Abstract
T cell activation in response to Ag is largely regulated by protein posttranslational modifications. Although phosphorylation has been extensively characterized in T cells, much less is known about the glycosylation of serine/threonine residues by O-linked N-acetylglucosamine (O-GlcNAc). Given that O-GlcNAc appears to regulate cell signaling pathways and protein activity similarly to phosphorylation, we performed a comprehensive analysis of O-GlcNAc during T cell activation to address the functional importance of this modification and to identify the modified proteins. Activation of T cells through the TCR resulted in a global elevation of O-GlcNAc levels and in the absence of O-GlcNAc, IL-2 production and proliferation were compromised. T cell activation also led to changes in the relative expression of O-GlcNAc transferase (OGT) isoforms and accumulation of OGT at the immunological synapse of murine T cells. Using a glycoproteomics approach, we identified >200 O-GlcNAc proteins in human T cells. Many of the identified proteins had a functional relationship to RNA metabolism, and consistent with a connection between O-GlcNAc and RNA, inhibition of OGT impaired nascent RNA synthesis upon T cell activation. Overall, our studies provide a global analysis of O-GlcNAc dynamics during T cell activation and the first characterization, to our knowledge, of the O-GlcNAc glycoproteome in human T cells.
Introduction
The activation of specific T lymphocytes by Ag is a critical step in the initiation of many immune responses. The binding of a TCR to its specific peptide-MHC ligand triggers an intracellular signaling cascade, culminating in the activation of T cell effector functions and the formation of long-lived memory cells. Similar to many other cell signaling pathways, the regulation of T cell activation is mediated largely through the regulation of protein posttranslational modifications. For instance, upon TCR ligation, lymphocyte-specific protein tyrosine kinase (LCK) phosphorylates the cytoplasmic tails of CD3, leading to the recruitment of ZAP-70, which potentiates the signaling cascade through phosphorylation of additional downstream substrates (1, 2). Eventually, these early signaling events translate into the expression of new genes that are necessary for T cell effector function. In addition to tyrosine phosphorylation, a variety of other posttranslational modifications are involved in the regulation of T cell activation, a process that must be strictly controlled to permit appropriate responses against foreign Ags while preventing inappropriate responses to self-antigens. Canonical examples include serine/threonine phosphorylation, lysine methylation and acetylation, lysine ubiquitylation, and cysteine palmitoylation, which can influence transcription factor activation, chromatin accessibility, protein stability, and membrane localization, respectively (3–19).
In contrast to some of these modifications, relatively little is known about the proteins targeted for glycosylation by O-linked N-acetylglucosamine (O-GlcNAc) during T cell activation and its effect on protein function. O-GlcNAc is the only known form of intracellular glycosylation, which involves the reversible attachment of a GlcNAc monosaccharide to serine and threonine residues (20–22). Remarkably, the opposing actions of just two enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), mediate the addition and removal of O-GlcNAc from the hundreds of protein substrates identified to date, including transcription factors, epigenetic regulators, and kinases (20). Mounting evidence suggests that O-GlcNAc functions in the regulation of protein activity much like phosphorylation and other modifications. Although the molecular mechanisms remain mostly unresolved, O-GlcNAc has clear biological significance because deletion of the OGT or OGA gene in mice leads to embryonic lethality (23) or perinatal death (24), respectively. Furthermore, conditional deletion of OGT results in cellular senescence and apoptosis in numerous cell types, including T cells (25). Considering that abnormal O-GlcNAc levels, possibly stemming from alterations in metabolism (26–29), may contribute to the pathology of several diseases, including cancer, diabetes, and neurodegeneration (29–36), a greater understanding of O-GlcNAc biology in T cells could help to explain the impact of metabolic health on the function of the immune system.
Although previous studies suggested a role for O-GlcNAc in T cell activation (37–39), they focused on immortalized cell lines, making the relevance to primary T cells unclear. Furthermore, only a selected few proteins were surveyed. In this study, we aimed to investigate the O-GlcNAc modification in primary human T cells so as to be as physiological as possible and to obtain a broader perspective of what proteins are modified using a proteomics approach. In this article, we show that activation of primary T cells directly ex vivo leads to increased staining for O-GlcNAc, which is accompanied by changes in OGT localization and splicing. Suppressing O-GlcNAc modification through inhibition of OGT activity reduced IL-2 production and proliferation, indicating that O-GlcNAc is important for T cell activation. To determine which proteins are modified by O-GlcNAc in T cells, we used a glycoproteomics approach and identified numerous proteins, including many involved in RNA metabolism. Consistent with this finding, the OGT inhibitor impaired nascent RNA synthesis in activated T cells. Overall, our results provide a global view of O-GlcNAc dynamics in response to T cell activation and constitute the first report, to our knowledge, of the O-GlcNAc glycoproteome in primary human T cells.
Materials and Methods
Abs
Anti–O-GlcNAc (clone RL-2, ab2739), anti–ubiquitin-associated protein 2-like (UBAP2L; ab138309), anti-NUP153 (ab24700), anti-NUP62 (ab96134), and anti-ELF1 (ab171813) were purchased from Abcam. Anti-NUP214 was purchased from Abcam (ab70497) or Abnova. Anti-IL-2–PE (559334) and anti-CD69–FITC (555530) were purchased from BD Biosciences. Anti–O-GlcNAc (clone CTD110.6, MMS-248R or 9875) was purchased from Covance and later Cell Signaling Technology. Anti-GAPDH (5174), anti-histone H2B (2934), anti-LCP1 (5350), anti-VIM (5741), anti–aryl hydrocarbon receptor nuclear translocator (ARNT; 5537), anti-HCLS1 (4503), anti-WNK1 (4979), anti-EWSR1 (11910), anti-ARID1A (12354), anti-MYPT1 (2634), anti-RUNX1 (4334), anti-SP1 (9389), and anti-NUP98 (2292) were purchased from Cell Signaling Technology. Anti-mouse IgM-FITC (11-5790-82) and IgM isotype control (14-4752-85) were purchased from eBioscience. Anti-mouse IgG–Alexa Fluor 647 (A21236) and anti-rabbit IgG–Alexa Fluor 488 (A11008) were purchased from Life Technologies. Anti–host cell factor 1 (HCFC1; ABD74) was purchased from Millipore. Anti-OGT (O6264) was purchased from Sigma.
Cell isolation and culture
Primary human T cells were isolated from leukocyte reduction shuttles obtained from anonymous donors at the Stanford Blood Center under institutional review board protocol. After purification by negative selection using RosetteSep (#15061; STEMCELL Technologies), T cells were cultured in RPMI 1640 (Life Technologies) supplemented with GlutaMAX, 10% FBS, and 1× penicillin/streptomycin in a humidified incubator at 37°C with 5% CO2. CH27 B cells were cultured similarly, with the exception that 2-ME was included in the culture medium at 0.1%.
Cell stimulation
Anti-CD3/CD28–coated beads were prepared by coating latex sulfate beads (5 μm, #S37227; Invitrogen) in PBS containing 0.1 mg/ml anti-CD3 (clone OKT3, #317304) and 0.2 mg/ml anti-CD28 (clone CD28.2, #302923; both from BioLegend) for 2 h at 37°C. Control beads were prepared in the same manner with isotype-control Abs. Primary human T cells were stimulated by incubating with beads at 2:1 ratio. For experiments involving intracellular cytokine staining, cells were stimulated in the presence of protein transport inhibitors (#00-4980; eBioscience). In the case of PMA/ionomycin stimulation, cells were treated with cell stimulation mixture plus protein transport inhibitors (#00-4975; eBioscience).
Cell lysis, subcellular fractionation, immunoprecipitation, and immunoblotting
For whole-cell lysates, cells were washed once in cold PBS, and cell pellets were immediately lysed or flash frozen in liquid N2 and stored at −80°C prior to lysis. Cells were lysed in cold RIPA buffer (1% Nonidet P-40 [NP-40], 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris [pH 8]) supplemented with protease inhibitors (#04-693-124-001; Roche complete with EDTA), 1 mM PMSF, and 2.5 μM PUGNAc (OGA inhibitor). IGEPAL CA-630 (Sigma-Aldrich) was used as a substitute for NP-40 in buffers in some cases.
For subcellular fractionation, cells were washed once in cold PBS and then resuspended in isotonic lysis buffer (10 mM Tris [pH 8], 2 mM MgCl2, 3 mM CaCl2, 300 mM sucrose). NP-40 was added to a final concentration of 0.5%, and plasma membrane lysis was allowed to proceed for 3 min on ice. Nuclei were pelleted for 5 min at 500 × g at 4°C. The supernatant, representing the crude cytosolic fraction, was supplemented with protease inhibitors and frozen at −80°C. Nuclei were washed once in isotonic lysis buffer and resuspended in extraction buffer (10 mM HEPES [pH 7.9], 0.75 mM MgCl2, 500 mM NaCl, 0.1 mM EDTA, 12.5% glycerol, 2.5 U/μl benzonase) for 30 min on ice with occasional vortexing. Nuclear extracts were centrifuged at 21,130 × g for 10 min at 4°C. The supernatant, representing the crude nuclear fraction, was supplemented with protease inhibitors, passed through a spin column filter (0.45 μm) to remove the beads, and frozen at −80°C. Upon thawing, both fractions were centrifuged at 21,130 × g to remove any insoluble material.
For immunoprecipitation, biotinylated proteins (see below) were resuspended in 1% SDS/50 mM Tris (pH 8) and diluted with four volumes of 1.25% NP-40/125 mM NaCl/50 mM Tris (pH 8). Anti-NUP214 was allowed to bind to magnetic protein A/G beads (Pierce) for 60 min at room temperature in TBST (50 mM Tris [pH 8], 150 mM NaCl, 0.1% Tween 20). The charged beads were washed with TBST and incubated with the biotinylated proteins overnight at 4°C. The beads were washed three times with TBST and eluted with 1× SDS-PAGE sample buffer containing 2.5% 2-ME for 10 min at room temperature.
