Abstract
Avicennia L. (Avicenniaceae), one of the most diverse mangrove genera, is distributed widely in tropical and subtropical intertidal zones worldwide. Five species of Avicennia in the Indo-West Pacific region have been previously described. However, their phylogenetic relationships were determined based on morphological and allozyme data. To enhance our understanding of evolutionary patterns in the clade, we carried out a molecular phylogenetic study using wide sampling and multiple loci. Our results support two monophyletic clades across all species worldwide in Avicennia: an Atlantic-East Pacific (AEP) lineage and an Indo-West Pacific (IWP) lineage. This split is in line with biogeographic distribution of the clade. Focusing on the IWP branch, we reconstructed a detailed phylogenetic tree based on sequences from 25 nuclear genes. The results identified three distinct subclades, (1) A. rumphiana and A. alba, (2) A. officinalis and A. integra, and (3) the A. marina complex, with high bootstrap support. The results strongly corresponded to two morphological traits in floral structure: stigma position in relation to the anthers and style length. Using Bayesian dating methods we estimated diversification of the IWP lineage was dated to late Miocene (c. 6.0 million years ago) and may have been driven largely by the fluctuating sea levels since that time.
Introduction
Avicennia L., an important component of forests worldwide in tropical and subtropical intertidal zones [1, 2], is currently thought to be derived from within the Acanthaceae based on recent phylogenetic studies (APG III, [3, 4]), although it has been placed in Verbenaceae or Avicenniaceae in some classifications. Avicennia has developed several unique features, such as salt glands, aerial and prop roots and finger-like pneumatophores, and cryptoviviparous fruits with the ability to float in salt water [2]. Members of the genus often occupy newly-formed seaward mangrove habitats on tropical coasts.
Avicennia L. is one of the most diverse mangrove genera, comprising eight species: A. germinans (L.) Stearn, A. schaueriana Stapf & Leechm. ex Moldenke, A. bicolor Standley, A. marina (Forssk.) Vierh, A. alba Blume, A. officinalis L., A. integra N. C. Duke, and A. rumphiana Hallier f [1]. Among them, the first three are endemic to the Atlantic-East Pacific (AEP) region; the remaining five are in the Indo-West Pacific region (IWP) [1]. One of the species in the IWP, A. marina, has the broadest distribution of all mangroves, ranging from approximately 25° N to 38° S. Three infraspecific taxa within A. marina have been recognized: A. marina var. marina, var. eucalyptifolia, and var. australasica [1, 5]. A recent systematic revision of Avicennia [1] based on morphological studies [6] divided the five IWP species into three groups: (i) A. marina; (ii) A. officinalis and A. integra; and (iii) A. rumphiana and A. alba [6]. However, detailed evolutionary relationships among the species of Avicennia, such as chronological speciation order and divergence time, have not been clearly described so far. Evidence from molecular markers [7–9] shed some light on the patterns of population genetics and phylogeny of Avicennia species in the AEP region. However, so far studies of the species from IWP have been relying on morphological and allozyme data [5].
Sequencing multiple loci has been a successful strategy used to resolve phylogenetic relationships among numerous species [10, 11]. Thus, we set out to re-evaluate the phylogenetic relationships among species in Avicennia based on a robust approach involving wide sampling and sequencing of a number of DNA segments. Specifically, we address the following questions: (1) Do molecular data support a phylogenetic grouping based on geographical distributions of AEP and IWP lineages? (2) What are the phylogenetic relationships of the five IWP Avicennia species and what do they tell us about evolution of flower characters? (3) When did the IWP Avicennia species diversify and what was the possible driving force behind this divergence? Our results shed fresh light on the evolution of this important mangrove clade and should enable further studies of this group’s remarkable adaptation to the intertidal environment.
Materials and Methods
Ethics statement
Avicennia species used in the manuscript are not included in the China, Thailand, Indonesia, Malaysia, Bangladesh, Kenya, Australia or New Zealand official lists of endangered plants. No specific permission was required for sampling these species in this study. The fieldwork was outside protected areas or natural reserves, and no specific permission was required for these collection activities. The field studies did not affect endangered or protected species.
Taxon sampling
We obtained samples from eight accessions of Avicennia, including A. germinans, A. officinalis, A. integra, A. alba, A. rumphiana, and three varieties of A. marina (A. marina var. marina, A. marina var. eucalyptifolia, and A. marina var. australasica). The sample of A. germinans was collected from La Paz, Mexico. The detailed sampling information is listed in Table 1. To infer the phylogeny of Avicennia from all available data, we retrieved four sequences from two taxa, A. bicolor and A. schaueriana from GenBank (Table 1)[7]. We included Thunbergia grandiflora as the outgroup, since a close relationship between Thunbergioideae and Avicennia has been suggested based on recent molecular and morphological data [3, 12, 13]. Leaf tissue for DNA extraction was stored and dried in plastic bags with silica gel. Voucher specimens were deposited in the Herbarium of Sun Yat-Sen University (SYS).
