Abstract
Crumbs 3 (CRB3) is a component of epithelial junctions, which has been implicated in apical-basal polarity, apical identity, apical stability, cell adhesion, and cell growth. CRB3 undergoes alternative splicing to yield two variants: CRB3a and CRB3b. Here, we describe novel data demonstrating that, as with previous studies on CRB3a, CRB3b also promotes the formation of tight junctions (TJs). However, significantly we demonstrate that the 4.1-ezrin–radixin–moesin-binding motif of CRB3b is required for CRB3b functionality and that ezrin binds to the FBM of CRB3b. Furthermore, we show that ezrin contributes to CRB3b functionality and the correct distribution of TJ proteins. We demonstrate that both CRB3 isoforms are required for the production of functionally mature TJs and also the localization of ezrin to the plasma membrane. Finally, we demonstrate that reduced CRB3b expression in head and neck squamous cell carcinoma (HNSCC) correlates with cytoplasmic ezrin, a biomarker for aggressive disease, and shows evidence that while CRB3a expression has no effect, low CRB3b and high cytoplasmic ezrin expression combined may be prognostic for HNSCC.
Keywords: CRB3, crumbs, FERM proteins, FBM, tight junctions, ezrin
Introduction
The maintenance of cell polarity and cell−cell adhesion is essential to the functional integrity of epithelial cell layers that line the surface of organs and assist in physiological mechanisms such as absorption and secretion (Zihni et al., 2014). Tight junctions (TJs) are apically localized intracellular structures that are part of the epithelial junctional complex. The basic structure of TJs consists of transmembrane proteins such as claudins, junctional adhesion molecules and occludins, along with regulatory proteins, which are predominantly PDZ domain-containing proteins (Gonzalez-Mariscal et al., 2003; Matter and Balda, 2003b; Guillemot et al., 2008; Zihni et al., 2014). Cell polarity and cell−cell adhesion are facilitated by TJs that allow tissues to regulate cell polarity within cells and to regulate selective permeability between cells (Anderson and Van Itallie, 2009). The barrier function of TJs was first illustrated by the retention of sphingomyelin at the apical surface of MDCK cells (Balda et al., 1996). Defects in cell polarity and cell−cell adhesion are implicated in mechanisms that range from an allergic response to inhaled antigens to metastatic cancer progression (Forster, 2008; Martin and Jiang, 2009; Sawada, 2013; Georas and Rezaee, 2014).
Crb is a transmembrane protein in Drosophila that mediates cell adhesion (Letizia et al., 2013). The intracellular domain (ICD) of Crb contains two recognizable motifs: a 4.1-ezrin–radixin–moesin-binding motif (FBM) and a PDZ-binding motif (Richard et al., 2006). In Drosophila, Crb interacts with Expanded (Ex), a FERM domain-containing protein (Ling et al., 2010; Robinson et al., 2010), via the intracellular FBM domain of Crb (Ribeiro et al., 2014), and with two FERM-containing proteins, Yurt and DMoesin (Medina et al., 2002; Laprise et al., 2006). In humans, there are three Crb orthologues: the most widely expressed orthologue in epithelial tissues is Crumbs 3 (CRB3) (Makarova et al., 2003), which is an important regulator of epithelial cell fate (Szymaniak et al., 2015). CRB3 has two isoforms: CRB3a contains a highly conserved C-terminus when compared with Drosophila Crb, while CRB3b is a splice variant resulting in a differing C-terminus. CRB3a has been shown to promote the formation of TJs and has been implicated in epithelial polarity (Roh et al., 2003; Lemmers et al., 2004; Fogg et al., 2005). The C-terminus of CRB3a contains three motifs, an FBM, a putative SH3-binding region (Zarrinpar et al., 2003), and a PDZ-binding motif (-ERLI) (Fogg et al., 2005). In contrast, the C-terminus of CRB3b retains the FBM but has a different C-terminus (ending ‘-CLPI’). The CRB3b isoform has been implicated in the generation of primary cilia, centrosomal localization during cell division and biochemically interacts with importin β1, which is inhibited by active Ran-GTP (Fan et al., 2007). To date, no interactions have been reported between CRB3b and PDZ-containing proteins.
Here, we describe novel data demonstrating that CRB3b promotes the formation of TJs. We demonstrate that the FBM of CRB3b is required for CRB3b functionality and that ezrin binds to the FBM of CRB3b. Furthermore, we show that ezrin contributes to CRB3b functionality and the correct distribution of TJ proteins. We demonstrate that Crb3 is required for the production of functionally mature TJs and the localization of ezrin to the plasma membrane. Finally, we demonstrate that in contrast to CRB3a, reduced CRB3b expression in human squamous cell carcinoma correlates with high cytoplasmic ezrin, a biomarker for poor survival outcome (Schlecht et al., 2012).
Results
CRB3a and CRB3b promote the formation of TJs in MCF10A cells via the FBM
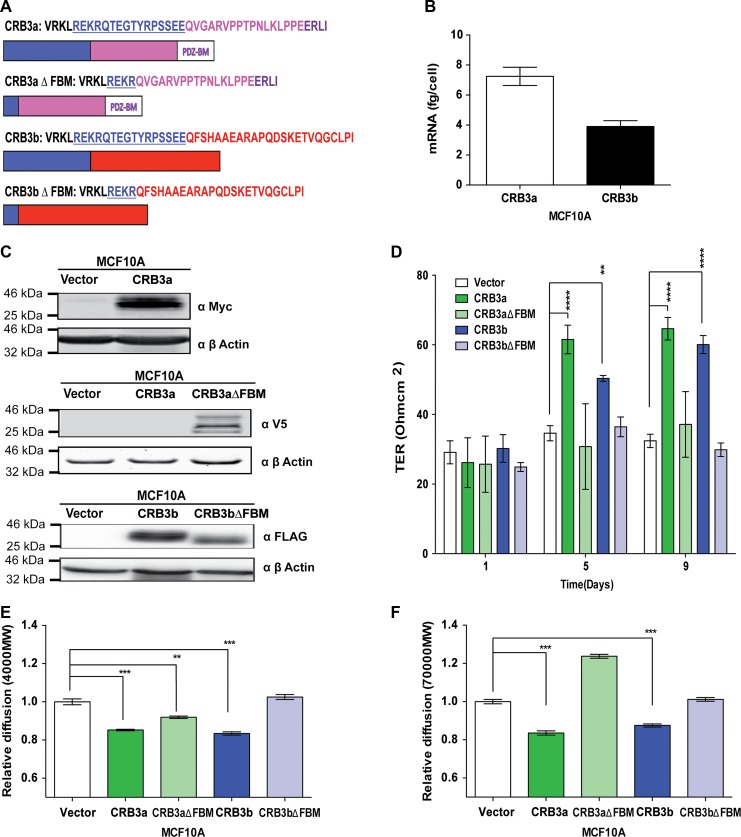
While CRB3a has been previously shown to promote the formation of TJs in MCF10A cells (Fogg et al., 2005; Elsum et al., 2013), little is known about the functional role of the CRB3b isoform. These isoforms show 100% homology across their extracellular domains and transmembrane domains. The two isoforms also share the FBM, but they differ in the last 20 amino acids of the ICD (Figure 1A). Absolute quantitative real-time polymerase chain reaction (qRT-PCR) was used to assess the basal levels of the different CRB3 mRNA isoforms in MCF10A cells, where CRB3a mRNA levels are higher than those of CRB3b (Figure 1B).
Figure 1.
CRB3b induces mature TJs in MCF10A cells. (A) Schematic diagram of the different CRB3 intracellular domains and deletion mutants used in this study: CRB3a with the FBM, C-terminal, and the PDZ-binding region; CRB3b with the FBM and the differing C-terminal. FBM deletion mutants of the CRB3 isoforms were engineered to disrupt CRB3−FERM protein interactions. (B) Absolute qRT-PCR analysis of the two isoforms of CRB3 in MCF10A cells shows higher levels of CRB3a mRNA than CRB3b. Error bars represent ±SD (n = 9). (C) Immunoblots show MCF10A-Vector, MCF10A-CRB3a, and MCF10A-CRB3aΔFBM cells probed with either MYC or V5 antibody or MCF10A-Vector, MCF10A-CRB3b, and MCF10A-CRB3bΔFBM cells probed with FLAG antibody for presence of ectopic proteins. β-actin was used as a loading control. (D) MCF10A-Vector, MCF10A-CRB3a, MCF10A-CRB3aΔFBM, MCF10A-CRB3b, and MCF10A-CRB3bΔFBM cells were grown on 0.4-μm PET membranes, and TER was recorded. Error bars represent ±SD (n = 9). (E and F) Size-selective assessment of TJ paracellular flux on MCF10A-Vector, MCF10A-CRB3a, MCF10A-CRB3aΔFBM, MCF10A-CRB3b, and MCF10A-CRB3bΔFBM cells using fluorescently labelled dextrans. Cells were grown for 5 days on 0.4-μm PET membranes prior to addition of fluorescent 4-kDa dextran–FITC (E) and 70-kDa dextran–Rhodamine (F). Error bars represent ±SD (n = 9). For all experiments, a two-way ANOVA was used with Bonferroni correction to compare against MCF10A-Vector. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Next, in order to investigate the functionality of CRB3a and CRB3b in MCF10A cells, stable cell lines were generated expressing Myc-tagged CRB3a (CRB3a), FLAG-tagged CRB3b (CRB3b) and deletions of the FBM domain in both isoforms (v5-tagged CRB3aΔFBM, FLAG-tagged CRB3bΔFBM), as shown schematically in Figure 1A. MCF10A cells expressing an empty vector (MCF10A-Vector) were used as control cells. Robust ectopic expression of full-length CRB3a (MCF10A-CRB3a), CRB3b (MCF10A-CRB3b), and the truncated mutants of CRB3a (MCF10A-CRB3aΔFBM) and CRB3b (MCF10A-CRB3bΔFBM) were confirmed by immunoblot analysis (Figure 1C). Confocal imaging of MCF10A cells revealed that both CRB3a and CRB3b localize to the regions of cell-to-cell contacts along with ezrin (Supplementary Figure S1A). These stable cell lines, MCF10A-Vector, MCF10A-CRB3a, MCF10A-CRB3aΔFBM, MCF10A-CRB3b, and MCF10A-CRB3bΔFBM were assessed by a series of different methodologies to investigate the potential physiological role of CRB3b and the potential role of the FBM in TJ formation.
