Abstract
The tumor microenvironment impacts tumor progression and individual cells, including CD4+ T cells, have been detected in bladder cancer (BCa) tissues. The detailed mechanism how these T cells were recruited to the BCa tumor and their impact on BCa progression, however, remains unclear. Using a human clinical BCa sample survey and in vitro co-culture system, we found that BCa has a greater capacity to recruit T cells than surrounding normal bladder tissues. The consequences of higher levels of recruited T cells in BCa included increased BCa metastasis. Mechanism dissection revealed that infiltrating T cells might function through secreting the cytokine IL-1 which increases the recruitment of T cells to BCa and enhances the BCa androgen receptor (AR) signaling that results in increased BCa cell invasion via up-regulation of HIF-1α/VEGFa expression. Interruption of the IL-1→AR→HIF-1α→VEGFa signals with inhibitors of HIF-1α or VEGFa partially reversed the enhanced-BCa cell invasion. Finally, in vivo mouse models of xenografted BCa T24 cells with CD4+ T cells confirmed in vitro co-culture studies and concluded that infiltrating CD4+ T cells can promote BCa metastasis via modulation of the IL-1→AR→HIF-1α→VEGFa signaling. Future clinical trials using small molecules to target this newly identified signaling pathway may facilitate the development of new therapeutic approaches to better suppress BCa metastasis.
Keywords: CD4+, T cell, bladder cancer, HIF-1α, VEGFa
Introduction
Bladder cancer (BCa) is the fourth most common cancer and eighth leading cause of death from cancer among men in the United States (1), with a recurrence rate up to 90%(2). The ratio of men to women that develop BCa is approximately 3:1 (1), suggesting that androgen/androgen receptor (AR) signals might play important roles in BCa progression. Early studies also demonstrated that AR is a key factor modulating the progression of many tumors, including BCa (3, 4).
The tumor microenvironment (TME), with its individual immune cells, may play key roles in tumor progression (5). Cancer could be due to the result of accumulation of random mutations with increasing dysregulation of several key pathways, which is named as the somatic mutation theory of carcinogenesis, SMT (6, 7). Yet many paradoxical aspects have been found under the SMT framework (8), and many questions remain about the lack of the computational strategies for dealing with those paradoxical aspects and interpreting the accumulated scientific data (8).
An alternative to SMT is the tissue organization field theory (TOFT), which defines a tumor as a tissue-based disease and assumes that cancer arises from the deregulated interplay among tumor cells and the infiltrating cells in the surrounding TME (9).
Several immune cells in the prostate TME have been demonstrated to have the capacity to impact prostate cancer progression (10) (11), and inflammation from TME may also play important roles in BCa progression (12), especially during therapy with Bacillus Calmutte Guerin (BCG) to suppress BCa progression (13). Another clinical study also linked a higher expression of Th17 cells, a subset of CD4+ T cells, in BCa tissues to the progression of BCa (14).
Here we demonstrate that infiltrating T cells promote BCa progression via increasing IL-1→AR→HIF-1α→VEGFa signaling.
Materials and Methods
Patients
We collected tissues from 20 patients that showed clinical cystoscopic evidence of BCa. All patients signed informed consent and received treatment by radical electrocision as their first line of therapy. Each patient's tissue was divided into 2 sections, the BCa tumor area and the adjacent normal tissue area. Both of these tissue types were identified by trained pathologists.
Cell culture
The T24 and J82 cell lines were purchased from the American Type Culture Collection (Rockville, MD) in December of 2008. After cells were received, the cell lines were frozen in liquid N2 after the first 3 passages with 50 ampules of cell stock. After an ampule was thawed, the cells were used for the designed experiments within 15 passages. They were grown in DMEM containing penicillin and streptomycin, supplemented with 10% fetal bovine serum (FBS). The normal immortalized non-transformed SV-HUC bladder urothelial cell line was purchased from American Type Culture Collection and grown in F-12k, supplemented with 10% FBS. In March 2012, the T-lymphocytic cell line HH (CD4+) was acquired from the American Type Culture Collection (ATCC# CRL-2105; CRL-1552) from ATCC and have been kept as 50 ampules of frozen cell stocks. HH cells were maintained in 10% RPMI (Invitrogen #A10491, Grand Island, NY) with 1% Pen/Strep and 10% FBS. After receipt from ATCC, all cell lines were used within 15 passages and were not re-authenticated by us. All cells were cultured in a 5% (v/v) CO2 humidified incubator at 37°C.
Reagents
Monoclonal anti-VEGFa neutralizing antibody was purchased from R&D Systems (Minneapolis, MN), and 500 μg/ml stock was reconstituted in phosphate buffered saline (PBS). HIF inhibitor FM19G11 was purchased from Sigma-Aldrich (St. Louis, MO), and a 500 μg/ml stock was reconstituted in DMSO. IL-1Ra and IL-8 neutralizing antibody were purchased from Peprotech (Rocky Hill, NJ).
Migration assay
The BCa and SV-HUC cells were plated into the lower chambers of the transwells with 8 μM pore polycarbonate membrane inserts precoated with fibronectin (Corning #3422, Corning, NY) at 1×105. 1×105 HH cells were plated into the upper chambers for the T cell migration assay. The membranes were removed after 6 hours, fixed in 75% ethanol and stained with toluidine blue.
Invasion assay
For the in vitro invasion assays, the upper chambers of the Transwell inserts (Corning; 8 μm pores) were pre-coated with diluted EGF-reduced matrigel (1:15 serum free RPMI) (BD Biosciences, Sparks, MD). Before invasion assays, BCa cells were co-cultured with T cells for 48 hrs in 6-well Transwell plates (Corning; 0.4 μm). The conditioned media (C.M.) and control media were collected diluted with 10% FBS RPMI, plated into the lower chambers of new Transwell plates and the parental untreated BCa cells were plated into the upper chambers at 1×105 cells/well. After 24 hours, the cells in the upper chambers were removed. The insert membranes were fixed in ice cold 75% alcohol, stained with crystal violet, and the positively stained cells were counted under a microscope. The numbers of cells were averaged by counting five random fields. Each sample was run in triplicate and in multiple experiments.
