Abstract
Purpose
Proinflammatory cytokines interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin-1 beta (IL-1β) secreted by infiltrating lymphocytes or macrophages may play a role in triggering RPE dysfunction associated with age-related macular degeneration (AMD). Binding of these proinflammatory cytokines to their specific receptors residing on the RPE cell surface can activate signaling pathways that, in turn, may dysregulate cellular gene expression. The purpose of the present study was to investigate whether IFN-γ, TNF-α, and IL-1β have an adverse effect on the expression of genes essential for RPE function, employing the RPE cell line ARPE-19 as a model system.
Methods
ARPE-19 cells were cultured for 3–4 months until they exhibited epithelial morphology and expressed mRNAs for visual cycle genes. The differentiated cells were treated with IFN-γ, TNF-α, and/or IL-1β, and gene expression was analyzed with real-time PCR analysis. Western immunoblotting was employed for the detection of proteins.
Results
Proinflammatory cytokines (IFN-γ + TNF-α + IL-1β) greatly increased the expression of chemokines and cytokines in cultured ARPE-19 cells that exhibited RPE characteristics. However, this response was accompanied by markedly decreased expression of genes important for RPE function, such as CDH1, RPE65, RDH5, RDH10, TYR, and MERTK. This was associated with decreased expression of the genes MITF, TRPM1, and TRPM3, as well as microRNAs miR-204 and miR-211, which are known to regulate RPE-specific gene expression. The decreased expression of the epithelial marker gene CDH1 was associated with increased expression of mesenchymal marker genes (CDH2, VIM, and CCND1) and epithelial–mesenchymal transition (EMT) promoting transcription factor genes (ZEB1 and SNAI1).
Conclusions
RPE cells exposed to proinflammatory cytokines IFN-γ, TNF-α, and IL-1β showed decreased expression of key genes involved in the visual cycle, epithelial morphology, and phagocytosis. This adverse effect of proinflammatory cytokines, which could be secreted by infiltrating lymphocytes or macrophages, on the expression of genes indispensable for RPE function may contribute to the RPE dysfunction implicated in AMD pathology.
Introduction
The RPE is an integral part of the human visual system and consists of a polarized monolayer of epithelial cells located in the eye between the retinal photoreceptors and the choroid [1,2]. The RPE forms the blood–retinal barrier, provides nutrients to the photoreceptor cells, and phagocytizes rod outer segment discs undergoing circadian shedding. The RPE plays an integral role in the visual cycle by regenerating 11-cis-retinal, the chromophore of the visual pigment rhodopsin. A normally functioning RPE is indispensable for vision, and dysfunction of the RPE may be associated with the pathogenesis of age-related macular degeneration (AMD). Inflammation is thought to be a major contributor to AMD pathogenesis, and inflammatory mediators secreted by lymphocytes and macrophages infiltrating the posterior compartment of the eye in this context could trigger RPE dysfunction [3-6]. The response of RPE cells to proinflammatory cytokines interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), and interleukin-1 beta (IL-1β) has been widely investigated. These cytokines can bind to their specific receptors present on RPE cells and highly increase the expression of several cytokines and chemokines [7-14]. This inflammatory response is associated with increased generation of reactive oxygen species (ROS) in the RPE cell [15]. The response of RPE cells to oxidant injury is also thought to play a role in the pathogenesis of AMD [16-20]. The activation of signaling pathways by IFN-γ, TNF-α, and IL-1β and the subsequent increase in ROS generation could potentially trigger RPE dysfunction by perturbing the expression of genes critical for the normal function of the RPE.
RPE cells are known to express a large number of genes to support their specialized function. CDH1 (Gene ID: 999; OMIM: 192090), which encodes cadherin-1 protein (CDH1, E-cadherin), is a gene that supports the epithelial function [21]. The RPE65 (Gene ID: 6121; OMIM: 180069) gene encodes the RPE-specific protein 65 kDa (RPE65), which is an essential visual cycle enzyme required for the conversion of all-trans-retinol to 11-cis-retinol [22,23]. The RDH5 (Gene ID: 5959; OMIM: 601617) gene encodes 11-cis-retinol dehydrogenase while the RDH10 (Gene ID: 157506; OMIM: 607599) gene encodes a retinol dehydrogenase that catalyzes the conversion of all-trans-retinol to all-trans-retinal [24,25]. The RLBP1 (Gene ID: 6017; OMIM: 180090) gene encodes the 11-cis-retinal binding protein retinaldehyde-binding protein 1 (RLBP1), which is also known as cellular retinaldehyde-binding protein (CRALBP) [26]. TYR (Gene ID: 7299; OMIM: 606933) encodes tyrosinase, the most important enzyme involved in the generation of melanin pigment from tyrosine [27]. The phagocytosis function of RPE cells is controlled by tyrosine-protein kinase MER encoded by the MERTK (Gene ID: 10461; OMIM: 604705) gene [28]. Microphthalmia-associated transcription factor (MITF) encoded by the MITF (Gene ID: 4286; OMIM: 156845) gene is a known regulator of RPE differentiation [29]. MITF keeps RPE cells at a differentiated stage by highly upregulating the expression of the RPE characteristic microRNAs miR-204 and miR-211 [30,31]. This transcription factor is also known to promote melanogenesis by inducing TYR and to increase the expression of the transient receptor potential cation channel, subfamily M, member 1 (TRPM1; Gene ID: 4308; OMIM: 603576) and transient receptor potential cation channel, subfamily M, member 3 (TRPM3; Gene ID: 80036; OMIM: 608961) genes [30]. We hypothesized that the interaction of IFN-γ, TNF-α, and IL-1β with RPE cells may adversely affect the expression of one or more genes required for the normal functioning of the RPE. Therefore, we investigated the effect of these inflammatory cytokines on the expression of several genes involved in RPE function. We employed ARPE-19, a widely used adult human RPE-derived cell culture system, for this purpose [32]. Here, we show that IFN-γ, TNF-α, and IL-1β decrease the expression of key genes involved in the visual cycle, epithelial morphology, melanogenesis, and phagocytosis in ARPE-19 cells, thus indicating that these proinflammatory cytokines could promote retinal pigment epithelial dysfunction.
Methods
STR analysis
Authentication of ARPE-19 cells [32] used in this study has been reported earlier from our laboratory [33]. Briefly, this validation of the established cell line using short tandem repeat analysis and amelogenin gender locus determination was performed by ATCC Cell Line Authentication Service, Promega, Madison, WI. Seventeen short tandem repeat (STR) loci plus the amelogenin gender determining locus were amplified using PowerPlex 18D kit. The cell line samples along with appropriate positive and negative controls were processed using an ABI Prism 3500xl Genetic Analyzer. Data were analyzed using GeneMapper ID-X v1.2 software (Applied Biosystems (Foster City, CA). The batch of the cells we had was found to be a perfect match for the ATCC human RPE cell line CRL-2302 (ARPE-19). The STR analysis result is shown in Appendix 1. These cells were used within 3 additional passages.
