Abstract
Precision medicine is an emerging approach to disease treatment and prevention that considers variability in patient genes, environment, and lifestyle. However, little has been written about how such research impacts emergency care. Recent advances in analytical techniques have made it possible to characterize patients in a more comprehensive and sophisticated fashion at the molecular level, promising highly individualized diagnosis and treatment. Among these techniques are various systematic molecular phenotyping analyses (e.g., genomics, transcriptomics, proteomics, and metabolomics). Although a number of emergency physicians use such techniques in their research, widespread discussion of these approaches has been lacking in the emergency care literature and many emergency physicians may be unfamiliar with them. In this article, we briefly review the underpinnings of such studies, note how they already impact acute care, discuss areas in which they might soon be applied, and identify challenges in translation to the emergency department. While such techniques hold much promise, it is unclear whether the obstacles to translating their findings to the emergency department will be overcome in the near future. Such obstacles include validation, cost, turnaround time, user interface, decision support, standardization, and adoption by end users.
Introduction
Precision medicine refers to creating individualized diagnostic and treatment plans for patients based on the way their unique characteristics may dictate disease response or progression1,2. Although there are many forms of precision medicine, one approach with particular relevance to emergency medicine is systematic molecular phenotyping—the comprehensive measurement of a category of molecules from a patient’s specimen in order to characterize their condition.
Technology in this area is advancing rapidly. In 2003, the Human Genome Project announced that it had completed its mission to sequence the entire human genome. That effort required 13 years, multiple research institutions processing samples from multiple subjects, and $2.7 billion3. In contrast, since 2014, more than 500,000 people have purchased a commercial product that provides identification of multiple DNA single nucleotide polymorphisms within weeks for only $99 each4. The United States Food and Drug Administration (FDA) has already approved marketing of one of the instruments underlying these products (Illumina MiSeqDx™)5 . Recently, a research team from one institution announced the application of whole genome sequencing to the care of a patient with a test turnaround time of 26 hours6. Direct-to-consumer advertising of whole genome sequencing is also available via pre-order with a physician’s authorization for $9994. In order to spur precision medicine’s role in the future of medicine, President Barack Obama announced a $215 million proposal to finance the Precision Medicine Initiative1 in January 2015, and the European Union has committed close to €1 billion for similar research since 20077.
Numerous proclamations have been made about the impact of precision medicine1,2,4,8–10. While some point to the current paucity of clinically applicable discoveries and other limitations of such research9,11, others point to its potential to alter the way doctors define, categorize, and diagnose diseases10,12,13. Despite numerous advances in this field, little has been written about how precision medicine may affect emergency care. In this paper, we highlight the impact of such research on the experience and care of emergency department patients. We further discuss the considerable challenges in applying these findings to the emergency department setting.
Background
Precision medicine refers to a broad field encompassing many techniques to more accurately deliver treatments. One common feature of these various techniques is a systematic analysis to more precisely characterize a subject’s phenotype. Precision medicine includes big-data analyses applied to socioeconomic or medical history data from the medical record to derive personalized recommendations; however, such approaches are beyond the scope of this paper14. Instead, we will focus on those analyses that focus on molecular phenotyping, which use analysis techniques often referred to as “-omics” (Table 1).
Table 1.