Quantitative PCR
Total RNA was extracted from each cell-line using Trizol (Invitrogen, Grand Island, NY), following the manufacturer's instructions. Reverse transcription was performed using the iScript Reverse Transcription Kit (Bio-Rad, Hercules, CA). Quantitative real-time PCR (qRT-PCR) was conducted using a Bio-Rad CFX96 system with SYBR green to determine the mRNA expression level of a gene of interest. Expression levels were normalized to the expression of GAPDH RNA.
Western Blot assay
Cells were washed twice in PBS and lysed with RIPA buffer containing 1% protease inhibitors (Amresco, Solon, OH). Protein concentrations in the cell lysate solutions were determined by BCA protein assay (Amresco). The cell lystates were mixed with 5× SDS-PAGE loading buffer (Amresco). Equivalent protein quantities were heated at 95°C for 10 min before separation on precast 7%-15% SDS-polyacrylamide gels (Bio-Rad). Proteins were electrotransferred to PVDF membranes (Millipore, Atlanta, GA) and blocked in Tris-buffered saline containing 0.05% Tween-20 (TBS-T) and 5% non-fat milk for 1 hr. The membranes were washed in TBS-T and incubated with primary monoclonal antibodies overnight at 4°C in TBS-T containing 1% non-fat milk. The following primary antibodies were used: rabbit anti-AR, anti-HIF-1α (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-VEGFa (1:1000; Abcam, Cambridge, MA) mouse anti-GAPDH (1:1000; Santa Cruz Biotechnology). After being washed in TBS-T buffer, membranes were incubated with goat anti-horseradish peroxidase-conjugated secondary antibody (1:1000; Invitrogen) for 1 hr at room temperature in TBS-T containing 1% non-fat milk. Membranes were then washed with TBS-T buffer, and signals were visualized by use of an enhanced chemiluminesence system (ThermoFisher Scientific, Waltham, MA).
Lentivirus packaging and transfection
We designed the AR siRNA sequence, inserted the oligo into the pLKO.1 vector, packaged with psPAX2 and pMD2.G plasmids. The plasmids were used to transfect 293T cells for 48 hr in order to generate the lentivirus soup. We collected the lentivirus soup and froze aliquots at −80°C for later use.
In vivo metastasis studies
Male 6-8 week old nude mice were purchased from NCI. Stably transfected T24 cells were engineered to express luciferase reporter gene (PCDNA3.0-luciferase) and the positive stable clones were selected and expanded in culture. Twenty-four (24) mice were injected with 1 × 106 T24 cells (mixed with Matrigel, 1:1) into the left sub-renal capsule; 8 mice received T24 cells only, 8 received T24 and 1 × 105 HH cells and 8 mice received T24 and HH cells and were treated with ASC-J9® (starting from the 5th week after xenografted implantation, 0.075 mg/g body weight; injected every other day for 3 weeks). Metastasis in live mice was measured using a Fluorescent Imager (IVIS Spectrum, Caliper Life Sciences, Hopkinton, MA) at 6 different time points (2, 3, 4, 5, 6 and 7 weeks after injection). After monitoring with the Imager, mice were sacrificed and the xenograft tumors were further examined by IHC staining.
Immunohistochemistry
Tumor samples from the BCa mice were fixed in 4% neutral buffered para-formaldehyde and embedded in paraffin. Rabbit anti-CD4+, rabbit (Thermo), rabbit anti-AR (Santa Cruz) and rabbit anti-VEGFa (Abcam) primary antibodies were used for staining. The primary antibodies were recognized by the biotinylated secondary antibody (Vector), and visualized using the VECTASTAIN ABC peroxidase system and peroxidase substrate DAB kit (Vector).
Statistical Analysis
Data are expressed as mean ± SEM from at least 3 independent experiments. Statistical analyses were performed by paired t-test with SPSS 17.0 (SPSS Inc., Chicago, IL). For in vivo studies, measurements of tumor metastasis among the three groups were analyzed through one-way ANOVA coupled with the Newman-Keuls test. P<0.05 was considered statistically significant.
Results
Clinical surveys using human BCa samples show BCa has better capacity than surrounding normal bladder cells to recruit T cells
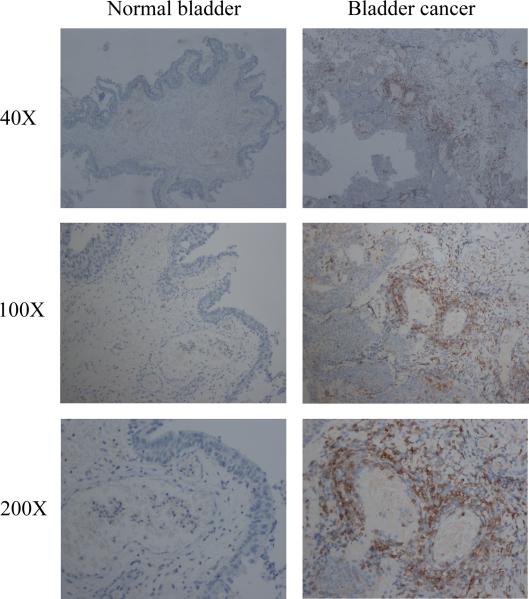
We first compared T cell infiltration in BCa with surrounding normal bladder in clinical specimens using IHC staining with T cell marker CD4+. The results revealed that more CD4+ T cells were recruited into BCa than surrounding normal bladder tissues (Fig. 1).
Figure 1.
Bladder cancer tissues recruit more T cells. IHC staining of clinical specimens from 20 BCa patients showed that more CD4+ T cells infiltrate the area surrounding BCa tissues than in the adjacent normal bladder tissues.