Equivalent amounts of protein were separated with 4–12% Bis-Tris gradient gels (Life Technologies) and blotted onto nitrocellulose. In the case of polyethylene glycol (PEG)-labeled proteins, 3–8% Tris-Acetate gels were used for enhanced resolution. After blocking in 3% milk or 3% BSA in TBST, primary Abs were added and incubated at room temperature for 1 h or overnight at 4°C in blocking buffer. Membranes were washed with TBST, and HRP-conjugated secondary reagents (goat anti-mouse IgG-HRP [Pierce 32430]; goat anti-rabbit IgG-HRP [Pierce 32460]; or streptavidin-HRP [Pierce 21134]) were added for 1 h at room temperature in blocking buffer. Membranes were washed again and developed with ECL reagents (Pierce). Where needed, membranes were stripped for 15 min at 65°C with mild stripping buffer (200 mM glycine [pH 2.2], 1% Tween-20, 0.1% SDS), washed in TBST, reblocked, and reprobed as above.
Derivatization of O-GlcNAc by β-elimination followed by Michael addition of DTT
To test the efficiency and specificity of β-elimination followed by Michael addition of DTT (BEMAD), 200 pmol of a synthetic O-GlcNAc peptide (gCREB, C33374) was spiked into a commercial mixture consisting of 200 pmol of four phosphopeptides and three unmodified peptides (P33357; both from Life Technologies). Peptides were subjected to mild performic acid oxidation, calf intestinal alkaline phosphatase (CIP; New England BioLabs) treatment, BEMAD, and thiol enrichment, as described below, with the exception that CIP was used at 0.2 U/μl. Half of the mixture was labeled with d0 DTT, whereas the other half was labeled with d6 DTT. Samples were analyzed by MALDI-TOF at the Beckman Protein and Nucleic Acid Facility.
To perform BEMAD on cell lysates, frozen cell pellets were resuspended in urea lysis buffer (8 M urea, 100 mM NaCl, 25 mM Tris [pH 8]) and incubated on ice for 20 min. Insoluble debris was pelleted by centrifugation, and the anti-CD3/CD28 or control beads were removed with 0.45-μm spin filters. After dilution with four volumes of 50 mM NH4HCO3, trypsin was added to 5 ng/ml, and proteins were digested overnight at 37°C in the presence of 1 μM PUGNAc. Digests were acidified with 0.5% trifluoroacetic acid, desalted with C18 columns (Waters), and dried in a speed-vac. To prevent labeling of cysteine residues, peptides were subjected to mild performic acid oxidation (3% formic acid/3% H2O2) (40) for 4 h at room temperature and then dried. In some cases, peptides were subjected to a crude fractionation by strong cation exchange (SCX), which was accomplished with SCX spin columns (Nest Group) and elution in stepwise gradients of 20, 50, 500, and 1000 mM KCl in loading buffer (5 mM KH2PO4/pH 3/20% acetonitrile), followed by desalting. To prevent labeling of phosphorylation sites, peptides were treated with CIP (0.4 U/μl in 100 mM NaCl, 10 mM MgCl2, 50 mM NH4HCO3, pH 8) at 37°C for 4 h followed by desalting. To label O-GlcNAc sites, peptides were incubated in BEMAD buffer (1% triethylamine, 0.1% NaOH) for 2 h at 56°C in the presence of 20 mM light (d0) DTT or heavy (d6) DTT (1,4-DTT-1,1,2,3,4,4-d6, CDN Isotopes) as described previously (41, 42). The labeling reaction was quenched with 1% trifluoroacetic acid and the heavy and light labeling reactions were combined into a single aliquot for desalting. To purify DTT-labeled peptides, total peptides were resuspended in 500 μl of binding buffer (PBS, 1 mM EDTA) and added to thiopropyl Sepharose (Sigma). After binding for 2 h at room temperature, the resin was washed three times with column buffer. Elution was accomplished with two treatments of column buffer containing 20 mM DTT for 30 min each at room temperature. The released peptides were passed through a 0.2 μm spin filter, acidified, and desalted prior to mass spectrometry analysis.
For the β-N-acetylglucosaminidase treatment, corresponding SCX fractions from control and stimulated conditions were combined to equalize any differences in protein expression levels and then split into two equivalent aliquots, one of which was treated with β-N-acetylglucosaminidase (16 U in 100 μl reaction; NEB) overnight at 37°C, according to the manufacturer’s protocol. β-N-acetylglucosaminidase was then heat inactivated at 65°C for 10 min, and the procedure was continued with CIP treatment, as described.
Biotin labeling, PEG labeling, and purification with streptavidin beads
For metabolic labeling, primary T cells were incubated with 40 μM tetraacetylated N-azidoacetylgalactosamine (Ac4GalNAz; Pierce) for 2 d, and then stimulated with beads in the presence of Ac4GalNAz for an additional 24 h. Cells were washed once in cold PBS and then flash frozen in liquid N2. Cell pellets were lysed in RIPA lysis buffer, as above. Cleared lysates were supplemented with additional SDS to 1%, and proteins (700 μg) were denatured for 10 min at 65°C, followed by biotinylation with 100 μM biotin phosphine (Pierce) overnight at room temperature.
To purify biotinylated proteins for mass spectrometry analysis, proteins were precipitated with methanol/chloroform to remove excess biotin reagent and then resolubilized in 1% SDS/50 mM Tris (pH 8). Proteins were diluted with four volumes of 1.25% NP-40, 50 mM Tris (pH 8), 125 mM NaCl and incubated with magnetic streptavidin beads (M-280 Dynabeads; Life Technologies) for 1.5 h at room temperature to capture biotinylated proteins. Beads were washed sequentially with 1% SDS/PBS, 8 M urea/100 mM NaCl/50 mM Tris (pH 8), 8 M urea/1 M NaCl/50 mM Tris (pH 8), 8 M urea/50 mM NH4HCO3, and PBS. An aliquot (10%) of the beads was treated with 6 M urea/2 M thiourea/2% SDS/15 mM biotin/PBS (pH 12) for 10 min at 95°C to release bound material for analysis by immunoblot to confirm capture of biotinylated protein. The remainder (90%) of the beads were resuspended in 8 M urea/50 mM NH4HCO3 and diluted with four volumes of 50 mM NH4HCO3 before adding ProteaseMAX surfactant to 0.03% and trypsin (both from Promega) to 5 ng/μl. After overnight digestion at 37°C, the supernatant containing tryptic peptides was recovered. Cysteines were reduced and alkylated by treatment with 5 mM DTT for 30 min at room temperature, 14 mM iodoacetamide for 30 min, and then an additional 5 mM DTT for 15 min. Trifluoroacetic acid was added to 0.5%, and the peptides were desalted with a C18 spin column (Nest Group) and dried. Peptides were resuspended in 5% formic acid/5% acetonitrile for analysis by liquid chromatography–tandem mass spectrometry.
For chemoenzymatic labeling, frozen cell pellets were resuspended in RIPA lysis buffer, and 200 μg of protein was labeled with the ClickIt O-GlcNAc Enzymatic Labeling Kit (Life Technologies), according to the manufacturer’s protocol. PNGase F (500 U; NEB) was added, and the labeling reaction was incubated for an additional 1 h at 37°C. Proteins were precipitated and resolubilized in 1% SDS/50 mM Tris (pH 8). Biotinylation was accomplished by treating proteins with 100 μM phosphine biotin (Pierce) at room temperature overnight. PEGylation was accomplished by treating proteins with 15 mM iodoacetamide for 30 min at room temperature, followed by 100 μM DBCO-5K PEG (Kerafast) overnight at room temperature. To purify biotinylated proteins with streptavidin beads for immunoblot analysis, proteins were separated from excess biotin reagent by precipitation and resolubilized in 1% SDS/50 mM Tris (pH 8). Proteins were diluted with four volumes of 1.25% NP-40/125 mM NaCl/50 mM Tris (pH 8) and allowed to bind to streptavidin beads for 1 h at room temperature or overnight at 4°C. Beads were washed sequentially with 1% SDS/PBS, 8 M urea/100 mM NaCl/50 mM Tris (pH 8), 1 M NaCl/1% NP-40/0.1% SDS/50 mM Tris (pH 8), and again with 1% SDS/PBS. Bound proteins were eluted with 6 M urea/2 M thiourea/2% SDS/15 mM biotin/PBS (pH 12) for 10 min at 95°C (43).
Mass spectrometry and data analysis
Desalted peptides were analyzed by liquid chromatography–tandem mass spectrometry on an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, Santa Clara, CA). Peak lists were generated with the msConvert algorithm23 (v. 3.0.45), and spectra were assigned to peptides using the SEQUEST (44) algorithm (v. 28.12) against the human proteome downloaded from the International Protein Index repository (ver 3.66). This sequence database also consisted of common protein contaminants, as well as reversed decoy sequences to facilitate false discovery rate estimation (45). Search parameters included precursor ion mass tolerance of 50 ppm and fragment ion mass tolerance of 1 Da. For BEMAD experiments, additional search parameters included static modification of cysteines (+47.98474387 Da, trioxidation) and methionines (+31.98982924, dioxidation) and variable modification of serines and threonines (+136.001659 for light DTT and +142.03931968 for heavy DTT) and tryptophans (+15.99491462, oxidation). For metabolic labeling experiments, additional search parameters included static modification of cysteines (+57.0214637236, carbamidomethylation), as well as variable modifications of methionines (+15.9949146221, oxidation), serines, threonines, and tyrosines (+79.9663304104, phosphorylation), and lysines (+42.0105646863, acetylation). Peptide identifications from each sample were filtered to a 1% peptide false-discovery rate (45).
Microscopy
For the analysis of OGT localization, CH27 B cells were pulsed with 10 μM MCC agonist peptide or 99E antagonist peptide for 4–6 h at 37°C. B cells were washed and incubated with 5CC7 T cell blasts on glass coverslips. In some cases, the coverslips were precoated with anti-CD16/32 to facilitate attachment of the CH27 cells. After 10–60 min, cells were fixed with 4% paraformaldehyde (PFA) in PBS, permeabilized in 0.3% Triton X-100/PBS, and stained with anti-OGT, followed by Alexa Fluor 488–labeled anti-rabbit IgG.
For experiments with primary human T cells, #1.5 glass coverslips were placed in 24-well plates and coated with 2.5 μg/ml anti-CD3 (OKT3) and 5 μg/ml anti-CD28 (CD28.2) in PBS at 4°C overnight, followed by washing with PBS. For the analysis of nascent RNA synthesis, T cells were cultured on coated coverslips in the presence of 200 μM 5-ethynyl uridine (5-EU; Life Technologies). Cells were fixed with 4% PFA in PBS for 15 min at room temperature. Cells were then permeabilized with 0.3% Triton X-100/PBS, and metabolically incorporated 5-EU was labeled with 10 μM azide-labeled Alexa Fluor 488 (Life Technologies) using click chemistry reagents (C10269; Life Technologies), according to the manufacturer’s instructions. After fluorescent labeling, coverslips were mounted on glass slides in VECTASHIELD mounting medium with DAPI and examined with a Zeiss LSM 710 META confocal microscope.
