Abstract
Fe-S cluster assembly is an essential process for all cells. Impairment of Fe-S cluster assembly creates diseases in diverse and surprising ways. In one scenario, the loss of function of lipoic acid synthase, an enzyme with Fe-S cluster cofactor in mitochondria, impairs activity of various lipoamide-dependent enzymes with drastic consequences for metabolism. In a second scenario, the heme biosynthetic pathway in red cell precursors is specifically targeted, and iron homeostasis is perturbed, but lipoic acid is unaffected. In a third scenario, tRNA modifications arising from action of the cysteine desulfurase and/or Fe-S cluster proteins are lost, which may lead to impaired protein synthesis. This can then result in cancer, neurologic dysfunction or type 2 diabetes.
Iron-sulfur (Fe-S) cluster assembly
Fe-S clusters are essential cofactors of proteins involved in numerous cellular processes. In eukaryotic cells, Fe-S proteins perform critical functions both inside and outside mitochondria, and their assembly is mediated by multi-subunit machineries termed the ISC (Iron Sulfur Cluster) and CIA (Cytoplasmic Iron-Sulfur Protein Assembly) [for recent reviews [1–3]. The ISC is found primarily in mitochondria in yeast, although in other eukaryotes including humans, ISC components are also found in the cytoplasm and nucleus [2]. The core ISC assembly complex contains the NFS1 cysteine desulfurase, an accessory protein ISD11, frataxin, and the scaffold protein ISCU. NFS1 binds the substrate cysteine and forms a persulfide on its active site. During this process, the enzyme likely undergoes at least two conformational changes for optimum activity. One change is mediated by frataxin interaction that exposes the “buried” substrate-binding sites and promotes substrate binding. A second change is mediated by ISD11 interaction that brings the bound substrate cysteine and the active site cysteine in proximity for persulfide formation [4]. Subsequently, the persulfide sulfur is transferred from NFS1 to ISCU, where it combines with iron to form [2Fe-2S] cluster intermediates. The NADPH-dependent ferredoxin reductase - ferredoxin redox couple supplies reducing equivalents that are needed for formation of the intermediate. The cluster intermediates are then transferred to apoproteins by the action of HSPA9-HSC20 chaperones. The transfer process also may involve a glutathione-bound intermediate on a monothiol glutaredoxin, GLRX5. Specific subsets of Fe-S cluster proteins are targeted by GLRX5, and allele effects have been noted on target selection [5]. A number of other proteins, including IBA57, NFU1 and BOLA3, are involved in modification of the [2Fe-2S] cluster intermediate into a [4Fe-4S] cluster intermediate and/or in targeting the preformed clusters to specific recipients. The mitochondrial ISC machinery must interface with the CIA machinery in the cytoplasm and nucleus. Interestingly, the CIA machinery bears some similarities to the mitochondrial ISC machinery. For example, scaffold and chaperone components (CFD1/NBP35) have been identified. Likewise, the NADPH-dependent NDOR1 and CIAPIN1 reductase complex is also involved in cytosolic Fe-S cluster synthesis [3]. This article focuses on the roles of Fe-S proteins in three important mitochondrial/cellular functions – biosynthesis of lipoamide that serves as a cofactor for several metabolic enzymes in mitochondria, maintenance of iron homeostasis in mitochondria, and modifications of mitochondrial and cytoplasmic transfer RNAs (tRNAs) required for efficient protein synthesis.
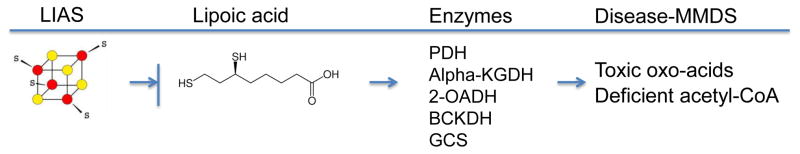
A. Lipoamide cofactor and its biosynthesis
The lipoamide cofactor consists of lipoic acid (a thiol derivative of octanoic acid) bound by an amide linkage to the epsilon amino group of a conserved lysine in a distinct lipoylation domain of some mitochondrial proteins. The cofactor can cycle between a reduced state in which the sulfhydryls are exposed and an oxidized state in which the thiols form a five member ring available for substrate binding. When bound to substrate, the lipoamide adduct acts as a 14Å swinging arm, enabling direct transfer of substrates between enzyme subunits [6]. The lipoamide-requiring enzymes include pyruvate dehydrogenase (PDH), α–ketoglutarate dehydrogenase (α–KGDH), 2-oxoadipate dehydrogenase (2-OADH), branched chain ketoacid dehydrogenase (BCKDH), and glycine cleavage system (GCS) [7] and Fig. 1.