BCa cells recruit more T cells than do normal bladder cells in the Transwell migration assay
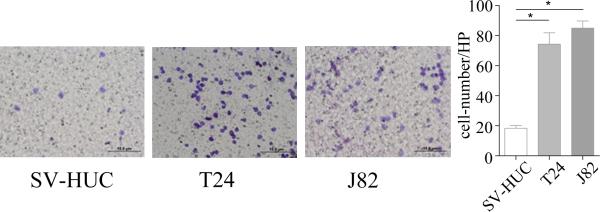
We then applied the in vitro T cell recruitment assay in the co-culture system to confirm the above human clinical results. In the Boyden chamber migration system, we placed the T24 BCa cells or SV-HUC normal bladder cells in the lower chambers and placed T cells (CD4+ HH cells) on the upper chambers for the migration assay. After 6 hours of co-culture, we counted the number of HH cells which had migrated through the membranes into the bottom chambers. The results revealed that T24 cells have a much better capacity to recruit T cells as compared to the non-malignant bladder SV-HUC cells (Fig. 2). Similar results were also obtained when we replaced T24 cells with BCa J82 cells (Fig. 2).
Figure 2.
Bladder cancer cells recruit more T cells in Transwell T cell migration assay. 1 ×105 of BCa cells or normal bladder cells were plated into the lower chambers of Transwell plates. 1 ×105 HH cells were plated into the upper chambers, with 8 μM pore polycarbonate membrane pre-coated with fibronectin, for T cell migration assay. After 6 hours, the upper chambers were removed, remaining T cells and fibronectin scraped off and then membranes fixed with 75% ethanol and stained with toluidine blue shown are representative images of T cells. Compared to the normal SV-HUC cells, BCa cell lines T24 and J82 recruit more HH cells.
Together, results from Fig. 2 suggest BCa cells have a better capacity than surrounding normal bladder cells to recruit T cells.
Recruited T HH cells increase BCa cell invasion
We next investigated the effects of T cells being recruited by BCa cells. Using a co-culture invasion assay with 6-well (0.4 μM membranes) Transwell plates, we found co-culturing BCa T24 cells with T HH cells for 3 days had better capacity to invade (Fig. 3). Similar results were also obtained when we replaced BCa T24 cells with J82 cells (Fig. 3, *P<0.05).
Figure 3.
Co-culture with HH cells promotes BCa cells invasion. For the Matrigel invasion assay, BCa/T cells were first co-cultured in 0.4 μM Transwell plates at a ratio of 2:1 for 3-4 days. Co-cultured/parental BCa cells were then trypsinized for 24 hrs prior to the Matrigel invasion assay. 10% FBS serum was put in the lower chambers and 5 ×104 BCa cells (co-cultured or parental) with serum-free media were plated into the Matrigel pre-coated upper chambers.
Together, results from Fig. 3 suggest that BCa cells have an improved capacity to invade after recruiting more T cells.
Mechanism dissection how recruited T cells can increase the BCa cell invasion
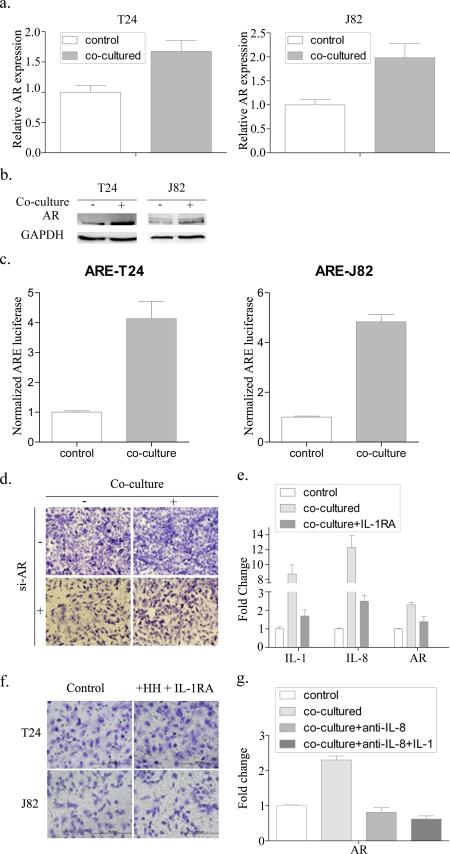
To dissect the mechanism of how recruited T cells promote BCa cell invasion, we focused on their impacts on AR signaling, as early studies suggested AR might have a positive role to increase BCa cell invasion (4, 15). As expected, after co-culturing BCa cells with T HH cells for 3-4 days, we found the expression of AR mRNA (Fig. 4a) and AR protein (Fig. 4b) was increased in BCa T24 cells after co-culture with T HH cells. Similar results were also obtained when we replaced BCa T24 cells with J82 cells (Fig. 4a-b). Importantly, further mechanism dissection using ARE-luciferase assay to examine their influence on AR transactivation revealed that AR transactivation was increased in BCa cells after co-culture with T HH cells (Fig. 4c). Knocking down AR with AR-siRNA also resulted in partially reversed BCa invasion promoted by infiltratingT cells (Fig. 4d).
Figure 4.
Mechanism of how recruited T cells increase BCa cell invasion. (a) After co-culture with HH cell for 3-4 days, AR mRNA levels and (b) protein levels were measured in T24 and J82 cells. (c) ARE-luciferase assay indicated that AR activity increased after BCa cells were co-cultured with T cells. (d) T24 and J82 invasion assays were performed after knocking down AR with AR-siRNA, and then with/without co-culture with HH cells. (e) IL-1, IL-8 and AR mRNA levels of T24 cells tested after co-culture with/without IL-1 antagonist IL-1RA. (f) Invasion assay performed after treating with IL-1 antagonist. (g) AR mRNA level of T24 cells tested after co-culture and addition of 1 mg/mL IL-8 neutralizing antibody.