Transepithelial resistance (TER) is a means to assess the integrity of a cell monolayer and whether cells are forming junctions by assaying the electrical resistance of cells (Balda et al., 1996). Interestingly, both MCF10A-CRB3a and MCF10A-CRB3b cells demonstrated significantly increased TER when compared with the MCF10A-Vector cells, indicating that both isoforms are able to promote the formation of TJs (Figure 1D) (Fogg et al., 2005; Elsum et al., 2013). However, the FBM deletion mutants of both isoforms did not show this phenotype, with no increase in TER compared with the MCF10A-Vector cells (Figure 1D), indicating that FBM of both isoforms is required for this function. These results were supported by size-selective assessment of TJ paracellular flux using fluorescently labelled dextrans (Matter and Balda, 2003a): both MCF10A-CRB3a and MCF10A-CRB3b cells displayed decreased paracellular flux as assessed by the passage of both 4-kDa fluorescein isothiocyanate (FITC) and 70-kDa Rhodamine dextrans when compared with the MCF10A-Vector cells (Figure 1E and F). In contrast, MCF10A-CRB3bΔFBM cells were not able to restrict the passage of dextrans along the paracellular space cells and maintained paracellular flux levels that were similar to MCF10A-Vector cells (Figure 1E and F). MCF10A-CRB3aΔFBM cells showed an increased passage of dextrans compared with MCF10A-CRB3a but less than MCF10A-Vector cells when using the 4-kDa FITC dextrans (Figure 1E).
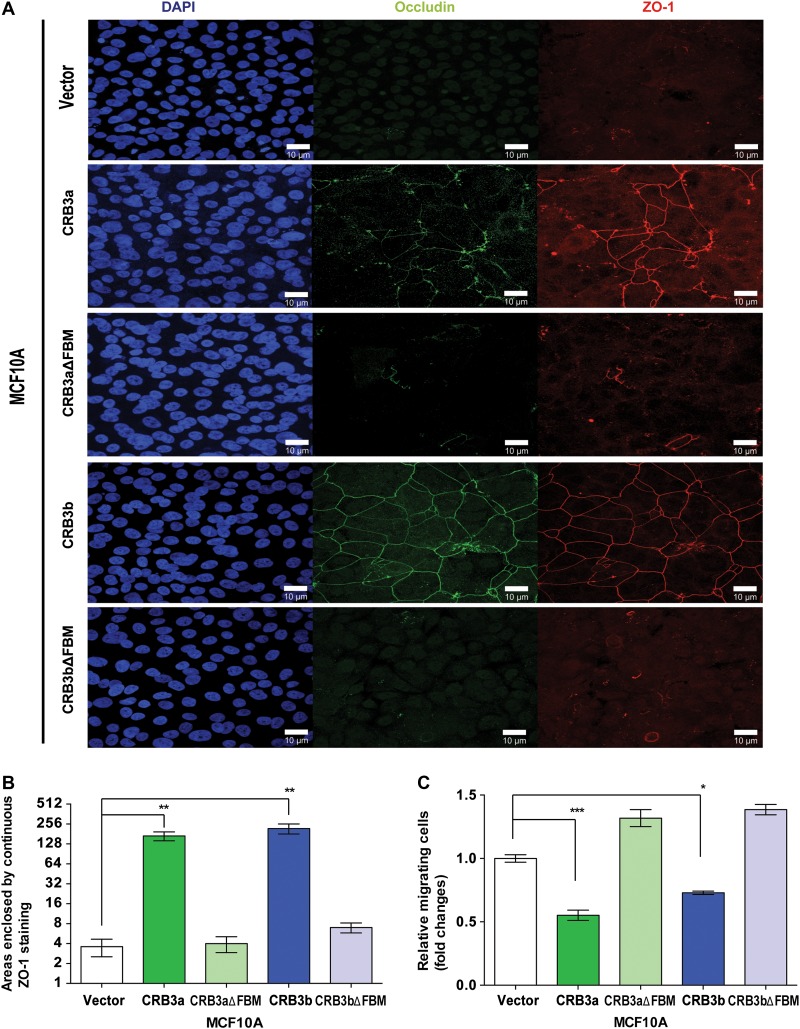
To further explore the role of these CRB3 isoforms and their effect on TJs, the presence of mature TJ proteins, occludin, and ZO-1, were assessed. While MCF10A-Vector cells lack the ability to form TJs and showed no expression of occludin and ZO-1, MCF10A-CRB3a cells promoted the formation of TJs in this cell type and displayed occludin and ZO-1 expression, as expected from published data (Fogg et al., 2005; Elsum et al., 2013). However, MCF10A-CRB3aΔFBM showed no expression of occludin or ZO-1 under the same conditions (Figure 2A). MCF10A-CRB3b cells displayed occludin and ZO-1 continuous patterning (Figure 2A), similar to that found in mature TJs and observed in MCF10A-CRB3a cells, while MCF10A-CRB3bΔFBM cells displayed an absence of ZO-1 and occludin expression under the same conditions (Figure 2A). To quantify the amount of TJ structures in MCF10A cells, we defined a TJ structure as an unbroken ring of ZO-1 staining. Using this method, we observed that the presence of CRB3a or CRB3b greatly increased the number of TJ structures present when compared with both the empty vector and the respective FBM deletion mutants (Figure 2B).
Figure 2.
CRB3a and CRB3b are able to promote TJ formation. (A) Immunofluorescent confocal imaging of MCF10A-Vector, MCF10A-CRB3a, MCF10A-CRB3aΔFBM, MCF10A-CRB3b, and MCF10A-CRB3bΔFBM cells were cultured for 4 days. Monolayers were stained for TJ markers occludin and ZO-1. Scale bar, 10 μm. (B) The areas completely enclosed by TJs were counted. Data are presented as the mean of 5 independent experiments. Error bars represent ±SD (n = 5). (C) MCF10A-Vector, MCF10A-CRB3a, MCF10A-CRB3aΔFBM, MCF10A-CRB3b, and MCF10A-CRB3bΔFBM cells were cultured in Boyden chambers across a serum gradient for 24 h and migration was assessed. Error bars represent ±SD (n = 9). For all experiments, a two-way ANOVA was used with Bonferroni correction to compare against MCF10A-Vector. *P < 0.05, **P < 0.01, ***P < 0.001.
To assess whether there were subsequent changes in migratory behaviour, the different cell lines were also assayed using an EGF-independent Boyden chamber assay. A serum gradient acted as a chemoattractant using a previously published protocol (Angus et al., 2012; Moleirinho et al., 2013a). Both MCF10A-CRB3a and MCF10A-CRB3b cells showed a decrease in migration when compared with MCF10A-Vector cells in a Boyden chamber assay (Figure 2C). However, MCF10A-CRB3aΔFBM and MCF10A-CRB3bΔFBM cells did not show a decrease in migration but displayed an increase in migration, as compared with MCF10A-Vector cells (Figure 2C). Therefore, we conclude that both isoforms of CRB3 induce TJ formation, and that this ability requires the presence of the intracellular FBM domain.
Ezrin binds to the FBM of CRB3b
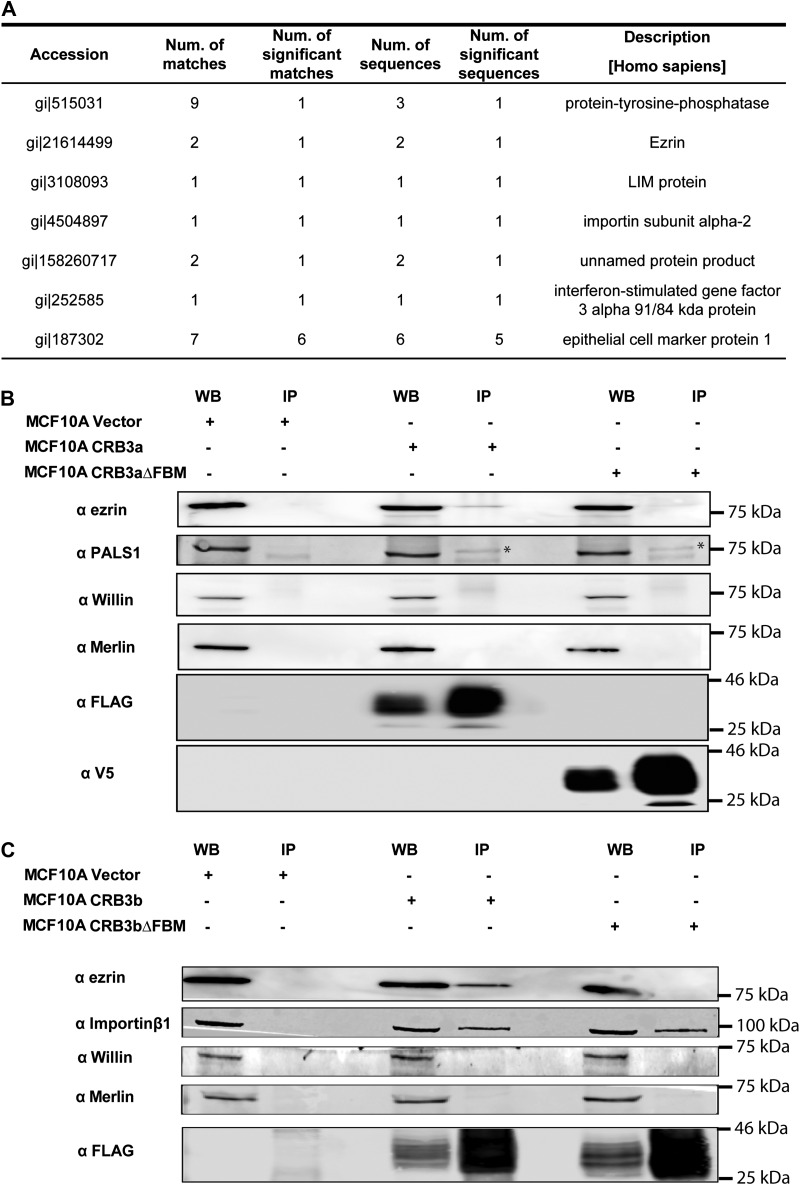
Since deletion of the FBM resulted in abrogation of CRB3b activity, we hypothesized that this deletion had disrupted an essential protein–protein interaction. To identify proteins that bind to the FBM of CRB3b, the MCF10A-CRB3b, MCF10A-CRB3bΔFBM, and MCF10A-Vector cell lines were subjected to co-immunoprecipitation (co-IP) using FLAG-mAb-conjugated magnetic bead followed by mass spectrometry analysis (Figure 3A). Potential interactions were identified, which bound to full-length CRB3b but not to CRB3bΔFBM (Supplementary Table S2). Of particular note, was the potential interaction with FERM domain-containing proteins, including ezrin (Figure 3A). To validate an interaction with ezrin, we performed co-IP on cell lysates from MCF10A-Vector, MCF10A-CRB3a, MCF10A-CRB3aΔFBM, MCF10A-CRB3b, and MCF10A-CRB3bΔFBM followed by immunoblotting using an ezrin-specific antibody. Interestingly, CRB3a–ezrin interaction was observed when full-length FLAG-tagged CRB3a was expressed (MCF10A-CRB3a cells) but was not observed when V5 tagged CRB3aΔFBM was expressed (MCF10A-CRB3aΔFBM cells) (Figure 3B). A CRB3b–ezrin interaction was observed when full-length FLAG tagged CRB3b was expressed (MCF10A-CRB3b cells) but was not observed when FLAG tagged CRB3bΔFBM was expressed (MCF10A-CRB3bΔFBM cells) (Figure 3C). To show this is not a general interaction with all FERM domain-containing proteins, the same co-IP lysates were probed for endogenous Willin/FRMD6 and Merlin/NF2 interaction. However, both endogenous Willin/FRMD6 and Merlin/NF2 did not bind to either the full-length CRB3a and CRB3b or their FBM deletion mutants, under these conditions (Figure 3B and C). PALS1, a previously known interaction partner of CRB3a was used to validate the co-IP, was observed to bind to both the full-length CRB3a and its FBM deletion mutant (Figure 3B). Importin β1 was observed to bind to CRB3b under these same conditions (Figure 3C), in agreement with a previously reported CRB3b-importin β1 interaction (Fan et al., 2007). Interestingly, we observed that importin β1 co-immunoprecipitated with both full-length CRB3b as well as the FBM deletion mutant (Figure 3C), suggesting importin β1 binds to CRB3b in a region outside the FBM.