Fig. 1.
Linkage of Fe-S cluster assembly defects and MMDS multiple mitochondrial dysfunction syndrome. Details are included in the text.
The biosynthesis of lipoic acid enzymes takes place in mitochondria in several steps. The precursor octanoic acid bound by the acyl carrier protein is delivered to recipient proteins and covalently attached by the lipoyltransferase 2 (LIPT2) enzyme to lysine residues on special lipoyl domains. The bound octanoic acid is then modified via the action of a unique mitochondrial Fe-S cluster enzyme, lipoic acid synthase or LIAS [7]. This enzyme carries two critical [4Fe-4S] clusters, an activating cluster and an auxiliary cluster [8]. The proposed mechanism involves interaction of the enzyme with the substrate, the lipoyl carrier protein with bound octanoic acid. The principal cluster mediates activation of adenosyl methionine to a free radical form able to sequentially abstract hydrogens from C6 and then C8 of the substrate. The auxiliary cluster is utilized as a sulfur donor, contributing two equivalents of S2− and “wasting” four equivalents of iron. The auxiliary [4Fe-4S] cluster must therefore be regenerated to process another molecule of substrate [6]. Thus even mild decrease in the activity or flux of the Fe-S cluster assembly system will target LIAS specifically, leading to LIAS deficiency and lipoamide deficiency with drastic metabolic consequences (see below and Fig. 1).
Multiple mitochondrial dysfunction syndrome
A number of devastating inherited metabolic diseases have recently been described that are characterized by lipoamide deficiency [7, 9]. These diseases are referred to as multiple mitochondrial dysfunction syndrome. The disease manifestations have considerable overlap, but interestingly, a number of genes/proteins involved in general Fe-S cluster assembly have been implicated, including IBA57 [10], NFU1 [11], BOLA3 [5] and GLRX5 [5]. Primary deficiencies of LIAS also share many of these features, such as non-ketotic hyperglycemia, seizures, brain damage, elevated plasma glycine and defective mitochondrial energy metabolism [5]. The constellation of symptoms and findings derive from accumulation of toxic oxo-acids on the one hand or energy deficiency on the other hand [7]. Mitochondria are also globally defective. Toxic oxo-acids, especially glycine, which must be handled by the glycine cleavage enzyme but also ketoacids and lactate, accumulate in the plasma and central nervous system. Here they may act as neurotoxins producing various neurologic symptoms such as intractable seizures, encephalopathy, hypotonia, brain malformations, and developmental regression. Non-ketotic hyperglycemia may result from pyruvate dehydrogenase deficiency. On the other hand, lack of energy as may result from decreased acetyl-CoA production and decreased metabolic flux through the mitochondrial electron transport chain may produce a broad spectrum of disease symptoms that alter functions of skeletal muscle (myopathy), heart (cardiomyopathy), and lungs (respiratory insufficiency) [7, 9].
B. Mitochondrial iron homeostasis - Defects in Fe-S cluster biogenesis causing sideroblastic anemia
Sideroblastic anemia is a hematologic syndrome characterized by iron accumulation in mitochondria of red cell precursors in the bone marrow leading to decreased viability of these precursors and anemia [12, 13]. Sideroblastic anemia reflects a problem with iron utilization for heme synthesis. In some cases, its pathogenesis has been linked to deficiency of Fe-S cluster synthesis. The manner in which Fe-S cluster deficiency causes mitochondrial iron accumulation is not entirely clear [14]. However, critical Fe-S cluster proteins have been implicated in mediating or regulating heme synthesis. For example, ferrochelatase, the enzyme that inserts iron into porphryin in mitochondria is a [2Fe-2S] cluster protein, and the cluster is necessary for the activity and stability of the protein, although it is not directly involved in the enzymatic activity [15]. The iron regulatory protein 1 (IRP1) in cytoplasm is another Fe-S cluster protein that acts as a regulatory switch for the heme/porphyrin biosynthetic pathway. When it assembles with a [4Fe-4S cluster] it functions as the cytosolic aconitase, but when Fe-S clusters (or iron) is scarce, the apoprotein binds to the 5′ regulatory stem loop in the amino terminal 5′ UTR of aminolevulinic acid synthase mRNA, repressing translation of this key rate-limiting enzyme for porphyrin synthesis [16]. Interference with Fe-S cluster biogenesis then may abrogate heme synthesis by blocking porphyrin synthesis at the first and last steps. Genes implicated in Fe-S cluster assembly and causing sideroblastic anemia include GLRX5 and ABCB7; SF3B1 has been linked to aberrant splicing of ABCB7 mRNA [17].