Table 1. Distribution of Avicennia and voucher specimens.
| Taxon | Locality | Voucher/Source |
|---|---|---|
| A. alba Blume | Bangkhunsai, Phetchaburi, Thailand | S.Shi 200908-AabBK01 |
| A. bicolor Standl. | Playa Panama, Costa Rica | Nettle and Dodd, 2008 |
| A. germinans (L.) Stearn | La Paz, Mexico | B. Liao 200901-AgeLP01 |
| A. integra N. C. Duke | South Alligator River, North Territory, Australia | N.C.Duke 201412-AinDW01 |
| N.C.Duke 201412-AinDW02 | ||
| A. marina (Forssk.) Vierh. | ||
| var. australasica | Northcote, Auckland, New Zealand | M.Zhang 201106-AmaNZ01 |
| var. eucalyptifolia | Cairns, Queensland, Australia | M.Zhang 201108-AmaCA01 |
| var. marina | Wenchang, Hainan, China | S.Shi 200908-AmaWC01 |
| Kinabalu, Sabah, Malaysia | S.Shi 201108-AmaSB03 | |
| Mida Creek, Kenya | Y.Deng 201104-AmaKY06 | |
| A. rumphiana Hallier f. | Kukup Johor, Malaysia | S.Shi 201108-AruKK01 |
| A. schaueriana Stapf & Leechm. ex Moldenke | Macao, Brazil | Nettle and Dodd, 2008 |
| A. officinalis L. | Laun, Ranong, Thailand | S.Shi 200908-AofLU01 |
| Sunderbans, Bangladesh | S.Shi 201102-AofSUN02 | |
| Cilacap, Central Java, Indonesia | S.Shi 201210-AofCL01 | |
| Thunbergia grandiflora (Rottl. ex Willd.) Roxb. | Sun Yat-Sen University, Guangzhou, China | S.Shi 201306-GZ01 |
Voucher specimens are in the Herbarium of Sun Yat-Sen University (SYSU).
DNA extraction, amplification and sequencing
The total genomic DNA of each individual was extracted by the modified CTAB method [14]. We used the trnT-trnD intergenic spacer region and the psbA gene for the phylogenetic reconstruction of all species of Avicennia because they are the only available sequence data from the two AEP species. Avicennia bicolor and A. schaueriana were not sampled in this study. For the detailed phylogenetic analyses of the IWP species, 25 pairs of primers were designed to amplify targeted nuclear genes from genomic DNA, based on the EST library of A. marina obtained from GenBank. The forward and reverse primers of each pair were located in different exons of the same gene to amplify a segment with introns. For most genes, only one band was amplified from all species. Two primer pairs produced PCR products with two bands, widely spaced in size. We re-designed allele-specific primers for these genes based on the sequences of isolated single bands that corresponded to the brightest-staining product that corresponded to the size of products from other taxa. The gene IDs, sequence lengths, gene descriptions, and primer sequences for the DNA markers are listed in S1 Table.
PCR was conducted with the following conditions: 94°C for 4 min, followed by 30 cycles of 94°C for 30 seconds, 48–58°C for 30 seconds, 72°C for 2 minutes, and a final extension of 8 minutes at 72°C. PCR products were purified using an agarose gel purification kit (QIAGEN, Hilden, Germany). Purified PCR products were subjected to direct sequencing. Sequencing reactions were conducted with amplification primers in an ABI 3730 DNA automated sequencer with BigDye chemistry (Applied Biosystems, Foster City, CA, USA). All sequences were deposited in GenBank. The accession numbers are listed in S2 Table.
Phylogenetic analyses
Raw sequences were aligned and manually edited using SeqMan 7.1 (Madison, WI, USA). Multiple-sequence alignments were performed using ClustalW 2.1 [15] by leaving the default parameters unchanged and visually adjusting as necessary. Each gene alignment was tested with a 5% level chi-squared test for stationary base composition at variable sites using TREE-PUZZLE-5.2 [16].
We first reconstructed the phylogeny of Avicennia, with Thunbergia grandiflora as the outgroup, using the combined sequence data from two chloroplast segments. Prior to combining the sequences, congruence was examined using the partition–homogeneity test by PAUP* 4b10 [17, 18]. We further concatenated the sequences of 25 nuclear genes to infer the phylogenetic relationship among the IWP species, with Avicennia germinans as the outgroup.
Phylogenetic trees were reconstructed using Bayesian inference (BI), maximum likelihood (ML) and maximum parsimony (MP) methods. Prior to using model-based analytical approaches (BI and ML), nucleotide substitution models were selected based on the Akaike information criterion, as determined by jModelTest 2.1.4 [19]. For the chloroplast matrix, the best-fit model was a general time reversible model with a gamma correction for rate variation among sites (GTR + G). The best model for the nuclear gene matrix was the Hasegawa-Kishino-Yano model, with a gamma correction for rate variation among sites (HKY+G). Bayesian inference was performed using MrBayes 3.2.6 [20] with the models mentioned above. We ran the Markov chains for 10,000,000 generations, with one cold and three heated chains, starting from random trees and saving one out of every 1,000 samples. The runs were repeated twice. The resulting log-likelihood and number of generations were plotted to determine the point after which the log-likelihoods had stabilized. The remaining trees were used to calculate the posterior probabilities (P) through the construction of a majority rule consensus tree. Internodes with P > 95% were considered to be statistically significant. Maximum likelihood (ML) estimation was conducted with PHYML [21] utilizing the HKY+G best-fit model. Bootstrapping of the datasets was performed with 500 replications, and all node values for the ML trees are represented as the proportion of replicates in which that clade was recovered.
Maximum parsimony (MP) analyses were conducted using PAUP* 4.0b10 [18] with a “heuristic search”: tree bisection–reconnection branch swapping, MULPARS option on, and 1,000 random taxon additions. All characteristics were weighted equally, and gaps were treated as missing data. Subsequently, bootstrap analyses were performed on 500 replicates [22] using the options described above.