To assay NFAT translocation, Jurkat cells stably expressing GFP-tagged NFAT (clone Cnc1g; generously provided by the R. Lewis Laboratory) were treated with 50 μM 5S-GlcNAc for 48 h. Cells were plated on #1.5 glass coverslips coated with 2.5 μg/ml OKT3 and 5 μg/ml CD28.2 in PBS at 37°C for 2 h. After stimulation, cells were fixed with 4% PFA in PBS for 15 min at room temperature, washed in PBS, and mounted on slides, as above, for confocal imaging.
Flow cytometry
After stimulation, T cells were washed in FACS buffer (2% FBS, 1 mM EDTA in PBS) and stained with Aqua LIVE/DEAD (Life Technologies). Cells were then fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) for intracellular staining to detect IL-2 and O-GlcNAc. FITC-labeled anti-mouse IgM secondary Ab was used to detect anti–O-GlcNAc Ab CTD110.6.
For the analysis of proliferation, T cells were labeled with CFSE (Life Technologies). Briefly, cells were resuspended in prewarmed complete RPMI 1640 medium diluted 1/100 in PBS at 5 × 106 cells/ml. CFSE was added to 1 μM, and cells were labeled for 8 min at 37°C. The labeling reaction was quenched by adding one volume of cold FBS to three volumes of cells. Cells were washed twice in cold complete medium and allowed to proliferate for 3–4 d at 37°C. Cells were harvested, washed in FACS buffer, and stained with Aqua LIVE/DEAD prior to analysis by flow cytometry. All cells were analyzed on an LSR II flow cytometer (BD Biosciences). Data were analyzed by FlowJo software (TreeStar).
For the analysis of 5-EU incorporation by flow cytometry, cells were treated the same as for microscopy except they were cultured in 96-well plates. Unless otherwise noted, plots are gated on total cells based on forward scatter versus side scatter.
Statistical analysis
Statistics were performed with GraphPad Prism and Microsoft Excel.
Results
Global levels of O-GlcNAc glycosylation increase after T cell activation
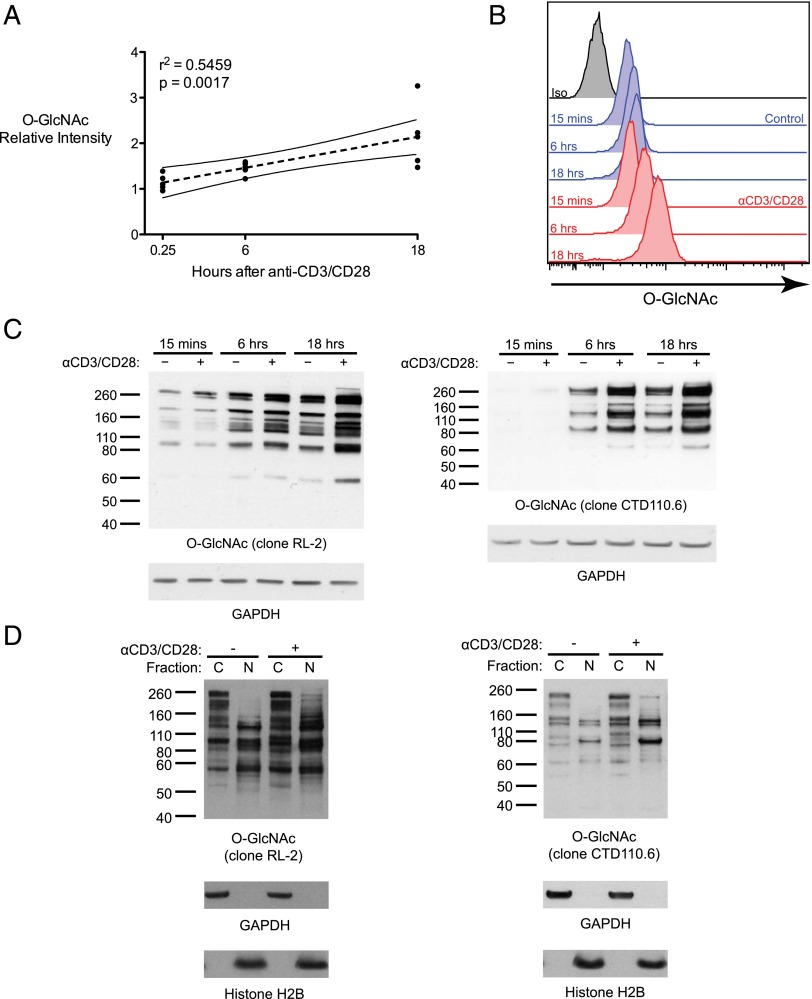
To determine whether TCR stimulation influences the modification of proteins with O-GlcNAc, we purified human T cells from blood bank donors and activated them with anti-CD3/CD28 beads. TCR stimulation resulted in increased staining for total O-GlcNAc levels over time, as assessed by flow cytometry (Fig. 1A, 1B), as well as by immunoblots of whole-cell lysates (Fig. 1C) and subcellular fractions (Fig. 1D).
FIGURE 1.
Global levels of O-GlcNAc glycosylation increase after T cell activation. (A) Primary human T cells were incubated with control beads or anti-CD3/CD28 beads and then analyzed for O-GlcNAc by flow cytometry. The staining intensity of stimulated cells relative to control cells at each time point is plotted (five donors over three independent experiments) along with the mean and 95% confidence interval of a linear regression analysis. (B) Representative line graphs are shown for one donor. (C) Whole-cell lysates from control and stimulated T cells were analyzed for O-GlcNAc levels by immunoblot with anti-O-GlcNAc Ab RL-2 or CTD110.6. GAPDH served as a loading control. A total of at least two donors over at least two independent experiments were analyzed with similar results for each blot. (D) Cytosolic (C) and nuclear (N) extracts from T cells incubated with control beads or anti-CD3/CD28 beads for 18 h were analyzed for O-GlcNAc levels by immunoblot with anti–O-GlcNAc Ab RL-2 or CTD110.6. GAPDH and histone H2B served as controls for loading and compartmentalization. A total of three donors over two independent experiments were analyzed with similar results for each blot.
Activity of OGT, but not OGA, is necessary for T cell effector function
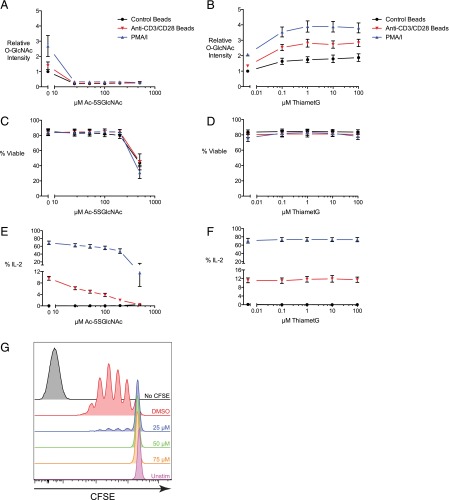
To verify that O-GlcNAc modification holds functional importance in primary human T cells, we stimulated cells in the presence of OGT or OGA inhibitors. Treatment with the OGT inhibitor 2-acetamido-1,3,4,6-tetra-O-acetyl-2-deoxy-5-thio-α-d-glucopyranose (Ac-5SGlcNAc) (46) diminished the production of IL-2 in response to anti-CD3/CD28 beads or PMA/ionomycin, without compromising cell viability, at concentrations up to 200 μM (Fig. 2A, 2C, 2E). The decrease in IL-2 production was likely not attributable to impaired NFAT translocation (Supplemental Fig. 1). Ac-5SGlcNAc also suppressed T cell proliferation (Fig. 2G). In contrast, treatment with the OGA inhibitor ThiametG (47) had no significant effect on IL-2 production (Fig. 2B, 2D, 2F). Together, these results indicate that the addition of O-GlcNAc, but not its removal, is important for T cell proliferation and cytokine production under the conditions used in this study.
FIGURE 2.
Activity of OGT, but not OGA, is necessary for T cell effector function. Primary human T cells were treated with the OGT inhibitor Ac-5SGlcNAc (A, C, and E) or the OGA inhibitor ThiametG overnight (B, D, and F) and then directly stimulated with anti-CD3/CD28 beads or PMA/ionomycin overnight in the continued presence of the inhibitors. Cells were analyzed for O-GlcNAc levels (A and B), viability (C and D), and IL-2 production (E and F) by flow cytometry. The plots of IL-2 and O-GlcNAc levels are gated on viable cells. Results are presented as the mean ± SEM of five donors across four experiments (A, C, and E) or four donors across three experiments (B, D, and F). O-GlcNAc staining intensity is expressed relative to unstimulated vehicle controls for each donor. (G) Human T cells were labeled with CFSE and then stimulated with anti-CD3/CD28 beads in the presence of increasing concentrations of Ac-5SGlcNAc. Proliferation was assessed after 3–4 d. Graphs are gated on viable cells. Results are representative of five donors across three experiments.
T cell activation leads to regulation of OGT localization and splicing
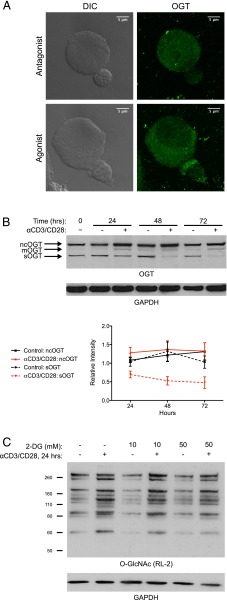
As shown in Fig. 1, T cell activation leads to a global increase in O-GlcNAc levels, which would require concomitant activation of OGT or repression of OGA. Thus, we investigated whether T cell activation is accompanied by changes in OGT localization or expression. A previous study noted that OGT binds to phosphatidylinositol-3,4,5-trisphosphate (PIP3) (48). Given that PIP3 is generated at the immunological synapse, we hypothesized that OGT is recruited to the synapse, where it would then have access to a different set of substrates. Indeed, we observed intense staining for OGT at the immunological synapse when transgenic murine T cells were incubated with B cells presenting cognate Ag (Fig. 3A).
FIGURE 3.