GLRX5 alleles, one gene and two diseases
An individual carrying inherited compound heterozygous mutations of GLRX5 L148S and K101Q was found to develop sideroblastic anemia [18]. A biochemical analysis of the alleles was performed by expressing them from plasmids in a K562 hematopoietic cell line with CRISPR/Cas9 inactivation of the native GLRX5 [19]. In the cell line, the L148S or coexpressed L148S and K101Q alleles were associated with Fe-S cluster deficient phenotypes: IRP1 was lacking its Fe-S cluster and c-aconitase activity was markedly down. The Fe-S cluster containing protein ferrochelatase was also decreased in amount. The iron-associated phenotypes apparent in these cells included an increase in transferrin receptor levels, increased cellular and mitochondrial iron, and decreased total heme levels [19], consistent with the sideroblastic anemia phenotype. However, not all Fe-S cluster proteins were deficient, and succinate dehydrogenase was normal in spite of the fact that it requires three different Fe-S cluster cofactors – [2Fe-2S], [3Fe-4S] and [4Fe-4S] – for its activity. In addition, lipoic acid dependent enzymes, pyruvate dehydrogenase and α-ketoglutarate dehydrogenase, were entirely normal, suggesting that lipoate formation via the FeS cluster-requiring enzyme LIAS was preserved [19].
A different homozygous mutant allele of GLRX5, K51del, was identified in an individual with an inherited metabolic syndrome. The clinical features included early age of onset at 3 years, non-ketotic hyperglycemia, psychomotor retardation, and glycine accumulation in blood and cerebral spinal fluid [5]. The latter probably resulted from deficient glycine cleavage enzyme activity, a lipoic acid-dependent function. Other lipoic acid-dependent enzymes, PDH and α–KGDH, were also very deficient, probably as a result of loss of activity of the Fe-S cluster enzyme, LIAS [7]. However, in the engineered K562 cells expressing only the K51del allele, cytoplasmic and mitochondrial aconitases and ferrochelatase were unaffected [19]. Accordingly in the patient with the K51del allele and inherited metabolic syndrome, the blood picture was entirely normal and sideroblastic changes were not seen [5].
ABCB7 involvement in inherited and acquired sideroblastic anemia
ABCB7 is an ATP-dependent mitochondrial transporter implicated in Fe-S cluster assembly [3]. The orientation of the transporter in the mitochondrial inner membrane with its ATP and substrate-binding sites in the mitochondrial interior suggests that it functions as an exporter [20]. The precise substrate is still unidentified, but likely candidates include sulfur species such as polysulfides or glutathione derivatives [21]. Biochemical studies suggest that NFS1/ISD11, the mitochondrial cysteine desulfurase, is required to generate the substrate from cysteine. Subsequent transfer of the substrate into the cytoplasm via ABCB7 may be necessary for Fe-S cluster assembly on cytoplasmic Fe-S cluster substrates such as IRP1. Inherited mutations in ABCB7 have been linked to XLSA-A, X-linked sideroblastic anemia with cerebellar ataxia [22]. The male predominance of the disease is consistent with the X-chromosomal location of the ABCB7 gene. The neurologic features are more striking than the hematologic features, and the cerebellar atrophy and motor impairment present early on, whereas the sideroblastic anemia presents later and is relatively mild [23]. The mechanism by which the ABCB7 deficit causes iron homeostatic changes leading to sideroblastic anemia probably involves defective formation of a regulatory Fe-S cluster outside mitochondria but the identity of this putative Fe-S cluster target is unknown.
