Abstract
BACKGROUND
Metabolic syndrome which underlies the increased prevalence of cardiovascular disease and Type 2 diabetes is considered as a group of metabolic abnormalities including central obesity, hypertriglyceridemia, glucose intolerance, hypertension, and dyslipidemia. Recently, artificial intelligence based health-care systems are highly regarded because of its success in diagnosis, prediction, and choice of treatment. This study employs machine learning technics for predict the metabolic syndrome.
METHODS
This study aims to employ decision tree and support vector machine (SVM) to predict the 7-year incidence of metabolic syndrome. This research is a practical one in which data from 2107 participants of Isfahan Cohort Study has been utilized. The subjects without metabolic syndrome according to the ATPIII criteria were selected. The features that have been used in this data set include: gender, age, weight, body mass index, waist circumference, waist-to-hip ratio, hip circumference, physical activity, smoking, hypertension, antihypertensive medication use, systolic blood pressure (BP), diastolic BP, fasting blood sugar, 2-hour blood glucose, triglycerides (TGs), total cholesterol, low-density lipoprotein, high density lipoprotein-cholesterol, mean corpuscular volume, and mean corpuscular hemoglobin. Metabolic syndrome was diagnosed based on ATPIII criteria and two methods of decision tree and SVM were selected to predict the metabolic syndrome. The criteria of sensitivity, specificity and accuracy were used for validation.
RESULTS
SVM and decision tree methods were examined according to the criteria of sensitivity, specificity and accuracy. Sensitivity, specificity and accuracy were 0.774 (0.758), 0.74 (0.72) and 0.757 (0.739) in SVM (decision tree) method.
CONCLUSION
The results show that SVM method sensitivity, specificity and accuracy is more efficient than decision tree. The results of decision tree method show that the TG is the most important feature in predicting metabolic syndrome. According to this study, in cases where only the final result of the decision is regarded significant, SVM method can be used with acceptable accuracy in decision making medical issues. This method has not been implemented in the previous research.
Keywords: Machine Learning, Metabolic Syndrome, Decision Tree, Support Vector Machine
Introduction
Nowadays, the health-care systems based on the intelligent systems have been drawing a lot of attention. Medical decision support systems which can be used for predicting diseases, and assisting physicians in the diagnosis and treatment decisions,1,2 and etc., can be mentioned as a kind of intelligent system. A metabolic syndrome which has an upward trend due to industrialization of countries is one of the medical disorders. Lifestyle and dietary changes tending toward fast food, salty, and fatty food have increased metabolic syndrome in the communities.3 Metabolic syndrome is a cluster of conditions including high blood pressure (BP), abdominal obesity and hyperlipidemia and insulin resistance. In many cases, these conditions are present simultaneously.4
It is estimated that about 20-25% of the world’s adult population suffer from the metabolic syndrome which doubles the risk of mortality in individuals. Moreover, risk of heart attack is 3 times as many and Type 2 diabetes is 5 times as many compared to people without the metabolic syndrome.4
In Asian countries, the metabolic syndrome has become an important issue due to changes in diet and lifestyle. In Iran, the prevalence of metabolic syndrome is spectacularly increasing that according to recent estimates equals generally 33 and 10-11% in normal weight adults.5,6
In recent years, artificial intelligence has been considered as a new approach of problem modeling. Nowadays, artificial intelligence has frequently been utilized to predict, diagnose and help medical diagnosis. Support vector machines (SVMs) and decision tree have been utilized for diagnosis and prediction of diseases in the medical domain. H. X. Liu et al. used the SVM to the diagnosis of breast cancer.7 SVM was also applied to predicting central nervous system (CNS) permeability of drug molecules.8 Decision tree algorithms have been used to prediction of pancreatic cancer9 and identification of metabolic syndrome.10 So far, this study has been accomplished to predict metabolic syndrome using SVM and decision tree. The purpose of this research is to predict the metabolic syndrome using decision tree and SVM decision methods.
In this study, decision tree and SVM methods are used to predict the metabolic syndrome since they are easy to understand and have a great efficiency, respectively.