Figure 3.
Interactions of CRB3b. MCF10A-Vector, MCF10A-CRB3b, and MCF10A-CRB3bΔFBM cells were cultured before being lysed and immunoprecipitation was carried out. The samples were reduced and separated by SDS-PAGE, following which bands were excised and sent for mass spectrometry analysis. The proteins that bound to the magnetic beads and CRB3bΔFBM were then removed from the list of proteins that bound to the full-length FLAG-CRB3b. (A) A list of potential interaction partners of CRB3b. (B) IP/western blot analysis of immunoprecipitation reactions involving both the full-length CRB3a and the CRB3aΔFBM, using anti-FLAG, anti-V5, anti-ezrin, anti-Willin, anti-Merlin, and anti-PALS1 antibodies. (C) IP/western blot analysis of immunoprecipitation reactions involving both the full-length CRB3b and the CRB3bΔFBM, using anti-FLAG, anti-ezrin, anti-Willin, anti-Merlin, and anti-importin β1 antibodies.
Ezrin contributes to CRB3b functionality
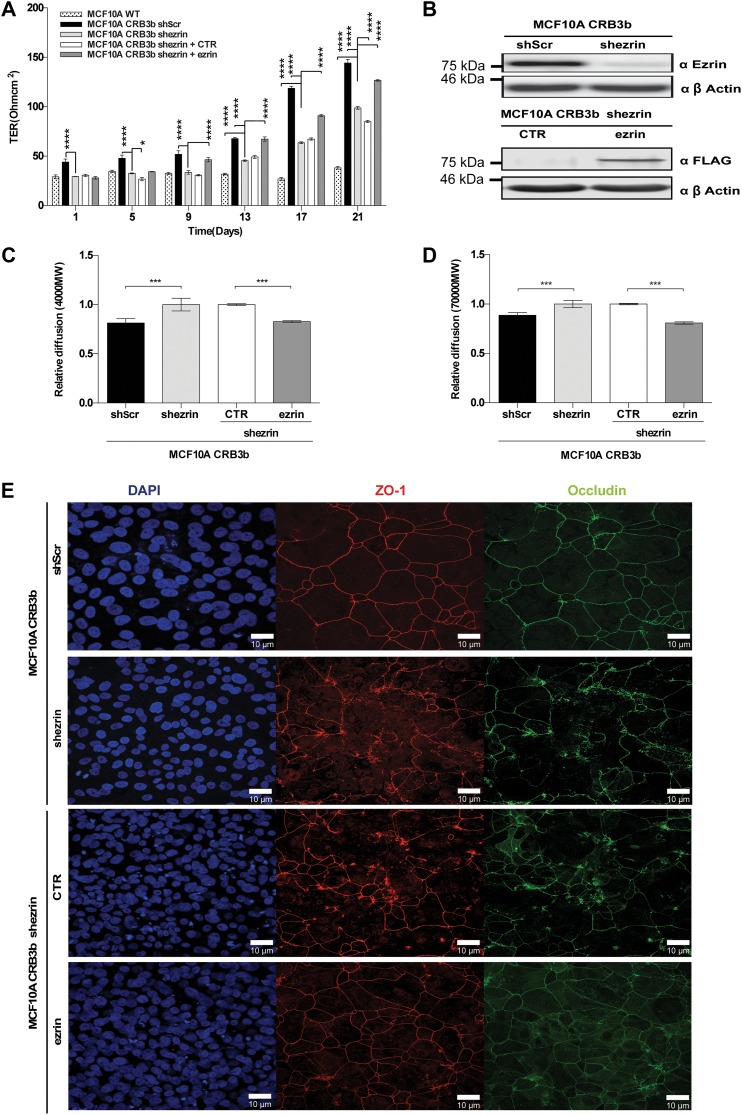
Having established that ezrin binds to CRB3b via its FBM, we determined whether this interaction is essential for CRB3b functionality. In order to do this, we stably expressed a short-hairpin RNA-targeting ezrin (shezrin) or a non-targeting scrambled control (shScr) in MCF10A-CRB3b cells. As predicted, MCF10A-CRB3b-shezrin cells displayed a significant decrease in TER when compared with MCF10A-CRB3b-shScr cells. However, the reduced expression of ezrin did not completely negate the ability of CRB3b to form TJs, as the TER reading was an intermediate reading between the wild-type MCF10A cells (MCF10A-WT) and the MCF10A-CRB3b-shScr (Figure 4A). Successful reduction in ezrin expression in MCF10A-CRB3b-shezrin cells when compared with MCF10A-CRB3b-shScr cells was confirmed by immunoblot analysis (Figure 4B). To demonstrate that the effects we observed were due to the depletion of ezrin, we overexpressed an empty vector control (MCF10A-CRB3b-shezrin+CTR) or full-length ezrin (MCF10A-CRB3b-shezrin+ezrin) that could not be targeted by the expressed shRNA in MCF10A-CRB3b-shezrin cells by the use of a lentiviral expression system. Only the reintroduction of ezrin (MCF10A-CRB3b-shezrin+ezrin) was able to rescue the observed TER phenotype, while the MCF10A-CRB3b-shezrin+CTR cells displayed TER similar to the MCF10A-CRB3b-shezrin cells (Figure 4A). The successful reintroduction of ezrin was confirmed by immunoblotting (Figure 4B).
Figure 4.
Depleted ezrin affects the functionality of CRB3b. Rescue of ezrin is able to restore the CRB3b phenotype. Short hairpin RNAs targeting ezrin (shezrin) and a non-targeting scramble control (shScr) were expressed in MCF10A-CRB3b cells. (A) MCF10A-CRB3b-shScr, MCF10A-CRB3b-shezrin, MCF10A-CRB3b-shezrin+CTR, MCF10A-CRB3b-shezrin+ezrin, and MCF10A cells were cultured on 0.4-μm PET membranes for 21 days, and TER was recorded at 4 day intervals. Error bars represent ±SD (n = 9). A two-way ANOVA was used to measure significance against MCF10A-CRB3b-shezrin cells. *P < 0.05, ****P < 0.0001. (B) Immunoblots show MCF10A-CRB3b-shScr and MCF10A-CRB3b-shezrin cells probed with an ezrin antibody or MCF10A-CRB3b-shezrin+CTR and MCF10A-CRB3b-shezrin+ezrin cells probed with a FLAG antibody to show levels of ezrin or protein expression, respectively. β-actin was used as a loading control. (C and D) Size-selective assessment of TJ paracellular flux on MCF10A-CRB3b-shScr, MCF10A-CRB3b-shezrin, MCF10A-CRB3b-shezrin+CTR, and MCF10A-CRB3b-shezrin+ezrin cells using fluorescently labelled dextrans. Cells were grown for 5 days on 0.4-μm PET membranes prior to addition of fluorescent 4-kDa dextran–FITC (C) and 70-kDa dextran–Rhodamine (D). Error bars represent ±SD (n = 9). The Student's t-test was used. ***P < 0.001. (E) Immunofluorescent confocal imaging of MCF10A-CRB3b-shScr, MCF10A-CRB3b-shezrin, MCF10A-CRB3b-shezrin+CTR, and MCF10A-CRB3b-shezrin+ezrin cells grown for 4 days. Monolayers were stained for TJ markers Occludin and ZO-1. Scale bar, 10 μm.
In addition, MCF10A-CRB3b-shezrin cells displayed an increase in paracellular flux as assessed by 4-kDa FITC and 70-kDa Rhodamine dextrans, when compared with MCF10A-CRB3b-shScr cells (Figure 4C and D), supporting the TER data. This phenotype was rescued by exogenous ezrin expression, and MCF10A-CRB3b-shezrin+ezrin cells displayed similar levels of paracellular flux as MCF10A-CRB3b-shScr cells (Figure 4C and D). Expression of markers of mature TJs was also assessed. MCF10A-CRB3b-shScr cells displayed both ZO-1 and occludin continuous patterning (Figure 4E), suggesting the presence of mature TJs. In contrast, MCF10A-CRB3b-shezrin cells displayed a disorganized pattern of both ZO-1 and occludin expression (Figure 4E). This phenotype was rescued by exogenous ezrin expression (Figure 4E). Taken together, these data suggest that ezrin contributes to CRB3b functionality by promoting the formation of mature TJs.