Definitions of Basic Science Techniques Utilized in Precision Medicine
| Genomics | The study of the entire complement or strategic sequences of genetic material. |
| Genome-Wide Association Studies (GWAS) | A form of study in which the researcher simultaneously probes all segments of the genome for evidence of association between single nucleotide polymorphisms and disease. |
| Exome Studies | Studies focused on the protein coding regions of the genome. Exome analyses focus on rare variants that are likely to be functional. This is in contrast to GWAS, which focuses on common variants that may or may not be related to functional proteins. |
| Epigenetics | The study of the regulation of gene activity by reversible modifications such as methylation of deoxyribonucleic acids (DNA). |
| Transcriptomics | The quantification of the relative levels of messenger ribonucleic acids (RNA) for a large number of genes in specific cells or tissues to measure differences in the expression levels of different genes. |
| Proteomics | The study of a comprehensive number of proteins, the products of gene transcription (including their expression and/or post-translational modifications). |
| Metabolomics | The study of a comprehensive number of small molecules (such as fatty acids, carbohydrates, and glycoproteins) that are the terminal downstream products of cellular processes. |
| Microbiomics | The study of the gut flora and their relative constituents by quantification of bacterial genomics. |
| Pharmaco-(genomics/proteomics/metabolomics) | The use of the aforementioned technologies to choose therapy, with the goal of maximizing efficacy and safety of therapy. |
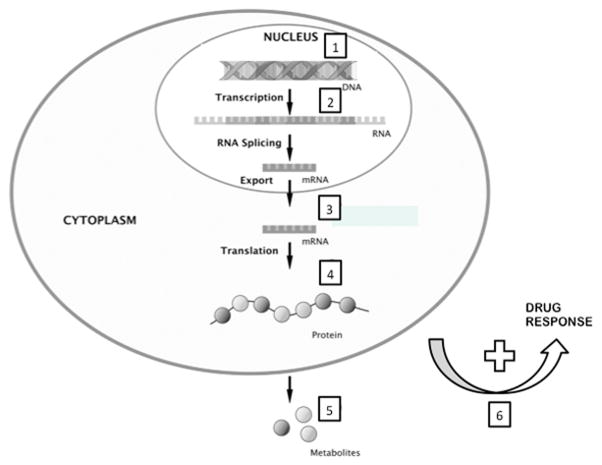
The combination of technological advances in molecular chemistry, assay automation, data storage and transmission, and computational mathematics now allow quantification of hundreds (e.g., metabolomics) to tens of thousands (e.g., transcriptomics) of analytes from a single patient specimen. In the case of genomics, billions of data points from a single patient sample can be generated. Some common techniques for systematic molecular phenotyping include genomics, genome-wide association studies (GWAS), epigenetics, transcriptomics, proteomics, metabolomics, and microbiomics, among others (Figure 1).
Figure 1.
Reproduced with permission from Skibsted et al. Crit Care 2013; 17: 231 and adapted. Numbers denote targets within cellular processes for different systems biology approaches: 1, Genetic analysis and Single Nucleotide Polymorphisms (SNPs) and 2, epigenomics (methylation variable positions) 3, transcriptomics (mRNA); 4, proteomics; and 5, metabolomics; 6, pharmaco-(genomics/metabolomics/proteomics).
Systematic molecular phenotyping approaches have helped us to better understand diseases, leading to improved diagnostic and treatment approaches. These techniques have also led to more efficient development of blood-based diagnostic tests. In some cases, such research has identified disease subtypes that may respond differentially to existing treatments. The vision of this approach is that patients will receive personalized treatments based on their unique genetic codes, metabolite, or other analyte profiles. As these techniques are increasingly being used to investigate acute care conditions, the practicing emergency physician will need to be familiar with their scientific basis.
At times, systematic molecular phenotyping identifies a single analyte associated with a diagnosis or treatment response. In such cases, these findings are translated into clinical care in traditional fashion. For example, if a genomic analysis identifies a single genetic variant that predicts a disease, then an assay for that single gene can be used as a diagnostic test. Such systematic studies are impacting many areas of acute care medicine.
However, what further distinguishes systematic molecular phenotyping as a field is the potential to create new approaches to diagnosis and treatment. Comprehensive analyte testing is one such new approach. Frequently, the inability to gather comprehensive data about a patient limits emergency physicians’ ability to make accurate diagnoses. At times, it is not only a lack of time or resources to obtain comprehensive data, but also a limitation regarding knowledge of which pieces of information to pursue. Most clinical tests assess a single analyte (e.g., troponin, creatinine) and often a single analyte is used to help determine whether an entire biological system is abnormal (e.g., white blood cell count). In contrast, systematic molecular phenotyping can quantify a large spectrum of analytes, which circumvents the piecemeal nature of ordering tests. Such comprehensive analysis of groups of analytes, and their relationship to one another, provide a more nuanced understanding of the patient’s physiology. For example, a panel of inflammatory system markers that analyzes many molecules might allow more precise identification of the type of infectious organism in a patient with fever.
Although for much of this article we will discuss these techniques as a group, there are important differences. For example, while a patient’s genome would not be expected to change over time, their metabolomic or proteomic profile might change rapidly according to their physiologic state. However, there are some commonalities in the ways in which they attempt to change our approach to diagnosis and treatment.