Materials and Methods
Data for this study included 2107 subjects participating in the research project of Isfahan Cohort Study (ICS)11. The first phase of the research project of ICS began in January 2001 and lasted until October of 2001. In the first phase, 6323 people who have not been diagnosed of heart disease were randomly selected. The second phase, in 2007, was re-conducted. In Phase II, the 3284 of them, after 7 years of follow-up, were re-evaluated. Among the participants in this project 2107 cases who did not suffer from metabolic syndrome in the first phase were chosen according to ATPIII. In the second phase of the study, 596 of them were diagnosed to suffer from metabolic syndrome. According to the criteria of ATPIII, there must exist at least three components of five following components12 for the diagnosis of metabolic syndrome:
Hypertriglyceridemia ≥ 150 mg/dl
Low- and high-density lipoprotein cholesterol (HDL-C) < 40 mg/dl in men and HDL < 50 mg/dl in women
Fasting glucose ≥ 100 or use of diabetes medication
BP ≥ 130/85 mmHg or treatment with BP medication
Waist circumference (WC) ≥ 95 cm in men and women
WC is considered 95 cm for the Iranian people.13
The features that have been used in this data set include: Gender, age, weight, body mass index (BMI), WC, waist-to-hip ratio (WHR), hip circumference (HC), physical activity, smoking history, hypertension, antihypertensive medication use, systolic BP, diastolic BP, fasting blood glucose, 2-hour blood glucose, triglycerides (TGs), total cholesterol, low-density lipoprotein (LDL), HDL-C, mean corpuscular volume (MCV), and mean corpuscular hemoglobin (MCH).
The decision tree is a supervised machine learning technique. The results of this method can be presented in the form of a tree or a set of if-then rules.14 The most significant feature in the decision tree method is the tree root. Other features are placed in the lower levels of tree in order of importance.15 Finally, the leaves of the tree represent the classification result. This technique is widely used in various fields including the medical field. Many studies show that decision tree is an effective method in the analysis of medical data.9,10,15-17 It has been applied in many studies because of its accurate and understandable results18 which make the decision tree more important than the other accurate methods.19 After learning tree by starting from the root, and following the conditions in the intermediate nodes of the tree, we can easily follow the process of decision tree. Decision tree is one of the methods that have been employed to predict the metabolic syndrome; with this method, we can extract features that are effective in predicting the metabolic syndrome. Decision tree C4.5 tools use the entropy equation for determining the tree nodes. If S contains positive and negative examples of a target concept, the entropy of S will be defined by relation (1) relative to this Boolean classification. P⊕ is the ratio of positive examples to all examples and P⊖ is the ratio of negative examples to all examples.
Formula 1
SVM
Another machine learning methods are SVM that can be used for classification and regression and recently has been widely used in the medical domain. Among them, studies in the following fields can be cited: diagnosis of heart diseases,20 predicting diabetes21 and diagnosis of breast cancer.7,22 Unlike the decision tree method, SVM is more a liked a black box, in other words, the knowledge derived from SVM is not directly sensible by professionals. However, SVM accuracy in classification has been caused its variety of applications. In this approach, each data is a vector where its and dimensions is equal to the number of features to be considered and SVM creates a hyperplane that separates samples of two categories. Such of the induced, created hyperplane has a maximum distance between the margin samples of between two classes. Figure 1 represents the solution for a two features problem.
Figure 1.
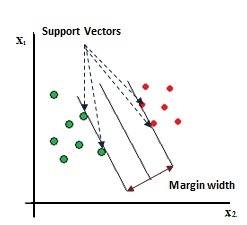
The induced line for the separation of positive and negative samples
Remember that SVM kernel maps the data that are linearly inseparable to a non-linear feature space and then induces a hyperplane between two classes.23 The advantage of this method is its implementation simplicity and, unlike the neural network does not get stuck in a local maximum.8 However, the only thing that should be carefully regarded is selecting the kernel type in this method.7 Type of selected kernel has a considerable impact on the prediction accuracy. The most commonly used kernels in SVM are the radial basis function (RBF), polynomial, sigmoid and linear. The C parameter must be set in all of them jointly. This parameter has an effect on the decision boundaries. As the value of C is increased, the distance between the boundaries of two classes is reduced. By changing C, a balance is established between the error and accuracy of the included hyperplane. Each kernel has its own parameters for setting which determines the degree of flexibility between the two classes. This study uses SVM method with polynomial kernel.