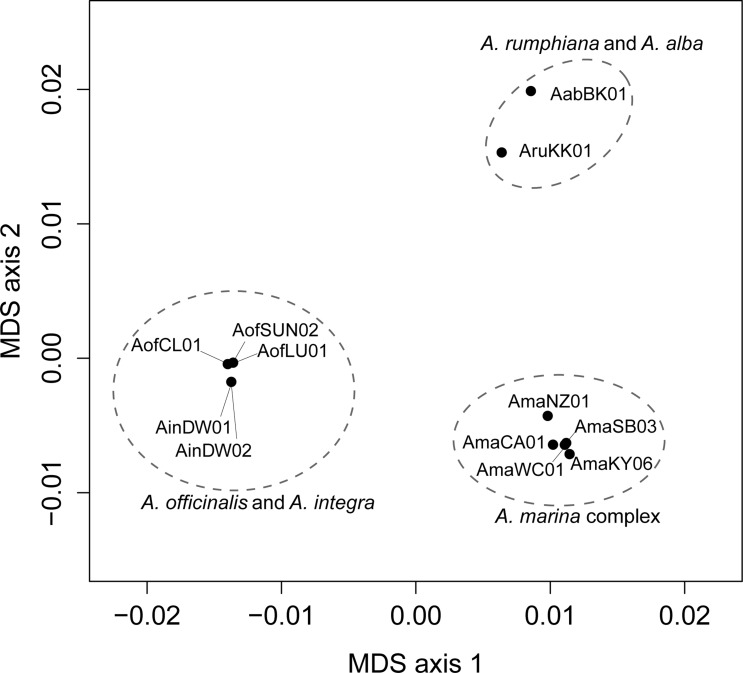
To evaluate the phylogenetic tree and to capture the major clusters of Avicennia species [23], a multi-dimensional scaling (MDS) analysis based on the matrix of the average pairwise genetic divergence value among all Avicennia species calculated using our sample of 2 chloroplast genes was conducted using the command "cmdscale" in R package stats [24]. The same analysis was conducted among IWP species calculated using our sample of 25 nuclear genes as well.
Estimation of divergence time
The concatenated sequences of 25 nuclear genes were used to estimate the divergence times within Avicennia, with gaps treated as missing data. A Bayesian dating method with a relaxed molecular clock was implemented using the mcmctree program in PAML 4.8a [25]. mcmctree employs two strategies to model the change in evolutionary rate among lineages: independent rates and auto-correlated rates [26]. The HKY85+G model was used with different transition/transversion rate ratios (kappa) and different shape parameters (alpha) among the loci. A gamma prior G (2.6, 2) was assigned for kappa and G (1, 1) was assigned for alpha. The calibration point at the basal position of Avicennia was set as B (0.1, 0.5), with 100 million years as one time unit. The assumed range of divergence time (from 10 to 50 million years ago, MYA) between the IWP and AEP lineages was estimated by a previous study based on fossil records and molecular dating [27].
The overall substitution rate (rgene_gamma) was assigned by a gamma distribution with prior G(14, 40), which is a mean of 0.35×10−8 substitutions per site per year, based on the substitution rate from nuclear genes in genus Avicennia (He et al., in review). To estimate substitution rates per year for dating purpose, we constructed a phylogenetic tree using 2,326 orthologous genes from the following species: Sesamum indicum, Avicennia officinalis, Avicennia marina, Acanthus leucostachyus and Acanthus ilicifolius (S1 Fig). These have been sequenced in our lab or have published in genomes or transcriptomes [28, 29], as well as a fossil record [30]. We estimated species divergence time with fossil calibration using coding sequences (CDS). Since our data set included introns as well as CDS, we adjusted the substitutions rate (μ) by inferring the μ intron/μCDS using Kintron/KCDS, where K is the number of segregating sites between A. marina and A. alba from 24 nuclear genes. We then calculated substitution rates for our sequences using weighted means of introns and CDS values. We saved 40,000 samples every two steps after discarding the initial 10,000 samples as the burn-in in mcmctree.
We next investigated the evolution of the flower characters among Avicennia species in IWP. We selected two morphologically important characters (stigma position and style length; S3 Table; [2, 31]) and reconstructed their ancestral states. The two characters were coded and scored following morphological descriptions in the literature [1, 2, 6, 32] and our field observations. The data matrixes used for the analyses are presented in the S4 Table. Based on the phylogenetic tree reconstructed with 25 nuclear genes, we performed our analyses using maximum parsimony in Mesquite v. 3.04 [33].
Results
The phylogeny of Avicennia based on combined chloroplast sequences
All gene segments we obtained satisfied the stationarity test (P > 5%) for base composition at the variable sites and were retained for further analysis. The partition–homogeneity test indicated that the trnT-trnD and psbA sequences could be combined for phylogenetic analyses (P = 1.0). Phylogenetic analyses of this dataset using the BI, ML and MP methods yielded identical topologies (Fig 1; see detail in S2 Fig).
Fig 1. Distribution and the phylogenetics of Avicennia.
Distributions of species are color coded on the map (modified from [34]). The map was modified from the 1:110m coastline map of Natural Earth (http://www.naturalearthdata.com). The phylogenetic relationship between species is based on the phylogenic analyses from chloroplast and nuclear genes. The divergence time for species in the Indo-Western Pacific (IWP) region was calibrated by mcmctree 4.8a [25].
In the combined chloroplast tree, species of Avicennia fell into two clades: five in the IWP region forming one clade and the remaining three in the AEP region forming the other (S2 and S3 Figs; Posterior Probability P = 1.00 for BI; Bootstraps BS = 100% and 99% for ML and MP, respectively). Long branches between the crown node and the roots of both the IWP and AEP clades indicated that the two clades had evolved independently over a long period since the initial split from the common ancestor. Within the AEP lineage, A. germinans was the sister to A. schaueriana (P = 0.99; BS = 78% and 72% for ML and MP, respectively). Within the IWP clade, A. officinalis was the sister to A. integra, with a high level of support (P = 1.00 and BS = 88% and 95%). Those two species were close to the three varieties of the A. marina complex (P = 0.99; BS = 70% and 65%). The phylogenetic positions of A. alba and A. rumphiana were still unclear in the chloroplast tree (P < 0.5 and BS < 50%).