SF3B1 involvement in sideroblastic anemia and myelodysplasia
SF3B1 encodes an essential component of the spliceosome [24]. The protein is involved in RNA splicing, mediating recognition of the branch point sequence and selection of the 3′ splice site. SF3B1 hotspot mutations are neomorphic and induce aberrant 3′ splice site selection [17]. Thus, it is the K700E allele of SF3B1, and not loss-of-function mutations, that have been strongly associated with the development of sideroblastic anemia [25]. Apparently this allele leads to altered branch point selection and altered splicing of a large number of target genes, including heme biosynthetic enzymes, cell cycle enzymes and DNA repair enzymes. Most interestingly, SF3B1 with the K700E allele results in the aberrant splicing of the ABCB7 mRNA, introducing a frame shift at the 3′ splice site. The frame shift in turn results in nonsense mediated decay of the mRNA and downregulation of expression [17]. The effects of ABCB7 downregulation on Fe-S cluster assembly outside mitochondria are likely to be similar to what occurs in the inherited XLSA-A syndrome. Various features of sideroblastic anemia result (e.g. heme deficiency, iron misregulation, mitochondrial iron accumulation and ineffective erythropoiesis), although the causal link with these outcomes has not yet been directly demonstrated.
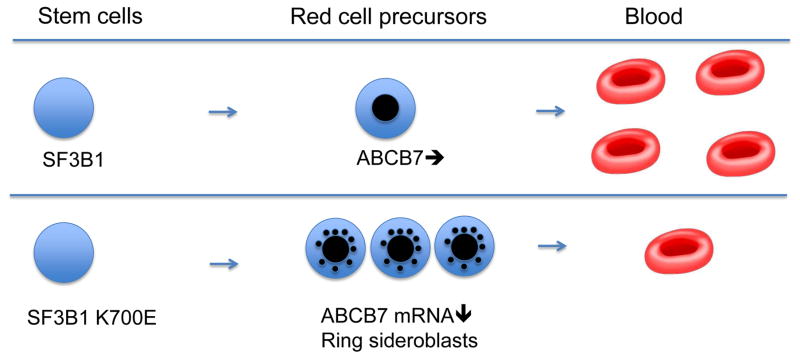
In the SF3B1 associated sideroblastic anemia, as opposed to the inherited form, the abnormalities are confined to hematopoietic cells [25]. Sideroblastic anemia is considered to be a variety of myelodysplasia, an acquired bone marrow disease caused by somatic genetic changes in a clone or clones of hematopoietic stem cells[26]. These clonally derived cells have a survival advantage in the bone marrow, but as they mature into precursors for the different blood lineages, they die prematurely (ineffective hematopoiesis) (Fig. 2). In some cases, SF3B1 mutations are associated with additional genetic changes that give rise to other cytopenias or myeloproliferative features (12).
Fig. 2.
Schematic showing possible causation of acquired sideroblastic anemia. Upper panel: normal hematopoiesis. Lower panel: SF3B1 K700E allele in a bone marrow stem cell causes altered splicing of ABCB7, non-sense mediated decay, and ring sideroblast formation in red cell precursors. Red cell precursors expand in the bone marrow but viable circulating red cells are decreased (ineffective erythropoiesis).
C. Fe-S clusters and tRNA modifications
tRNAs are fundamental components of the protein-synthesizing machinery. Many tRNAs contain conserved nucleotide modifications, and these modifications are often required for accurate and efficient recognition of codons in mRNA, maintenance of proper reading frame, and/or tRNA structural stability. It is therefore not surprising that defects in tRNA modifications are associated with numerous diseases including cancer, neurological disorders, and type 2 diabetes [27–29].
Many tRNAs have modifications at position 34 corresponding to the wobble position of the anticodon, and at position 37 that is adjacent to the 3′-position of the anticodon. For example, cytoplasmic tRNAs are usually modified by the covalent addition of methoxycarbonylmethyl (mcm) or carbamoylmethyl (ncm) to the 5′ carbon of the wobble base uridine (U34). A subset of these tRNAs - tRNALys (UUU), tRNAGlu (UUC), and tRNAGln (UUG) – the U34 base is further decorated with a thio group, replacing oxygen at 2′ position [30]. In Saccharomyces cerevisiae, loss of thiolation does not eliminate mcm5 modification, and cells lacking either mcm5 or s2 are viable. However, simultaneous loss of mcm5 and s2 is lethal [31]. Other derivatives such as 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U) and 5-taurinomethyl-2-thiouridine (τm5s2U) are found in yeast and mammalian mitochondrial tRNAs, respectively [30].