Balance in data set is another subject that impacts the performance of machine learning methods. In the real world, especially in the medical field, the number of positive samples is much less than the negative samples.24,25 On the other hand, in this field, accurate prediction and diagnosis of positive samples is very important. In medical issues, a high sensitivity is more important than high specificity. The lack of positive data causes the learned model is based on negative data which means they learn the classes containing negative data and ignore the minority positive samples. For example, in a dataset, if the number of positive samples would be 1%, the specificity and accuracy will be 100 and 99%, respectively, while it is the negative class that is learned. Instead of the positive class,25 one-way to fix this problem is to balance the given data set can be used. In this research, due to the unbalanced given data set Synthetic Minority Over-sampling Technique (SMOTE) method is used to balance the given data set. In this method, with the help of minima samples, a new synthetic data set is generated using the nearest neighbor.24
In general, there are several criteria to evaluate the success rate of a learning model. In this study, the criteria of sensitivity, specificity and accuracy are used. Furthermore, in this study, we have used 10-fold cross validation for performance evaluation of prediction metabolic syndrome.26
These criteria are calculated according to the values of the confusion matrix. The confusion matrix displays the results of predicting the classify problem samples compared to their original classes. In a problem with two classes including patient and healthy, there is a 2 × 2 confusion matrix. Four members of this matrix are numbers that show the true positive (TP), false positive (FP), true negative (TN), and false negative (FN). Criteria of sensitivity, specificity and accuracy are calculated using elements of the confusion matrix. Sensitivity as is shown in relation3 is an accuracy rate of the right diagnosis in patients with metabolic syndrome. Specificity as is shown in relation4 is the accuracy rate of right diagnosis of normal subjects. Accuracy as is shown in relation5 is the rate of the right diagnosis of metabolic syndrome and healthy subjects.
Formula 2
Formula 3
Formula 4
We use 10-fold cross validation approach to validate the performance of predictive models.14
The experiments of research are conducted using the WEKA open source software (Waikato Environment for Knowledge Analysis, University of Waikato, New Zealand).
Results
We used ICS data set to determine the input factors (gender, age, weight, BMI, WC, WHR, HC, physical activity, smoking history, hypertension, antihypertensive medication use, systolic BP, diastolic BP, fasting blood glucose, 2-hour blood glucose, TGs, total cholesterol, LDL, HDL-C, MCV and MCH) (Table 1). Details of ICS study were reported previously.27
Table 1.
Baseline subject characteristics in this study
| Characteristics | Total (n = 2107) Mean (Min-Max) |
|---|---|
| Age | 48.07 (34.0-86.0) |
| Weight | 67.97 (39.0-120.0) |
| BMI | 25.67 (14.96-39.9) |
| HC | 99.77 (53.0-143.0) |
| WC | 90.96 (52.0-131.0) |
| WHR | 91.16 (0.7-1.2) |
| SBP | 115.52 (75.0-200.0) |
| DBP | 75.27 (20.0-150.0) |
| FBS | 79.66 (41.0-298.0) |
| 2-HP | 97.22 (60.0-383.0) |
| HDL | 48.48 (25.0-79.0) |
| LDL | 125.70 (15.0-316.0) |
| TG | 164.11 (47.0-726.0) |
| TCH | 206.68 (76.0-450.0) |
| MCV | 86.63 (44.0-106.0) |
| MCH | 28.45 (0.0-42.3) |
BMI: Body mass index; HC: Hip circumference; WC: Waist circumference; WHR: Waist-to-hip ratio; SBP: Systolic blood pressure; DBP: Diastolic blood pressure; FBS: Fasting blood sugar; HDL: High density lipoprotein; LDL: Low-density lipoprotein; TG: Triglycerides; TCH: Total cholesterol; MCV: Mean corpuscular volume; MCH: Mean corpuscular hemoglobin; 2-HP: 2-hour postprandial blood sugar
The results of the experimenting of the decision tree algorithm and SVM on ISC data set is presented in table 2. According to table 2, the sensitivity of the decision tree and SVM are 0.337 and 0.320, respectively. These sensitivity values are very low for predicting, so the result is not acceptable.