Cell culture
ARPE-19 cells were grown on tissue culture plates in Dulbecco’s modified Eagle’s medium (DMEM) with 4.5 g/l glucose, L-glutamine, and 1 mM sodium pyruvate supplemented with 1% fetal bovine serum, and penicillin and streptomycin (Thermo Fisher Scientific, Grand Island, NY). The cells were maintained at 37 °C in a humidified environment of 5% CO2, with media exchange performed twice a week. The ARPE-19 cells grown for 3 to 4 months appeared differentiated as indicated by epithelial morphology, pigmentation, and expression of RPE characteristic genes [data not shown]. The differentiated cells were treated with the proinflammatory cytokines IFN-γ, IL-1β, and/or TNF-α for either 20 h in the absence of serum or 4 days in the presence of serum. The concentrations of the cytokines used were IFN-γ, 10 u/ml; IL-1β, 1 ng/ml; and TNF-α, 1 ng/ml. The cytokine concentration was increased tenfold in certain experiments. Human IL-1β was purchased from R&D Systems (Minneapolis, MN) while TNF-α and IFN-γ were from Roche Applied Science (Indianapolis, IN). The effect of proinflammatory cytokines on gene expression was also tested using an RPE cell culture system established from the eyes of adult human donors (70–85 years old) by Nagineni et al. [12,34]. These cells were grown to confluence in minimum essential medium supplemented with 10% fetal bovine serum, non-essential amino acids, penicillin (100 u/ml), streptomycin (100 μg/ml), and amphotericin B (25 ng/ml) and then exposed to IFN-γ (100 u/ml), IL-1β (10 ng/ml), and TNF-α (10 ng/ml) for 20 h in the absence of serum.
Real-time PCR
The reagents for RNA isolation, cDNA synthesis, and real-time PCR were obtained from Life Technologies Corporation (Grand Island, NY). The total RNA fraction isolated from the ARPE-19 cells with the Ambion mirVana miRNA Isolation Kit was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit. The cDNA preparation was then used as the template for quantitative real-time PCR analysis of the gene transcripts IL-8 (Gene ID: 3576; OMIM: 146930), CCL2 (Gene ID: 6347; OMIM: 158105), CCL5 (Gene ID: 6352; OMIM: 187011), CSF2 (Gene ID: 1437; OMIM: 138960), CXCL11 (Gene ID: 6373; OMIM: 604852), CXCL10 (Gene ID: 3627; OMIM: 147310), CDH1, RLBP1, RPE65, RDH5, RDH10, TYR, MERTK, VIM (Gene ID: 7431; OMIM: 193060), CCND1 (Gene ID: 595; OMIM: 168461), CDH2 (Gene ID: 1000; OMIM: 114020), ZEB1 (Gene ID: 6935; OMIM: 189909), SNAI1 (Gene ID: 6615; OMIM: 604238), MITF, TRPM1, and TRPM3. Briefly, 96-well reaction plate containing the PCR samples was heated at 95 ⁰C for 20 s to activate the polymerase. Amplification condition consisted of 40 cycles of 1 sec at 95 ⁰C and 20 s at 60 ⁰C. Each PCR reaction (20 µl) was set up using validated TaqMan probes (labeled with reporter dye FAM at the 5′ end), primers specific for the gene of interest, and TaqMan Fast Advanced Master Mix. Human 18S rRNA was used as the endogenous control. An Applied Biosystems ViiA 7 Real-Time PCR System was employed for gene amplification analysis following the manufacturer’s default thermal cycling conditions. The real-time PCR results were expressed as n-fold induction in gene expression calculated using the relative quantification (ΔΔCT) method. The expression of miRNAs was also analyzed with real-time PCR using Individual TaqMan MicroRNA Assays (hsa-miR-204, hsa-miR-211, and hsa-miR-155), the TaqMan MicroRNA Reverse Transcription Kit, TaqMan Fast Advanced Master Mix, and RNU48 (endogenous control).
Western immunoblot analysis
Following treatment with proinflammatory cytokines, the cells were suspended in RIPA buffer (Sigma-Aldrich, St. Louis, MO) containing a protease/phosphatase inhibitor cocktail (Cell Signaling Technology, Danvers, MA) at 4 °C, sonicated, and then centrifuged at 12,000 ×g for 10 min. Equal amounts of the supernatants (corresponding to 20 µg protein) were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and then blotted on to nitrocellulose membranes using the iBlot dry blotting system (Invitrogen, Carlsbad, CA). The blots were then probed using a rabbit anti-CDH1 antibody (1:1,000 dilution; Cell Signaling Technology, Danvers, MA) or rabbit anti-RLBP1 antibody (1:10,000 dilution; gift from Dr. John Saari, Emeritus Professor of Biochemistry/Ophthalmology, University of Washington, Seattle, WA ). Mouse anti-α-tubulin (1:10,000 dilution) was used as the primary antibody to detect α-tubulin as the loading control. IRDye 800CW goat anti-rabbit immunoglobulin G (IgG; 1:15,000) and IRDye 680LT goat-anti-mouse IgG (1:15,000) were used as the secondary antibodies. Odyssey blocking buffer (PBS), mouse anti-α-tubulin antibody, and IRDye-labeled secondary antibodies were purchased from LI-COR Biosciences (Lincoln, NE). The blots were scanned using a LI-COR Odyssey Clx Infrared Imaging System for the detection of immunoreactive bands and to estimate the fluorescence intensity.
Statistical analysis
A paired Student t test was used for the analysis of statistical significance. The alpha value assigned for significance was a p value of less than 0.05. Representative experiments are shown in the figures, and the values are shown as mean ± standard deviation (SD).