Phylogenetic relationships among the IWP species based on 25 nuclear genes
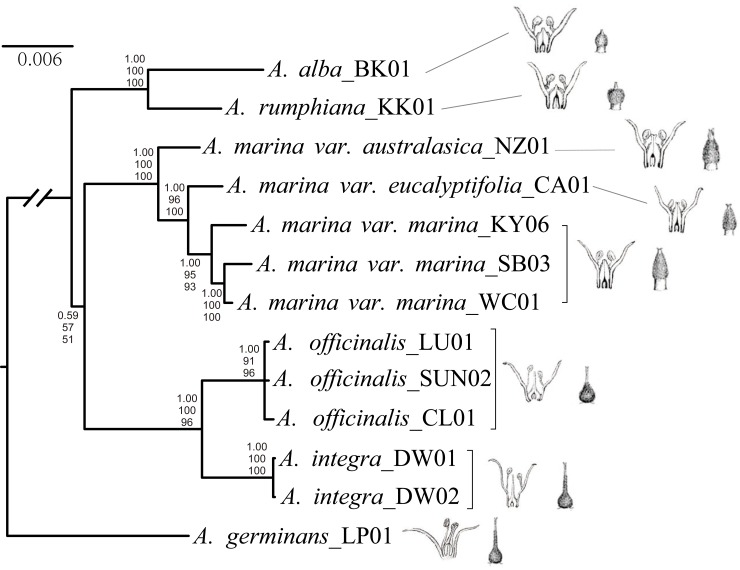
We conducted further phylogenetic analyses among the IWP species using concatenated sequences of 25 nuclear genes, with Avicennia germinans as an outgroup. The BI, ML, and MP methods yielded identical topologies (only the BI tree is shown in Fig 2). The results suggest that the IWP clade could be further split into three distinct subclades (Fig 2). Avicennia alba and A. rumphiana were grouped into one subclade (P = 1.00 and BS = 100% for both ML and MP). The three varieties of A. marina formed another subclade (P = 1.00, BS = 100% for ML and MP). Among the three varieties, A. marina, var. marina is closer to A. marina var. eucalyptifolia than to A. marina var. australasica (P = 1.00, BS = 96% for ML and BS = 100% MP). Avicennia integra and A. officinalis were grouped into the third subclade (P = 1.00, BS = 100% for ML and BS = 96% for MP), which was consistent with results based on the chloroplast segments (S2 Fig). We had low power to resolve the sub-clade relationships, but our results suggest that the A. officinalis/A. integra subclade is more closely related to the A. marina complex than to the A. alba/A. rumphiana subclade (P = 0.59; BS = 57% and 51% for ML and MP, respectively). If we concatenate the chloroplast and nuclear genes and repeat phylogenetic reconstruction using three methods as before, the clustering pattern is the same, with increased support rate for this node (P = 0.72; BS = 69% and 58% for ML and MP, respectively; P = 1.00 for partition–homogeneity test. See the S4 Fig) MDS analysis of the mean pairwise genetic divergence among IWP species, again based on our sample of 25 nuclear genes, revealed a consistent pattern, with A. officinalis and A. integra, A. alba and A. rumphiana, and the three varieties of A. marina forming clear groups identical to those revealed by our phylogenetic analyses (Fig 3).
Fig 2. Bayesian Inference tree of Avicennia species in the Indo-West Pacific (IWP) region.
Based on concatenated sequences of 25 nuclear genes, with A. germinans as the outgroup. Bayesian posterior probabilities for Bayesian Inference (BI), Likelihood bootstrap values from the maximum likelihood analysis (ML) and Parsimony bootstrap values from maximum parsimony analysis (MP) are indicated at nodes (BI/ML/MP). Two selected morphological characters of taxonomic importance (source from [1]) have been mapped on the tree.
Fig 3. Multidimensional scaling plot.
Plot of the first and second axis of a multidimensional scaling matrix based on pairwise genetic divergence value among IWP species of 25 nuclear genes. The numbers and letters refer to sample identity is listed in Table 1.
Fig 1 shows divergence time estimates. Within the IWP clade, the A. alba/A. rumphiana subclades first diverged c. 6.0 MYA, while the other two subclades diverged from each other c. 5.3 MYA. Within the subclades, the initial split between A. alba and A. rumphiana was estimated to be c. 3.6 MYA, whereas the split between A. officinalis and A. integra was estimated to have occurred c. 2.4 MYA. The two divergence events within the A. marina complex were estimated to have occurred at 2.7 and 1.8 MYA, respectively.
Molecular phylogenies can shed light on evolution of morphological traits. Therefore, we selected two important flower structure phenotypes: stigma position and style length. Ancestral states reconstruction for stigma position is shown in the S5 Fig, while the results for style length are in the S6 Fig. Our analyses indicate that the ancestral stigma position in Avicennia from IWP is between anthers, and the ancestral style length is over 1.0 mm. These results suggest that A. officinalis retains ancestral state in flower structure, while A. alba/A. rumphiana lineage and the A. marina complex exhibit independent degeneration.