Together, results from Fig. 4a-d suggest recruited T cells may increase the BCa cell invasion via enhancing AR expression and AR transactivation.
Investigation of the mechanism of infiltrating T cell promoted increase of AR expression in BCa cells
Next we dissected the mechanism(s) by which infiltrated T cells promote AR expression in the co-cultured BCa cells. We hypothesized that infiltrated T cells might function through releasing cytokines to influence BCa AR expression and focused on the interleukin family members, since early studies suggested these cytokines can influence cancer invasion (16-18), and AR expression (18, 19).
In our initial screen, we found the expression of the IL-1 and IL-8 was consistently higher in infiltrating T cells after co-culture with T24, and an IL-1 antagonist can partially inhibit IL-8 and AR. (Fig. 4e, *P<0.05). We utilized an interruption assay to confirm these observations and the results revealed that treatment with an IL-1 antagonist in the co-culture system partially reversed the T cell-enhanced BCa cell (T24 and J82) invasion (Fig. 4f, *P<0.05). Since IL-1 can promote IL-8 expression (20-22), and IL-8 could promote AR mRNA expression (19, 23), we further explored if IL-1 can promote AR expression through altering the IL-8 expression. As expected, we found that adding IL-8 neutralizing antibody can block IL-1 recombinant protein-increased AR expression in the co-culture system (Fig. 4g), suggesting that infiltrating T cells might be able to function through modulation of IL-1 to increase BCa AR expression and enhancing BCa cell invasion and that IL-1 promotion of AR expression is IL-8 dependent.
Examination of the mechanism by which infiltrating T cell-enhanced AR expression increases BCa cell invasion
Among many genes related to BCa metastasis (24), we were able to confirm a few, including MTA1, MMP2, and VEGFa in our co-culture system (Fig. 5a). We focused on VEGFa, because early studies suggested VEGFa expression might be important in BCa metastasis (25). We first verified our screening in the co-cultured T24 and J82 cells with HH cells, and the results confirmed the expression of VEGFa expression was increased in both T24 and J82 cells (Fig. 5b). Importantly, when we applied the interruption approach using VEGFa antagonist Su-5416 to suppress VEGFa, we found suppression of VEGFa can partially reverse infiltrating T cell-increased invasion (Fig. 5c).
Figure 5.
Mechanism by which infiltrating T cell-enhanced AR expression increases BCa cell invasion. (a) Changes in metastasis related genes after BCa cells were co-cultured with T cells. (b) VEGFa mRNA expression (left panel) and qPCR (right panel) in T24 and J82 cells after co-culture with HH cells. (c) Invasion assay after treatment with VEGFa antagonist in the co-culture system. (d) HIFα expression after AR knockdown in T24 and J82 cells. si-AR, AR siRNA; si-luc, AR luciferase. (e) Impact of AF degradation enhancer ASC-J9® on T cell- induced BCa invasion. (f) HIF-1α expression after over expressing AR (OE-AR). (g-h) After inhibiting HIF-1α using the HIF-1α inhibitor FM-19G11, (g) VEGFa expression and (h) invasion induced by T cells was tested.
Together, results from Fig. 5a-c suggest that infiltrating T cells may promote BCa cell invasion via increasing VEGFa expression.
To link the above findings as a newly identified signaling pathway from infiltrating T cells→AR→VEGFa to the increase BCa cell invasion, we dissected the mechanism by which AR modulated VEGFa expression in BCa cells. As early studies suggest VEGFa can be modulated via HIF-1α (26, 27), we then knocked-down AR with AR-siRNA and the results revealed that expression of both VEGFa and HIF-1α was suppressed in T24 and J82 cells (Fig. 5d). Similar results were also obtained when we replaced AR-siRNA with the AR degradation enhancer ASC-J9® (4, 15, 28-30), suggesting reversed T cell-induced invasion via inhibition of the AR pathway (Fig. 5e).
Using a different approach, overexpression of AR also slightly increased HIF-1α expression in T24 and J82 cells (Fig. 5f). Using an interruption approach with HIF inhibitor FM19G11, we found that suppressed HIF-1α inhibited VEGFa expression, and this interruption also partially reversed AR-enhanced VEGFa expression (Fig. 5g) as well as infiltratingT cells-enhanced invasion (Fig. 5h).
Together, results from Fig. 5a-h suggest that infiltrating T cells may be able to function through modulating the IL-1→AR→HIF-1α→VEGFa signaling to increase BCa cell invasion.