Results
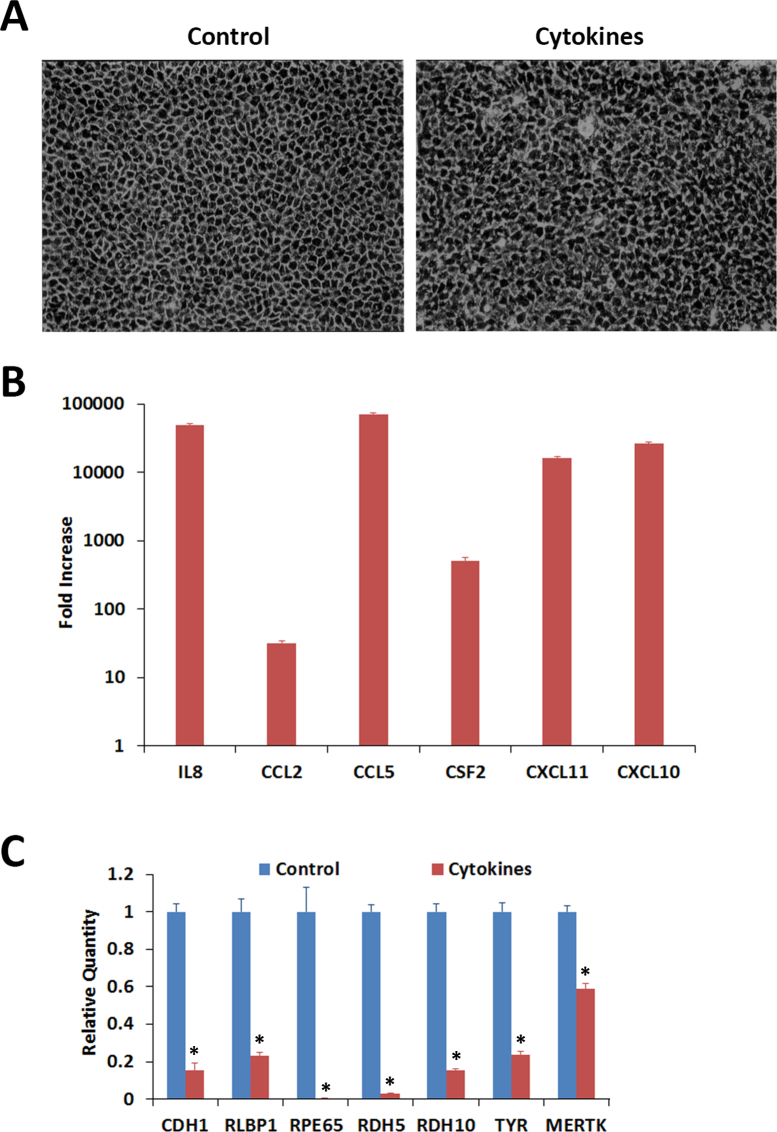
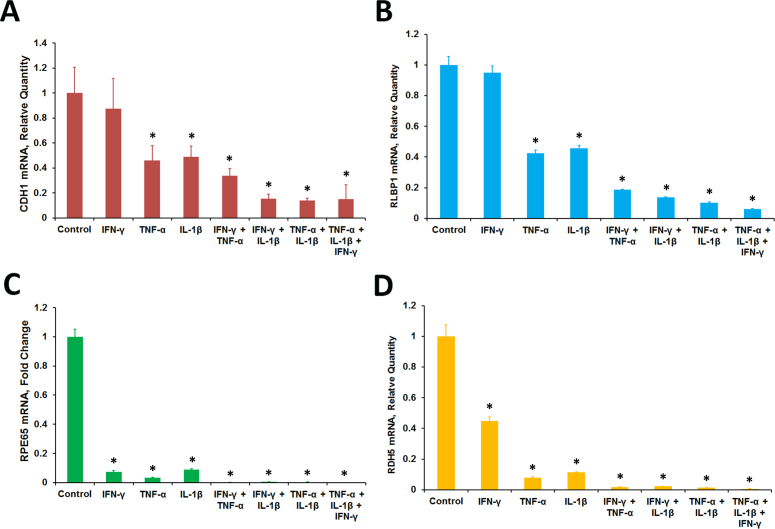
The response of the RPE cells to the proinflammatory cytokines was investigated. The ARPE-19 cells maintained in culture for 4 months acquired RPE characteristics, such as epithelial morphology and visual cycle gene expression [data not shown]. The cells were treated with IFN-γ (10 u/ml), TNF-α (1 ng/ml), and IL-1β (1 ng/ml) for 20 h in the absence of serum; cytokines were omitted from the controls. The control cells showed typical epithelial morphology characteristic of RPE cells while the treated cells exhibited an irregular shape and thickened cell junctions (Figure 1A). This was accompanied by the increased expression of several cytokines and chemokines. Real-time PCR analysis of the control and treated cells showed that the expression of transcripts for IL8, CCL2, CCL5, CSF2, CXCL10, and CXCL11 was highly increased by the treatment (Figure 1B). We then analyzed the expression of several genes essential for RPE function with real-time PCR in the control and treated cells (Figure 1C). The proinflammatory cytokines considerably decreased the expression of mRNA for CDH1. This gene encodes the CDH1 (E-cadherin) protein that is essential for epithelial function. Interestingly, the treatment also markedly decreased the expression of genes involved in the visual cycle (RLBP1, RPE65, RDH5, and RDH10), melanin pigment synthesis (TYR), and phagocytosis (MERTK). The effect of proinflammatory cytokines on the expression of transcripts for CDH1, RLBP1, RPE65, and RDH5 in ARPE-19 cells was investigated further. The cells were treated with IFN-γ (10 u/ml), TNF-α (1 ng/ml), and IL-1β (1 ng/ml) either individually or in combination for 20 h in the absence of serum and gene expression analyzed with real-time PCR (Figure 2). TNF-α and IL-1β when tested individually noticeably decreased the expression of the transcripts for CDH1, RLBP1, RPE65, and RDH5. IFN-γ by itself failed to decrease the expression of CDH1 and RLBP1 but decreased the expression of RPE65 and RDH5. Combinations of two cytokines (IFN-γ + TNF-α, IFN-γ + IL-1β, or TNF-α + IL-1β) or all three cytokines (IFN-γ + TNF-α + IL-1β) were highly effective in decreasing the expression of CDH1, RLBP1, RPE65, and RDH5. Thus, the interaction of proinflammatory cytokines with RPE cells may result in the decreased expression of genes necessary for RPE function.
Figure 1.
Effect of proinflammatory cytokines on cultured ARPE-19 cells exhibiting RPE characteristics. The cells were treated with interferon gamma (IFN-γ; 10 u/ml), tumor necrosis factor alpha (TNF-α; 1 ng/ml), and interleukin-1 beta (IL-1β; 1 ng/ml) for 20 h in the absence of serum and their gene expression analyzed with real-time PCR. A: Control cells exhibited typical epithelial morphology when examined with phase contrast microscopy. Cells treated with cytokines appeared distorted with thickened cell junctions. Magnification: 100X. B: The cells responded to the proinflammatory cytokines by highly increasing the expression of cytokines and chemokines. Gene expression in the treated and control cells was analyzed with real-time PCR. The fold increases shown are statistically significant (p<0.05 compared to control, n = 4). C: The expression of the RPE characteristic genes when analyzed with real-time PCR decreased substantially in the differentiated ARPE-19 cells exposed to the proinflammatory cytokines. *p<0.5 when compared to control, n = 4.
Figure 2.
Proinflammatory cytokines decreased the expression of RPE characteristic genes. Differentiated ARPE-19 cells were treated with interferon gamma (IFN-γ; 10 u/ml), tumor necrosis factor alpha (TNF-α; 1 ng/ml), interleukin-1 beta (IL-1β; ng/ml) either alone or in combination for 20 h in the absence of 1% serum. The gene transcripts were analyzed with real-time PCR. A: CDH1. B: RLBP1. C: RPE65. D: RDH5. *p<0.5 when compared to control, n = 4.