The modifications at position 37 often form hyper-modified nucleosides such as N6-isopentenyladenosine (i6A) and its 2-methylthio derivative (ms2i6A), N6-threonyl-carbamoyladenosine (t6A) and its 2-methylthio derivative (ms2t6A), and wybutosine (yW). Interestingly, several enzymes involved in these modifications contain Fe-S clusters (see below and Table 1). Furthermore, like in the case for Fe-S clusters and lipoic acid, the source of sulfur for tRNA thiolation is also the persulfide sulfur formed on the NFS1 cysteine desulfurase. For tRNA thiolation, methylthiotransferases are responsible for the s2 modification. However, unlike in the case for lipoic acid biosynthesis, the [4Fe-4S] clusters of methylthiotransferases do not appear to be sacrificed as sulfur donors [32].
Table 1.
tRNA modifications mediated by Fe-S cluster enzymes, and defects associated with impaired modifications
| Enzyme | [4Fe-4S] clusters | Localization | tRNA | Substrate | Product | Abnormalities associated with impaired tRNA modifications | Refs. |
|---|---|---|---|---|---|---|---|
| TYW1 | Two | Cytoplasm | tRNAPhe (position 37) | m1G | imG-14 | More prone to frame-shifting. Impaired modifications found in HIV-infected cells, Ehrlich ascites tumor, and neuroblastoma cells | 32–36 |
| CDKAL1 | Two | Associated with ER | tRNALys (position 37) | t6A | ms2t6A | Pancreatic islet hypertrophy, impaired blood glucose control, and increased risk of type 2 diabetes in humans | 37, 38 |
| CDK5RAP1 | Two | Mitochondria | Various (position 37) | i6A | ms2i6A | Defective mitochondrial protein synthesis, respiratory deficiency, and myopathy | 39 |
| ELP3 | One | Cytoplasm | Various (position 34) | U | cm5U | Neuronal defects observed in mouse, zebrafish, Drosophila, and C. elegans; increased risk of ALS in humans | 26–28, 41–43 |
TYW1 (tRNA-yW synthesizing protein 1)
TYW1 is found in cytoplasm; it contains two [4Fe-4S] clusters and is involved in wybutosine (yW) biosynthesis. yW biosynthesis occurs by a series of post-transcriptional modifications of the genetically encoded guanine base at position 37 of eukaryotic tRNAPhe. The initial steps involve methylation of N1 at G37 to form N-methylguanosine (m1G), which is then converted to 4-demethylwyosine (imG-14) by TYW1. Further modifications of imG-14 to yW require several other enzymes [33, 34] (Table 1).
yW is a tricyclic nucleoside that contributes to translational fidelity. It stabilizes the codon anticodon pairing through an increased base-stacking interaction that helps in maintaining the reading frame [35]. Lack of yW may therefore cause increased frame-shifting during translation [36]. For example, yW is absent in tRNAPhe in cells infected with human immunodeficiency virus (HIV). The lack of yW in these cells causes a greatly enhanced -1 frame-shifting, allowing the HIV virus to produce the reverse transcriptase that is essential for virus replication and activity [37]. The lack of yW in tRNAPhe could be specifically due to malfunction of TYW1 in HIV-infected cells, although this remains to be established.
CDKAL1 (Cdk5 regulatory subunit associated protein 1-like 1)
CDKAL1 is a methylthiotransferase that contains two [4Fe-4S] clusters. The enzyme catalyzes the 2-methylthio (ms2) modification of N6-threonyl-carbamoyladenosine (t6A) in tRNA to 2-methylthio-N6-threonyl-carbamoyladenosine (ms2t6A) at position 37 of tRNALys (UUU). Lack of this ms2 modification in tRNALys (UUU) leads to abnormal proinsulin synthesis, ultimately causing impaired glucose metabolism and development of type 2 diabetes [38, 39] (Table 1).