Table 2.
The sensitivity, specificity and accuracy of the learning methods applied on the unbalanced Isfahan Cohort Study (ICS) data set
| Method | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
| Decision tree | 0.337 | 0.917 | 0.75273 |
| SVM | 0.320 | 0.934 | 0.76032 |
SVM: Support vector machine
The lack of sensitivity is due to the ICS unbalanced data set. Following the lack of positive samples compared to with the negative samples, the model learns healthy subjects instead of patient subjects. To vanish this problem we used, SMOTE method to balance ICS data set. We applied decision tree and SVM method on the balanced ICS data set. The results are presented in table 3.
Table 3.
The sensitivity, specificity and accuracy of the learning methods applied on the balanced Isfahan Cohort Study (ICS) data set
| Method | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
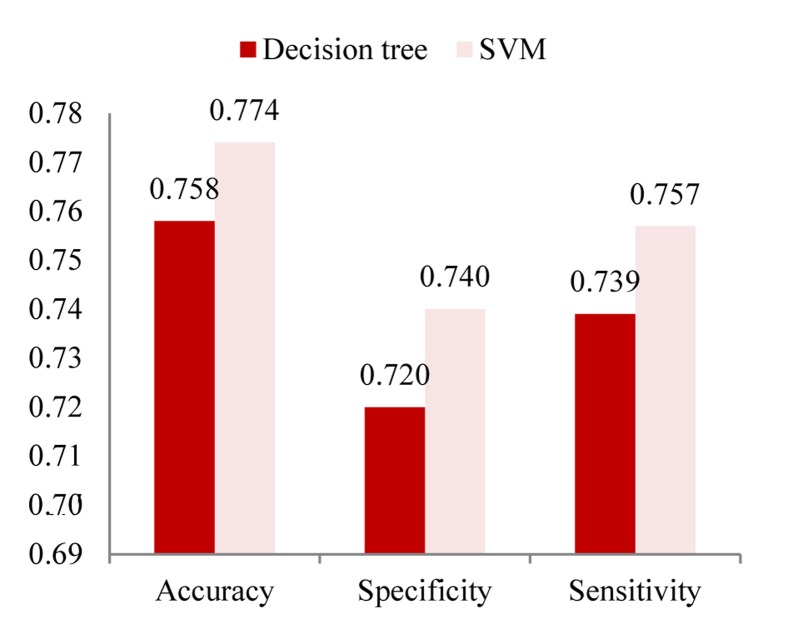
| Decision tree | 0.758 | 0.72 | 0.739 |
| SVM | 0.774 | 0.74 | 0.757 |
SVM: Support vector machine
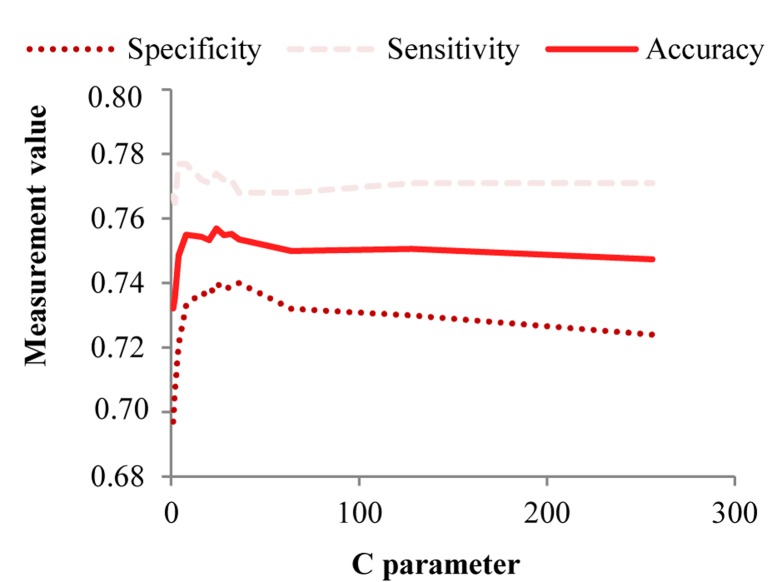
The value of the parameter C [2(−5), 2(−3), …, 210] with polynomial degree 2, 3 and 4 are used in SVM method. To determine the optimal value C, values between two consecutive C are tested again, after obtaining the highest value of C. Finally, the highest accuracy is obtained at C = 24. This accuracy is obtained with polynomial kernel of degree 4. Changes in accuracy, sensitivity and specificity based on C, is presented in figure 2.