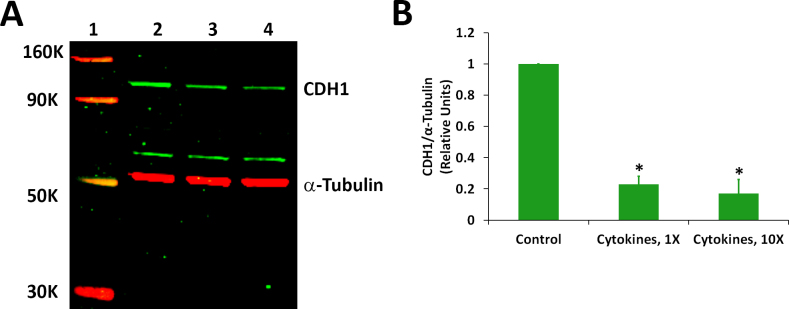
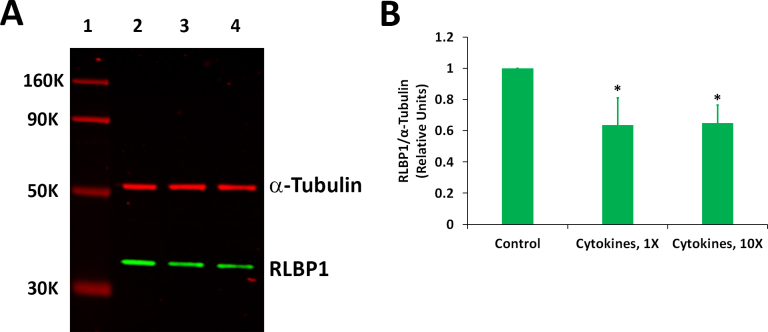
We analyzed whether proinflammatory cytokines can decrease the expression of proteins encoded by the CDH1 and RLBP1 genes in ARPE-19 cells. The cells treated with a mixture of IFN-γ, TNF-α, and IL-1β for 20 h under serum-free conditions, as described for the gene expression studies, were initially employed for the western blot analysis of the CDH1 and RLBP1 proteins. However, we did not observe any significant decrease in the expression of these proteins between the control and treated samples under this condition. This is not quite unexpected as proteins in a living cell are thought to turn over much more slowly compared to mRNAs. Therefore, we decided to increase the duration of the treatment of the cytokines to 4 days. However, it was necessary to use serum-containing medium in this case as the cells could not tolerate serum-free conditions for this duration. The differentiated ARPE-19 cells treated with proinflammatory cytokines for 4 days in the presence of the serum also exhibited distorted morphology similar to those treated under serum-free conditions for 20 h (data not shown). The cells were treated with a mixture of IFN-γ (10 u/ml), TNF-α (1 ng/ml), and IL-1β (1 ng/ml) for 4 days in the presence of serum for the western blot analysis. The results showed that the amount of CDH1 protein in the cells exposed to the cytokines was about 25% of that present in the control (Figure 3). A similar decrease was also observed when the cells were treated with ten times the indicated cytokine concentrations. Proinflammatory cytokines also decreased the expression of the RLBP1 protein in ARPE-19 cells (Figure 4). The RLBP1 protein content in the treated cells was approximately 65% of that present in the control cells.
Figure 3.
Proinflammatory cytokines decreased the expression of the CDH1 protein. Differentiated ARPE-19 cells were treated with the cytokines (1X = interferon gamma (IFN-γ), 10 u/ml + tumor necrosis factor alpha (TNF-α), 1 ng/ml + interleukin-1 beta (IL-1β), 1 ng/ml) for 4 days in the presence of 1% serum. A: Western immunoblot analysis of CDH1 expression. Lane 1: molecular weight markers; lane 2: control; lane 3: cytokines, 1X; and lane 4: cytokines, 10X. A representative blot from three similar experiments is shown. B: Histogram showing CDH1 expression. The CDH1 band intensity was normalized with corresponding α-tubulin band intensity; *p<0.05 when compared to control, n = 3.
Figure 4.
Proinflammatory cytokines decreased the expression of the RLBP1 protein. Differentiated ARPE-19 cells were treated with the cytokines (1X = interferon gamma (IFN-γ), 10 u/ml + tumor necrosis factor alpha (TNF-α), 1 ng/ml + interleukin-1 beta (IL-1β), 1 ng/ml) for 4 days in the presence of 1% serum. A: Western immunoblot analysis of RLBP1 expression. Lane 1: molecular weight markers; lane 2: control; lane 3: cytokines, 1X; and lane 4: cytokines, 10X. A representative blot from three similar experiments is shown. B: Histogram showing RLBP1 expression. The RLBP1 band intensity was normalized with corresponding α-tubulin band intensity; *p<0.05 when compared to control, n = 3.
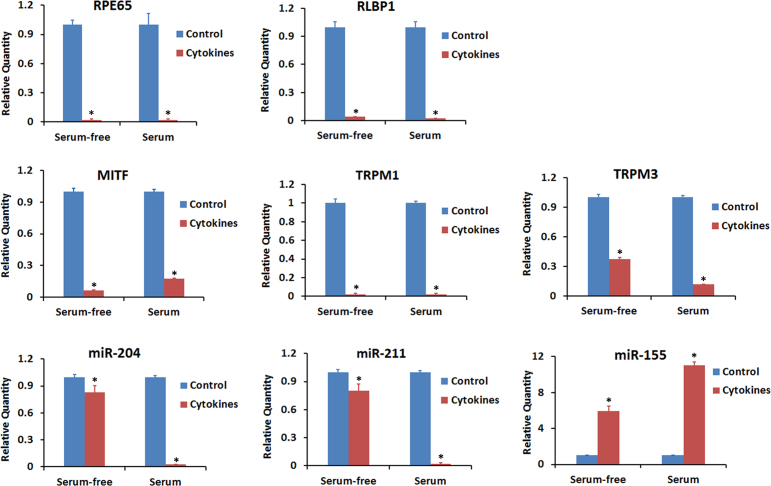
The genes MITF, TRPM1, and TRPM2 and microRNAs miR-204 and miR-211 are highly expressed in RPE cells, and the expression of these genes and microRNAs is necessary for maintaining RPE characteristics [30,31]. The effect of proinflammatory cytokines on the expression of these genes and microRNAs was investigated in relation to that of RPE65 and RLBP1. Differentiated ARPE-19 cells were treated with a mixture of IFN-γ (10 u/ml), TNF-α (1 ng/ml), and IL-1β (1 ng/ml) either for 20 h under serum-free conditions or for 4 days in the presence of 1% serum, and the gene expression was analyzed with real-time PCR (Figure 5). A decrease in the expression of the transcripts for RPE65 and RLBP1 was observed under the two different conditions that we employed to expose the cells to the cytokines. The expression of MITF, TRPM1, and TRPM2 was also considerably decreased following exposure to proinflammatory cytokines under both conditions. The expression of miR-204 and miR-211 was decreased by more than 95% when the cells were exposed to the cytokines for a longer duration (4 days with serum). However, the decrease in miR-204 and miR-211 was less than 15% when the cells were treated with cytokines for a shorter time (20 h, without serum). The expression of miR-155 was considerably increased in the cells exposed to proinflammatory cytokines, and a higher increase was observed with longer exposure.