T cell activation leads to regulation of OGT localization and splicing. (A) Transgenic 5CC7 T cell blasts were incubated with CH27 B cells pulsed with MCC agonist or 99E antagonist peptide. After 50 min, cells were fixed, permeabilized, and stained for OGT for analysis by confocal microscopy. Results are representative of two independent experiments and incubation times from 10 to 60 min. (B) Primary human T cells were stimulated with control beads or anti-CD3/CD28 beads, and OGT protein levels were measured by immunoblot. A total of four donors over two independent experiments were analyzed. One blot is pictured. The ncOGT and sOGT bands on all blots were analyzed by densitometry and plotted. Data represent the mean band intensity ± SEM relative to GAPDH loading controls and normalized to the ncOGT or sOGT band intensity at the initial time point (0 h). (C) Primary human T cells were treated with increasing concentrations of 2-DG for 0–2 h prior to overnight stimulation with control beads or anti-CD3/CD28 beads in the continued presence of 2-DG. Whole-cell lysates were analyzed for O-GlcNAc levels by immunoblot. Similar results were obtained with two additional donors across two independent experiments using anti–O-GlcNAc Ab CTD110.6.
To assess the expression levels of OGT, we performed immunoblots on whole-cell lysates from control or activated human T cells. As evident from the three bands in Fig. 3B, OGT exists as three differentially spliced isoforms, termed nucleocytoplasmic OGT (ncOGT), mitochondrial OGT (mOGT), and short OGT (sOGT). Each isoform is identical except for the number of tetratricopeptide repeats in the N terminus and, in the case of mOGT, an N-terminal mitochondrial-targeting sequence (49). Although there was little, if any, increase in ncOGT expression in activated T cells, a striking difference was seen in the relative levels of sOGT, which were consistently higher in control cells (Fig. 3B). In some cases, an increase in mOGT expression was observed in activated T cells, but the intensity of this band was not always strong enough for reliable measurements.
In addition to direct regulation of its localization or expression, OGT activity may also be regulated indirectly by the availability of its nucleotide sugar donor substrate, UDP-GlcNAc, which, in turn, is heavily influenced by cell metabolism (26–28). In response to activation, T cells exhibit a remarkable metabolic shift from fatty acid oxidation to glycolysis (50–52). Thus, increased sugar metabolism may lead to higher levels of UDP-GlcNAc, which would promote OGT activity. To test whether changes in T cell metabolism contribute to changes in O-GlcNAc levels, we stimulated cells in the presence of 2-deoxyglucose (2-DG), a glucose analog that inhibits glycolysis. As shown in Fig. 3C, 2-DG treatment reduced the intensity of a subset of O-GlcNAc bands, but most were unaffected. Thus, we speculate that the increased O-GlcNAc levels in activated T cells are primarily due to changes in OGT localization and splicing and, in some cases, glucose metabolism.
Metabolic labeling identifies O-GlcNAc proteins from human T cells
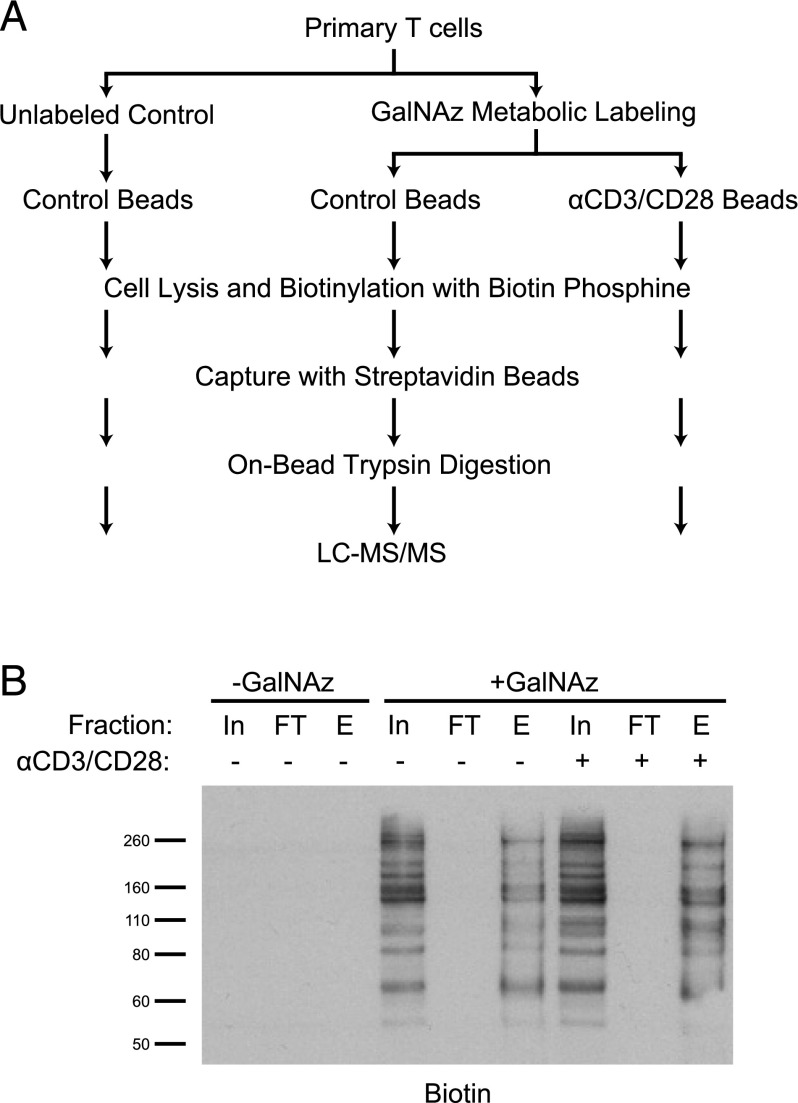
Next, we focused on the comprehensive identification of proteins that are subject to O-GlcNAc glycosylation in activated T cells. To this end, we used a proteomics approach based on metabolic incorporation of Ac4GalNAz, which is processed intracellularly to UDP-GlcNAz for use as the donor nucleotide sugar of OGT (53). O-GlcNAz groups are subsequently labeled with a biotin phosphine reagent through Staudinger ligation (54). O-GlcNAc proteins from activated and control T cells were metabolically labeled and purified according to the workflow outlined in Fig. 4A. Cells incubated without Ac4GalNAz served as a control for nonspecific purification. We confirmed efficient capture of the labeled proteins by immunoblot analysis (Fig. 4B). Comparison of the peptides present in the unlabeled control sample with those in the labeled samples yielded a list of 133 specifically purified proteins identified with high confidence, which were taken to be probable O-GlcNAc–modified proteins (Supplemental Table I). An additional 81 proteins, classified as potential O-GlcNAc proteins based on their enrichment in the labeled samples but with fewer peptides sequenced, are also listed in Supplemental Table I. The list of purified O-GlcNAc proteins includes nuclear pore proteins, transcription factors, and other proteins of diverse functions, many of which were reported in a study of HEK293 cells (55). However, our list also included novel O-GlcNAc proteins, such as HCLS1, ZAP-70, SHIP1 (INPP5D), LCK, and PI3K (PI3KCD). Although it is possible for GalNAz to label glycoproteins other than O-GlcNAc glycoproteins, the latter can be distinguished based on their intracellular localization. Pathway analysis (56, 57) of the 133 higher-confidence glycoproteins showed enrichment of proteins involved in the metabolism of RNA, including transcription, processing, and transport, most likely driven by the significant presence of transcription factors, splicing factors, and nuclear pore proteins (Supplemental Table II). The glycoproteins were also enriched for proteins subject to modification by phosphorylation and acetylation, indicating the potential for cross-talk among multiple posttranslational modifications.
FIGURE 4.
Metabolic labeling approach identifies O-GlcNAc proteins from human T cells. (A) Diagram illustrating the workflow for identification of O-GlcNAc proteins from T cells using metabolic labeling. (B) Primary human T cells from one donor were metabolically labeled with GalNAz and stimulated with control beads or anti-CD3/CD28 beads for 24 h. Azide-labeled proteins were then biotinylated for subsequent enrichment by streptavidin beads. Samples of the input (In) and flow-through (FT) material were analyzed by immunoblot to detect the biotin tag. Biotinylated protein was eluted (E) from an aliquot of the streptavidin beads to confirm capture.
Chemical proteomics identifies O-GlcNAc sites enriched in activated T cells
Although the metabolic-labeling approach enables efficient identification of O-GlcNAc proteins given the opportunity to detect many unique peptides from the same protein, it does not provide information regarding the site of modification. Furthermore, because of the possibility of multiple O-GlcNAc sites on the same protein, it is necessary to perform quantitative analyses at the site level rather than the protein level. Thus, to identify O-GlcNAc sites that undergo changes in occupancy during T cell activation, we turned to a second labeling technique, BEMAD, which involves alkaline cleavage of the sugar from peptides with subsequent addition of a DTT adduct (41, 42). Unlike the native O-GlcNAc linkage, which is labile and tends to undergo neutral loss from the peptide backbone, the DTT adduct remains stable during conventional collision-induced dissociation mass spectrometry, allowing detection of the modification site. Additionally, the use of DTT isotopes permits the relative quantification of O-GlcNAc sites between experimental conditions. Using a synthetic O-GlcNAc peptide spiked into a mixture of irrelevant peptides, we confirmed that our BEMAD reaction conditions resulted in efficient and specific labeling (data not shown).
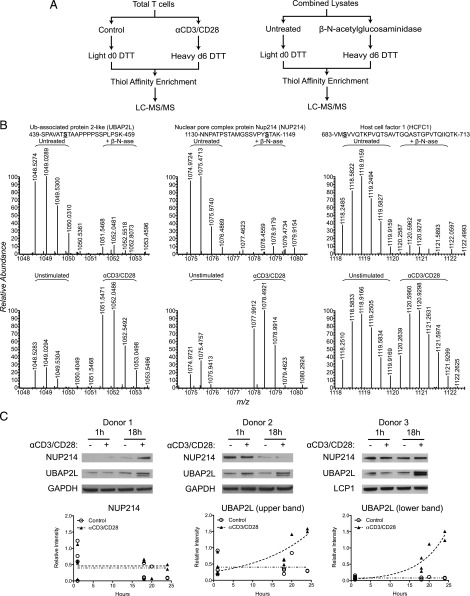
We proceeded by performing BEMAD on tryptic peptides obtained from activated and control T cells. The use of DTT isotopes (outlined in Fig. 5A) facilitated the relative quantification of any differences between activated and control T cells. In addition, we used stable isotope tagging to control for labeling specificity by analyzing samples with and without enzymatic removal of O-GlcNAc by treatment with β-N-acetylglucosaminidase. Using this approach, O-GlcNAc sites were detected on Ser445 of UBAP2L and Ser1146 of nuclear pore complex protein Nup214 (NUP214), as previously described (55). Both of these sites represent true positives, as demonstrated by their sensitivity to β-N-acetylglucosaminidase (Fig. 5B, upper panels). Notably, these O-GlcNAc–modified peptides were more prevalent in activated T cells (Fig. 5B, lower panels).