Discussion
Phylogenetic relationships within Avicennia
The classification and phylogeny of Avicennia were first studied by Tomlinson (1986) [2], Duke (1990, 1991) [1, 6] and Duke et al., (1998) [5] based on morphological and allozyme data. Some studies on phylogeographic patterns of this genus based on a few nuclear and chloroplast DNA markers have been conducted for three AEP species of Avicennia [8, 9]. The present study is the first to provide a phylogenetic reconstruction of Avicennia based on multiple loci and comprehensive sampling (Fig 1 & S2 and S3 Figs). Our phylogeny reveals with high confidence that Avicennia is divided into two distinct clades, consistent with our expectations based on geography and systematics.
Using nuclear gene sequences sampled in this study, we were able to resolve phylogenetic relationships among five species of the IWP region. Our analyses suggest three major subclades within the IWP clade: (1) A. rumphiana and A. alba, (2) A. officinalis and A. integra, and (3) the A. marina complex (Figs 2 and 3). Interestingly, these subclades can be clearly differentiated using two morphological traits in floral structure, i.e. stigma position in relation to the anthers, and the length of styles (Fig 2). In previous studies, species of Avicennia in the IWP region were divided into two lineages, the Officinalis lineage and the Marina lineage, based on the main types of corolla [2]. The Officinalis lineage includes A. officinalis and A. integra, while the latter includes all other species. Our molecular phylogenetic analyses strongly support a subclade with A. officinalis and A. integra (Fig 2). In fact, those two species share some taxonomically important morphological characters (e.g., stamens markedly unequal; one pair is short and the one pair is long). Both species have an elongate flask- or ampulla-shape style exerted above or equal to the upper edge of anthers. The length of style is about 2mm (Fig 2, [1]).
The A. marina complex is the sister to the A. officinalis/A. integra subclade and is not grouped with the other two species, A. rumphiana and A. alba, which were included in the Marina lineage by Tomlinson (1986) [2]. Structural attributes of the floral organs of A. rumphiana and A. alba differ from those of the A. marina complex. The style of A. rumphiana and A. alba is minute and narrow, ending well below the anthers. The length of style is from 0.2–0.4mm [1]. In contrast, the stigma is positioned level with the middle or the lower edge of the anthers in the A. marina complex. The style is about 0.5mm [1].
Our phylogenetic tree suggests that A. rumphiana and A. alba are closely related to each other and form a basal subclade in the IWP lineage. Previous morphological analyses produced conflicting results regarding their affinity to other species of Avicennia. For example, an analysis of flower, fruit and leaf data [6] showed A. alba to be closely related to the A. marina complex, but positioned A. rumphiana as an independent group [32, 35]. However, an analysis of 22 morphological attribute means, such as width and length of the leaves, flowers, and fruit, as well as the inflorescence, stigma position, and propagule shape, suggested that A. rumphiana and A. alba were most closely related to each other [6], which is supported by the present study.
We used our molecular phylogeny to study evolution of flower morphology in Avicennia. We infer the ancestral of two important traits: stigma position and style length (S5 and S6 Figs). Our results indicated the ancestral flower structure is preserved in the A. officinalis/A. integra lineage, with style length about 2mm and stigma level in the middle of the variety anthers (Fig 2). The style degenerated and the stigma became positioned under the anthers in the other IWP Avicennia species, especially for A. abla and A. rumphiana. Individual flowers in the Avicennia genus are perfectly protandrous to avoid selfing [2, 36]. The change of relative positions of anthers and stigma indicates that these species might have evolved herkogamy as a supplemental mechanism for selfing avoidance [37, 38]. Li et al., (2013) [37] suggest that herkogamy is more effective than dichogamy for avoiding reduction of fitness through selfing. Additional field observations and investigations of flower phenology in Avicennia species are needed to establish evolutionary significance of this observation.
Diversification and Evolution within Avicennia in IWP
Although the first fossil record of Avicennia in the IWP region dates back to the late Eocene of southwest Australia, significant flourishing of Avicennia in this region has occurred since the Miocene [39–41]. The distribution of ancestral Avicennia was likely to have been similar to its present location, extending from Japan to Borneo and from the Marshall Islands to the Red Sea [40, 41]. In this study, diversification of the species of Avicennia in the IWP region was dated at approximately 6.0 MYA (95% HPD: 2.8~9.7 MYA; Fig 2). The distribution of the major biogeographic barriers and the shallow seas in the IWP region has not changed significantly since that time [42]. Sea levels, however, began to fluctuate because of climate change [43]. Lowered sea levels resulted in the emergence of land barriers in the IWP region [44], particularly within the Indo-Australia archipelago, which may have facilitated isolation of the Avicennia populations and the subsequent diversification of this clade throughout the region. A number of studies of marine species have also suggested a diversification surrounding the IWP oceans in that period (late Miocene-Pliocene), such as wrasse (Halichoeres in 3.5–8 MYA [45]), Pomacentrus coelestis species complex (within 2.8 MYA [46]) and mangrove snails (Cerithidea, within Plio-Pleistocene [47]).
As the most dominant Avicennia species in this area, A. marina displayed a similar evolutionary history based on the present study. In our phylogenetic tree, the crown age of the A. marina subclades was dated at 2.8 MYA (95% HPD: 1.2~4.6 MYA; Fig 2), which is consistent with the age estimated for this subclade based on allozyme data (approximately 2 MYA, [5]). These estimates support the conclusion that varieties of A. marina diverged more recently, in the Pleistocene, possibly during periods of lowered sea levels during periods of glaciation [43]. Based on genetic diversity of three A. marina varieties, Duke et al., (1998) [5] suggested that the split occurred during the ice age, when the emerged land barrier between New Guinea and Australia isolated the populations on the west coast (var. marina) from those on the east (var. eucalyptifolia). Population genetic data have revealed the role of land barriers in shaping genetic structure across IWP in many species, such as Kandelia candel [48, 49], Ceriops spps., [50–52], Bruguiera gymnorhiza [53], Rhizophora apiculata [54], reef fish [55] and starfish [56]. However, the divergence between var. australasica and var. eucalyptifolia along the coast of East Australia is difficult to explain without positing an additional barrier in this region. This barrier has not been identified in these prior studies [5]. Hewitt (2000) [57] proposed that sea level oscillations would lead to many changes in distribution of marine and coastal species in IWP. Considering repeated oscillations of sea level, Avicennia marina populations, especially var. eucalyptifolia near the Strait of Torres, may experienced several rounds of isolation, dispersal and admixture. Wide sampling and population-genomic studies are necessary to reveal the demography of Avicennia marina and to understand the causes of evolution and diversification of Avicennia species.