CRB3 is essential for TJ maintenance
To analyse the effects of the loss of CRB3 on TJs, we next tested the mouse mammary epithelial cell-line EPH4. Unlike MCF10A cells, which do not readily form TJs, EPH4 cells readily form TJs (Nagaoka et al., 2012; Yano et al., 2013), therefore providing a supplementary model to study the contribution of Crb3 to TJ formation.
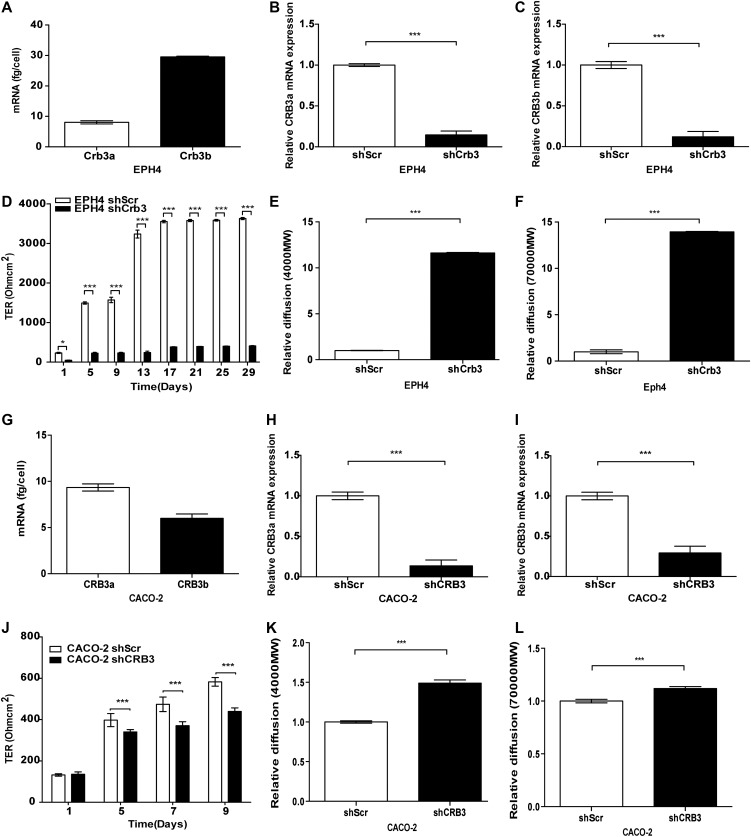
As a baseline, we assessed levels of Crb3a and Crb3b mRNA in EPH4 cells by absolute qRT-PCR: Crb3b mRNA expression was higher compared with Crb3a mRNA (Figure 5A). To investigate the effects of the loss of either Crb3 isoform expression, we designed shRNAs targeting CRB3a and CRB3b isoforms separately. Isoform-specific knock-down of CRB3 in EPH4 cells was attempted and assessed by qPCR. However, the use of isoform-specific shRNAs caused the up-regulation of the non-targeted CRB3 isoform (Supplementary Figure S2A, B). The knock-down of either Crb3a or Crb3b in EPH4 cells demonstrated a modest reduction in TER (Supplementary Figure S2C) and an increased reduction in paracellular flux as assessed by 4-kDa FITC (Supplementary Figure S2D) and 70-kDa Rhodamine dextrans (Supplementary Figure S2E), when compared with the EPH4-shScr cells. As the observed phenotypes in the single isoform knockdowns could not be singled out to be an effect of the loss of the targeted isoform or the up-regulation of the non-targeted isoform, we simultaneously knocked down expression of both Crb3a and Crb3b (shCrb3) and showed that both isoforms were reduced in expression by 90% in EPH4-shCrb3 cells, relative to their mRNA levels in EPH4-shScr cells (Figure 5B and C). EPH4-shCrb3 cells demonstrated a significant decrease in TER when compared with EPH4-shScr cells, throughout the 29-day time course (Figure 5D). EPH4-shCrb3 cells also displayed an increase in paracellular flux as assessed by 4-kDa FITC and 70-kDa Rhodamine dextrans, when compared with EPH4-shScr cells at day 5 (Figure 5E and F). In order to demonstrate that this phenotype was not cell-type specific, we additionally used CACO-2 cells, which have been previously used to study TJs (Anderson et al., 1989; Li et al., 2004). Absolute qRT-PCR analysis showed that endogenous levels of CRB3a mRNA are higher compared with those of CRB3b (Figure 5G). The knock-down of CRB3 in CACO-2 cells was performed using shRNA targeting a region common to both isoforms. Depletion of CRB3a and CRB3b mRNA in CACO-2 shCRB3 cells compared with CACO-2-shScr cells was confirmed by qPCR (Figure 5H and I). CACO-2 shCRB3 cells showed reduced TER as compared with CACO-2-shScr cells (Figure 5J) and increased paracellular flux (Figure 5K and L), when compared with CACO-2-shScr cells.
Figure 5.
The knock-down of CRB3 affects TJ behaviour. (A and G) Absolute qRT-PCR analysis of the endogenous levels of the two CRB3 isoforms in EPH4 (A) and CACO-2 cells (G). Error bars represent ±SD (n = 9). (B and H) Relative qPCR analysis of CRB3a in EPH4-shScr and EPH4-shCrb3 (B) and CACO-2-shScr and CACO-2-shCRB3 (H) cells. β-actin was used to normalize for variations in input cDNA. Error bars represent ±SD (n = 9). (C and I) Relative qPCR analysis of CRB3b in EPH4-shScr and EPH4-shCrb3 (C) and CACO-2-shScr and CACO-2-shCRB3 (I) cells. β-actin was used to normalize for variations in input cDNA. Error bars represent ±SD (n = 9). (D and J) EPH4-shScr and EPH4-shCrb3 (D) and CACO-2-shScr and CACO-2-shCRB3 (J) cells were grown on 0.4-μm PET membranes, and TER was recorded. Error bars represent ±SD (n = 9). (E, F, K, and L) Size-selective assessment of TJ paracellular flux on EPH4-shScr and EPH4-shCrb3 cells (E and F) and CACO-2-shScr and CACO-2-shCRB3 cells (K and L) using fluorescently labelled dextrans 4-kDa dextran–FITC (E, K) and 70-kDa dextran–Rhodamine (F, L). Error bars represent ±SD (n = 9). For all experiments, the Student's t-test was used. *P < 0.05, ***P < 0.001.
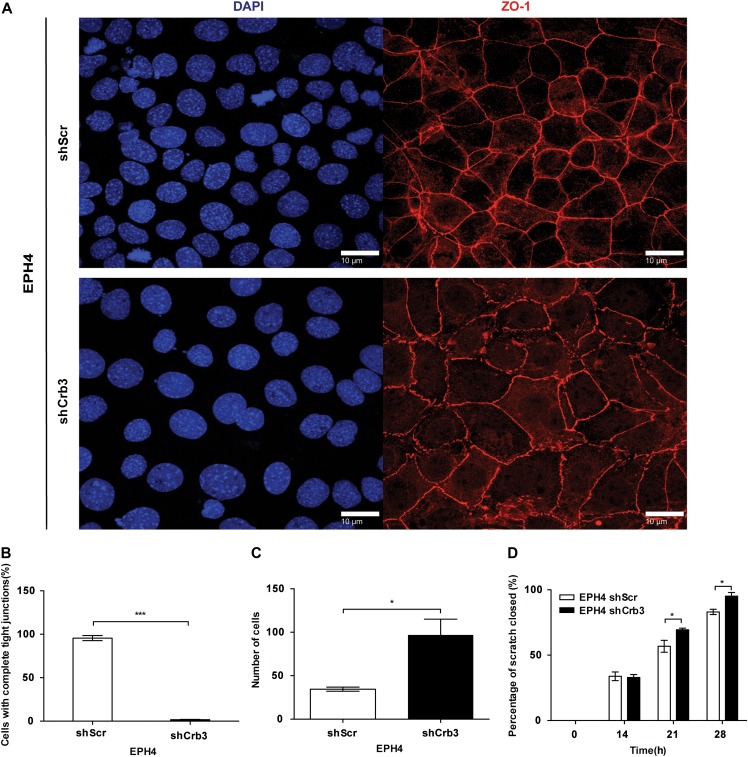
While EPH4-shScr cells displayed mature TJs as reflected by well-defined continuous patterning of ZO-1 around each individual cell, EPH4-shCrb3 cells displayed an impaired ability to form continuous regions of ZO-1 (Figure 6A). To quantify these TJ structures, we counted nuclei that were surrounded by a complete ZO-1 ring. We observed that in the control cells, nearly every nuclei had a continuous ZO-1 expression pattern around it while almost every cell with depleted Crb3 had an incomplete ZO-1 expression pattern around each nuclei (Figure 6B). EPH4-shCrb3 cells also displayed a 3-fold increase in migration in a Boyden chamber assay, when compared with EPH4-shScr cells (Figure 6C). As expected, EPH4-shCrb3 cells also displayed a significant increase in scratch closure in a wound-healing assay, when compared with EPH4-shScr cells (Figure 6D). Therefore, CRB3 expression is required to maintain mature TJs in EPH4 cells.
Figure 6.
CRB3 suppresses migration in EPH4 cells. (A) Immunofluorescent confocal imaging of EPH4-shScr and EPH4-shCrb3 cells grown on glass coverslips. Monolayers were stained for TJ marker, ZO-1. Scale bar, 10 μm. (B) The areas completely enclosed by TJs were counted. Data are presented as the mean of 5 independent experiments. Error bars represent ±SD (n = 5). (C) Wound closure ability of stable cells was assessed by a wound-healing assay. Error bars represent ±SD (n = 9). (D) EPH4-shScr and EPH4-shCrb3 cells were cultured in Boyden chambers across a serum gradient for 24 h, and migration was assessed. Error bars represent ±SD (n = 9). For all experiments, the Student's t-test was used. *P < 0.05, ***P < 0.001.