FIGURE 5.
Chemical labeling identifies O-GlcNAc sites enriched in activated T cells. (A) Diagrams illustrating the workflow for DTT isotope labeling to detect differences between control and activated T cells (left panel) or between samples with and without enzymatic removal of O-GlcNAc using β-N-acetylglucosaminidase (right panel). (B) MS1 spectra of peptides from UBAP2L, NUP214, and HCFC1. DTT-labeled residues are underlined in bold. Experiment comparing samples with and without enzymatic removal of O-GlcNAc (upper panels). Experiment comparing control and activated T cells after 18 h of stimulation (lower panels). For this experiment, lysates from three donors were pooled and labeled together. (C) Whole-cell lysates from control or activated T cells (six donors across four independent experiments, one or two time points each) were analyzed for the expression of NUP214 and UBAP2L by immunoblot. The individual results of three donors are shown (upper panels). The intensities of the NUP214 band and the two UBAP2L bands relative to loading controls (GAPDH or lymphocyte cytosolic protein 1 (LCP1)) are plotted below for all donors and time points. Dashed lines represent nonlinear regression analyses for the control and stimulated samples. The plots of the upper and lower UBAP2L bands have significantly nonzero slopes (p < 0.01, p < 0.0001) with r2 values of +0.63 and +0.89, respectively.
The increased abundance of these O-GlcNAc sites could arise from increased expression of the protein or increased levels of O-GlcNAc modification. Analysis of protein expression levels revealed that the UBAP2L O-GlcNAc site is more prevalent in activated T cells, probably because UBAP2L is upregulated after activation (Fig. 5C). Unlike the consistently increased expression of UBAP2L, the expression of NUP214 appeared variable from donor to donor, making it unclear whether the increased abundance of the O-GlcNAc site at Ser1146 in activated T cells is due to increased O-GlcNAc modification or increased NUP214 expression. O-GlcNAc was also detected at Ser685 of HCFC1, as previously reported (55, 58), although this site was equally present in activated and control T cells. Unfortunately, although nearly 2000 unique DTT-labeled peptides were detected, their insensitivity to β-N-acetylglucosaminidase precluded their assignment as O-GlcNAc modified. A low percentage of true hits from BEMAD was described in a previous study that similarly used OGA treatment as a negative control when mapping O-GlcNAc sites on proteasome subunits (59). Nonetheless, the metabolic and chemical labeling approaches together enabled us to identify numerous O-GlcNAc proteins and some O-GlcNAc sites in activated and resting human T cells.
Blotting techniques verify mass spectrometry results
To validate our mass spectrometry findings, we used chemoenzymatic labeling (60) to transfer an azide-tagged galactose residue onto O-GlcNAc groups. Following biotinylation of the azide, O-GlcNAc proteins can be enriched with streptavidin beads as before, and the enriched fraction can be probed for proteins of interest by blotting. We confirmed that several candidates are true O-GlcNAc proteins in primary T cells (Fig. 6A). We also confirmed O-GlcNAc modification of ARNT and vimentin. O-GlcNAc glycosylation of the former protein was suggested by our metabolic labeling experiment on Jurkat T cells (data not shown), whereas the latter was reported previously (61).
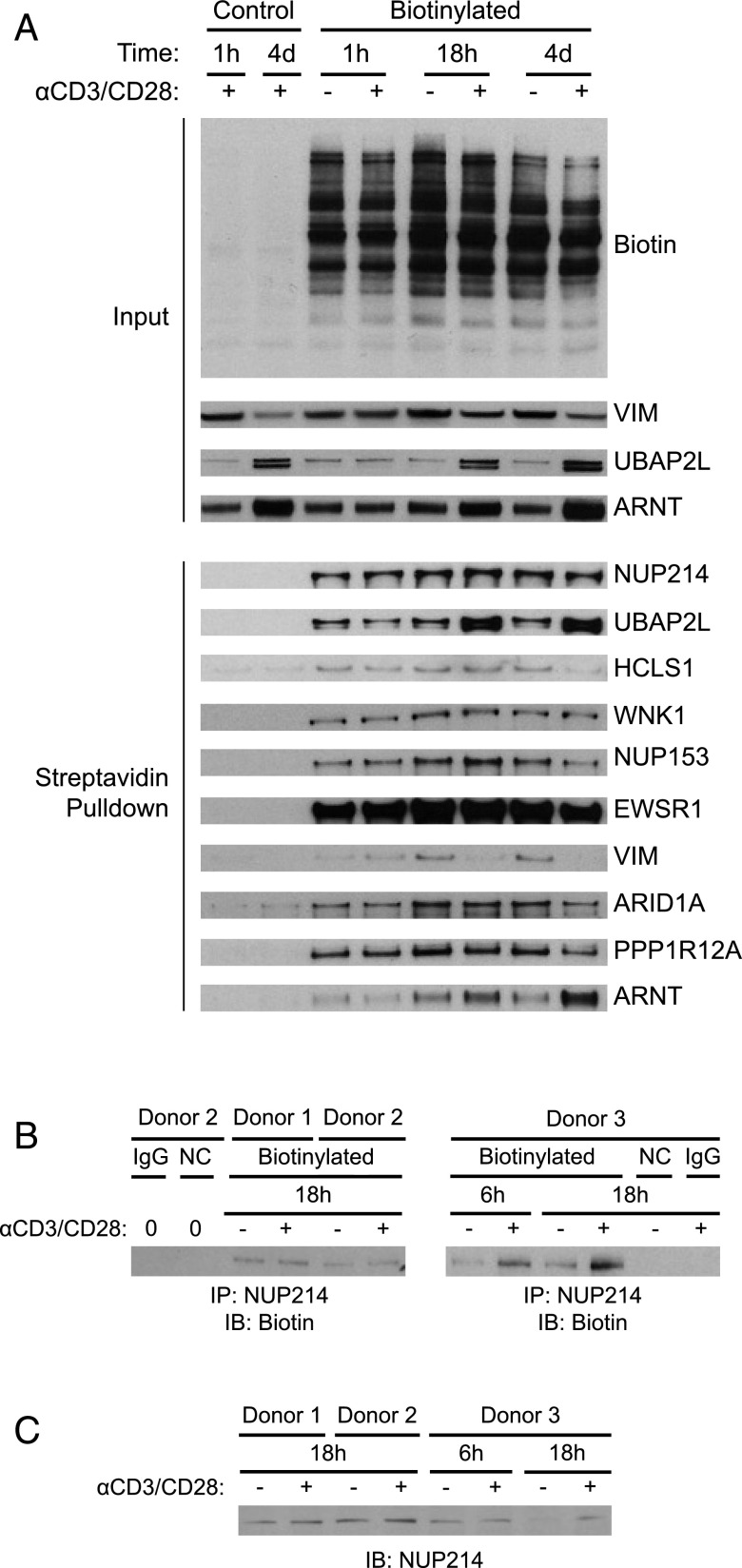
FIGURE 6.
Blotting techniques verify mass spectrometry results. (A) O-GlcNAc proteins from control or activated T cells from one donor were enzymatically labeled, biotinylated, and enriched with streptavidin beads. Samples of unlabeled protein were also prepared by omitting the azide label to control for nonspecific labeling and enrichment. The enriched fractions were analyzed for the presence of the indicated proteins by immunoblot. (B) O-GlcNAc proteins from control or activated T cells (three donors across two independent experiments) were biotinylated as in (A). NUP214 was immunoprecipitated and analyzed for biotinylation by immunoblotting. An unlabeled sample served as a negative control (NC) for biotinylation, and a labeled sample immunoprecipitated with normal rabbit IgG served as a negative control for immunoprecipitation. Zero (0) indicates cells that were not treated with any beads. (C) Cell lysates from the three donors used in (B) were analyzed for NUP214 protein levels by immunoblotting with a different NUP214 Ab.
As seen in Fig. 6A, the abundance of O-GlcNAc–glycosylated NUP214 appears equivalent in control and activated T cells, whereas the isotope-labeling data in Fig. 5B suggest increased glycosylation at Ser1146 of NUP214 in activated T cells. We attribute this discrepancy to the presence of multiple O-GlcNAc sites on NUP214, leading to equivalent enrichment by streptavidin across all conditions. To circumvent this issue, we first purified NUP214 by immunoprecipitation and then detected the biotin label by blotting with streptavidin. Now, the signal intensity is directly proportional to the number of biotinylated O-GlcNAc groups. Although we observed an obvious increase in O-GlcNAc glycosylation on NUP214 at 6 h after activation in one sample, the biotin signal was only slightly increased in other samples or was artificially increased as a result of changes in NUP214 protein levels (Fig. 6B, 6C). It is possible that this biotin-detection method is not sensitive enough to detect minor changes in O-GlcNAc in highly glycosylated proteins, which might be better analyzed by reliable mass spectrometry–based methods.
A variation of the enzymatic labeling method involves the addition of a PEG mass tag instead of a biotin tag to the azide group (62). Based on the number of attached mass tags, labeled proteins undergo discrete shifts in electrophoretic mobility, providing information regarding how many O-GlcNAc groups exist per molecule and what fraction of the total protein is modified. Using this approach, we analyzed a variety of our candidate O-GlcNAc proteins in activated and control T cells. Ten of the candidate proteins that we tested underwent gel shifts, as expected (Supplemental Fig. 2). Stoichiometries ranged from a single O-GlcNAc modification per protein to as many as five. A gel shift was not observed in the case of CDKN2AIP (data not shown), which may be due to a low stoichiometry of modification and, thus, a gel shift is not evident in the absence of enrichment techniques. Unexpectedly, many proteins appeared to have higher O-GlcNAc stoichiometries in control T cells. One potential explanation is that these proteins are phosphorylated upon activation, and phosphorylation antagonizes O-GlcNAc glycosylation. Furthermore, protein molecules modified at the same stoichiometry may appear identical by this approach when in reality they may be modified at different residues. Based on the results of complementary labeling and detection techniques presented in Fig. 6 and Supplemental Fig. 2, we conclude that the O-GlcNAc proteins that we identified by mass spectrometry represent true hits and that analysis of O-GlcNAc at the protein level may fail to capture changes at individual modification sites.