Figure 2.
The sensitivity, specificity and accuracy of support vector machine, based on C in polynomial kernel of degree 4 applied on the balanced Isfahan cohort study data set
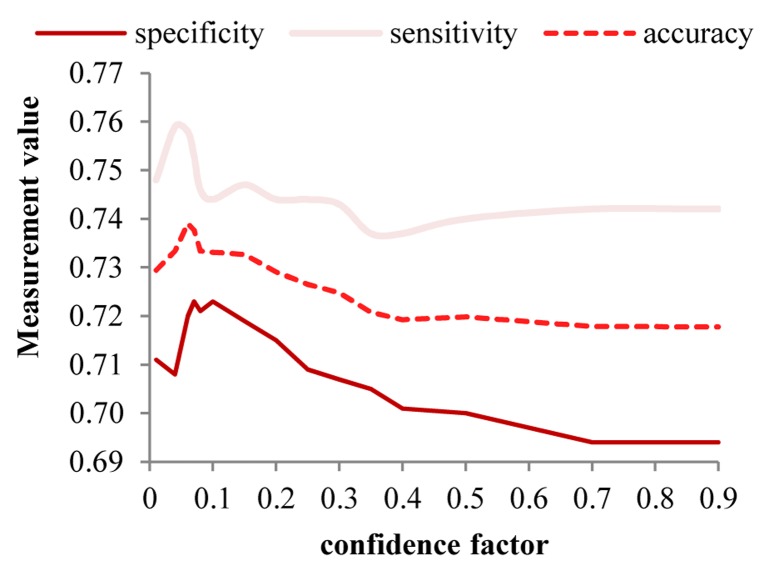
The decision tree method is studied for the confidence factor between 0.01 and 0.99. The best accuracy is obtained with a confidence factor equal to 0.06. Figure 3 shows the effectiveness of in sensitivity, specificity and accuracy of decision tree C4.5 with respect to changes of the confidence factor.
Figure 3.
The sensitivity, specificity and accuracy of the decision tree C4.5 with different on faience factor applied on the balanced Isfahan Cohort Study data set
In this study, we have used four methods for performance evaluation of prediction metabolic syndrome. These methods are classification accuracy, analysis of sensitivity and specificity, and 10-fold cross validation.
Comparison of the decision tree and SVM methods based on figure 4 shows that SVM has better accuracy and efficiency than the decision tree. However, the advantages of decision tree in comparison with SVM are a high speed and low-cost learning and prediction. The decision tree generated in this study shows that TG is the main prognostic factor of metabolic syndrome. A combination of TGs + BMI is an accurate predictor for predicting. Moreover, according to the induced decision tree, a person with TGs > 223 and BMI ≥ 24.35, with probability 85%, will suffer the metabolic syndrome.
Figure 4.
The sensitivity, specificity and accuracy of the learning methods applied on the balanced Isfahan cohort study data set SVM: Support vector machine
Discussion
The purpose of this study is comparing the prediction of metabolic syndrome using decision tree and SVM methods. The decision tree method is chosen because the induced tree is understandable and shows the process of prediction. According to studies, the method of SVM also has a high accuracy. The most significant feature in the decision tree method is the tree root. In the constructed decision tree, TGs represent the root. Therefore, TGs is the most important feature for classifying the metabolic syndrome. The derived results are corresponded with research by Worachartcheewan et al.10 as “identification of metabolic syndrome using decision tree analysis.” According to this study, participants had BMI ≥ 25. The results of this study show that TGs is an important feature for the diagnosis of metabolic syndrome and combining (TG + BP) and (TG + BP + glucose) are as an accurate predictor of metabolic syndrome. In another study by Lemieux et al.,28 it is found that TGs is the most important feature for predicting the metabolic syndrome.