Figure 5.
Proinflammatory cytokines decreased the expression of genes that control RPE function. Differentiated ARPE-19 cells were treated with the cytokines (interferon gamma (IFN-γ), 100 u/ml + tumor necrosis factor alpha (TNF-α), 10 ng/ml + interleukin-1 beta (IL-1β), 10 ng/ml) either for 20 h under serum-free conditions or for 4 days in the presence of 1% serum. Real-time PCR was employed to analyze the expression of the indicated transcripts and miRNAs. *p<0.05 when compared to respective control, n = 3.
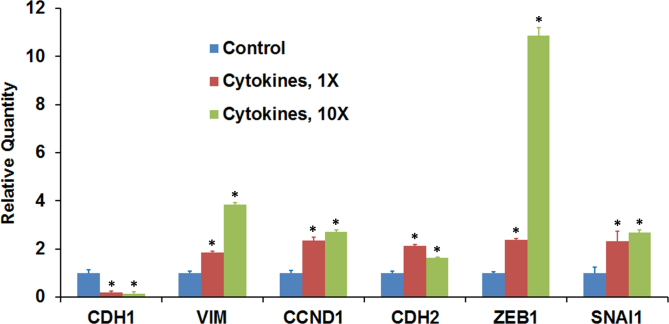
The decrease in the expression of CDH1, a known epithelial marker, in ARPE-19 cells on exposure to proinflammatory cytokines could indicate EMT-like changes. Therefore, we analyzed the expression of mesenchymal marker genes in cells treated with a mixture of IFN-γ (10 u/ml), TNF-α (1 ng/ml), and IL-1β (1 ng/ml) for 20 h under serum-free conditions. The cells were also exposed to cytokine concentrations ten times higher than those indicated here. The treatment increased the expression of mesenchymal marker genes VIM, CCND1, and CDH2 while decreasing the expression of CDH1 (Figure 6). The expression of ZEB1 and SNAI1, two transcription factors known to promote EMT, was also increased by exposing the cells to proinflammatory cytokines. Thus, a decrease in the expression of the epithelial marker gene is associated with increased expression of several mesenchymal genes.
Figure 6.
Proinflammatory cytokines modulate the expression of genes that regulate the EMT. Differentiated ARPE-19 cells were treated with the cytokines (1X = interferon gamma (IFN-γ), 10 u/ml + tumor necrosis factor alpha (TNF-α), 1 ng/ml + interleukin-1 beta (IL-1β), 1 ng/ml) for 20 h in the absence of serum, and the expression transcripts for the epithelial marker gene (CDH1), mesenchymal marker genes (VIM, CCND1, and CDH2), and transcription factor genes (ZEB1 and SNAI1) was analyzed with real-time PCR. *p<0.5 when compared to control, n = 4.
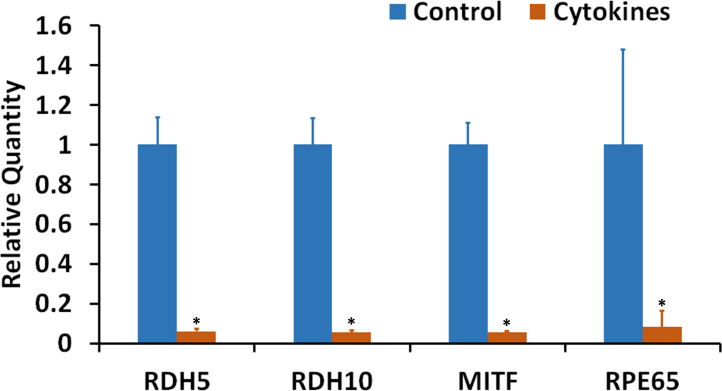
The effect of proinflammatory cytokines on the expression of RPE characteristic genes was tested using an RPE cell culture established from adult human donor eyes by Nagineni et al. [12,34]. These cells were exposed to IFN-γ (100 u/ml), IL-1β (10 ng/ml), and TNF-α (10 ng/ml) for 20 h in the absence of serum. The expression of the transcripts for RDH5, RDH10, MITF, and RPE65 was highly decreased in response to the cytokine treatment (Figure 7). Thus, these cultured RPE cells also exhibited decreased expression of RPE characteristic genes, similar to the differentiated ARPE-19 cells, in response to treatment with proinflammatory cytokines.
Figure 7.
Proinflammatory cytokines decreased the expression of RPE characteristic genes in an RPE cell culture system established from adult human donor eyes. The cultured cells were treated with the cytokines (interferon gamma (IFN-γ), 100 u/ml + tumor necrosis factor alpha (TNF-α), 10 ng/ml + interleukin-1 beta (IL-1β), 10 ng/ml) for 20 h under serum-free conditions. Real-time PCR analysis of the expression of mRNAs for RDH5, RDH10, MITF, and RPE65 is shown. *p<0.05 when compared to respective control, n = 4.
Discussion
Inflammation is known to contribute to the pathogenesis of AMD [4,35-38]. Infiltrating lymphocytes and macrophages during ocular inflammation may facilitate RPE dysfunction associated with AMD [5,6]. RPE cells in culture respond to the proinflammatory cytokines IFN-γ, TNF-α, and IL-1β by increasing the expression of a large number of cytokines and chemokines [7-10]. This is associated with increased production of cytotoxic levels of reactive oxygen species in the cell [15]. In the present study, we showed that the expression of several genes important in RPE function are decreased in RPE cells exposed to IFN-γ, TNF-α, and IL-1β. The proinflammatory cytokines decreased the expression of transcripts for visual cycle genes RLBP1, RPE65, RDH5, and RDH10; epithelial gene CDH1; melanogenesis gene TYR; and phagocytosis gene MERTK in treated ARPE-19 cells in culture. We also observed that the decrease in mRNA expression in the case of CDH1 and RLBP1 resulted in decreased expression of their encoded proteins. Thus, proinflammatory cytokines have the potential to trigger RPE dysfunction by downregulating the expression of important functional genes in RPE.