Inhibition of OGT blocks RNA synthesis
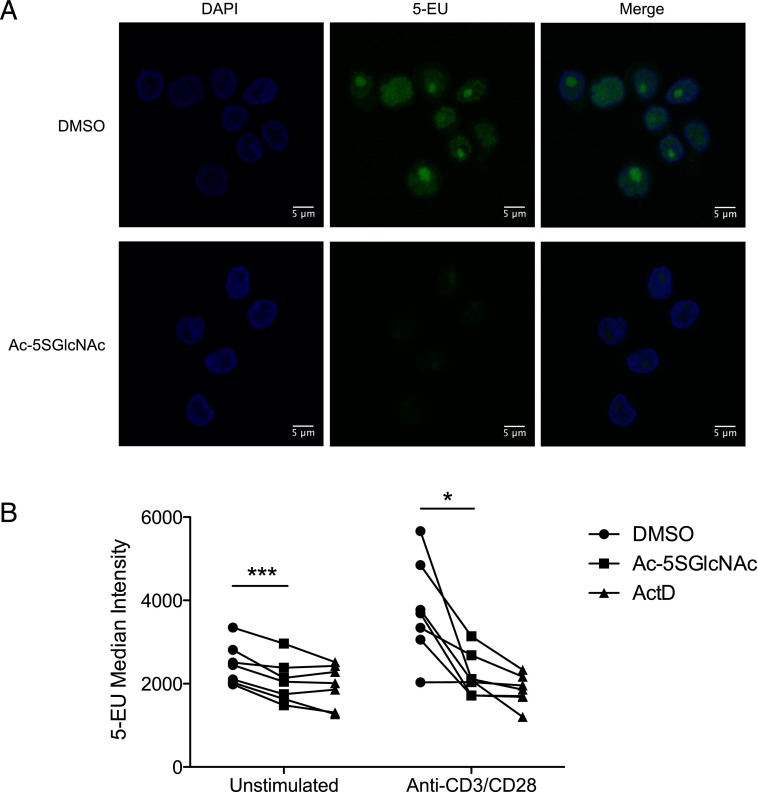
As mentioned above, many proteins from our proteomics data had functional relationships to RNA metabolism. Therefore, we investigated whether O-GlcNAc is important for RNA synthesis. As illustrated in Fig. 7A, the OGT inhibitor suppressed nascent RNA synthesis in T cells cultured on anti-CD3/CD28–coated coverslips, as measured by metabolic incorporation of a uridine analog. Although this result is consistent with the notion that O-GlcNAc directly affects the activity of RNA polymerase, transcription factors, or epigenetic regulators (63, 64), the observed decrease in labeling may also arise indirectly from a defect in T cell activation, which normally leads to increased RNA synthesis (65). Thus, we repeated this experiment using flow cytometry to enable the analysis of resting T cells. Activated T cells showed dramatically increased levels of RNA synthesis, which was again suppressed by the OGT inhibitor (Fig. 7B). Resting T cells treated with the OGT inhibitor showed a smaller, but statistically significant, decrease in RNA synthesis as well. Therefore, we conclude that the lack of RNA synthesis observed upon OGT inhibition is likely a combination of defective T cell activation and diminished transcription.
FIGURE 7.
Inhibition of OGT blocks RNA synthesis. (A) Human T cells were cultured on glass coverslips coated with anti-CD3/CD28 for 24 h in the presence of 50 μM Ac-5SGlcNAc. 5-EU was added during the final 7 h of culture, and nascent RNA synthesis was subsequently visualized with Alexa Fluor 488 azide. Similar results were obtained with a second donor in an independent experiment using a 24-h 5-EU pulse. (B) Human T cells were treated with 50 μM Ac-5SGlcNAc overnight (24–28 h) and then stimulated with anti-CD3/CD28 beads or control beads overnight (18–22.5 h) in the presence of 5-EU. 5-EU was then labeled with Alexa Fluor 488, and cells were analyzed by flow cytometry. The median intensity of 5-EU staining is plotted for seven donors across three experiments with lines connecting data points from each donor. Cells treated with 2 μg/ml of the RNA polymerase inhibitor actinomycin D (ActD) during the 5-EU pulse served as negative controls and indicate the level of background staining *p < 0.05, ***p < 0.001, two-tailed, paired t test.
Discussion
The regulation of intracellular signaling pathways downstream of cell surface receptors is governed largely by the posttranslational modification of proteins. Ultimately, these signal-transduction cascades dictate cellular behavior, a prime example of which is the activation of T cells during an adaptive immune response to a pathogen. Compared with other types of posttranslational modifications, the glycosylation of intracellular proteins by O-GlcNAc in T cells remains largely unexplored, despite several studies indicating that it possesses functional significance (23, 25, 37–39).
In these studies, we sought to further delineate the role of O-GlcNAc in T cell activation. As opposed to previous studies, which investigated O-GlcNAc on specific targets, such as NF-κB and NFAT (38, 39), we elected to broadly profile O-GlcNAc targets using multiple labeling methods in conjunction with mass spectrometry. Additionally, we conducted nearly all of our experiments with primary human T cells to complement prior work that relied mostly on immortalized cell lines and, thus, may not constitute an accurate representation of normal cell physiology. We found that the activation of human T cells through the TCR induces a gradual increase in overall O-GlcNAc levels over the course of several hours, similar to previous work characterizing a T cell hybridoma activated with PMA and ionomycin (37). However, in that study, cytosolic O-GlcNAc levels underwent a transient decrease with a corresponding increase in nuclear O-GlcNAc levels. In contrast, we observed that primary human T cells stimulated with anti-CD3/CD28 beads undergo a sustained increase in O-GlcNAc levels in both cellular compartments (Fig. 1). Notably, stimulation of the T cell hybridoma eventually compromised cell viability, which complicates the interpretation of those findings and may explain, in part, our contrasting results. Our observation of higher O-GlcNAc levels in activated T cells, which undergo rapid division, suggests that O-GlcNAc levels are positively correlated with cell proliferation. Indeed, elevated O-GlcNAc levels also were reported in cancer (31, 66, 67). Consistent with previous work using genetic approaches (25, 38), we show in this study with a specific inhibitor that OGT is important for T cell function, because blocking its activity suppressed IL-2 production and cell proliferation. Notably, evidence suggests that the transcription factor NFAT, a major driver of IL-2 expression, is regulated by O-GlcNAc (38, 68). However, we failed to detect any functional differences in T cells treated with an OGA inhibitor, unlike another study that reported enhanced expression of the activation marker CD69 (38). The reason for this discrepancy is unclear, but because the earlier study was conducted with PBMCs as opposed to purified T cells, the enhanced expression of CD69 in T cells could represent an indirect effect of inhibiting OGA in other cell types.
OGT is the one enzyme that transfers O-GlcNAc to proteins. Because O-GlcNAc sites lack a well-defined consensus sequence (69), many questions remain regarding the regulation of OGT. One potential regulatory mechanism is the control of its localization. OGT features a patch of basic residues near the C terminus that mediate binding to PIP3 (48), although this was disputed (70). Given that PIP3 is generated at the immunological synapse (71, 72), we hypothesized that OGT would undergo recruitment to the synapse, which we confirmed by immunofluorescence microscopy. At the synapse, which is rife with signaling molecules like ZAP-70, OGT may have access to a distinct set of substrates. Another mechanism of regulation relies on changes in enzyme expression. Although the expression of full-length ncOGT was not significantly altered in activated T cells, we noted a striking difference in the expression levels of sOGT and possibly mOGT. Because OGT is thought to function as a heterotrimer composed of two ncOGT subunits and one sOGT subunit (73), changes in the relative expression of these subunits may influence substrate specificity or enzymatic activity. Studies with recombinant OGT showed that certain substrates are only modified by specific isoforms (74). Additionally, owing to its mitochondrial localization, mOGT presumably interacts with a different set of substrates than its nucleocytoplasmic counterparts. We also tested whether glucose metabolism contributes to the regulation of OGT. Alterations in cell metabolism may indirectly influence OGT activity by affecting the availability of UDP-GlcNAc, which serves as the nucleotide sugar donor for OGT (26–28). After activation, T cells exhibit a dramatic increase in the consumption of glucose and glutamine (75). Both glucose and glutamine provide important precursors for the hexosamine biosynthesis pathway, which is responsible for the generation of UDP-GlcNAc. Although the bands of some O-GlcNAc proteins appeared sensitive to treatment with 2-DG, we were surprised to see that most were unaffected, which suggests that the increase in O-GlcNAc levels after T cell activation is not simply due to increased glucose metabolism promoting OGT activity. OGT activity may also depend on its interaction partners (20, 22), especially because the N terminus of OGT contains multiple tetratricopeptide repeats, which usually mediate protein–protein interactions.
To discover proteins modified by O-GlcNAc across the entire proteome, we used various techniques based on mass spectrometry, which resulted in the identification of 214 probable or potential O-GlcNAc proteins. We believe that this constitutes the first broad proteomic analysis of O-GlcNAc–modified proteins in primary human T cells. Many of the proteins that we identified appeared in prior proteomics studies (55, 76–78), supporting the validity of our results. However, we also identified novel O-GlcNAc substrates, including ARNT, HCLS1, ZAP-70, SHIP1, LCK, and PI3K. Furthermore, this global analysis suggested a functional role for O-GlcNAc in RNA metabolism. Subsequent experiments revealed that inhibiting OGT activity suppressed RNA synthesis (Fig. 7), which may contribute to the decrease in proliferation and IL-2 production (Fig. 2). Decreased proliferation may also stem from inhibiting the protease activity of OGT, which is responsible for cleaving HCFC1, a regulator of the cell cycle (79, 80). Through the use of stable isotope labeling, we detected a greater abundance of O-GlcNAc–modified peptides from UBAP2L and NUP214 after T cell activation. Expression of UBAP2L was greatly increased upon activation, which likely accounts for its increase in O-GlcNAc glycosylation. We found that nearly all UBAP2L is modified by O-GlcNAc. Recent work indicated a potential role for UBAP2L in the epigenetic regulation of cell proliferation (81, 82), and another study reported that UBAP2L is phosphorylated upon T cell activation (83), warranting future investigation of UBAP2L and its regulation by posttranslational modifications in T cells.