However, it is noted in other studies, using SVM is one of the most successful methods of classification. In our study, SVM method shows that it is more efficient than a decision tree method based on the criteria of sensitivity, specificity and accuracy. Note that, previously, SVM method is not used to predict metabolic syndrome.
Conclusion
One of the main objectives in the field of health is diseases prevention. One of the most effective actions to prevent diseases is prediction of disease in susceptible individuals. One of the effective methods for prediction of diseases is the use of methods of statistical inference and artificial intelligence. Statistical methods do not provide acceptable results in nonlinear and complex problems where features are dependent.3 In recent years, machine learning techniques are frequently used in medical issues, and they acceptable results in predicting and diagnosing of diseases. Since to the metabolic syndrome is a precursor to heart disease and diabetes and increases the risk of these diseases so Predicting and analyzing the characteristics of this disorder, it can be an effective step in preventing these diseases. According to the results obtained in our study, increasing of TGs, BP, and BMI are regarded as the most important causes of this disorder. These factors are easily modifiable by changes in the lifestyle and the use of healthy eating.
Furthermore, due to the high efficiency of SVM method, this method can be used for prediction and generality issues related to the classification in the medical domain. According to the results of the two learning candidate methods, in other to have a more accurate prediction for new instances of the disorder we recommend to use SVM method. Prediction of ATPIII based metabolic syndrome using machine learning.
Acknowledgments
This article is the result of a master’s thesis on the topic of “Prediction of ATPIII based metabolic syndrome using machine learning” in 2014. The authors are grateful to the Cardiovascular Research Center, Isfahan University of Medical Sciences for providing the ICS data set, Dr. Mohammad Talaei, Head cohort unit, for collaboration.
Footnotes
Conflicts of Interest
Authors have no conflict of interests.
REFERENCES
- 1.Karabatak M, Ince MC. An expert system for detection of breast cancer based on association rules and neural network. Expert Syst Appl. 2009;36(2, Part 2):3465–9. [Google Scholar]
- 2.Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7(6):673–9. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirose H, Takayama T, Hozawa S, Hibi T, Saito I. Prediction of metabolic syndrome using artificial neural network system based on clinical data including insulin resistance index and serum adiponectin. Comput Biol Med. 2011;41(11):1051–6. doi: 10.1016/j.compbiomed.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 4.International Diabetes Federation. The IDF consensus worldwide definition of the metabolic syndrome [Online]. 2006. Available from: URL: http://www.idf.org/webdata/docs/MetS_def_update2006.pdf.
- 5.Zabetian A, Hadaegh F, Azizi F. Prevalence of metabolic syndrome in Iranian adult population, concordance between the IDF with the ATPIII and the WHO definitions. Diabetes Res Clin Pract. 2007;77(2):251–7. doi: 10.1016/j.diabres.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Esteghamati A, Ashraf H, Rashidi A, Meysamie A. Waist circumference cut-off points for the diagnosis of metabolic syndrome in Iranian adults. Diabetes Res Clin Pract. 2008;82(1):104–7. doi: 10.1016/j.diabres.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Liu HX, Zhang RS, Luan F, Yao XJ, Liu MC, Hu ZD, et al. Diagnosing breast cancer based on support vector machines. J Chem Inf Comput Sci. 2003;43(3):900–7. doi: 10.1021/ci0256438. [DOI] [PubMed] [Google Scholar]
- 8.Doniger S, Hofmann T, Yeh J. Predicting CNS permeability of drug molecules: comparison of neural network and support vector machine algorithms. J Comput Biol. 2002;9(6):849–64. doi: 10.1089/10665270260518317. [DOI] [PubMed] [Google Scholar]