We employed the adult human RPE derived ARPE-19 cell line for this study. This cell culture system developed by Dunn et al. [32] has been employed in more than 1,000 publications to study RPE function. We have previously shown that ARPE-19 cells respond to IFN-γ, TNF-α, and IL-1β by increasing the expression of cytokines and chemokines [10]. However, ARPE-19 cells grown under normal culture conditions did not show many RPE characteristics required for the present study. Therefore, we maintained these cells in culture for the long term employing culture conditions similar to those reported by Ahmado et al. [39]. ARPE-19 cells grown under this condition for 4 months exhibited typical epithelial morphology and pigmentation, and expressed many RPE functional genes [data not shown]. These differentiated cells responded to IFN-γ, TNF-α, and IL-1β by decreasing the expression of many RPE characteristic genes as reported in the present study. This important observation must be verified using other RPE cell culture systems. We tested the effect of proinflammatory cytokines on an RPE cell culture system established by Nagineni et al. [34] from human donor eyes. A combination of IFN-γ, TNF-α, and IL-1β effectively decreased the expression of RPE characteristic genes RDH5, RDH10, RPE65, and MITF in this cell culture system that has served as a valuable model system for studying the RPE inflammatory response [9,11-14,34,40,41]. It remains to be elucidated whether the adverse effect of inflammatory cytokines on the expression of genes important in critical RPE functions, such as the visual cycle, is applicable to in vivo situations. However, Camelo et al. [42] reported that the RPE dysfunction observed in aged or light-challenged mice is associated with a decrease in the expression of the Rpe65 gene. Camelo et al. employed these mice model systems to study age-related retinal degeneration and found that the RPE dysfunction is triggered by the inflammatory response resulting from T lymphocyte infiltration into the retina.
We observed that proinflammatory cytokines decreased the expression of MITF in ARPE-19 cells. This gene encodes MITF, a basic helix–loop–helix leucine zipper transcription factor that regulates RPE development [29]. Recently, this transcription factor was reported to regulate the expression of two visual cycle genes, RLBP1 and RDH5, in RPE cells [43]. Adijanto et al. [30] showed that MITF keeps the human fetal RPE cells in a differentiated state by upregulating the expression of miR-204 and miR-211. These two microRNAs are highly expressed in RPE cells and control RPE characteristics [31]. Fetal human RPE cells transfected with MITF siRNA showed decreased expression of miR-204 and miR-211 that was associated with the decreased expression of the RPE characteristic genes RLBP1 and RPE65 [30]. Antimers to miR-204 and miR-211 also decreased the expression of visual cycle genes [31]. MITF also regulated the expression of TRPM1 and TRPM3 along with that of miR-204 and miR-211 [30]. The present results clearly showed that the decrease in the expression of RLBP1, RPE65, and other RPE characteristic genes in ARPE-19 cells exposed to IFN-γ, TNF-α, and IL-1β was associated with a decrease in the expression of MITF along with that of miR-204, miR-211, TRPM1, and TRPM3. Therefore, downregulation of MITF may potentially mediate the adverse effect of proinflammatory cytokines on the expression of RPE characteristic genes due to the inherent ability of MITF to regulate miR-204/211 expression.
Morphometric analysis of RPE from donor eyes has indicated that AMD progression is associated with the presence of RPE cells with irregular size and shape [44]. Interestingly, we observed a disruption in the typical epithelial morphology of cultured ARPE-19 cells on exposure to the proinflammatory cytokines IFN-γ, TNF-α, and IL-1β. This disruption was associated with gene expression changes indicative of EMT. The decrease in the expression of the epithelial marker gene CDH1 in cells treated with the proinflammatory cytokines was accompanied by increased expression of the mesenchymal marker genes VIM, CDH2, and CCND1. This was associated with an increase in the expression of ZEB1 and SNAI1. These two transcription factor genes are known promoters of EMT [45-48]. Inhibiting the expression of miR-204 and miR-211 has been shown to induce EMT-like changes in human fetal RPE cells [30,31]. In addition, miR-204 and miR-211 are known to inhibit EMT in cancer cells [49,50]. Liu et al. [51] reported that Zeb1 downregulated Mitf expression in mouse RPE cells causing EMT-like changes and the loss of RPE characteristics. We observed in the ARPE-19 cells that the increase in ZEB1 expression is associated with decreases in the expression of MITF, miR-204, and miR-211. Thus, ZEB1 may mediate the proinflammatory cytokine induced RPE dysfunction by decreasing the expression of MITF and miR-204/miR-211. The possibility that apoptosis plays a role in the proinflammatory cytokine-induced RPE dysfunction that we observed remains to be elucidated. A review of the literature showed that the study of apoptosis in RPE cells in response to treatment with proinflammatory cytokines is limited. No apoptosis was detected when human RPE cells in culture were exposed to TNF-α (20 ng/ml), IL-1β (2 ng/ml), or lipopolysaccharide (1,000 ng/ml) for 24 h [52], but the effect of a combination of all these proinflammatory agents on apoptosis in human RPE cells was not tested. However, human retinal capillary endothelial cells treated with a mixture of 10 ng/ml TNF-α, 10 ng/ml IL-1β, and 50 units/ml IFN-γ for 48 h were reported to exhibit apoptosis in about 14% cells compared to about 7% in the case of untreated cells [53].
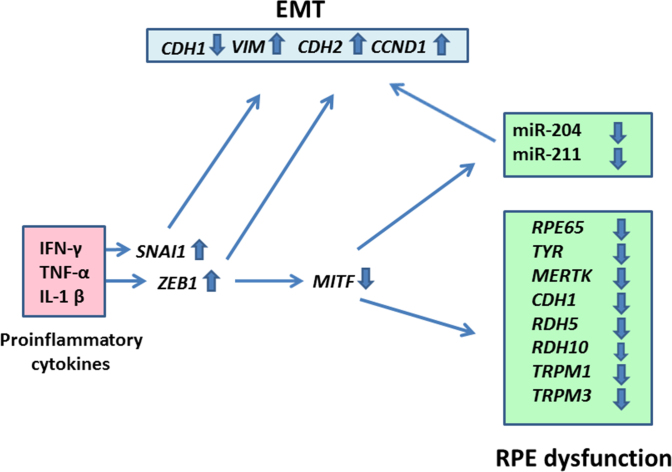
In summary, the present results showed that the proinflammatory cytokines IFN-γ, TNF-α, and IL-1β decrease the expression of key genes involved in visual cycle, epithelial morphology, melanogenesis, and phagocytosis in cultured ARPE-19 RPE cells (Figure 8). Proinflammatory cytokines may elicit this effect by increasing the expression of ZEB1 and SNAI1, pro-EMT transcription factor genes. The increase in ZEB1 expression may, in turn, decrease the expression of MITF, a key regulator of RPE differentiation, causing a decrease in the expression of miR-204 and miR-211, two key microRNAs regulating RPE-characteristics. Thus, a decrease in the expression of critical functional genes due to targeting of RPE cells by IFN-γ, TNF-α, and IL-1β secreted by infiltrating inflammatory cells may potentially contribute to the RPE dysfunction implicated in AMD pathology.
Figure 8.