In contrast to the consistent upregulation of UBAP2L, the expression of NUP214 was more variable after T cell activation. Although NUP214 likely contains multiple O-GlcNAc sites, modification at Ser1146 was more prevalent in activated T cells, although this may vary between donors as a result of variations in NUP214 expression levels (Figs. 5B, 6B, 6C). Nuclear pore proteins were among the first proteins to be identified as O-GlcNAc modified (84); given their high degree of modification (Supplemental Fig. 2), the O-GlcNAc modification likely holds functional importance. Indeed, recent studies showed that O-GlcNAc glycosylation of nuclear pore proteins prevents their proteasomal degradation (85) and facilitates the transport of protein cargo (86). Interestingly, phosphorylation of nuclear pore proteins, including NUP214, also increases after T cell activation (83), which raises the possibility of multiple posttranslational modifications working synergistically to regulate nuclear pore function in activated T cells.
After submission of this manuscript, we note that a newly published study of murine T cells reinforces many of our findings and shows their generality across these species. Specifically Swamy et al. (87) reported that the T cells of inbred mice also undergo an increase in total protein O-GlcNAc glycosylation in the hours after activation and require OGT for efficient clonal expansion. The investigators also demonstrated that activated T cells consume more glucose and glutamine, leading to higher levels of UDP-GlcNAc and, therefore, higher levels of O-GlcNAc glycosylation. However, the effects of glucose and glutamine on protein O-GlcNAc levels, although apparent, were less dramatic compared with their effects on UDP-GlcNAc levels, which is in line with our results showing that inhibition of glycolysis only affected the glycosylation status of a small subset of proteins (Fig. 3C). Thus, the regulation of protein O-GlcNAc levels likely consists of multiple mechanisms that could be substrate specific and may also involve changes in OGT localization or isoform expression (Fig. 3A, 3B). The transcription factor c-myc is clearly essential for increased O-GlcNAc levels in activated T cells (85), probably due, in part, to its role in driving expression of nutrient transporters (75). At the same time, OGT activity is critical for stabilizing c-myc expression (35, 87). Our results showing reduced RNA synthesis in human T cells treated with the OGT inhibitor (Fig. 7) are fully consistent with decreased c-myc expression, because c-myc activates transcription of rRNA (88, 89), and a large fraction of the labeled RNA likely represents rRNA based on its nucleolar staining pattern. Although c-myc is undoubtedly one important OGT substrate, we identified hundreds of other potentially relevant substrates (Supplemental Table I).
In summary, we believe that a full understanding of the signaling pathways that underlie T cell activation and other cellular processes requires knowledge of the different types and sites of posttranslational modifications because they have the potential to dramatically modulate the properties of proteins. In this study, we focused on the O-GlcNAc modification and identified numerous molecules that are modified in activated human T cells. We also found clear effects on proliferation and IL-2 production, indicating the functional importance of this posttranslational modification for T cell biology.
Supplementary Material
Acknowledgments
We thank members of the Elias Lab, especially Dominique Figueroa, Josh Lichtman, Niclas Olsson, and Kavya Swaminathan, for discussions and assistance with mass spectrometry; Karin Kealoha of the Stanford Blood Center for help in obtaining blood samples; Andrew Olson of the Stanford Neuroscience Microscopy Service for assistance with confocal imaging; Shripa Patel and Dick Winant of the Beckman Protein and Nucleic Acid Facility for performing MALDI-TOF analysis; the Stanford Shared FACS Facility for assistance with flow cytometry; Nelida Prado for animal care; Christina Woo, Ian Blong, Lissette Andres, and other members of the Bertozzi Lab for discussions and the Ac-5SGlcNAc inhibitor; and Justin Sonnenburg, Yueh-hsiu Chien, Naresha Saligrama, Jun Huang, Feng Wang, and other members of the Davis Lab for helpful discussions.
This work was supported by National Institutes of Health Grants AI090019 and AI057229, the Howard Hughes Medical Institute, a George D. Smith Stanford Graduate Fellowship, and a National Science Foundation Graduate Research Fellowship. The Stanford Neuroscience Microscopy Service is supported by National Institutes of Health Grant NS069375.
The online version of this article contains supplemental material.
- Ac4GalNAz
- tetraacetylated N-azidoacetylgalactosamine
- Ac-5SGlcNAc
- 2-acetamido-1,3,4,6-tetra-O-acetyl-2-deoxy-5-thio-α-d-glucopyranose
- BEMAD
- β-elimination followed by Michael addition of DTT
- CIP
- calf intestinal alkaline phosphatase
- 2-DG
- 2-deoxyglucose
- OGA
- O-GlcNAcase
- O-GlcNAc
- O-linked N-acetylglucosamine
- OGT
- O-GlcNAc transferase
- PEG
- polyethylene glycol
- PIP3
- phosphatidylinositol-3,4,5,-trisphosphate.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Acuto O., Di Bartolo V., Michel F. 2008. Tailoring T-cell receptor signals by proximal negative feedback mechanisms. Nat. Rev. Immunol. 8: 699–712. [DOI] [PubMed] [Google Scholar]
- 2.Smith-Garvin J. E., Koretzky G. A., Jordan M. S. 2009. T cell activation. Annu. Rev. Immunol. 27: 591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabouridis P. S., Magee A. I., Ley S. C. 1997. S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes. EMBO J. 16: 4983–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avni O., Lee D., Macian F., Szabo S. J., Glimcher L. H., Rao A. 2002. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 3: 643–651. [DOI] [PubMed] [Google Scholar]
- 5.Anandasabapathy N., Ford G. S., Bloom D., Holness C., Paragas V., Seroogy C., Skrenta H., Hollenhorst M., Fathman C. G., Soares L. 2003. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity 18: 535–547. [DOI] [PubMed] [Google Scholar]
- 6.Duan L., Reddi A. L., Ghosh A., Dimri M., Band H. 2004. The Cbl family and other ubiquitin ligases: destructive forces in control of antigen receptor signaling. Immunity 21: 7–17. [DOI] [PubMed] [Google Scholar]
- 7.Mueller D. L. 2004. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 5: 883–890. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y. C., Penninger J., Karin M. 2005. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5: 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fann M., Godlove J. M., Catalfamo M., Wood W. H., III, Chrest F. J., Chun N., Granger L., Wersto R., Madara K., Becker K., et al. 2006. Histone acetylation is associated with differential gene expression in the rapid and robust memory CD8(+) T-cell response. Blood 108: 3363–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J., Paul W. E. 2008. CD4 T cells: fates, functions, and faults. Blood 112: 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araki Y., Wang Z., Zang C., Wood W. H., III, Schones D., Cui K., Roh T. Y., Lhotsky B., Wersto R. P., Peng W., et al. 2009. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30: 912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T. Y., Watford W. T., et al. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30: 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dispirito J. R., Shen H. 2010. Histone acetylation at the single-cell level: a marker of memory CD8+ T cell differentiation and functionality. J. Immunol. 184: 4631–4636. [DOI] [PubMed] [Google Scholar]
- 14.Müller M. R., Rao A. 2010. NFAT, immunity and cancer: a transcription factor comes of age. Nat. Rev. Immunol. 10: 645–656. [DOI] [PubMed] [Google Scholar]
- 15.Nurieva R. I., Zheng S., Jin W., Chung Y., Zhang Y., Martinez G. J., Reynolds J. M., Wang S. L., Lin X., Sun S. C., et al. 2010. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity 32: 670–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiting C. C., Su L. L., Lin J. T., Fathman C. G. 2011. GRAIL: a unique mediator of CD4 T-lymphocyte unresponsiveness. FEBS J. 278: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Shea J. J., Lahesmaa R., Vahedi G., Laurence A., Kanno Y. 2011. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat. Rev. Immunol. 11: 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yount J. S., Zhang M. M., Hang H. C. 2013. Emerging roles for protein S-palmitoylation in immunity from chemical proteomics. Curr. Opin. Chem. Biol. 17: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray S. M., Kaech S. M., Staron M. M. 2014. The interface between transcriptional and epigenetic control of effector and memory CD8+ T-cell differentiation. Immunol. Rev. 261: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hart G. W., Housley M. P., Slawson C. 2007. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446: 1017–1022. [DOI] [PubMed] [Google Scholar]
- 21.Zeidan Q., Hart G. W. 2010. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J. Cell Sci. 123: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. 2011. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80: 825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shafi R., Iyer S. P., Ellies L. G., O’Donnell N., Marek K. W., Chui D., Hart G. W., Marth J. D. 2000. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97: 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y. R., Song M., Lee H., Jeon Y., Choi E. J., Jang H. J., Moon H. Y., Byun H. Y., Kim E. K., Kim D. H., et al. 2012. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11: 439–448. [DOI] [PubMed] [Google Scholar]
- 25.O’Donnell N., Zachara N. E., Hart G. W., Marth J. D. 2004. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24: 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W. H., Park S. Y., Nam H. W., Kim D. H., Kang J. G., Kang E. S., Kim Y. S., Lee H. C., Kim K. S., Cho J. W. 2008. NFkappaB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. USA 105: 17345–17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanover J. A., Krause M. W., Love D. C. 2010. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 1800: 80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanover J. A., Krause M. W., Love D. C. 2012. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13: 312–321. [DOI] [PubMed] [Google Scholar]
- 29.Springhorn C., Matsha T. E., Erasmus R. T., Essop M. F. 2012. Exploring leukocyte O-GlcNAcylation as a novel diagnostic tool for the earlier detection of type 2 diabetes mellitus. J. Clin. Endocrinol. Metab. 97: 4640–4649. [DOI] [PubMed] [Google Scholar]
- 30.Liu F., Shi J., Tanimukai H., Gu J., Gu J., Grundke-Iqbal I., Iqbal K., Gong C. X. 2009. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer’s disease. Brain 132: 1820–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y., Tomic J., Wen F., Shaha S., Bahlo A., Harrison R., Dennis J. W., Williams R., Gross B. J., Walker S., et al. 2010. Aberrant O-GlcNAcylation characterizes chronic lymphocytic leukemia. Leukemia 24: 1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whelan S. A., Dias W. B., Thiruneelakantapillai L., Lane M. D., Hart G. W. 2010. Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes. J. Biol. Chem. 285: 5204–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi W., Clark P. M., Mason D. E., Keenan M. C., Hill C., Goddard W. A., III, Peters E. C., Driggers E. M., Hsieh-Wilson L. C. 2012. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang X., Pan Q., Sun D., Chen W., Shen A., Huang M., Ding J., Geng M. 2013. O-GlcNAcylation of cofilin promotes breast cancer cell invasion. J. Biol. Chem. 288: 36418–36425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itkonen H. M., Minner S., Guldvik I. J., Sandmann M. J., Tsourlakis M. C., Berge V., Svindland A., Schlomm T., Mills I. G. 2013. O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res. 73: 5277–5287. [DOI] [PubMed] [Google Scholar]