CRB3 is required for membrane localization of ezrin
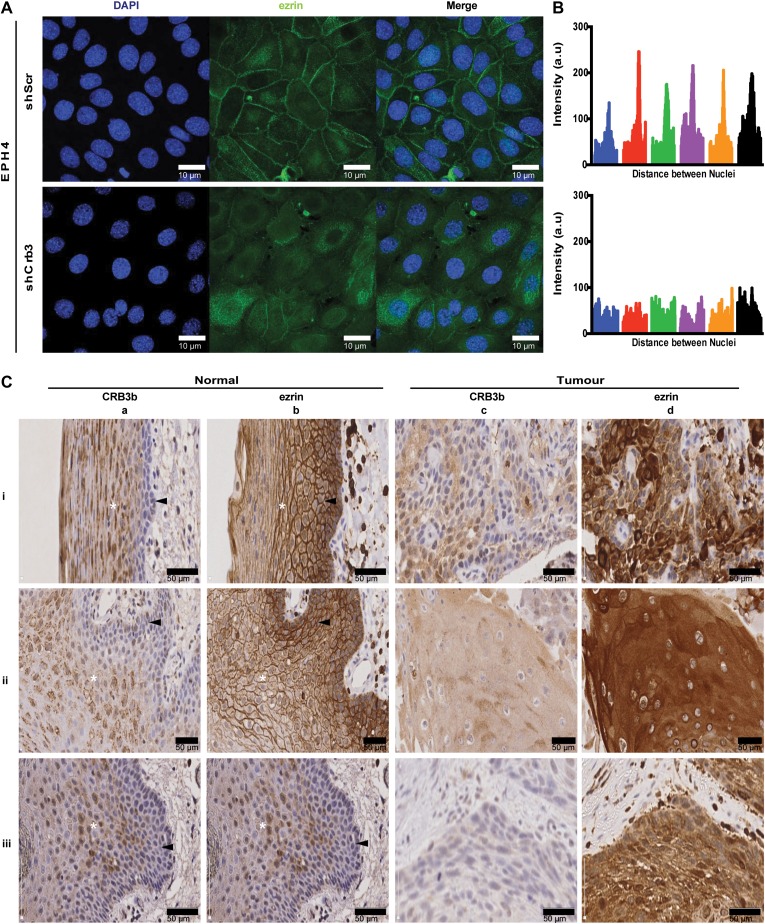
Previous data have demonstrated that the intracellular distribution of ezrin changes location from the cell membrane in normal squamous mucosa to the cytoplasm in squamous carcinoma cells (Madan et al., 2006). A subsequent report showed that high levels of cytoplasmic ezrin in head and neck squamous cell carcinoma (HNSCC) correlate with poor clinical outcome for patients, along with reduced CRB3 mRNA expression (Schlecht et al., 2012). Given these results, we hypothesized that a decrease in CRB3 expression would cause a shift in the subcellular localization of ezrin. To test this hypothesis, we used the EPH4-shScr and the EPH4-shCrb3 cells, which were grown on coverslips to allow cell−cell contact. EPH4-shScr cells displayed ezrin localization at the plasma membrane at regions of cell−cell contact, while EPH4-shCrb3 cells displayed a shift in the localization of ezrin from the membrane to the cytoplasm (Figure 7A). Using line intensity scan profiles of lines drawn from nuclei to nuclei, a peak that corresponds to the membrane accumulation of ezrin in the control cells, which is not present within the EPH4-shCrb3 cells is observed, suggesting a change in ezrin subcellular localization (Figure 7B).
Figure 7.
CRB3 affects the localization of ezrin. (A) Immunofluorescent confocal imaging of EPH4-shScr and EPH4-shCrb3 cells grown on glass coverslips. Cell monolayers were assessed for expression of endogenous ezrin. Scale bar, 10 μm. (B) The line scan intensity profiles of ezrin across cell boundaries from a line drawn between the nuclei of adjacent cells. Enrichment at the cell border is seen as a tall spike in the scan profile. Tracings from 6 cells are shown. (C) Expression of CRB3b and ezrin in squamous cell carcinoma and normal adjacent squamous epithelium (n = 25), as assessed by IHC and DAB detection. Three representative cases (i, ii, iii) show images of normal squamous epithelium (40× objective magnification) detecting expression of CRB3b (a-i, a-ii, a-iii) or ezrin (b-i, b-ii, b-iii) and images of squamous cell carcinoma (40× objective magnification) detecting expression of CRB3b (c-i, c-ii, c-iii) or ezrin (d-i, d-ii, d-iii). A white asterisk (*) denotes the stratum spinosum, while the black arrowhead denotes the stratum basale. Scale bar, 50 μm.
In order to determine whether the reduced expression of CRB3b correlates with cytoplasmic ezrin in a more physiological and clinical context, HNSCC tissue and corresponding normal adjacent healthy tissue were assessed by immunohistochemistry using anti-CRB3b and anti-ezrin antibodies followed by DAB to detect expression of either endogenous CRB3b or ezrin. Twenty-five cases of squamous cell carcinoma tissue and corresponding normal adjacent healthy tissue were selected based on cytoplasmic ezrin expression levels in the squamous carcinoma cells (Schlecht et al., 2012). In normal squamous epithelium, both CRB3b and ezrin were expressed in the stratum spinosum (n = 21). Expression of CRB3b was observed at the cell membrane (n = 15), and also occasionally perinuclear (n = 15) (Figure 7C-aii, aiii) and within the nucleus (n = 11) (Figure 7C-aiii). Similarly, CRB3b was observed to be present at cell-to-cell junctions and in the nucleus of MCF10A cells (Supplementary Figure S1B). Notably, CRB3b was strongly expressed in the stratum spinosum, but little or no expression was observed within the stratum basale in the normal tissue (n = 21) (Figure 7C-ai, aii, aiii). In adjacent serial sections, expression of ezrin was localized at the cell membrane in the stratum spinosum and stratum basale (n = 19) (Figure 7C-bi, bii, biii). In contrast, in squamous cell carcinoma tissue, CRB3b expression was absent in the majority of cells (n = 18) (Figure 7C-ci, cii, ciii) or displayed heterogeneity in expression throughout the tumour but never at the cell membrane (n = 8) (Figure 7C-ci). However, high cytoplasmic ezrin was observed within squamous carcinoma cells with little or no expression at the cell membrane (n = 16) (Figure 7C-di, dii, diii). To address the specificity of the antibody used to detect CRB3b, validation by immunoblotting was performed, showing detection of CRB3b but not of CRB3a (Supplementary Figure S3). Therefore, we conclude that reduced CRB3b expression correlates with an increase in cytoplasmic ezrin within HNSCC.
High CRB3b, but not CRB3a, improves high cytoplasmic ezrin survival prediction in HNSCC
A significant association between increased cytoplasmic ezrin and poor survival in HNSCC has previously been reported (Schlecht et al., 2012). Therefore, we assessed whether CRB3 expression added prognostic information to cytoplasmic ezrin status. CRB3 expression was measured using the Illumina HumanHT-12 v3 Expression BeadChip platform from total RNA extracted from 55 fresh and frozen primary HNSCC tumour samples that were assessed for ezrin protein expression (Schlecht et al., 2012). Three Illumina probes detected CRB3; one targeting a precursor RNA of both CRB3a and CRB3b isoform variants (ILMN_2405680), one targeting CRB3b (ILMN_1723092), and one targeting CRB3a (ILMN_1793932).
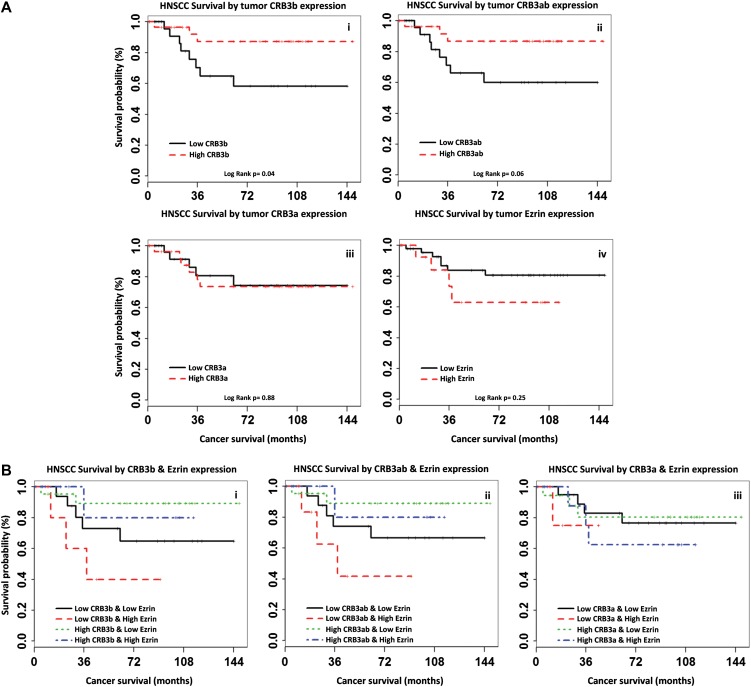
Differences in cancer survival probabilities were estimated for each CRB3 probe using Kaplan–Meier analyses and Log-rank tests comparing HNSCC stratified by CRB3 status defined using median cut-off levels in CRB3 RNA expression in the tumour (Figure 8A). Poorer survival outcome was observed with decreased CRB3 expression as measured using probes that detected CRB3b RNA (ILMN_1723092, Figure 8A-i; ILMN_2405680, Figure 8A-ii), but not for the CRB3a variant (ILMN_1793932, Figure 8A-iii). Poorer survival outcome was observed in patients with tumours with high levels of cytoplasmic ezrin (Figure 8A-iv). Compared with patients with tumours displaying high cytoplasmic ezrin and CRB3 expression, patients with tumours displaying low CRB3b RNA expression (ILMN_2405680 P = 0.03, Figure 8B-i; ILMN_1723092 log-rank P = 0.04, Figure 8B-ii) and high cytoplasmic ezrin expression had significantly poorer cancer survival outcome, but patients with tumours displaying low CRB3a RNA expression (ILMN_1793932 P = 0.45, Figure 8B-iii) and high cytoplasmic ezrin expression did not.
Figure 8.