Infiltrating T cells enhance BCa metastasis in mouse models
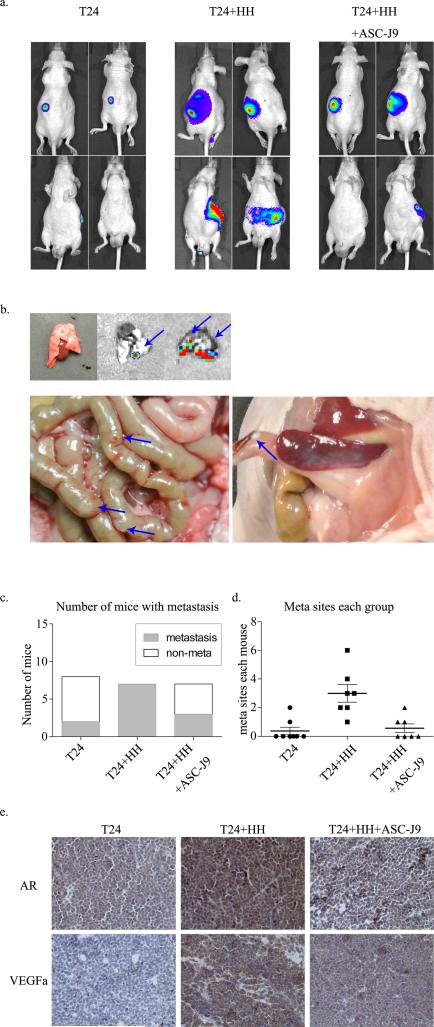
To confirm the in vitro results using various cell line studies in mouse models, we investigated BCa T24 cells with or without co-cultured T (HH) cells at a 9:1 ratio and their therapeutic potential by blocking AR signal with ASC-J9® using mouse BCa xenograft models. T24 cells were transfected with pCDNA3-luciferase for monitoring tumor growth and metastasis using fluorescent imaging (IVIS Spectrum, Caliper Life Sciences) (11). 7 weeks after implantation, we found that the T24/HH cell co-implanted group mice had more metastatic luminescent signals located at distant organs when compared with the T24-only group, and this promoted metastasis could be partially reversed by ASC-J9® (Fig. 6a). The mice were sacrificed for tumor examination (Fig. 6b), and results confirmed higher metastastic rates (100% in 7 of 7 mice) with metastatic tumors found in the lungs and/or gut in the co-implanted T24+HH cell group and 25% (2 of 8 mice) in the co-implanted T24+HH group. Notably, only 42.9% (3 of 7 mice) were found to have meta sites in the ASC-J9® treated T24+HH group. Total meta sites of each group have also been calculated. We found that the co-implanted T cell group has more meta sites (21 in total) than the T24-only group (3 in total) (P<0.05) (Fig. 6c), and the ASC-J9® treated T24+HH group has significantly fewer meta sites than the co-implant group (4 in total) (P<0.05) (Fig. 6d). The expression of AR and VEGFa were also examined using IHC methods. We confirmed that co-implanted specimens have higher AR and VEGFa expression, and this promoted expression can be partially reversed by the AR degradation enhancer ASC-J9® (Fig. 6e).
Figure 6.
AR signal in CD4+ T cells as conducted in bladder BCa TME in the mouse models. (a) IVIS performed 7 weeks after xenograft implantation into male nude mice sub-renal capsules. (b) Meta sites after distal metastases. Blue arrows point to metastases. (c) Number of mice with metastasis in each group was counted; (d) calculation of total meta sites of each group . (e) IHC performed on xenograft tumors of each group; AR and VEGFa expression levels were tested.
Discussion
Early studies linked CD4+ lymphocytes in the prostate TME to poor outcomes in PCa patients (31). Similar clinical reports also revealed that increased CD4+ T-lymphocyte infiltration is associated with decreased survival in patients with kidney cancer (32). Furthermore, recruited CD4+ T cells in mammary tumors might also be able to enhance their metastasis (33). Interestingly, infiltrating T cells were also found in BCa (34), yet the impact of each subtype on BCa remained controversial (34-36). Here we demonstrated that infiltrating T lymphocytes could enhance BCa invasion via increasing the IL-1→ AR→HIF-1α→VEGFa signaling pathway. This is in agreement with previous studies suggesting that AR can increase BCa metastasis (37, 38).
IL-1 has been reported to play more important roles than IL-8 to recruit more CD4+ T lymphocytes into skin tissues (39). Results shown in Fig 4g indicate that IL-1 may function through modulating IL-8 to alter AR expression. Comparing Lane 1, 2 and 3, we see that anti-IL-8 can reverse T cell-promoted AR up-regulation, while comparing Lane 3 and 4 it is seen that, when IL-8 as blocked using an IL-8 neutralizing antibody, adding more IL-1 failed to promote further AR expression. These results are consistent with earlier studies showing that IL-1 can promote IL-8 expression (20-22), and that IL-8 promotes AR expression (19, 23). A previous study also indicated that IL-1 regulates IL-8 at the transcriptional level via inducing a nuclear factor that binds to the cis-element between −80 and −71 bp of the IL-8 gene (21). A study by Hoffman et al also suggested that IL-1 might stimulate the recruitment of p65 NF-κB and RNA polymerase II to the endogenous IL-8 promoter (40), and that IL-8 can regulate AR via both transcriptional and non-transcriptional mechanisms that involve the altering the NF-kappaB, Hsp90, phosphorylation and co-activators (19, 41-43).
Early studies found that VEGF expression is associated with BCa stages and recurrence (44), and that HIF-1α has been linked to the progression of renal cell carcinoma (45, 46) and BCa (47-49). Here we further linked this modulation to the AR expression showing AR could positively regulate HIF1α→VEGFa signaling in BCa. Interestingly, using androgen deprivation therapy to prevent androgen binding to AR, Ragnum et al found that targeting the androgen/AR signals suppressed the HIF-1α signaling in prostate cancer (50), and Mitani et al suggested that AR and HIF-1α might be able to act together as co-regulators to modulate AR signals (51).
Furthermore, we found ASC-J9®, a novel AR degradation enhancer (52), functions in a manner similar to AR-siRNA to partially reverse the T cell-induced BCa cell invasion by reducing AR expression, which is in agreement with early studies showing ASC-J9® decreases BCa progression via targeting AR signaling (4, 15, 53).
The TCGA database is a valuable resource which can be used to predict relationships among genes in many cancers (54, 55). In order to further investigate the expression of mRNA in bladder cancers, and to confirm the relationship of the markers in the pathway studied in this research, we mined the TCGA RNA-seq dataset for BLCA (bladder cancer; available online at https://tcga-data.nci.nih.gov/docs/publications/blca_2013/) to determine the association between the biomarkers. In this dataset, 129 records contained information regarding bladder cancer subtype (Subtypes 1-4), and these were selected for our analysis so that we could later stratify the results by subtype. A correlation analysis of all 129 samples in this dataset determined a high degree of correlation between HIF-1α and IL-8 (Pearson correlation = 0.415) and between IL-1a and IL-8 (Pearson correlation = 0.545) (Supplementary Fig. S1). We analyzed CD4, AR, HIF-1α and VEGFa in the BLCA dataset as a whole, but pairwise analyses did not produce strong correlations. However, when we analyzed the dataset by individual subtype (Subtypes 1-4), pairwise correlation analyses produced dramatically different results. Subtype 4 had the strongest correlations with the CD4/AR (Pearson correlation = 0.444), CD4/HIF1α (0.416), IL-1a/VEGFa (0.473), IL-8/VEGFa (0.473) and IL-1a/IL-8 (0.675) pairs being more strongly correlated than in the other subtypes (Supplementary Table S2). For example, in Subtype 3, only IL-1a/IL-8 showed a strong correlation (Pearson correlation = 0.655), while the other marker pairs were weakly or almost entirely un-correlated (correlation coefficient < 0.4). Additionally, the mRNA expression levels covered a broad range, with some markers displaying a 1000-fold or greater change (Supplementary Fig. S1). These results demonstrate the heterogeneity in cancers of this type. The subtypes may present with differential expression of biomarkers, potentially correlating with different pathways or mechanisms for invasion, migration and proliferation.