- 36.Jin F. Z., Yu C., Zhao D. Z., Wu M. J., Yang Z. 2013. A correlation between altered O-GlcNAcylation, migration and with changes in E-cadherin levels in ovarian cancer cells. Exp. Cell Res. 319: 1482–1490. [DOI] [PubMed] [Google Scholar]
- 37.Kearse K. P., Hart G. W. 1991. Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc. Natl. Acad. Sci. USA 88: 1701–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golks A., Tran T. T., Goetschy J. F., Guerini D. 2007. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 26: 4368–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramakrishnan P., Clark P. M., Mason D. E., Peters E. C., Hsieh-Wilson L. C., Baltimore D. 2013. Activation of the transcriptional function of the NF-κB protein c-Rel by O-GlcNAc glycosylation. Sci. Signal. 6: ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pesavento J. J., Garcia B. A., Streeky J. A., Kelleher N. L., Mizzen C. A. 2007. Mild performic acid oxidation enhances chromatographic and top down mass spectrometric analyses of histones. Mol. Cell. Proteomics 6: 1510–1526. [DOI] [PubMed] [Google Scholar]
- 41.Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. 2002. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1: 791–804. [DOI] [PubMed] [Google Scholar]
- 42.Vosseller K., Hansen K. C., Chalkley R. J., Trinidad J. C., Wells L., Hart G. W., Burlingame A. L. 2005. Quantitative analysis of both protein expression and serine/threonine post-translational modifications through stable isotope labeling with dithiothreitol. Proteomics 5: 388–398. [DOI] [PubMed] [Google Scholar]
- 43.Rybak J., Scheurer S. B., Neri D., Elia G. 2004. Purification of biotinylated proteins on streptavidin resin: A protocol for quantitative elution. Proteomics 4: 2296–2299. [DOI] [PubMed] [Google Scholar]
- 44.Eng J. K., McCormack A. L., Yates J. R. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5: 976–989. [DOI] [PubMed] [Google Scholar]
- 45.Elias J. E., Gygi S. P. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4: 207–214. [DOI] [PubMed] [Google Scholar]
- 46.Gloster T. M., Zandberg W. F., Heinonen J. E., Shen D. L., Deng L., Vocadlo D. J. 2011. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol. 7: 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., Vocadlo D. J. 2008. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4: 483–490. [DOI] [PubMed] [Google Scholar]
- 48.Yang X., Ongusaha P. P., Miles P. D., Havstad J. C., Zhang F., So W. V., Kudlow J. E., Michell R. H., Olefsky J. M., Field S. J., Evans R. M. 2008. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451: 964–969. [DOI] [PubMed] [Google Scholar]
- 49.Butkinaree C., Park K., Hart G. W. 2010. O-linked beta-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800: 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox C. J., Hammerman P. S., Thompson C. B. 2005. Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 5: 844–852. [DOI] [PubMed] [Google Scholar]
- 51.Jacobs S. R., Herman C. E., Maciver N. J., Wofford J. A., Wieman H. L., Hammen J. J., Rathmell J. C. 2008. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 180: 4476–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maciver N. J., Jacobs S. R., Wieman H. L., Wofford J. A., Coloff J. L., Rathmell J. C. 2008. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J. Leukoc. Biol. 84: 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyce M., Carrico I. S., Ganguli A. S., Yu S. H., Hangauer M. J., Hubbard S. C., Kohler J. J., Bertozzi C. R. 2011. Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. USA 108: 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saxon E., Bertozzi C. R. 2000. Cell surface engineering by a modified Staudinger reaction. Science 287: 2007–2010. [DOI] [PubMed] [Google Scholar]
- 55.Hahne H., Sobotzki N., Nyberg T., Helm D., Borodkin V. S., van Aalten D. M., Agnew B., Kuster B. 2013. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J. Proteome Res. 12: 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang D. W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 57.Huang D. W., Sherman B. T., Lempicki R. A. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Myers S. A., Daou S., Affar B., Burlingame A. 2013. Electron transfer dissociation (ETD): the mass spectrometric breakthrough essential for O-GlcNAc protein site assignments-a study of the O-GlcNAcylated protein host cell factor C1. Proteomics 13: 982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Overath T., Kuckelkorn U., Henklein P., Strehl B., Bonar D., Kloss A., Siele D., Kloetzel P. M., Janek K. 2012. Mapping of O-GlcNAc sites of 20 S proteasome subunits and Hsp90 by a novel biotin-cystamine tag. Mol. Cell. Proteomics 11: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khidekel N., Arndt S., Lamarre-Vincent N., Lippert A., Poulin-Kerstien K. G., Ramakrishnan B., Qasba P. K., Hsieh-Wilson L. C. 2003. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J. Am. Chem. Soc. 125: 16162–16163. [DOI] [PubMed] [Google Scholar]
- 61.Slawson C., Lakshmanan T., Knapp S., Hart G. W. 2008. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol. Biol. Cell 19: 4130–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rexach J. E., Rogers C. J., Yu S. H., Tao J., Sun Y. E., Hsieh-Wilson L. C. 2010. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 6: 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ranuncolo S. M., Ghosh S., Hanover J. A., Hart G. W., Lewis B. A. 2012. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem. 287: 23549–23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hardivillé S., Hart G. W. 2014. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20: 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asmal M., Colgan J., Naef F., Yu B., Lee Y., Magnasco M., Luban J. 2003. Production of ribosome components in effector CD4+ T cells is accelerated by TCR stimulation and coordinated by ERK-MAPK. Immunity 19: 535–548. [DOI] [PubMed] [Google Scholar]
- 66.Caldwell S. A., Jackson S. R., Shahriari K. S., Lynch T. P., Sethi G., Walker S., Vosseller K., Reginato M. J. 2010. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene 29: 2831–2842. [DOI] [PubMed] [Google Scholar]
- 67.Lynch T. P., Ferrer C. M., Jackson S. R., Shahriari K. S., Vosseller K., Reginato M. J. 2012. Critical role of O-linked β-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J. Biol. Chem. 287: 11070–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Facundo H. T., Brainard R. E., Watson L. J., Ngoh G. A., Hamid T., Prabhu S. D., Jones S. P. 2012. O-GlcNAc signaling is essential for NFAT-mediated transcriptional reprogramming during cardiomyocyte hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 302: H2122–H2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pathak S., Alonso J., Schimpl M., Rafie K., Blair D. E., Borodkin V. S., Schüttelkopf A. W., Albarbarawi O., van Aalten D. M. 2015. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 22: 744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lazarus M. B., Nam Y., Jiang J., Sliz P., Walker S. 2011. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469: 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costello P. S., Gallagher M., Cantrell D. A. 2002. Sustained and dynamic inositol lipid metabolism inside and outside the immunological synapse. Nat. Immunol. 3: 1082–1089. [DOI] [PubMed] [Google Scholar]
- 72.Harriague J., Bismuth G. 2002. Imaging antigen-induced PI3K activation in T cells. Nat. Immunol. 3: 1090–1096. [DOI] [PubMed] [Google Scholar]
- 73.Kreppel L. K., Hart G. W. 1999. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 274: 32015–32022. [DOI] [PubMed] [Google Scholar]
- 74.Lazarus B. D., Love D. C., Hanover J. A. 2006. Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 16: 415–421. [DOI] [PubMed] [Google Scholar]
- 75.Wang R., Dillon C. P., Shi L. Z., Milasta S., Carter R., Finkelstein D., McCormick L. L., Fitzgerald P., Chi H., Munger J., Green D. R. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35: 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G., II, Shabanowitz J., Stanley P., et al. 2012. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. USA 109: 7280–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., Burlingame A. L. 2012. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11: 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woo C. M., Iavarone A. T., Spiciarich D. R., Palaniappan K. K., Bertozzi C. R. 2015. Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 12: 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Capotosti F., Guernier S., Lammers F., Waridel P., Cai Y., Jin J., Conaway J. W., Conaway R. C., Herr W. 2011. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144: 376–388. [DOI] [PubMed] [Google Scholar]
- 80.Lazarus M. B., Jiang J., Kapuria V., Bhuiyan T., Janetzko J., Zandberg W. F., Vocadlo D. J., Herr W., Walker S. 2013. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 342: 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bordeleau M. E., Aucagne R., Chagraoui J., Girard S., Mayotte N., Bonneil E., Thibault P., Pabst C., Bergeron A., Barabé F., et al. 2014. UBAP2L is a novel BMI1-interacting protein essential for hematopoietic stem cell activity. Blood 124: 2362–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li D., Huang Y. 2014. Knockdown of ubiquitin associated protein 2-like inhibits the growth and migration of prostate cancer cells. Oncol. Rep. 32: 1578–1584. [DOI] [PubMed] [Google Scholar]
- 83.Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. 2009. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2: ra46. [DOI] [PubMed] [Google Scholar]
- 84.Holt G. D., Snow C. M., Senior A., Haltiwanger R. S., Gerace L., Hart G. W. 1987. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J. Cell Biol. 104: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu Y., Liu T. W., Madden Z., Yuzwa S. A., Murray K., Cecioni S., Zachara N., Vocadlo D. J. 2016. Post-translational O-GlcNAcylation is essential for nuclear pore integrity and maintenance of the pore selectivity filter. J. Mol. Cell Biol. 8: 2–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Labokha A. A., Gradmann S., Frey S., Hülsmann B. B., Urlaub H., Baldus M., Görlich D. 2013. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 32: 204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swamy M., Pathak S., Grzes K. M., Damerow S., Sinclair L. V., van Aalten D. M., Cantrell D. A. 2016. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 17: 712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arabi A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., et al. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7: 303–310. [DOI] [PubMed] [Google Scholar]
- 89.Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., White R. J. 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7: 311–318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.