Conclusions
Avicennia consists of two monophyletic clades: Atlantic-East Pacific (AEP) lineage and Indo-West Pacific (IWP) lineage. Three distinct subclades were identified within the IWP group with high bootstrap support: (1) A. rumphiana and A. alba, (2) A. officinalis and A. integra, and (3) the A. marina complex. The result was also supported by two morphological traits in floral structure: stigma position in relation to the anthers and the length of the styles. The diversification of the IWP lineage was dated to late Miocene (c. 6 MYA) and may have been driven largely by fluctuating sea levels since that time.
Supporting Information
Bars show the error bar of 95% confidential interval of divergence time. Red rectangle shows the earliest fossil records of mangrove lineages while gray one shows that of inland relatives. The time range of fossils are the earliest and most conformed fossil was from Spain and dated to Middle Bartonia (38.3–39.4 MYA, [30]).
(TIF)
Bayesian posterior probabilities for Bayesian Inference (BI), Likelihood bootstrap values for maximum likelihood (ML) and Parsimony bootstrap values for maximum parsimony (MP) are indicated at nodes (BI/ML/MP), respectively. IWP: Indo-West Pacific region; AEP: Atlantic-East Pacific region.
(TIF)
Plot of the first and second axis of a multidimensional scaling matrix based on pairwise genetic divergence value among all Avicennia species of two chloroplast genes.
(TIF)
Based on concatenated sequences of chloroplast gene and 25 nuclear genes, with A. germinans as the outgroup. Bayesian posterior probabilities for Bayesian Inference (BI), Likelihood bootstrap values from the maximum likelihood analysis (ML) and Parsimony bootstrap values from maximum parsimony analysis (MP) are indicated at nodes (BI/ML/MP).
(TIF)
The coding and its meaning were showed in legend.
(TIF)
The coding and its meaning were showed in legend.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Chung-I Wu for his critical evaluation of the manuscript. We are grateful to David E. Bufford and Anthony Greenberg for English proofreading.
Data Availability
All sequences data for this study are available from the GenBank database (accession numbers is listed in S2 Table).
Funding Statement
This study was supported by grants from the National Natural Science Foundation of China (Grant Numbers: 41130208 and 91331202 to SS, 41276107 to YH. http://www.nsfc.gov.cn/), the Science Foundation of State Key Laboratory of Biocontrol (SKLBC16A35 to SS), and the Chang Hungta Science Foundation of Sun Yat-sen University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Duke NC. A systematic revision of the mangrove genus Avicennia (Avicenniaceae) in Australasia. Aust Syst Bot. 1991;4(2):299–324. 10.1071/sb9910299 [DOI] [Google Scholar]
- 2.Tomlinson P. The botany of mangrove. London: Cambridge Univ; 1986. [Google Scholar]
- 3.Schwarzbach AE, McDade LA. Phylogenetic relationships of the mangrove family Avicenniaceae based on chloroplast and nuclear ribosomal DNA sequences. Syst Bot. 2002;27(1):84–98. [Google Scholar]
- 4.The Angiosperm Phylogeny G. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161(2):105–21. 10.1111/j.1095-8339.2009.00996.x [DOI] [Google Scholar]
- 5.Duke NC, Benzie JA, Goodall JA, Ballment ER. Genetic structure and evolution of species in the mangrove genus Avicennia (Avicenniaceae) in the Indo-West Pacific. Evolution. 1998;52(6):1612–26. 10.2307/2411335 [DOI] [PubMed] [Google Scholar]
- 6.Duke NC. Morphological variation in the mangrove genus Avicennia in Australasia: Systematic and ecological considerations. Aust Syst Bot. 1990;3(2):221–39. 10.1071/sb9900221 [DOI] [Google Scholar]
- 7.Nettel A, Dodd RS. Drifting propagules and receding swamps: Genetic footprints of mangrove recolonization and dispersal along tropical coasts. Evolution. 2007;61(4):958–71. 10.1111/j.1558-5646.2007.00070.x [DOI] [PubMed] [Google Scholar]
- 8.Nettel A, Dodd RS, Afzal-Rafii Z, Tovilla-Hernandez C. Genetic diversity enhanced by ancient introgression and secondary contact in East Pacific black mangroves. Mol Ecol. 2008;17(11):2680–90. 10.1111/j.1365-294X.2008.03766.x [DOI] [PubMed] [Google Scholar]
- 9.Mori GM, Zucchi MI, Sampaio I, Souza AP. Species distribution and introgressive hybridization of two Avicennia species from the Western Hemisphere unveiled by phylogeographic patterns. BMC Evol Biol. 2015;15 10.1186/s12862-015-0343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015. 10.1038/nature15697 [DOI] [PubMed] [Google Scholar]
- 11.Zou XH, Zhang FM, Zhang JG, Zang LL, Tang L, Wang J, et al. Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol. 2008;9(3). 10.1186/gb-2008-9-3-r49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borg AJ, McDade LA, Schönenberger J. Molecular phylogenetics and morphological evolution of Thunbergioideae (Acanthaceae). Taxon. 2008;57(3):811–22. [Google Scholar]
- 13.McDade LA, Daniel TF, Kiel CA. Toward a comprehensive understanding of phylogenetic relationships among lineages of Acanthaceae s.l. (Lamiales). Am J Bot. 2008;95(9):1136–52. 10.3732/ajb.0800096 [DOI] [PubMed] [Google Scholar]
- 14.Doyle J, Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19(11):11–5. [Google Scholar]
- 15.Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 16.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18(3):502–4. 10.1093/bioinformatics/18.3.502 [DOI] [PubMed] [Google Scholar]
- 17.Farris JS, Källersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1995;10(3):315–9. 10.1006/clad.1994.1021 [DOI] [Google Scholar]
- 18.Swofford D. PAUP*: phylogenetic analysis using parsimony, version 4.0 b10. 2003.
