jbc.M110.156380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ott A., Oehme F., Keller H., Schuster S.C. Osmotic stress response in Dictyostelium is mediated by cAMP. EMBO J. 2000;19:5782–5792. doi: 10.1093/emboj/19.21.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schuster S.C., Noegel A.A., Oehme F., Gerisch G., Simon M.I. The hybrid histidine kinase DokA is part of the osmotic response system of Dictyostelium. EMBO J. 1996;15:3880–3889. [PMC free article] [PubMed] [Google Scholar]
- 60.Loomis W.F. Cell signaling during development of Dictyostelium. Dev. Biol. 2014;391:1–16. doi: 10.1016/j.ydbio.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDonough K.A., Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: From signal to sword. Nat. Rev. Microbiol. 2012;10:27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krell T., Lacal J., Busch A., Silva-Jimenez H., Guazzaroni M.E., Ramos J.L. Bacterial sensor kinases: Diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 2010;64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 63.Kaiser D. Signaling in myxobacteria. Annu. Rev. Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 64.Heidel A., Lawal H., Felder M., Schilde C., Helps N., Tunggal B., Rivero F., John U., Schleicher M., Eichinger L., Platzer M., Noegel A., Schaap P., Glockner G. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 2011;21:1882–1891. doi: 10.1101/gr.121137.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loomis W.F. Comparative genomics of the dictyostelids. Methods Mol. Biol. 2013;983:39–58. doi: 10.1007/978-1-62703-302-2_3. [DOI] [PubMed] [Google Scholar]
- 66.Eichinger L., Pachebat J.A., Glockner G., Rajandream M.A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Tunggal B., Kummerfeld S., Madera M., Konfortov B.A., Rivero F., Bankier A.T., Lehmann R., Hamlin N., Davies R., Gaudet P., Fey P., Pilcher K., Chen G., Saunders D., Sodergren E., Davis P., Kerhornou A., Nie X., Hall N., Anjard C., Hemphill L., Bason N., Farbrother P., Desany B., Just E., Morio T., Rost R., Churcher C., Cooper J., Haydock S., van Driessche N., Cronin A., Goodhead I., Muzny D., Mourier T., Pain A., Lu M., Harper D., Lindsay R., Hauser H., James K., Quiles M., Madan Babu M., Saito T., Buchrieser C., Wardroper A., Felder M., Thangavelu M., Johnson D., Knights A., Loulseged H., Mungall K., Oliver K., Price C., Quail M.A., Urushihara H., Hernandez J., Rabbinowitsch E., Steffen D., Sanders M., Ma J., Kohara Y., Sharp S., Simmonds M., Spiegler S., Tivey A., Sugano S., White B., Walker D., Woodward J., Winckler T., Tanaka Y., Shaulsky G., Schleicher M., Weinstock G., Rosenthal A., Cox E.C., Chisholm R.L., Gibbs R., Loomis W.F., Platzer M., Kay R.R., Williams J., Dear P.H., Noegel A.A., Barrell B., Kuspa A. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sucgang R., Kuo A., Tian X., Salerno W., Parikh A., Feasley C.L., Dalin E., Tu H., Huang E., Barry K., Lindquist E., Shapiro H., Bruce D., Schmutz J., Salamov A., Fey P., Gaudet P., Anjard C., Babu M.M., Basu S., Bushmanova Y., van der Wel H., Katoh-Kurasawa M., Dinh C., Coutinho P.M., Saito T., Elias M., Schaap P., Kay R.R., Henrissat B., Eichinger L., Rivero F., Putnam N.H., West C.M., Loomis W.F., Chisholm R.L., Shaulsky G., Strassmann J.E., Queller D.C., Kuspa A., Grigoriev I.V. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol. 2011;12:R20. doi: 10.1186/gb-2011-12-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Urushihara H., Kuwayama H., Fukuhara K., Itoh T., Kagoshima H., Shin I.T., Toyoda A., Ohishi K., Taniguchi T., Noguchi H., Kuroki Y., Hata T., Uchi K., Mohri K., King J.S., Insall R.H., Kohara Y., Fujiyama A. Comparative genome and transcriptome analyses of the social amoeba Acytostelium subglobosum that accomplishes multicellular development without germ-soma differentiation. BMC Genomics. 2015;16:80. doi: 10.1186/s12864-015-1278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clarke M., Lohan A.J., Liu B., Lagkouvardos I., Roy S., Zafar N., Bertelli C., Schilde C., Kianianmomeni A., Burglin T.R., Frech C., Turcotte B., Kopec K.O., Synnott J.M., Choo C., Paponov I., Finkler A., Heng Tan C.S., Hutchins A.P., Weinmeier T., Rattei T., Chu J.S., Gimenez G., Irimia M., Rigden D.J., Fitzpatrick D.A., Lorenzo-Morales J., Bateman A., Chiu C.H., Tang P., Hegemann P., Fromm H., Raoult D., Greub G., Miranda-Saavedra D., Chen N., Nash P., Ginger M.L., Horn M., Schaap P., Caler L., Loftus B.J. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013;14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ritchie A.V., van Es S., Fouquet C., Schaap P. From drought sensing to developmental control: Evolution of cyclic AMP signaling in social amoebas. Mol. Biol. Evol. 2008;25:2109–2118. doi: 10.1093/molbev/msn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawabe Y., Schilde C., Du Q., Schaap P. A conserved signalling pathway for amoebozoan encystation that was co-opted for multicellular development. Sci. Rep. 2015;5:9644. doi: 10.1038/srep09644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du Q., Schilde C., Birgersson E., Chen Z.H., McElroy S., Schaap P. The cyclic AMP phosphodiesterase RegA critically regulates encystation in social and pathogenic amoebas. Cell. Signal. 2014;26:453–459. doi: 10.1016/j.cellsig.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fritz-Laylin L.K., Prochnik S.E., Ginger M.L., Dacks J.B., Carpenter M.L., Field M.C., Kuo A., Paredez A., Chapman J., Pham J., Shu S., Neupane R., Cipriano M., Mancuso J., Tu H., Salamov A., Lindquist E., Shapiro H., Lucas S., Grigoriev I.V., Cande W.Z., Fulton C., Rokhsar D.S., Dawson S.C. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 74.Saran S., Meima M.E., Alvarez-Curto E., Weening K.E., Rozen D.E., Schaap P. cAMP signaling in Dictyostelium—Complexity of cAMP synthesis, degradation and detection. J. Muscle Res. Cell Motil. 2002;23:793–802. doi: 10.1023/a:1024483829878. [DOI] [PubMed] [Google Scholar]
- 75.Alvarez-Curto E., Rozen D.E., Ritchie A.V., Fouquet C., Baldauf S.L., Schaap P. Evolutionary origin of cAMP-based chemoattraction in the social amoebae. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6385–6390. doi: 10.1073/pnas.0502238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kawabe Y., Morio T., James J.L., Prescott A.R., Tanaka Y., Schaap P. Activated cAMP receptors switch encystation into sporulation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7089–7094. doi: 10.1073/pnas.0901617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Louis J.M., Saxe C.L., III, Kimmel A.R. Two transmembrane signaling mechanisms control expression of the cAMP receptor gene cAR1 during Dictyostelium development. Proc. Natl. Acad. Sci. U. S. A. 1993;90:5969–5973. doi: 10.1073/pnas.90.13.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kawabe Y., Weening K.E., Marquay-Markiewicz J., Schaap P. Evolution of self-organisation in Dictyostelia by adaptation of a non-selective phosphodiesterase and a matrix component for regulated cAMP degradation. Development. 2012;139:1336–1345. doi: 10.1242/dev.077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossier C., Gerisch G., Malchow D., Eckstein F. Action of a slowly hydrolysable cyclic AMP analogue on developing cells of Dictyostelium discoideum. J. Cell Sci. 1978;35:321–338. doi: 10.1242/jcs.35.1.321. [DOI] [PubMed] [Google Scholar]
- 80.Faure M., Franke J., Hall A.L., Podgorski G.J., Kessin R.H. The cyclic nucleotide phosphodiesterase gene of Dictyostelium discoideum contains three promoters specific for growth, aggregation, and late development. Mol. Cell. Biol. 1990;10:1921–1930. doi: 10.1128/mcb.10.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galardi-Castilla M., Garciandia A., Suarez T., Sastre L. The Dictyostelium discoideum acaA gene is transcribed from alternative promoters during aggregation and multicellular development. PLoS One. 2010;5:e13286. doi: 10.1371/journal.pone.0013286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sarwar Z., Garza A.G. The Nla28S/Nla28 two-component signal transduction system regulates sporulation in Myxococcus xanthus. J. Bacteriol. 2012;194:4698–4708. doi: 10.1128/JB.00225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Petters T., Zhang X., Nesper J., Treuner-Lange A., Gomez-Santos N., Hoppert M., Jenal U., Sogaard-Andersen L. The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol. Microbiol. 2012;84:147–165. doi: 10.1111/j.1365-2958.2012.08015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schaap P. Evolutionary crossroads in developmental biology: Dictyostelium discoideum. Development. 2011;138:387–396. doi: 10.1242/dev.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fellmann C., Lowe S.W. Stable RNA interference rules for silencing. Nat. Cell Biol. 2014;16:10–18. doi: 10.1038/ncb2895. [DOI] [PMC free article] [PubMed] [Google Scholar]