CDK5RAP1 (Cdk5 regulatory subunit-associated protein 1)
It is a methylthiotransferase in mammalian mitochondria. It is homologous to CDKAL1, and like CDKAL1, CDK5RAP1 also contains two [4Fe-4S] clusters required for ms2 group insertion. CDK5RAP1 specifically converts N6-isopentenyladenosine (i6A) to 2-methylthio-N6-isopentenyladenosine (ms2i6A) at A37 of tRNAs for Phe, Ser, Trp, and Tyr in mammalian mitochondria (Table 1). In CDK5RAP1 knockout mice, a deficiency in the ms2 modification of i6A resulted in impaired mitochondrial protein synthesis and respiratory defects. Furthermore, the knockout mice exhibited accelerated myopathy and cardiac dysfunction particularly under stressed conditions [40]. Interestingly, the ms2 modification was also found to be reduced in patients with mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) carrying the A3243G mutation in mitochondrial tRNALeu. However, mt-tRNALeu does not contain an ms2 modification. Instead, tRNALeu carrying the A3243G mutation is deficient in taurine modification and this defect appears to be the primary cause of MELAS [41]. The myopathy in MELAS could be due to combined effects of decoding errors occurring at multiple codons.
ELP3 (Elongator protein 3)
The Elongator complex has been shown to associate with actively transcribing RNA polymerase II. The eukaryotic Elongator complex consists of six Elongator protein subunits (ELP1–ELP6), and the complex is conserved from yeast to humans. Defects in the ELP complex elicit pleiotropic phenotypes involving a variety of cellular processes that include histone acetylation, transcription, and exocytosis. Remarkably, all of these phenotypes appear to originate primarily from the lack of modifications at position 34 of some tRNAs. In fact, all of the six subunits of the ELP complex (together with the Killer toxin-insensitive proteins) are required for formation of mcm5U, mcm5s2U, and ncm5U at the wobble position [27–29] (Table 1).
Among the components of the Elongator complex, ELP3 is generally considered the catalytic subunit. Only ELP3 has a [4Fe-4S] cluster, and it has two enzymatic domains – a radical S-adenosylmethionine (SAM) near the N-terminus, and a histone acetyltransferase (HAT) at the C-terminus of the protein. Interestingly, both enzymatic activities are necessary for ELP3 functions [27, 42]. In an association study, allelic variants of ELP3 identified in three human populations were found to be associated with amyotrophic lateral sclerosis (ALS) [43]. Likewise, neuronal defects have also been observed in mouse, flies, zebrafish, and C. elegans with impaired ELP3 function [27, 29, 44].
Acknowledgments
The research in our laboratories is supported by National Institutes of Health Grant R37DK53953 to AD, and National Institutes of Health Grant R01GM107542 and a grant from the Friedreich’s Ataxia Research Alliance to AD and DP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1•.Pain D, Dancis A. In: Fe-S cluster assembly and regulation in yeast In Iron-Sulfur Clusters in Chemistry and Biology. Rouault TA, editor. de Gruyter; Berlin: 2014. pp. 367–410. Review of yeast pathways. [Google Scholar]
- 2•.Rouault TA. Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat Rev Mol Cell Biol. 2015;16:45–55. doi: 10.1038/nrm3909. Review of mammalian Fe-S biogenesis. [DOI] [PubMed] [Google Scholar]
- 3•.Lill R, Dutkiewicz R, Freibert SA, Heidenreich T, Mascarenhas J, Netz DJ, Paul VD, Pierik AJ, Richter N, Stumpfig M, Srinivasan V, Stehling O, Muhlenhoff U. The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins. Eur J Cell Biol. 2015;94:280–291. doi: 10.1016/j.ejcb.2015.05.002. Review of role of mitochondria. [DOI] [PubMed] [Google Scholar]
- 4.Pandey A, Gordon DM, Pain J, Stemmler TL, Dancis A, Pain D. Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly. J Biol Chem. 2013;288:36773–36786. doi: 10.1074/jbc.M113.525857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Baker PR, 2nd, Friederich MW, Swanson MA, Shaikh T, Bhattacharya K, Scharer GH, Aicher J, Creadon-Swindell G, Geiger E, MacLean KN, Lee WT, Deshpande C, Freckmann ML, Shih LY, Wasserstein M, Rasmussen MB, Lund AM, Procopis P, Cameron JM, Robinson BH, Brown GK, Brown RM, Compton AG, Dieckmann CL, Collard R, Coughlin CR, 2nd, Spector E, Wempe MF, Van Hove JL. Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain. 2014;137:366–379. doi: 10.1093/brain/awt328. Characterization of novel metabolic syndrome and its association with global defects in Fe-S cluster assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanz ND, Booker SJ. The role of iron-sulfur clusters in the biosynthesis of the lipoyl cofactor. In: Rouault TA, editor. Iron-Sulfur Clusters in Chemistry and Biology. de Gruyter; Berlin: 2014. pp. 211–238. [Google Scholar]