- 19.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772–. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–91. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- 23.Brazil M, Thomas DA, Nielsen BK, Winter P, Wulff‐Nilsen C, Zachariasen M. A novel approach to phylogenetic trees: d‐Dimensional geometric Steiner trees. Networks. 2009;53(2):104–11. 10.1002/net.20279 [DOI] [Google Scholar]
- 24.Team RC. R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 25.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 26.Rannala B, Yang Z. Inferring speciation times under an episodic molecular clock. Syst Biol. 2007;56(3):453–66. 10.1080/10635150701420643 [DOI] [PubMed] [Google Scholar]
- 27.Tripp EA, McDade LA. A rich fossil record yields calibrated phylogeny for Acanthaceae (Lamiales) and evidence for marked biases in timing and directionality of intercontinental disjunctions. Syst Biol. 2014:syu029. 10.1093/sysbio/syu029 [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Yang S, Li J, Deng Y, Zhang Z, Xu S, et al. Transcriptome analysis of the Holly mangrove Acanthus ilicifolius and its terrestrial relative, Acanthus leucostachyus, provides insights into adaptation to intertidal zones. BMC Genomics. 2015;16(1). 10.1186/s12864-015-1813-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Miao H, Wang L, Qu L, Liu H, Wang Q, et al. Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biol. 2013;14(1):401 10.1186/gb-2013-14-1-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavagnetto C, Anadon P. Preliminary palynological data on floristic and climatic changes during the Middle Eocene-Early Oligocene of the eastern Ebro Basin, northeast Spain. Rev Palaeobot Palynol. 1996;92(3):281–305. 10.1016/0034-6667(95)00096-8 [DOI] [Google Scholar]
- 31.Nadia TDL, De Menezes NL, Machado IC. Floral traits and reproduction of Avicennia schaueriana Moldenke (Acanthaceae): a generalist pollination system in the Lamiales. Plant Species Biol. 2013;28(1):70–80. 10.1111/j.1442-1984.2011.00361.x [DOI] [Google Scholar]
- 32.Duke NC. The mangrove genus Avicennia (Avicenniaceae) in Australasia Townsville Queensland: James Cook University; 1988. [Google Scholar]
- 33.Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2001.
- 34.Duke NC. World Mangrove ID. Australia: MangroveWatch; 2013. [Google Scholar]
- 35.Duke NC. Genetic diversity, distributional barriers and rafting continents–more thoughts on the evolution of mangroves. Hydrobiologia. 1995;295(1–3):167–81. 10.1007/978-94-011-0289-6_21 [DOI] [Google Scholar]
- 36.Clarke PJ, Myerscough PJ. Floral biology and reproductive phenology of Avicennia marina in south-eastern Australia. Aust J Bot. 1991;39:283–93. 10.1071/bt9910283 [DOI] [Google Scholar]
- 37.Li XX, Zou Y, Xiao CL, Gituru RW, Guo YH, Yang CF. The differential contributions of herkogamy and dichogamy as mechanisms of avoiding self-interference in four self-incompatible Epimedium species. J Evol Biol. 2013;26(9):1949–58. 10.1111/jeb.12193 [DOI] [PubMed] [Google Scholar]
- 38.Webb CJ, Lloyd DG. The avoidance of interference between the presentation of pollen and stigmas in angiosperms II. Herkogamy. N Z J Bot. 1986;24(1):163–78. 10.1080/0028825x.1986.10409726 [DOI] [Google Scholar]
- 39.Renema W, Bellwood D, Braga J, Bromfield K, Hall R, Johnson K, et al. Hopping hotspots: global shifts in marine biodiversity. Science. 2008;321(5889):654–7. 10.1126/science.1155674 [DOI] [PubMed] [Google Scholar]
- 40.Saenger P. Mangrove vegetation: an evolutionary perspective. Marine and Freshwater Research. 1998;49(4):277–86. [Google Scholar]
- 41.Plaziat J-C, Cavagnetto C, Koeniguer J-C, Baltzer F. History and biogeography of the mangrove ecosystem, based on a critical reassessment of the paleontological record. Wetlands Ecol Manage. 2001;9(3):161–79. [Google Scholar]
- 42.Hall R. Southeast Asia's changing palaeogeography. Blumea. 2009;54(1–3):148–61. 10.3767/000651909x475941 [DOI] [Google Scholar]
- 43.Miller KG, Kominz MA, Browning JV, Wright JD, Mountain GS, Katz ME, et al. The Phanerozoic record of global sea-level change. Science. 2005;310(5752):1293–8. 10.1126/science.1116412 [DOI] [PubMed] [Google Scholar]
- 44.Duke NC, Lo E, Sun M. Global distribution and genetic discontinuities of mangroves–emerging patterns in the evolution of Rhizophora. Trees. 2002;16(2–3):65–79. 10.1007/s00468-001-0141-7 [DOI] [Google Scholar]
- 45.Barber PH, Bellwood DR. Biodiversity hotspots: evolutionary origins of biodiversity in wrasses (Halichoeres: Labridae) in the Indo-Pacific and new world tropics. Mol Phylogenet Evol. 2005;35(1):235–53. 10.1016/j.ympev.2004.10.004 [DOI] [PubMed] [Google Scholar]