A schematic depiction of RPE cell dysfunction induced by IFN-γ, TNF-α, and IL-1β. The proinflammatory cytokines increased the expression of proepithelial–mesenchymal transition (EMT) transcription factor genes SNAI1 and ZEB1 in the RPE cells. This could result in the downregulation of the epithelial marker gene CDH1 and upregulation of the mesenchymal marker genes VIM, CDH2, and CCND1. Increased ZEB1 expression also caused a decrease in the expression of the MITF gene that encodes a transcription factor that controls RPE differentiation. This, in turn, may decrease the expression of the RPE functional genes RPE65, TYR, MERTK, RDH5, RDH10, TRPM1, and TRPM3. The decrease in MITF expression may also reduce the expression of miR-204 and miR-211, two microRNAs necessary for maintaining epithelial physiology and RPE function.
Acknowledgments
The authors thank Dr. C. Vijayasarathy (National Institute on Deafness and Other Communication Disorders) for his contribution and critical reading of the manuscript. This study was supported by the Intramural Research Program, NIH, NEI (1ZIAEY000444-09).
Appendix 1. STR analysis.
To access the data, click or select the words “Appendix 1.”
References
- 1.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–81. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 2.Sparrow JR, Hicks D, Hamel CP. The retinal pigment epithelium in health and disease. Curr Mol Med. 2010;10:802–23. doi: 10.2174/156652410793937813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buschini E, Piras A, Nuzzi R, Vercelli A. Age related macular degeneration and drusen: neuroinflammation in the retina. Prog Neurobiol. 2011;95:14–25. doi: 10.1016/j.pneurobio.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Ambati J, Atkinson JP, Gelfand BD. Immunology of age-related macular degeneration. Nat Rev Immunol. 2013;13:438–51. doi: 10.1038/nri3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cousins SW, Espinosa-Heidmann DG, Csaky KG. Monocyte activation in patients with age-related macular degeneration: a biomarker of risk for choroidal neovascularization? Arch Ophthalmol. 2004;122:1013–8. doi: 10.1001/archopht.122.7.1013. [DOI] [PubMed] [Google Scholar]
- 6.Hooks JJ, Chan CC, Detrick B. Identification of the lymphokines, interferon-gamma and interleukin-2, in inflammatory eye diseases. Invest Ophthalmol Vis Sci. 1988;29:1444–51. [PubMed] [Google Scholar]
- 7.Shi G, Maminishkis A, Banzon T, Jalickee S, Li R, Hammer J, Miller SS. Control of chemokine gradients by the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2008;49:4620–30. doi: 10.1167/iovs.08-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elner SG, Delmonte D, Bian ZM, Lukacs NW, Elner VM. Differential expression of retinal pigment epithelium (RPE) IP-10 and interleukin-8. Exp Eye Res. 2006;83:374–9. doi: 10.1016/j.exer.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Nagineni CN, Kutty RK, Detrick B, Hooks JJ. Inflammatory cytokines induce intercellular adhesion molecule-1 (ICAM-1) mRNA synthesis and protein secretion by human retinal pigment epithelial cell cultures. Cytokine. 1996;8:622–30. doi: 10.1006/cyto.1996.0083. [DOI] [PubMed] [Google Scholar]
- 10.Kutty RK, Samuel W, Abay R, Cherukuri A, Nagineni CN, Duncan T, Jaworski C, Vijayasarathy C, Redmond TM. Resveratrol attenuates CXCL11 expression induced by proinflammatory cytokines in retinal pigment epithelial cells. Cytokine. 2015;74:335–8. doi: 10.1016/j.cyto.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagineni CN, Kommineni VK, William A, Detrick B, Hooks JJ. Regulation of VEGF expression in human retinal cells by cytokines: implications for the role of inflammation in age-related macular degeneration. J Cell Physiol. 2012;227:116–26. doi: 10.1002/jcp.22708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagineni CN, Raju R, Nagineni KK, Kommineni VK, Cherukuri A, Kutty RK, Hooks JJ, Detrick B. Resveratrol Suppresses Expression of VEGF by Human Retinal Pigment Epithelial Cells: Potential Nutraceutical for Age-related Macular Degeneration. Aging Dis. 2014;5:88–100. doi: 10.14366/AD.2014.050088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kutty RK, Nagineni CN, Samuel W, Vijayasarathy C, Hooks JJ, Redmond TM. Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochem Biophys Res Commun. 2010;402:390–5. doi: 10.1016/j.bbrc.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kutty RK, Nagineni CN, Samuel W, Vijayasarathy C, Jaworski C, Duncan T, Cameron JE, Flemington EK, Hooks JJ, Redmond TM. Differential regulation of microRNA-146a and microRNA-146b-5p in human retinal pigment epithelial cells by interleukin-1beta, tumor necrosis factor-alpha, and interferon-gamma. Mol Vis. 2013;19:737–50. [PMC free article] [PubMed] [Google Scholar]
- 15.Yang D, Elner SG, Bian ZM, Till GO, Petty HR, Elner VM. Pro-inflammatory cytokines increase reactive oxygen species through mitochondria and NADPH oxidase in cultured RPE cells. Exp Eye Res. 2007;85:462–72. doi: 10.1016/j.exer.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mettu PS, Wielgus AR, Ong SS, Cousins SW. Retinal pigment epithelium response to oxidant injury in the pathogenesis of early age-related macular degeneration. Mol Aspects Med. 2012;33:376–98. doi: 10.1016/j.mam.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Zhu D, Wu J, Spee C, Ryan SJ, Hinton DR. BMP4 mediates oxidative stress-induced retinal pigment epithelial cell senescence and is overexpressed in age-related macular degeneration. J Biol Chem. 2009;284:9529–39. doi: 10.1074/jbc.M809393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollyfield JG, Bonilha VL, Rayborn ME, Yang X, Shadrach KG, Lu L, Ufret RL, Salomon RG, Perez VL. Oxidative damage-induced inflammation initiates age-related macular degeneration. Nat Med. 2008;14:194–8. doi: 10.1038/nm1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarrett SG, Boulton ME. Consequences of oxidative stress in age-related macular degeneration. Mol Aspects Med. 2012;33:399–417. doi: 10.1016/j.mam.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khandhadia S, Lotery A. Oxidation and age-related macular degeneration: insights from molecular biology. Expert Rev Mol Med. 2010;12:e34. doi: 10.1017/S146239941000164X. [DOI] [PubMed] [Google Scholar]
- 21.Burke JM, Cao F, Irving PE, Skumatz CM. Expression of E-cadherin by human retinal pigment epithelium: delayed expression in vitro. Invest Ophthalmol Vis Sci. 1999;40:2963–70. [PubMed] [Google Scholar]
- 22.Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK, Pfeifer K. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat Genet. 1998;20:344–51. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 23.Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B, Redmond TM. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J Biol Chem. 1993;268:15751–7. [PubMed] [Google Scholar]