Because BCa is heterogeneous, low grade and high grade tumors may have different signaling pathways (56). For example, recent studies indicate that muscle-invasive bladder cancers (MIBCs) could be divided into two (57) or four (58) intrinsic subtypes, similar to those found in breast cancer (59-61). The biomarkers, prognosis and response to chemo- or radio-therapy could be significantly different among the subtypes (60, 61). We therefore believe our findings do not represent every case of BCa found in all patients, but we believe our study provides a rationale for exploring novel therapeutic strategies for the treatment of BCa. For example, in the development of Precision Medicine, especially with the continuous updating of TCGA's RNAseq dataset (54, 55), our findings may be applicable in some cases of BCa for the development of better therapeutic strategies.
A seen in Figure 1, we found T cell infiltration in different degrees among the specimens, which is consistent with the study by Loskog et al (34). McConey et al (61) reported that T cell infiltration is highly enriched in one of the intrinsic subtypes, according to Choi et al's (60) work, and that different BCa subtypes may yield different conclusions. However, we believe there are additional factors contributing to the discrepancy. First, Choi et al (60) and McConey et al (61) used mRNAseq to detect lymphocyte biomarkers, and interpreted the data using bioinfomatics methods. Meanwhile, our study used IHC to detect the CD4+ T cells. Different methodologies may be part of the reason for the differing results. Secondly, we compared BCa tissue with adjacent normal urothelial tissues, while all of the specimens in the previous studies were MIBC tissues. Thus, the control groups are different. Finally, we used an anti-CD4 antibody to detect the CD4+ T cells, while, according to Figure 2 of McConkey et al's work (61), CD4 was not evaluated in the previous study. The biomarkers listed might reflect the presence of all kinds of lympohcytes, but it is difficult to conclude CD4+ T cells. Importantly, our clinical results were supported by our subsequent in vitro cell line study.
In summary, our results concluded that infiltrating T cells increase BCa metastasis via modulation of the IL-1→AR→HIF-1α→VEGFa signaling pathway, which may provide a new therapeutic approach to battle BCa metastasis via targeting this newly identified pathway, beginning with infiltrating T cells.
Supplementary Material
Acknowledgments
This work was supported by NIH grants (CA155477 and CA156700; to C. Chang), the George Whipple Professorship Endowment (to C. Chang), and the Taiwan Department of Health Clinical Trial, Research Center of Excellence (DOH99-TD-B-111-004 to China Medical University, Taichung, Taiwan; to C. Chang).
Footnotes
The authors declare that no conflicts of interest exist.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Piergentili R, Carradori S, Gulia C, De Monte C, Cristini C, Grande P, et al. Bladder cancer: innovative approaches beyond the diagnosis. Current medicinal chemistry. 2014;21:2219–36. doi: 10.2174/0929867321666140304110231. [DOI] [PubMed] [Google Scholar]
- 3.Chang C, Lee SO, Yeh S, Chang TM. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene. 2014;33:3225–34. doi: 10.1038/onc.2013.274. [DOI] [PubMed] [Google Scholar]
- 4.Miyamoto H, Yang Z, Chen YT, Ishiguro H, Uemura H, Kubota Y, et al. Promotion of bladder cancer development and progression by androgen receptor signals. Journal of the National Cancer Institute. 2007;99:558–68. doi: 10.1093/jnci/djk113. [DOI] [PubMed] [Google Scholar]
- 5.Bizzarri M, Cucina A. Tumor and the microenvironment: a chance to reframe the paradigm of carcinogenesis? BioMed research international. 2014;2014:934038. doi: 10.1155/2014/934038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michor F, Iwasa Y, Nowak MA. Dynamics of cancer progression. Nature reviews Cancer. 2004;4:197–205. doi: 10.1038/nrc1295. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Baker SG, Kramer BS. Paradoxes in carcinogenesis: new opportunities for research directions. BMC cancer. 2007;7:151. doi: 10.1186/1471-2407-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnenschein C, Soto AM. The death of the cancer cell. Cancer research. 2011;71:4334–7. doi: 10.1158/0008-5472.CAN-11-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yates C. Prostate tumor cell plasticity: a consequence of the microenvironment. Advances in experimental medicine and biology. 2011;720:81–90. doi: 10.1007/978-1-4614-0254-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu S, Li L, Yeh S, Cui Y, Li X, Chang HC, et al. Infiltrating T cells promote prostate cancer metastasis via modulation of FGF11-->miRNA-541-->androgen receptor (AR)-->MMP9 signaling. Molecular oncology. 2014 doi: 10.1016/j.molonc.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Chin AI. Role of Rip2 in development of tumor-infiltrating MDSCs and bladder cancer metastasis. PloS one. 2014;9:e94793. doi: 10.1371/journal.pone.0094793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boccafoschi C, Montefiore F, Pavesi M, Pastormerlo M, Annoscia S, Lozzi C, et al. Immunophenotypic characterization of the bladder mucosa infiltrating lymphocytes after intravesical BCG treatment for superficial bladder carcinoma. European urology. 1992;21:304–8. doi: 10.1159/000474862. [DOI] [PubMed] [Google Scholar]