High CRB3b improves high cytoplasmic ezrin survival prediction in HNSCC. (A) Differences in cancer survival probabilities are shown in the Kaplan–Meier plots grouping HNSCC patients by tumour CRB3 isoform precursor RNA or cytoplasmic ezrin protein expression status alone. Whereas poorer cancer survival was seen for HNSCC patients with low CRB3b RNA or high cytoplasmic ezrin protein expression, this was significant only for CRB3b expression (ILMN_1723092 log-rank P = 0.04) (i), but not for CRB3ab (ILMN_2405680 P = 0.06) (ii) or cytoplasmic ezrin expression (P = 0.25) (iv). And no association was seen for CRB3a expression alone (ILMN_1793932 P = 0.88) (iii). (B) Differences in cancer survival probabilities are shown in the Kaplan–Meier plots grouping HNSCC patients by combined CRB3 and cytoplasmic ezrin protein expression defined using median cut-off levels in CRB3b (ILMN_1723092) (i), CRB3ab (ILMN_2405680) (ii), and CRB3a (ILMN_1793932) (iii) isoform precursor RNA expression in the tumour. Compared with HNSCC patients with either high CRB3 or low cytoplasmic ezrin expression (i.e. combined as one group), HNSCC patients with low tumour CRB3b RNA and high cytoplasmic ezrin protein expression had significantly poorer cancer survival for both CRB3b variant probes (ILMN_1723092 P = 0.04; ILMN_2405680 P = 0.03), but not for CRB3a (ILMN_1793932 P = 0.45).
Discussion
TJs influence both cell polarity and cell−cell adhesion and are important for epithelial tissue physiology (Shin et al., 2006; Matter and Balda, 2007). MCF10A cells have been used by numerous groups to test the ability of proteins to promote the formation of TJs, due to the absence of mature TJs in these cells (Fogg et al., 2005; Elsum et al., 2013; Rousseau et al., 2013). While previous studies on the CRB3b isoform have implicated it in the generation of primary cilia, in cell division and as an interaction partner with importin β1 in a Ran-GTP-dependent fashion (Fan et al., 2007), here we show that CRB3b promotes the formation of TJs in MCF10A cells. This occurs even though the C-terminus of CRB3b lacks a conserved PDZ-binding motif and a conserved SH3 domain, when compared with the C-terminus of CRB3a. Functionally, CRB3b expression increases TER and reduces paracellular flux, and this is dependent on the presence of the FBM. CRB3b expression also induces both ZO-1 and occludin expression similar to published data in MCF10A cells expressing the CRB3a isoform (Fogg et al., 2005; Elsum et al., 2013). The absence of the SH3 domain and the PDZ-binding motif in CRB3b suggests that the FBM of CRB3b is indispensable for the formation of mature TJs. Our data suggest that both isoforms of CRB3 are able to promote the formation of TJs, and that the FBM is required for this function in both Crb3a and CRB3b.
Surprisingly, no proteins from known polarity complexes of CRB3-PALS1-PATJ or PAR6-PAR3-aPKC were identified by mass spectrometry in our CRB3b immunoprecipitation, suggesting that this isoform of CRB3 may use a mechanism for mature TJ formation independent of these protein complexes. This is supported by data showing that PALS1 and PAR6 bind to the “-ERLI” PDZ-binding motif in CRB3a (Lemmers et al., 2004; Fogg et al., 2005; Elsum et al., 2013). In contrast with Drosophila Crb, which interacts with Ex (Ling et al., 2010; Robinson et al., 2010), we find that under physiologically relevant conditions, mammalian CRB3a and CRB3b bind to ezrin and not to Willin/FRMD6, the reported human homologue of Ex (Hamaratoglu et al., 2006; Angus et al., 2012). Our study has been unable to replicate the interaction between the mammalian orthologues of Crb and Ex; however, this could be due to the difference between the sizes and structural conformations of Ex (1429 aa) and Willin/FRMD6 (614 aa) (Moleirinho et al., 2013b). Furthermore, we show that the FERM domain protein ezrin interacts with the FBM of CRB3b and importantly reduced ezrin expression results in diminished TER and increased paracellular flux in CRB3b-expressing cell monolayers. Similarly, we find no interaction of CRB3 with Merlin/NF2. Our data suggest that the FBM of Crb/CRB3 selectively binds FERM domain-containing proteins, but there are fundamental differences between Drosophila and mammals. Recent work has suggested that the Drosophila Crb-C-terminus is able to crystalize with the dMoesin FERM domain, where the FBM of Crb interacts with the F3 lobe of the dMoesin FERM domain (Wei et al., 2015). The same group also demonstrated that the interaction between the Drosophila Crb-C-terminus and PALS1 via the PDZ motif is a much stronger interaction than Crb-dMoesin (Li et al., 2014). Two binding motifs may have allowed for the evolution of gain of function activities in some tissues (Le Bivic, 2013), and CRB3b may have evolved to allow for a more stable FBM interaction as compared with a PDZ interaction. Nevertheless, we would argue that the FBM of both isoforms is required for the formation of TJs.
In MCF10A cells, both CRB3 isoforms are able to promote the formations of TJ junctions and colocalize with ezrin at cell-to-cell contacts. The precise mechanism of how CRB3 and ezrin promote the formation of TJs is currently unknown. One of the technical issues encountered during knocking down the individual isoforms of Crb3 in EPH4 cells was the resultant up-regulation of the non-targeted isoform. However, the single knock-down of either Crb3a or Crb3b demonstrated that TJ formation was affected when assessed by TER and dextran diffusion, suggesting functional redundancy between Crb3a and Crb3b. It is important to note that due to up-regulation of the non-targeted isoform, no firm conclusions can be made as to the precise roles of the individual isoforms of Crb3 in the formation of TJs. Data from the double knock-down of Crb3 in EPH4 demonstrated that both isoforms are required for the formation of TJs and the membrane localization of ezrin.
Down-regulation or loss of function of Crb3 contributes to tumour progression (Karp et al., 2008). When kidney cells of immortalized baby mouse were injected into nude mice, tumours formed, which could be reintroduced into tissue culture. Further analysis revealed that these tumour-derived cell lines (TDCL) harboured a loss of Crb3, and they lost the ability to form epithelial junctions when compared with the parent cells. The reintroduction of Crb3a into these TDCLs caused the cells to regain their epithelial phenotype and regain contact inhibited growth, reduced migration, and allowed for the formation of TJs (Karp et al., 2008). This suggests that CRB3 acts as a tumour suppressor by promoting epithelial characteristics. Interestingly, CRB3a and ezrin are both considered being apical determinants in cell culture models (Makarova et al., 2003; Lemmers et al., 2004; Viswanatha et al., 2012; Mojallal et al., 2014). Ezrin interacts with PALS1 (Cao et al., 2005), which also binds to the ‘-ERLI’ PDZ-binding motif in CRB3a (Lemmers et al., 2004; Fogg et al., 2005; Elsum et al., 2013). Recently, in vitro GST binding studies have suggested that the FBM of CRB3a interacts with the FERM domain of ezrin (Whiteman et al., 2014), although no functional studies were carried out. However, this raises the possibility that mammalian CRB3 isoforms may dimerize via FBM-mediated ezrin binding, since we show that both CRB3a and CRB3b bind to ezrin. It is interesting that the intestines of Crb3− /− mouse (Whiteman et al., 2014) share defect similarities with those of the Ezr− /− mouse (Casaletto et al., 2011), suggesting that they are involved in the same processes, supporting this notion. Paradoxically, ezrin may contribute to the tumour-suppressive function of CRB3 under normal physiological conditions, and may only display oncogenic properties when this interaction is disrupted (Karp et al., 2008). Recent work from the Varelas Lab has demonstrated that Crb3 controls cell fate by the retention of YAP in the cytoplasm allowing for the differentiation of airway epithelial cells (Szymaniak et al., 2015). The loss of Crb3 resulted in the nuclear localization of YAP in lung epithelial airway progenitor cells resulting in the failure of these cells to differentiate into mature airway epithelial cells, which also exhibited the loss of apicobasal polarity and TJs (Szymaniak et al., 2015). Although the expression of YAP in our HNSCC cohort has not been investigated, high expression of nuclear YAP has been reported in HNSCC (Ge et al., 2011).
Interestingly, the subcellular localization of ezrin has been implicated with aggressiveness in HNSCC (Schlecht et al., 2012). In our cohort of HNSCC, we observed that ezrin localized to the cytoplasm of the carcinoma cells, which correlated with poor outcomes for the patients (Schlecht et al., 2012). In this same cohort, we now show high CRB3b expression in normal mucosa and reduced CRB3b expression in squamous cell carcinoma, which supports the idea that the loss of CRB3 plays a role in epithelial tumours. While the expression levels of CRB3a did not stratify the outcome for patients in our study, high levels of CRB3b mRNA correlated with increased clinical survival outcome, indicating that CRB3b and significantly not CRB3a, is correlated with clinical outcome. Also, combined CRB3b mRNA expression and cytoplasmic ezrin protein expression data were able to stratify patient survival outcome better than either CRB3b mRNA expression or cytoplasmic ezrin protein expression alone.
Loss of CRB3b may be a potential mechanistic explanation for high cytoplasmic ezrin. Intense cytoplasmic ezrin has been observed and may predict poor survival in colorectal cancer, endometrioid carcinomas, uterine cervical cancer and pancreatic ductal adenocarcinomas (Elzagheid et al., 2008; Park et al., 2010; Kong et al., 2013). With cytoplasmic ezrin being reported in a variety of carcinomas, it would be relevant to determine if the loss of CRB3b correlated with this localization shift within these subsets of tumours. Currently, one study has reported loss of CRB3 expression in human tumours, in which in clear cell renal cell carcinoma patients, low CRB3-expressing tumours correlate with a poor clinical outcome (Mao et al., 2015). Our data support reported literature describing CRB3 tumour-suppressive behaviour and raises the question of whether CRB3 loss is common in cancers that have high cytoplasmic ezrin. Our data suggest that reduced or absent CRB3b expression should be investigated as a prognostic marker in squamous cell carcinoma.