- 46.Sorenson L, Allen GR, Erdmann MV, Dai C-F, Liu S-YV. Pleistocene diversification of the Pomacentrus coelestis species complex (Pisces: Pomacentridae): historical biogeography and species boundaries. Mar Biol. 2014;161(11):2495–507. 10.1007/s00227-014-2521-8 [DOI] [Google Scholar]
- 47.Reid DG, Claremont M, Smith L, Shamoto M, Glaubrecht M, Ozawa T. Mosaics in the mangroves: allopatric diversification of tree-climbing mudwhelks (Gastropoda: Potamididae: Cerithidea) in the Indo-West Pacific. Biol J Linn Soc. 2013;110(3):564–80. 10.1111/bij.12151 [DOI] [Google Scholar]
- 48.Chiang TY, Chiang YC, Chen Y, Chou CH, Havanond S, Hong T, et al. Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Mol Ecol. 2001;10(11):2697–710. 10.1046/j.0962-1083.2001.01399.x [DOI] [PubMed] [Google Scholar]
- 49.Giang LH, Geada GL, Hong PN, Tuan MS, Lien NTH, Ikeda S. Genetic Variation of Two Mangrove Species in Kandelia (Rhizophoraceae) in Vietnam and Surrounding Area Revealed by Microsatellite Markers. Int J Plant Sci. 2015. [Google Scholar]
- 50.Liao P, Havanond S, Huang S. Phylogeography of Ceriops tagal (Rhizophoraceae) in Southeast Asia: the land barrier of the Malay Peninsula has caused population differentiation between the Indian Ocean and South China Sea. Conserv Genet. 2006;8(1):89–98. 10.1007/s10592-006-9151-8 [DOI] [Google Scholar]
- 51.Huang Y, Zhu C, Li X, Li X, Hu L, Tan F, et al. Differentiated population structure of a genetically depauperate mangrove species Ceriops tagal revealed by both Sanger and deep sequencing. Aquat Bot. 2012;101:46–54. 10.1016/j.aquabot.2012.04.001 [DOI] [Google Scholar]
- 52.Huang YJ, Tan F, Su G, He H, Shi S. Population genetic structure of three tree species in the mangrove genus Ceriops (Rhizophoraceae) from the Indo West Pacific. Genetica. 2007;133(1):47 10.1007/s10709-007-9182-1 [DOI] [PubMed] [Google Scholar]
- 53.Minobe S, Saiki R, Kajita T, Changtragoon S, Shukor NAA, Latiff A, et al. Highly differentiated population structure of a Mangrove species, Bruguiera gymnorhiza (Rhizophoraceae) revealed by one nuclear GapCp and one chloroplast intergenic spacer trnF–trnL. Conserv Genet. 2010;11(1):301–10. 10.1007/s10592-009-9806-3 [DOI] [Google Scholar]
- 54.Guo Z, Huang YJ, Chen Y, Duke NC, Zhong C, Shi S. Genetic discontinuities in a dominant mangrove Rhizophora apiculata (Rhizophoraceae) in the Indo‐Malesian region. J Biogeogr. 2016. 10.1111/jbi.12770 [DOI] [Google Scholar]
- 55.Gaither MR, Bowen BW, Bordenave TR, Rocha LA, Newman SJ, Gomez JA, et al. Phylogeography of the reef fish Cephalopholis argus (Epinephelidae) indicates Pleistocene isolation across the indo-pacific barrier with contemporary overlap in the coral triangle. BMC Evol Biol. 2011;11 10.1186/1471-2148-11-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.John AHB. Major Genetic Differences between Crown-of-Thorns Starfish (Acanthaster planci) Populations in the Indian and Pacific Oceans. Evolution. 1999;53(6):1782–95. 10.2307/2640440 [DOI] [PubMed] [Google Scholar]
- 57.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405(6789):907–13. 10.1038/35016000 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bars show the error bar of 95% confidential interval of divergence time. Red rectangle shows the earliest fossil records of mangrove lineages while gray one shows that of inland relatives. The time range of fossils are the earliest and most conformed fossil was from Spain and dated to Middle Bartonia (38.3–39.4 MYA, [30]).
(TIF)
Bayesian posterior probabilities for Bayesian Inference (BI), Likelihood bootstrap values for maximum likelihood (ML) and Parsimony bootstrap values for maximum parsimony (MP) are indicated at nodes (BI/ML/MP), respectively. IWP: Indo-West Pacific region; AEP: Atlantic-East Pacific region.
(TIF)
Plot of the first and second axis of a multidimensional scaling matrix based on pairwise genetic divergence value among all Avicennia species of two chloroplast genes.
(TIF)
Based on concatenated sequences of chloroplast gene and 25 nuclear genes, with A. germinans as the outgroup. Bayesian posterior probabilities for Bayesian Inference (BI), Likelihood bootstrap values from the maximum likelihood analysis (ML) and Parsimony bootstrap values from maximum parsimony analysis (MP) are indicated at nodes (BI/ML/MP).
(TIF)
The coding and its meaning were showed in legend.
(TIF)
The coding and its meaning were showed in legend.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All sequences data for this study are available from the GenBank database (accession numbers is listed in S2 Table).