- 14.Chugh S, Anand V, Swaroop L, Sharma M, Seth A, Sharma A. Involvement of Th17 cells in patients of urothelial carcinoma of bladder. Human immunology. 2013;74:1258–62. doi: 10.1016/j.humimm.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 15.Hsu JW, Hsu I, Xu D, Miyamoto H, Liang L, Wu XR, et al. Decreased tumorigenesis and mortality from bladder cancer in mice lacking urothelial androgen receptor. The American journal of pathology. 2013;182:1811–20. doi: 10.1016/j.ajpath.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar J, Ward AC. Role of the interleukin 6 receptor family in epithelial ovarian cancer and its clinical implications. Biochimica et biophysica acta. 2014;1845:117–25. doi: 10.1016/j.bbcan.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Voronov E, Carmi Y, Apte RN. The role IL-1 in tumor-mediated angiogenesis. Frontiers in physiology. 2014;5:114. doi: 10.3389/fphys.2014.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:6735–41. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 19.Seaton A, Scullin P, Maxwell PJ, Wilson C, Pettigrew J, Gallagher R, et al. Interleukin-8 signaling promotes androgen-independent proliferation of prostate cancer cells via induction of androgen receptor expression and activation. Carcinogenesis. 2008;29:1148–56. doi: 10.1093/carcin/bgn109. [DOI] [PubMed] [Google Scholar]
- 20.Unger BL, McGee DW. Hepatocyte growth factor and keratinocyte growth factor enhance IL-1-induced IL-8 secretion through different mechanisms in Caco-2 epithelial cells. In vitro cellular & developmental biology Animal. 2011;47:173–81. doi: 10.1007/s11626-010-9365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukaida N, Mahe Y, Matsushima K. Cooperative interaction of nuclear factor-kappa B- and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. The Journal of biological chemistry. 1990;265:21128–33. [PubMed] [Google Scholar]
- 22.Bae JY, Kim EK, Yang DH, Zhang X, Park YJ, Lee DY, et al. Reciprocal Interaction between Carcinoma-Associated Fibroblasts and Squamous Carcinoma Cells through Interleukin-1alpha Induces Cancer Progression. Neoplasia (New York, NY) 2014;16:928–38. doi: 10.1016/j.neo.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Yang J, Gao Y, Dong LJ, Liu S, Yao Z. Reciprocal regulation of 5alpha-dihydrotestosterone, interleukin-6 and interleukin-8 during proliferation of epithelial ovarian carcinoma. Cancer biology & therapy. 2007;6:864–71. doi: 10.4161/cbt.6.6.4093. [DOI] [PubMed] [Google Scholar]
- 24.Mitra AP, Bartsch CC, Cote RJ. Strategies for molecular expression profiling in bladder cancer. Cancer metastasis reviews. 2009;28:317–26. doi: 10.1007/s10555-009-9196-5. [DOI] [PubMed] [Google Scholar]
- 25.Gakis G. The role of inflammation in bladder cancer. Advances in experimental medicine and biology. 2014;816:183–96. doi: 10.1007/978-3-0348-0837-8_8. [DOI] [PubMed] [Google Scholar]
- 26.Deniz H, Karakok M, Yagci F, Guldur ME. Evaluation of relationship between HIF-1alpha immunoreactivity and stage, grade, angiogenic profile and proliferative index in bladder urothelial carcinomas. International urology and nephrology. 2010;42:103–7. doi: 10.1007/s11255-009-9590-5. [DOI] [PubMed] [Google Scholar]
- 27.Tang N, Wang L, Esko J, Giordano FJ, Huang Y, Gerber HP, et al. Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer cell. 2004;6:485–95. doi: 10.1016/j.ccr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Wen S, Chang HC, Tian J, Shang Z, Niu Y, Chang C. Stromal Androgen Receptor Roles in the Development of Normal Prostate, Benign Prostate Hyperplasia, and Prostate Cancer. The American journal of pathology. 2015;185:293–301. doi: 10.1016/j.ajpath.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He D, Li L, Zhu G, Liang L, Guan Z, Chang L, et al. ASC-J9 suppresses renal cell carcinoma progression by targeting an androgen receptor-dependent HIF2alpha/VEGF signaling pathway. Cancer research. 2014;74:4420–30. doi: 10.1158/0008-5472.CAN-13-2681. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita S, Lai KP, Chuang KL, Xu D, Miyamoto H, Tochigi T, et al. ASC-J9 suppresses castration-resistant prostate cancer growth through degradation of full-length and splice variant androgen receptors. Neoplasia (New York, NY) 2012;14:74–83. doi: 10.1593/neo.111436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McArdle PA, Canna K, McMillan DC, McNicol AM, Campbell R, Underwood MA. The relationship between T-lymphocyte subset infiltration and survival in patients with prostate cancer. British journal of cancer. 2004;91:541–3. doi: 10.1038/sj.bjc.6601943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bromwich EJ, McArdle PA, Canna K, McMillan DC, McNicol AM, Brown M, et al. The relationship between T-lymphocyte infiltration, stage, tumour grade and survival in patients undergoing curative surgery for renal cell cancer. British journal of cancer. 2003;89:1906–8. doi: 10.1038/sj.bjc.6601400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan W, Zhang W, Strasner A, Grivennikov S, Cheng JQ, Hoffman RM, et al. Tumour-infiltrating regulatory T cells stimulate mammary cancer metastasis through RANKL-RANK signalling. Nature. 2011;470:548–53. doi: 10.1038/nature09707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loskog A, Ninalga C, Paul-Wetterberg G, de la Torre M, Malmstrom PU, Totterman TH. Human bladder carcinoma is dominated by T-regulatory cells and Th1 inhibitory cytokines. The Journal of urology. 2007;177:353–8. doi: 10.1016/j.juro.2006.08.078. [DOI] [PubMed] [Google Scholar]