- 24.Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL, Dryja TP. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat Genet. 1999;22:188–91. doi: 10.1038/9707. [DOI] [PubMed] [Google Scholar]
- 25.Wu BX, Chen Y, Chen Y, Fan J, Rohrer B, Crouch RK, Ma JX. Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Invest Ophthalmol Vis Sci. 2002;43:3365–72. [PubMed] [Google Scholar]
- 26.Sparkes RS, Heinzmann C, Goldflam S, Kojis T, Saari JC, Mohandas T, Klisak I, Bateman JB, Crabb JW. Assignment of the gene (RLBP1) for cellular retinaldehyde-binding protein (CRALBP) to human chromosome 15q26 and mouse chromosome 7. Genomics. 1992;12:58–62. doi: 10.1016/0888-7543(92)90406-i. [DOI] [PubMed] [Google Scholar]
- 27.Reinisalo M, Putula J, Mannermaa E, Urtti A, Honkakoski P. Regulation of the human tyrosinase gene in retinal pigment epithelium cells: the significance of transcription factor orthodenticle homeobox 2 and its polymorphic binding site. Mol Vis. 2012;18:38–54. [PMC free article] [PubMed] [Google Scholar]
- 28.D’Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, LaVail MM, Vollrath D. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet. 2000;9:645–51. doi: 10.1093/hmg/9.4.645. [DOI] [PubMed] [Google Scholar]
- 29.Bharti K, Gasper M, Ou J, Brucato M, Clore-Gronenborn K, Pickel J, Arnheiter H. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet. 2012;8:e1002757. doi: 10.1371/journal.pgen.1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adijanto J, Castorino JJ, Wang ZX, Maminishkis A, Grunwald GB, Philp NJ. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem. 2012;287:20491–503. doi: 10.1074/jbc.M112.354761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S, Miller SS. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–71. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–69. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 33.Poliakov E, Strunnikova NV, Jiang JK, Martinez B, Parikh T, Lakkaraju A, Thomas C, Brooks BP, Redmond TM. Multiple A2E treatments lead to melanization of rod outer segment-challenged ARPE-19 cells. Mol Vis. 2014;20:285–300. [PMC free article] [PubMed] [Google Scholar]
- 34.Nagineni CN, Detrick B, Hooks JJ. Synergistic effects of gamma interferon on inflammatory mediators that induce interleukin-6 gene expression and secretion by human retinal pigment epithelial cells. Clin Diagn Lab Immunol. 1994;1:569–77. doi: 10.1128/cdli.1.5.569-577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kauppinen A, Paterno JJ, Blasiak J, Salminen A, Kaarniranta K. Inflammation and its role in age-related macular degeneration. Cell Mol Life Sci. 2016;73:1765–86. doi: 10.1007/s00018-016-2147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin T, Walker GB, Kurji K, Fang E, Law G, Prasad SS, Kojic L, Cao S, White V, Cui JZ, Matsubara JA. Parainflammation associated with advanced glycation endproduct stimulation of RPE in vitro: implications for age-related degenerative diseases of the eye. Cytokine. 2013;62:369–81. doi: 10.1016/j.cyto.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nussenblatt RB, Lee RW, Chew E, Wei L, Liu B, Sen HN, Dick AD, Ferris FL. Immune responses in age-related macular degeneration and a possible long-term therapeutic strategy for prevention. Am J Ophthalmol. 2014;158:5–11. doi: 10.1016/j.ajo.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parmeggiani F, Romano MR, Costagliola C, Semeraro F, Incorvaia C, D'Angelo S, Perri P, De Palma P, De Nadai K, Sebastiani A. Mechanism of inflammation in age-related macular degeneration. Mediators Inflamm. 2012;2012:546786. doi: 10.1155/2012/546786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmado A, Carr AJ, Vugler AA, Semo M, Gias C, Lawrence JM, Chen LL, Chen FK, Turowski P, da Cruz L, Coffey PJ. Induction of differentiation by pyruvate and DMEM in the human retinal pigment epithelium cell line ARPE-19. Invest Ophthalmol Vis Sci. 2011;52:7148–59. doi: 10.1167/iovs.10-6374. [DOI] [PubMed] [Google Scholar]
- 40.Kutty RK, Nagineni CN, Kutty G, Hooks JJ, Chader GJ, Wiggert B. Increased expression of heme oxygenase-1 in human retinal pigment epithelial cells by transforming growth factor-beta. J Cell Physiol. 1994;159:371–8. doi: 10.1002/jcp.1041590221. [DOI] [PubMed] [Google Scholar]
- 41.Nagineni CN, Pardhasaradhi K, Martins MC, Detrick B, Hooks JJ. Mechanisms of interferon-induced inhibition of Toxoplasma gondii replication in human retinal pigment epithelial cells. Infect Immun. 1996;64:4188–96. doi: 10.1128/iai.64.10.4188-4196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camelo S, Calippe B, Lavalette S, Dominguez E, Hur J, Devevre E, Guillonneau X, Raoul W, Sennlaub F. Thinning of the RPE and choroid associated with T lymphocyte recruitment in aged and light-challenged mice. Mol Vis. 2015;21:1051–9. [PMC free article] [PubMed] [Google Scholar]
- 43.Wen B, Li S, Li H, Chen Y, Ma X, Wang J, Lu F, Qu J, Hou L. Microphthalmia-associated transcription factor regulates the visual cycle genes Rlbp1 and Rdh5 in the retinal pigment epithelium. Sci Rep. 2016;6:21208. doi: 10.1038/srep21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashid A, Bhatia SK, Mazzitello KI, Chrenek MA, Zhang Q, Boatright JH, Grossniklaus HE, Jiang Y, Nickerson JM. RPE Cell and Sheet Properties in Normal and Diseased Eyes. Adv Exp Med Biol. 2016;854:757–63. doi: 10.1007/978-3-319-17121-0_101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–88. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 48.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–85. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 49.Qiu YH, Wei YP, Shen NJ, Wang ZC, Kan T, Yu WL, Yi B, Zhang YJ. miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol Biochem. 2013;32:1331–41. doi: 10.1159/000354531. [DOI] [PubMed] [Google Scholar]
- 50.Yu H, Yang W. MiR-211 is epigenetically regulated by DNMT1 mediated methylation and inhibits EMT of melanoma cells by targeting RAB22A. Biochem Biophys Res Commun. 2016;476:400–5. doi: 10.1016/j.bbrc.2016.05.133. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Ye F, Li Q, Tamiya S, Darling DS, Kaplan HJ, Dean DC. Zeb1 represses Mitf and regulates pigment synthesis, cell proliferation, and epithelial morphology. Invest Ophthalmol Vis Sci. 2009;50:5080–8. doi: 10.1167/iovs.08-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bian ZM, Elner SG, Elner VM. Dual involvement of caspase-4 in inflammatory and ER stress-induced apoptotic responses in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2009;50:6006–14. doi: 10.1167/iovs.09-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nahomi RB, Palmer A, Green KM, Fort PE, Nagaraj RH. Pro-inflammatory cytokines downregulate Hsp27 and cause apoptosis of human retinal capillary endothelial cells. Biochim Biophys Acta. 2014;1842:164–74. doi: 10.1016/j.bbadis.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]