In normal squamous mucosa, CRB3b showed reduced expression in basal and parabasal layers that are responsible for populating the stratum spinosum (Birajdar et al., 2014). Our CRB3b expression data in the stratum spinosum are strikingly similar to the expression patterns of differentiation markers involucrin, SPRR2, and SPRR3 observed in buccal mucosa (Gibbs and Ponec, 2000) and to the expression patterns of TJ proteins ZO-1, Claudin-1, Claudin-4, and Claudin-7 in buccal mucosa and lip epithelium (Lourenco et al., 2010). This suggests that CRB3b is preferentially expressed in maturing and differentiated cells that form TJs, and these cells display protective characteristics in the uppermost layers of oral squamous epithelia. The reasons for CRB3b nuclear localization in the MCF10A cells and HNSCC tumours are unclear at present. Previous literature and data from this study show the CRB3b interaction with Importin β1, which is responsible for both nuclear and ciliary import of proteins (Kee and Verhey, 2013). One possible explanation for this nuclear localization of CRB3b in both cell culture and HNSCC tumours could be the occurrence of high levels of nuclear import of proteins dependent on a Ran-GTP/Ran-GDP gradient. For example, nuclear EGFR is observed in HNSCC p16-negative tumours, and this nuclear transport can be facilitated by Importin β1 (Lo et al., 2006; Husain et al., 2012). In our cohort of highly aggressive and invasive tumours, the reduced levels of CRB3b may result in reduced epithelial characteristics, allowing for the tumour cells to be more proliferative. Indeed, reduced expression of TJ proteins occurs in the invasive fronts of oral squamous cell carcinomas, and there is no expression of TJ proteins in poorly differentiated tumours (Lourenco et al., 2010). This loss of TJ proteins has also been observed in breast cancer (Martin et al., 2004). As our model suggests that CRB3b is able to promote the formation of TJs, the loss of CRB3b in HNSCC may result in the inability of the tumour cells to form and maintain TJs allowing for the delamination of tumour cells from an epithelial sheet thereby undergoing single cell–mediated invasion rather than collective invasion. The absence of cell junctions is a classical feature of amoeboid/mesenchymal invasion, while the presence of cell junctions is widely observed in collective invasion of tumour cells (Friedl et al., 2012). In oral squamous cell carcinoma, poor survival outcomes correlate with a pattern of invasion where tumour satellites of single cells occur 1 mm away from the site of tumour islands, indicative of amoeboid/mesenchymal invasion (Brandwein-Gensler et al., 2005).
To conclude, we demonstrate that CRB3b promotes the formation of functional TJs in a FBM-dependent manner. Furthermore, we show that ezrin interacts with the FBM of CRB3b. These findings further elucidate the mechanism of TJ formation, implicating ezrin in a process that is independent or parallel to CRB3-PALS1-PATJ complex function. In addition, in contrast to CRB3a, reduced CRB3b expression correlates with high cytoplasmic ezrin in HNSCC, and further investigation of CRB3b as a potential prognostic marker in HNSCC is warranted in conjunction with cytoplasmic ezrin.
Materials and methods
Cell culture
MCF10A cells were maintained as described previously (Moleirinho et al., 2013a). EPH4 cells were maintained as described previously (Montesano et al., 1998). CACO-2 cells were maintained as described previously (Natoli et al., 2012). All cultures were maintained at 37°C in 5% CO2.
Retroviral and lentiviral plasmids
The FLAG tag was incorporated into the full-length and truncated CRB3b constructs by the use of the reverse primer 5′-CCCCGAATTCCTACTTGTCATCGTCATCCTTGTAATCGATGGGCAGGCAGCCCTG-3′. For pBabe plasmids, the respective inserts were cloned into the BamHI and EcoRI sites. pBabe plasmids were packaged in Phoenix A cells for viral production, and retroviral infection of MCF10A and EPH4 cells was performed according to standard protocols. Puromycin selection (2 μg/ml) or hygromycin selection (300 μg/ml) was applied 48 hours after retroviral infection and continued for 5 days (puromycin) or 8 days (hygromycin), after which cells were harvested or used for further experiments. For shRNA plasmids, the following oligonucleotides were synthesized and cloned into AgeI and EcoRI sites of pLKO.1 puro (Addgene; plasmid 8453). The shRNA duplex hairpin sequences for mouse Crb3a is 5′-TCATCACTACCAACGCCTGG-3′, mouse Crb3b 5′-TGAGGAGCAGTTTTCCCACG-3′, and human CRB3 5′-GGGCAAATACAGACCACTTCT-3′. The shRNA duplex hairpin sequence for human ezrin 5′-GCCTGATTCTCGCGATTAT-3′ has been previously published (Hsu et al., 2012). A scramble control (shScr) was obtained from Addgene (plasmid 1864). The ViraPower™ HiPerform™ Promoterless Gateway® Expression System was used in rescue experiments. The full-length human ezrin cDNA was inserted into Gateway® pENTR™ 11 Dual Selection Vector and recombined into pLenti6.4/R4R2/V5-DEST, according to manufacturers’ instructions, to create the ezrin rescue construct pLenti6.4/EF-1α/ezrin (ezrin). pLenti6.4/CMV V5-MSGW/lacZ was used as the control for the rescue experiments (CTR). pLenti6.4 constructs were packaged in HEK293FT cells. The lentiviral supernatant was added to MCF10A-CRB3b-shezrin cells. Blasticidin selection (3.5 μg/ml) was applied 48 hours after retroviral infection and continued for 15 days after which cells were harvested or used for further experiments. All constructs were verified by sequencing.
RNA isolation and qRT-PCR
Isolation of RNA from cell pellets was carried out using an RNeasy Mini Kit. First Strand cDNA Synthesis Kit for RT-PCR (AMV) was used to synthesize cDNA from 1 μg of the DNase-digested RNA. Relative qPCR was performed using Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix on a Rotor-GeneQ. All samples were normalized to β-actin, and fold change between samples was calculated using the comparative C(T) method. qPCR experiments were performed in triplicate from three biological replicates. Statistics were calculated on results from 3–6 runs using GraphPad Prism data software (GraphPad version 5.0b). Absolute qRT-PCR was performed using a one-step RT-PCR using Brilliant III Ultra-Fast QRT-PCR Master Mix on a Rotor-GeneQ. A standard curve generated using either Stratagene QPCR Human Reference Total RNA or Stratagene QPCR Mouse Reference Total RNA was run along side unknown RNA samples. qRT-PCR experiments were performed in triplicate from three biological replicates. Information about the standard curves and primer efficiencies from the absolute qPCR is listed in Supplementary Figure S4. Primers used for qRT-PCR are listed in Supplementary Table S1.
Transepithelial electrical resistance measurements
MCF10A, EPH4, and CACO-2 cells were seeded at 2 × 105 per well onto Millicell 24-well Cell Culture Assembly plates and readings recorded at regular intervals using a Millicell-ERS according to the manufacturer's instructions. Sample readings were measured in triplicate for each reading. Media was replaced following each reading.
Size-selective assessment of TJ paracellular flux
MCF10A, EPH4, and CACO-2 cells were seeded at 2 × 105 cells per well on polyester transwell filters (CLS3460; Sigma Aldrich) for 13 days. Media was replaced every 2 days. Fluorescently labelled dextrans were added to the top well, 25 μl of 4 kDa FITC-Dextran (Sigma, FD-4) and 25 μl of 70 kDa Rhodamine-Dextran (Sigma, R-9379) and incubated at 37°C for ~3 hours. Media from the bottom plate was aliquoted to three wells in a black-walled 96-well plate and the following wavelengths; FITC (Exc: 485 nm, Em: 544 nm) and Rhodamine (Exc: 520 nm, Em: 590 nm) on a FLUOstar microplate reader (Matter and Balda, 2003a). The average of the readings was taken as technical replicates, and the experiments were performed in three biological repeats.
Supplementary Material
Acknowledgements
We thank Catherine Sarta for her help enrolling patients and with data entry, Gregory Rosenblatt for his assistance with data management and analysis, and Nicole Kawachi for her help handling samples and the RNA expression analyses.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
A.M.T.-L. is supported by the School of Biology, University of St Andrews. A.M.T.-L., P.A.R., and F.J.G.-M. were funded by the Anonymous Trust, University of St Andrews. P.A.R. is supported by the Melville Trust for the Care and Cure of Cancer. The mass spectrometry work was supported by the Wellcome Trust (grant number 094476/Z/10/Z), which funded the purchase of the TripleTOF 5600 mass spectrometer at the BSRC Mass Spectrometry and Proteomics Facility, University of St Andrews. The clinical study was supported by the Department of Pathology, Albert Einstein College of Medicine/Montefiore Medical Center.
Conflict of interest: none declared.