- 35.Coe D, Begom S, Addey C, White M, Dyson J, Chai JG. Depletion of regulatory T cells by anti-GITR mAb as a novel mechanism for cancer immunotherapy. Cancer immunology, immunotherapy : CII. 2010;59:1367–77. doi: 10.1007/s00262-010-0866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krpina K, Babarovic E, Dordevic G, Fuckar Z, Jonjic N. The association between the recurrence of solitary non-muscle invasive bladder cancer and tumor infiltrating lymphocytes. Croatian medical journal. 2012;53:598–604. doi: 10.3325/cmj.2012.53.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jing Y, Cui D, Guo W, Jiang J, Jiang B, Lu Y, et al. Activated androgen receptor promotes bladder cancer metastasis via Slug mediated epithelial-mesenchymal transition. Cancer letters. 2014;348:135–45. doi: 10.1016/j.canlet.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Jitao W, Jinchen H, Qingzuo L, Li C, Lei S, Jianming W, et al. Androgen receptor inducing bladder cancer progression by promoting an epithelial-mesenchymal transition. Andrologia. 2014;46:1128–33. doi: 10.1111/and.12203. [DOI] [PubMed] [Google Scholar]
- 39.Colditz IG, Watson DL. The effect of cytokines and chemotactic agonists on the migration of T lymphocytes into skin. Immunology. 1992;76:272–8. [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. Journal of leukocyte biology. 2002;72:847–55. [PubMed] [Google Scholar]
- 41.Chen SY, Cai C, Fisher CJ, Zheng Z, Omwancha J, Hsieh CL, et al. c-Jun enhancement of androgen receptor transactivation is associated with prostate cancer cell proliferation. Oncogene. 2006;25:7212–23. doi: 10.1038/sj.onc.1209705. [DOI] [PubMed] [Google Scholar]
- 42.Gioeli D, Ficarro SB, Kwiek JJ, Aaronson D, Hancock M, Catling AD, et al. Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. The Journal of biological chemistry. 2002;277:29304–14. doi: 10.1074/jbc.M204131200. [DOI] [PubMed] [Google Scholar]
- 43.Lu S, Ren C, Liu Y, Epner DE. PI3K-Akt signaling is involved in the regulation of p21(WAF/CIP) expression and androgen-independent growth in prostate cancer cells. International journal of oncology. 2006;28:245–51. [PubMed] [Google Scholar]
- 44.Kopparapu PK, Boorjian SA, Robinson BD, Downes M, Gudas LJ, Mongan NP, et al. Expression of VEGF and its receptors VEGFR1/VEGFR2 is associated with invasiveness of bladder cancer. Anticancer research. 2013;33:2381–90. [PubMed] [Google Scholar]
- 45.Keefe SM, Nathanson KL, Rathmell WK. The molecular biology of renal cell carcinoma. Seminars in oncology. 2013;40:421–8. doi: 10.1053/j.seminoncol.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Rhim T, Lee DY, Lee M. Hypoxia as a target for tissue specific gene therapy. Journal of controlled release : official journal of the Controlled Release Society. 2013;172:484–94. doi: 10.1016/j.jconrel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 47.Badr S, Salem A, Yuosif AH, Awadallah H, Awed N, Bakr A. Hypoxia inducible factor-1alpha and microvessel density as angiogenic factors in bilharzial and non-bilharzial bladder cancer. Clinical laboratory. 2013;59:805–12. doi: 10.7754/clin.lab.2012.120605. [DOI] [PubMed] [Google Scholar]
- 48.Xue M, Li X, Li Z, Chen W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1alpha-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:6901–12. doi: 10.1007/s13277-014-1925-x. [DOI] [PubMed] [Google Scholar]
- 49.Chen MC, Lee CF, Huang WH, Chou TC. Magnolol suppresses hypoxia-induced angiogenesis via inhibition of HIF-1alpha/VEGF signaling pathway in human bladder cancer cells. Biochemical pharmacology. 2013;85:1278–87. doi: 10.1016/j.bcp.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 50.Ragnum HB, Roe K, Holm R, Vlatkovic L, Nesland JM, Aarnes EK, et al. Hypoxia-independent downregulation of hypoxia-inducible factor 1 targets by androgen deprivation therapy in prostate cancer. International journal of radiation oncology, biology, physics. 2013;87:753–60. doi: 10.1016/j.ijrobp.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 51.Mitani T, Harada N, Nakano Y, Inui H, Yamaji R. Coordinated action of hypoxia-inducible factor-1alpha and beta-catenin in androgen receptor signaling. The Journal of biological chemistry. 2012;287:33594–606. doi: 10.1074/jbc.M112.388298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Z, Chang YJ, Yu IC, Yeh S, Wu CC, Miyamoto H, et al. ASC-J9 ameliorates spinal and bulbar muscular atrophy phenotype via degradation of androgen receptor. Nature medicine. 2007;13:348–53. doi: 10.1038/nm1547. [DOI] [PubMed] [Google Scholar]
- 53.Soh SF, Huang CK, Lee SO, Xu D, Yeh S, Li J, et al. Determination of androgen receptor degradation enhancer ASC-J9((R)) in mouse sera and organs with liquid chromatography tandem mass spectrometry. Journal of pharmaceutical and biomedical analysis. 2014;88:117–22. doi: 10.1016/j.jpba.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. European urology. 2011;59:671–81. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 57.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3110–5. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi W, Czerniak B, Ochoa A, Su X, Siefker-Radtke A, Dinney C, et al. Intrinsic basal and luminal subtypes of muscle-invasive bladder cancer. Nature reviews Urology. 2014;11:400–10. doi: 10.1038/nrurol.2014.129. [DOI] [PubMed] [Google Scholar]
- 60.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer cell. 2014;25:152–65. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McConkey DJ, Choi W, Dinney CP. Genetic subtypes of invasive bladder cancer. Current opinion in urology. 2015;25:449–58. doi: 10.1097/MOU.0000000000000200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.