- 9.Yu Y, Chen S, Wang LS, Chen WL, Guo WJ, Yan H, et al. Prediction of pancreatic cancer by serum biomarkers using surface-enhanced laser desorption/ionization-based decision tree classification. Oncology. 2005;68(1):79–86. doi: 10.1159/000084824. [DOI] [PubMed] [Google Scholar]
- 10.Worachartcheewan A, Nantasenamat C, Isarankura-Na-Ayudhya C, Pidetcha P, Prachayasittikul V. Identification of metabolic syndrome using decision tree analysis. Diabetes Res Clin Pract. 2010;90(1):e15–e18. doi: 10.1016/j.diabres.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Talaei M, Sarrafzadegan N, Sadeghi M, Oveisgharan S, Marshall T, Thomas GN, et al. Incidence of cardiovascular diseases in an Iranian population: the Isfahan Cohort Study. Arch Iran Med. 2013;16(3):138–44. [PubMed] [Google Scholar]
- 12.Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112(17):2735–52. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 13.Azizi F, Khalili D, Aghajani H, Esteghamati A, Hosseinpanah F, Delavari A, et al. Appropriate waist circumference cut-off points among Iranian adults: the first report of the Iranian National Committee of Obesity. Arch Iran Med. 2010;13(3):243–4. [PubMed] [Google Scholar]
- 14.Witten LH, Frank E. Data mining: practical machine learning tools and techniques. Burlington, MA: Morgan Kaufmann; 2005. [Google Scholar]
- 15.Jerez-Aragones JM, Gomez-Ruiz JA, Ramos-Jimenez G, Munoz-Perez J, Alba-Conejo E. A combined neural network and decision trees model for prognosis of breast cancer relapse. Artif Intell Med. 2003;27(1):45–63. doi: 10.1016/s0933-3657(02)00086-6. [DOI] [PubMed] [Google Scholar]
- 16.Palaniappan S, Awang R. Intelligent heart disease prediction system using data mining techniques.; Proceedings of the IEEE/ACS International Conference on Computer Systems and Applications; 2008 Mar 31-Apr 4; Doha, Qatar. [Google Scholar]
- 17.Anbananthen KS, Sainarayanan G, Chekima A, Teo J. Artificial neural network tree approach in data mining. Malaysian Journal of Computer Science. 2007;20(1):51–62. [Google Scholar]
- 18.Murthy SK. Automatic Construction of Decision Trees from Data: A Multi-Disciplinary Survey. Data Min Knowl Discov. 1998;2(4):345–89. [Google Scholar]
- 19.Alpaydin E. Introduction to machine learning. 2nd. Cambridge, MA: MIT Press; 2010. [Google Scholar]
- 20.Yan WW, Shao HH. Application of support vector machines and least squares support vector machines to heart disease diagnoses. Control and Decision. 2003;18(3):358–60. [Google Scholar]
- 21.Yu W, Liu T, Valdez R, Gwinn M, Khoury MJ. Application of support vector machine modeling for prediction of common diseases: the case of diabetes and pre-diabetes. BMC Med Inform Decis Mak. 2010;10:16. doi: 10.1186/1472-6947-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akay MF. Support vector machines combined with feature selection for breast cancer diagnosis. Expert Syst Appl. 2009;36(2):3240–7. [Google Scholar]
- 23.Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16(10):906–14. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- 24.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: Synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–57. [Google Scholar]
- 25.Zhang D, Liu W, Gong X, Jin H. A Novel Improved SMOTE Resampling Algorithm Based on Fractal. Journal of Computational Information Systems. 2011;7(6):2204–11. [Google Scholar]
- 26.Alpaydin E. Introduction to machine learning. 3rd. Cambridge, MA: MIT Press; 2014. [Google Scholar]
- 27.Sarrafzadegan N, Talaei M, Sadeghi M, Kelishadi R, Oveisgharan S, Mohammadifard N, et al. The Isfahan cohort study: rationale, methods and main findings. J Hum Hypertens. 2011;25(9):545–53. doi: 10.1038/jhh.2010.99. [DOI] [PubMed] [Google Scholar]
- 28.Lemieux I, Poirier P, Bergeron J, Almeras N, Lamarche B, Cantin B, et al. Hypertriglyceridemic waist: a useful screening phenotype in preventive cardiology? Can J Cardiol. 2007;23(Suppl B):23B–31B. doi: 10.1016/s0828-282x(07)71007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]