References
- Anderson J.M., and Van Itallie C.M. (2009). Physiology and function of the tight junction. Cold Spring Harb. Perspect. Biol. 1, a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.M., Van Itallie C.M., Peterson M.D., et al. (1989). ZO-1 mRNA and protein expression during tight junction assembly in Caco-2 cells. J. Cell Biol. 109, 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus L., Moleirinho S., Herron L., et al. (2012). Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene 31, 238–250. [DOI] [PubMed] [Google Scholar]
- Balda M.S., Whitney J.A., Flores C., et al. (1996). Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol. 134, 1031–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birajdar S.S., Radhika M., Paremala K., et al. (2014). Expression of Ki-67 in normal oral epithelium, leukoplakic oral epithelium and oral squamous cell carcinoma. J. Oral Maxillofac. Pathol. 18, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandwein-Gensler M., Teixeira M.S., Lewis C.M., et al. (2005). Oral squamous cell carcinoma: histologic risk assessment, but not margin status, is strongly predictive of local disease-free and overall survival. Am. J. Surg. Pathol. 29, 167–178. [DOI] [PubMed] [Google Scholar]
- Cao X., Ding X., Guo Z., et al. (2005). PALS1 specifies the localization of ezrin to the apical membrane of gastric parietal cells. J. Biol. Chem. 280, 13584–13592. [DOI] [PubMed] [Google Scholar]
- Casaletto J.B., Saotome I., Curto M., et al. (2011). Ezrin-mediated apical integrity is required for intestinal homeostasis. Proc. Natl Acad. Sci. USA 108, 11924–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsum I.A., Martin C., and Humbert P.O. (2013). Scribble regulates an EMT polarity pathway through modulation of MAPK-ERK signaling to mediate junction formation. J. Cell Sci. 126, 3990–3999. [DOI] [PubMed] [Google Scholar]
- Elzagheid A., Korkeila E., Bendardaf R., et al. (2008). Intense cytoplasmic ezrin immunoreactivity predicts poor survival in colorectal cancer. Hum. Pathol. 39, 1737–1743. [DOI] [PubMed] [Google Scholar]
- Fan S., Fogg V., Wang Q., et al. (2007). A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J. Cell Biol. 178, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg V.C., Liu C.J., and Margolis B. (2005). Multiple regions of Crumbs3 are required for tight junction formation in MCF10A cells. J. Cell Sci. 118, 2859–2869. [DOI] [PubMed] [Google Scholar]
- Forster C. (2008). Tight junctions and the modulation of barrier function in disease. Histochem. Cell Biol. 130, 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Locker J., Sahai E., et al. (2012). Classifying collective cancer cell invasion. Nat. Cell Biol. 14, 777–783. [DOI] [PubMed] [Google Scholar]
- Ge L., Smail M., Meng W., et al. (2011). Yes-associated protein expression in head and neck squamous cell carcinoma nodal metastasis. PLoS One 6, e27529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georas S.N., and Rezaee F. (2014). Epithelial barrier function: at the frontline of asthma immunology and allergic airway inflammation. J. Allergy Clin. Immunol. 134, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs S., and Ponec M. (2000). Intrinsic regulation of differentiation markers in human epidermis, hard palate and buccal mucosa. Arch. Oral Biol. 45, 149–158. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L., Betanzos A., Nava P., et al. (2003). Tight junction proteins. Prog. Biophys. Mol. Biol. 81, 1–44. [DOI] [PubMed] [Google Scholar]
- Guillemot L., Paschoud S., Pulimeno P., et al. (2008). The cytoplasmic plaque of tight junctions: a scaffolding and signalling center. Biochim. Biophys. Acta 1778, 601–613. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., et al. (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36. [DOI] [PubMed] [Google Scholar]
- Hsu Y.Y., Shi G.Y., Kuo C.H., et al. (2012). Thrombomodulin is an ezrin-interacting protein that controls epithelial morphology and promotes collective cell migration. FASEB J. 26, 3440–3452. [DOI] [PubMed] [Google Scholar]
- Husain H., Psyrri A., Markovic A., et al. (2012). Nuclear epidermal growth factor receptor and p16 expression in head and neck squamous cell carcinoma. Laryngoscope 122, 2762–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp C.M., Tan T.T., Mathew R., et al. (2008). Role of the polarity determinant crumbs in suppressing mammalian epithelial tumor progression. Cancer Res. 68, 4105–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee H.L., and Verhey K.J. (2013). Molecular connections between nuclear and ciliary import processes. Cilia 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J., Li Y., Liu S., et al. (2013). High expression of ezrin predicts poor prognosis in uterine cervical cancer. BMC Cancer 13, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P., Beronja S., Silva-Gagliardi N.F., et al. (2006). The FERM protein Yurt is a negative regulatory component of the Crumbs complex that controls epithelial polarity and apical membrane size. Dev. Cell 11, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bivic A. (2013). Evolution and cell physiology. 4. Why invent yet another protein complex to build junctions in epithelial cells. Am. J. Physiol. Cell Physiol. 305, C1193–C1201. [DOI] [PubMed] [Google Scholar]
- Lemmers C., Michel D., Lane-Guermonprez L., et al. (2004). CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol. Biol. Cell 15, 1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letizia A., Ricardo S., Moussian B., et al. (2013). A functional role of the extracellular domain of Crumbs in cell architecture and apicobasal polarity. J. Cell Sci. 126, 2157–2163. [DOI] [PubMed] [Google Scholar]
- Li N., Lewis P., Samuelson D., et al. (2004). Glutamine regulates Caco-2 cell tight junction proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G726–G733. [DOI] [PubMed] [Google Scholar]
- Li Y., Wei Z., Yan Y., et al. (2014). Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex. Proc. Natl Acad. Sci. USA 111, 17444–17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C., Zheng Y., Yin F., et al. (2010). The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl Acad. Sci. USA 107, 10532–10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.W., Ali-Seyed M., Wu Y., et al. (2006). Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J. Cell. Biochem. 98, 1570–1583. [DOI] [PubMed] [Google Scholar]
- Lourenco S.V., Coutinho-Camillo C.M., Buim M.E., et al. (2010). Oral squamous cell carcinoma: status of tight junction claudins in the different histopathological patterns and relationship with clinical parameters. A tissue-microarray-based study of 136 cases. J. Clin. Pathol. 63, 609–614. [DOI] [PubMed] [Google Scholar]
- Madan R., Brandwein-Gensler M., Schlecht N.F., et al. (2006). Differential tissue and subcellular expression of ERM proteins in normal and malignant tissues: cytoplasmic ezrin expression has prognostic significance for head and neck squamous cell carcinoma. Head Neck 28, 1018–1027. [DOI] [PubMed] [Google Scholar]
- Makarova O., Roh M.H., Liu C.J., et al. (2003). Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1). Gene 302, 21–29. [DOI] [PubMed] [Google Scholar]
- Mao X., Li P., Ren Y., et al. (2015). Cell polarity protein CRB3 is an independent favorable prognostic factor for clear cell renal cell carcinoma. Int. J. Oncol. 46, 657–666. [DOI] [PubMed] [Google Scholar]
- Martin T.A., and Jiang W.G. (2009). Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta 1788, 872–891. [DOI] [PubMed] [Google Scholar]
- Martin T.A., Watkins G., Mansel R.E., et al. (2004). Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur. J. Cancer 40, 2717–2725. [DOI] [PubMed] [Google Scholar]
- Matter K., and Balda M.S. (2003. a). Functional analysis of tight junctions. Methods 30, 228–234. [DOI] [PubMed] [Google Scholar]
- Matter K., and Balda M.S. (2003. b). Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4, 225–236. [DOI] [PubMed] [Google Scholar]
- Matter K., and Balda M.S. (2007). Epithelial tight junctions, gene expression and nucleo-junctional interplay. J. Cell Sci. 120, 1505–1511. [DOI] [PubMed] [Google Scholar]
- Medina E., Williams J., Klipfell E., et al. (2002). Crumbs interacts with moesin and beta(Heavy)-spectrin in the apical membrane skeleton of Drosophila. J. Cell Biol. 158, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojallal M., Zheng Y., Hultin S., et al. (2014). AmotL2 disrupts apical-basal cell polarity and promotes tumour invasion. Nat. Commun. 5, 4557. [DOI] [PubMed] [Google Scholar]
- Moleirinho S., Chang N., Sims A.H., et al. (2013. a). KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene 32, 1821–1830. [DOI] [PubMed] [Google Scholar]
- Moleirinho S., Tilston-Lunel A., Angus L., et al. (2013. b). The expanding family of FERM proteins. Biochem. J. 452, 183–193. [DOI] [PubMed] [Google Scholar]
- Montesano R., Soriano J.V., Fialka I., et al. (1998). Isolation of EpH4 mammary epithelial cell subpopulations which differ in their morphogenetic properties. In Vitro Cell. Dev. Biol. Anim. 34, 468–477. [DOI] [PubMed] [Google Scholar]
- Nagaoka K., Udagawa T., and Richter J.D. (2012). CPEB-mediated ZO-1 mRNA localization is required for epithelial tight-junction assembly and cell polarity. Nat. Commun. 3, 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli M., Leoni B.D., D'Agnano I., et al. (2012). Good Caco-2 cell culture practices. Toxicol. In Vitro 26, 1243–1246. [DOI] [PubMed] [Google Scholar]
- Park H.R., Min S.K., Min K., et al. (2010). Differential expression of ezrin in epithelial skin tumors: cytoplasmic ezrin immunoreactivity in squamous cell carcinoma. Int. J. Dermatol. 49, 48–52. [DOI] [PubMed] [Google Scholar]
- Ribeiro P., Holder M., Frith D., et al. (2014). Crumbs promotes expanded recognition and degradation by the SCFSlimb/beta-TrCP ubiquitin ligase. Proc. Natl Acad. Sci. USA 111, E1980–E1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Roepman R., Aartsen W.M., et al. (2006). Towards understanding CRUMBS function in retinal dystrophies. Hum. Mol. Genet. 15(Spec No 2), R235–R243. [DOI] [PubMed] [Google Scholar]
- Robinson B.S., Huang J., Hong Y., et al. (2010). Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr. Biol. 20, 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh M.H., Fan S., Liu C.J., et al. (2003). The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J. Cell Sci. 116, 2895–2906. [DOI] [PubMed] [Google Scholar]
- Rousseau A., McEwen A.G., Poussin-Courmontagne P., et al. (2013). TRAF4 is a novel phosphoinositide-binding protein modulating tight junctions and favoring cell migration. PLoS Biol. 11, e1001726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada N. (2013). Tight junction-related human diseases. Pathol. Int. 63, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht N.F., Brandwein-Gensler M., Smith R.V., et al. (2012). Cytoplasmic ezrin and moesin correlate with poor survival in head and neck squamous cell carcinoma. Head Neck Pathol. 6, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K., Fogg V.C., and Margolis B. (2006). Tight junctions and cell polarity. Annu. Rev. Cell Dev. Biol. 22, 207–235. [DOI] [PubMed] [Google Scholar]
- Szymaniak A.D., Mahoney J.E., Cardoso W.V., et al. (2015). Crumbs3-mediated polarity directs airway epithelial cell fate through the Hippo pathway effector Yap. Dev. Cell 34, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R., Ohouo P.Y., Smolka M.B., et al. (2012). Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J. Cell Biol. 199, 969–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Li Y., Ye F., et al. (2015). Structural basis for the phosphorylation-regulated interaction between the cytoplasmic tail of cell polarity protein crumbs and the actin-binding protein moesin. J. Biol. Chem. 290, 11384–11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman E.L., Fan S., Harder J.L., et al. (2014). Crumbs3 is essential for proper epithelial development and viability. Mol. Cell. Biol. 34, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T., Matsui T., Tamura A., et al. (2013). The association of microtubules with tight junctions is promoted by cingulin phosphorylation by AMPK. J. Cell Biol. 203, 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A., Bhattacharyya R.P., and Lim W.A. (2003). The structure and function of proline recognition domains. Sci. STKE 2003, RE8. [DOI] [PubMed] [Google Scholar]
- Zihni C., Balda M.S., and Matter K. (2014). Signalling at tight junctions during epithelial differentiation and microbial pathogenesis. J. Cell Sci. 127, 3401–3413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.