- 7.Mayr JA, Feichtinger RG, Tort F, Ribes A, Sperl W. Lipoic acid biosynthesis defects. J Inherited Metab Dis. 2014;37:553–563. doi: 10.1007/s10545-014-9705-8. [DOI] [PubMed] [Google Scholar]
- 8.Harmer JE, Hiscox MJ, Dinis PC, Fox SJ, Iliopoulos A, Hussey JE, Sandy J, Van Beek FT, Essex JW, Roach PL. Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem J. 2014;464:123–133. doi: 10.1042/BJ20140895. [DOI] [PubMed] [Google Scholar]
- 9.Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Ajit Bolar N, Vanlander AV, Wilbrecht C, Van der Aa N, Smet J, De Paepe B, Vandeweyer G, Kooy F, Eyskens F, De Latter E, Delanghe G, Govaert P, Leroy JG, Loeys B, Lill R, Van Laer L, Van Coster R. Mutation of the iron-sulfur cluster assembly gene IBA57 causes severe myopathy and encephalopathy. Hum Mol Genet. 2013;22:2590–2602. doi: 10.1093/hmg/ddt107. [DOI] [PubMed] [Google Scholar]
- 11•.Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA, Robinson BH. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am J Hum Genet. 2011;89:486–495. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcindor T, Bridges KR. Sideroblastic anaemias. Br J Haematol. 2002;116:733–743. doi: 10.1046/j.0007-1048.2002.03378.x. [DOI] [PubMed] [Google Scholar]
- 13•.Cazzola M, Malcovati L. Diagnosis and treatment of sideroblastic anemias: from defective heme synthesis to abnormal RNA splicing. Hematology Am Soc Hematol Educ Program. 2015;2015:19–25. doi: 10.1182/asheducation-2015.1.19. Nice description of pathology and causes of sideroblastic anemias. [DOI] [PubMed] [Google Scholar]
- 14.Rouault TA, Tong WH. Tangled up in red: intertwining of the heme and iron-sulfur cluster biogenesis pathways. Cell Metab. 2009;10:80–81. doi: 10.1016/j.cmet.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC. The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Biol. 2001;8:156–160. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 16.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 17•.Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, Corson L, Feala J, Fekkes P, Ichikawa K, Keaney GF, Lee L, Kumar P, Kunii K, MacKenzie C, Matijevic M, Mizui Y, Myint K, Park ES, Puyang X, Selvaraj A, Thomas MP, Tsai J, Wang JY, Warmuth M, Yang H, Zhu P, Garcia-Manero G, Furman RR, Yu L, Smith PG, Buonamici S. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. First clear connection of SF3B1 allele and ABCB7 splicing. [DOI] [PubMed] [Google Scholar]
- 18.Liu G, Guo S, Anderson GJ, Camaschella C, Han B, Nie G. Heterozygous missense mutations in the GLRX5 gene cause sideroblastic anemia in a Chinese patient. Blood. 2014;124:2750–2751. doi: 10.1182/blood-2014-08-598508. [DOI] [PubMed] [Google Scholar]
- 19•.Liu G, Wang Y, Anderson GJ, Camaschella C, Chang Y, Nie G. Functional Analysis of GLRX5 Mutants Reveals Distinct Functionalities of GLRX5 Protein. J Cell Biochem. 2016;117:207–217. doi: 10.1002/jcb.25267. Rigorous characterization of two distinct phenotypes from different GLRX5 alleles. [DOI] [PubMed] [Google Scholar]
- 20.Leighton J, Schatz G. An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. The EMBO J. 1995;14:188–195. doi: 10.1002/j.1460-2075.1995.tb06989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaedler TA, Thornton JD, Kruse I, Schwarzlander M, Meyer AJ, van Veen HW, Balk J. A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly. J Biol Chem. 2014;289:23264–23274. doi: 10.1074/jbc.M114.553438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A) Hum Mol Genet. 1999;8:743–749. doi: 10.1093/hmg/8.5.743. [DOI] [PubMed] [Google Scholar]
- 23.Pagon RA, Bird TD, Detter JC, Pierce I. Hereditary sideroblastic anaemia and ataxia: an X linked recessive disorder. J Med Genet. 1985;22:267–273. doi: 10.1136/jmg.22.4.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Effenberger KA, Urabe VK, Prichard BE, Ghosh AK, Jurica MS. Interchangeable SF3B1 inhibitors interfere with pre-mRNA splicing at multiple stages. RNA. 2016 doi: 10.1261/rna.053108.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, Mudie LJ, Stephens PJ, McLaren S, Massie CE, Tarpey PS, Varela I, Nik-Zainal S, Davies HR, Shlien A, Jones D, Raine K, Hinton J, Butler AP, Teague JW, Baxter EJ, Score J, Galli A, Della Porta MG, Travaglino E, Groves M, Tauro S, Munshi NC, Anderson KC, El-Naggar A, Fischer A, Mustonen V, Warren AJ, Cross NC, Green AR, Futreal PA, Stratton MR, Campbell PJ. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jadersten M, Jansson M, Elena C, Galli A, Walldin G, Della Porta MG, Raaschou-Jensen K, Travaglino E, Kallenbach K, Pietra D, Ljungstrom V, Conte S, Boveri E, Invernizzi R, Rosenquist R, Campbell PJ, Cazzola M, Hellstrom Lindberg E. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126:233–241. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Kimura S, Suzuki T. Iron-sulfur proteins responsible for RNA modifications. Biochim Biophys Acta. 2015;1853:1272–1283. doi: 10.1016/j.bbamcr.2014.12.010. Excellent review. [DOI] [PubMed] [Google Scholar]
- 28••.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. Excellent review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20:306–314. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Shigi N. Biosynthesis and functions of sulfur modifications in tRNA. Front Genet. 2014;5:67. doi: 10.3389/fgene.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjork GR, Huang B, Persson OP, Bystrom AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forouhar F, Arragain S, Atta M, Gambarelli S, Mouesca JM, Hussain M, Xiao R, Kieffer-Jaquinod S, Seetharaman J, Acton TB, Montelione GT, Mulliez E, Hunt JF, Fontecave M. Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat Chem Biol. 2013;9:333–338. doi: 10.1038/nchembio.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young AP, Bandarian V. Radical mediated ring formation in the biosynthesis of the hypermodified tRNA base wybutosine. Curr Opin Chem Biol. 2013;17:613–618. doi: 10.1016/j.cbpa.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, Nureki O. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J Mol Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Carlson BA, Kwon SY, Chamorro M, Oroszlan S, Hatfield DL, Lee BJ. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255:2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- 37.Hatfield D, Feng YX, Lee BJ, Rein A, Levin JG, Oroszlan S. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology. 1989;173:736–742. doi: 10.1016/0042-6822(89)90589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei FY, Tomizawa K. Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes. Endocr J. 2011;58:819–825. doi: 10.1507/endocrj.ej11-0099. [DOI] [PubMed] [Google Scholar]
- 39.Zhou B, Wei FY, Kanai N, Fujimura A, Kaitsuka T, Tomizawa K. Identification of a splicing variant that regulates type 2 diabetes risk factor CDKAL1 level by a coding-independent mechanism in human. Hum Mol Genet. 2014;23:4639–4650. doi: 10.1093/hmg/ddu184. [DOI] [PubMed] [Google Scholar]
- 40.Wei FY, Zhou B, Suzuki T, Miyata K, Ujihara Y, Horiguchi H, Takahashi N, Xie P, Michiue H, Fujimura A, Kaitsuka T, Matsui H, Koga Y, Mohri S, Oike Y, Tomizawa K. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab. 2015;21:428–442. doi: 10.1016/j.cmet.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 41.Kirino Y, Yasukawa T, Ohta S, Akira S, Ishihara K, Watanabe K, Suzuki T. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc Natl Acad Sci U S A. 2004;101:15070–15075. doi: 10.1073/pnas.0405173101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Huang B, Eliasson M, Ryden P, Bystrom AS. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011;7:e1002258. doi: 10.1371/journal.pgen.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson CL, Lemmens R, Miskiewicz K, Broom WJ, Hansen VK, van Vught PW, Landers JE, Sapp P, Van Den Bosch L, Knight J, Neale BM, Turner MR, Veldink JH, Ophoff RA, Tripathi VB, Beleza A, Shah MN, Proitsi P, Van Hoecke A, Carmeliet P, Horvitz HR, Leigh PN, Shaw CE, van den Berg LH, Sham PC, Powell JF, Verstreken P, Brown RH, Jr, Robberecht W, Al-Chalabi A. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet. 2009;18:472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, Tuck S, Bystrom AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5:e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]