How Systematic Molecular Phenotyping is Poised to Impact Clinical Care in the ED
Consider the following case, taken from the actual clinical experience of one of the authors.
Case Presentation:
A 31-year-old male presents to the emergency department with left calf pain for 2 days. He has not noticed any swelling, skin changes, or prominence of the leg veins. He denies chest pain, shortness of breath, or dyspnea. He has no significant past medical history, no history of recent travel, and an unremarkable family history. His exam and vitals are normal, and his Wells deep vein thrombosis (DVT) score is 0.
The patient is diagnosed with muscle strain and prepared for discharge without further testing. It is explained that this is unlikely to be a DVT given the clinical data. The patient replies that he came to the emergency department because he was concerned his pain might be caused by a DVT, since a commercial DNA analysis service reported a genetic predisposition to blood clots. He shares the report, which states that he is heterozygous for Factor V Leiden.
In light of the new information, a duplex ultrasound was ordered and is negative for DVT and the patient was discharged home.
Did the physician do the right thing by allowing the patient’s genetic data to influence the decision to order an ultrasound? Does the patient require further testing or referral for Factor V Leiden?
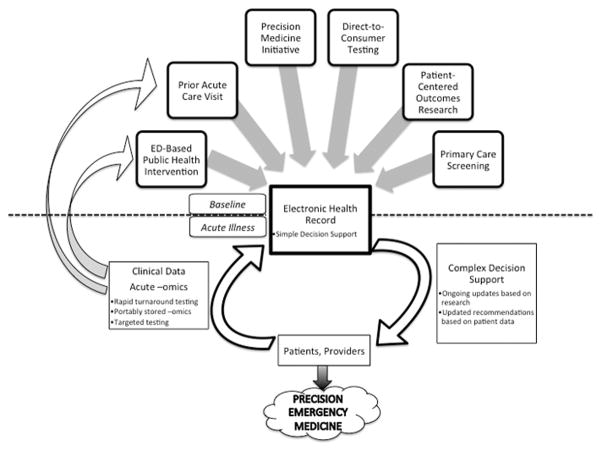
This case demonstrates both the promise and possible dangers of widespread systematic molecular phenotyping. There are a number of ways that systematic molecular phenotyping can be integrated into emergency department care (Figure 2, for one example). Some institutions have existing prototypes in place15,16. Many of the key features of such a system already exist or are beginning to take shape: universal electronic health records with underlying decision support, multiple venues to access systematic molecular phenotyping, and technical analytic advances that allow rapid turnaround and application of results.
Figure 2. A Model for Incorporating Systematic Molecular Phenotyping into Emergency Care.
There are multiple potential sources for baseline systematic molecular phenotyping data to be entered into an acute care patient’s electronic health record. During the visit itself, clinical data can be combined with this data to help provide complex decision support. In the future, with more rapid turnaround times, systematic molecular phenotyping might be performed primarily in the ED and incorporated into such systems. Alternatively, patients may arrive with portably stored systematic molecular phenotyping data, or targeted testing of genotypes might be posited based on the patient’s clinical scenario. The results of any testing done in the acute care setting would be retained for future visits, or a patient might have systematic molecular phenotyping done as part of an ED-based public health intervention and stored for future use.
In such a model, the electronic health record represents a key tool for transfer of outpatient information to the emergency department and vice versa15. This creates a feedback loop where emergency care visits could lead to baseline systematic molecular phenotypes (similar to other laboratory blood tests) which can then be used for future acute episodes or chronic disease management. Alternatively, data may be stored in portable formats that move with the patient between systems.
Increasingly, patients can have such systematic analyses performed in multiple environments. For example, the aforementioned Precision Medicine Initiative will provide for patients to have their genomic information linked with 8 their electronic health records1. There are a rising number of validated clinical applications for molecular phenotyping; this leads to more patients having such analyses available from their usual clinical care. Furthermore, a burgeoning private industry seeks to provide patients genetic analysis information directly. Therefore, it will be increasingly likely that a patient presenting to an emergency department has already had a complete genomic or other systematic molecular analysis performed prior to the ED visit.
Finally, it is possible that technical advances in analyte analysis, reporting, and integration will allow for real-time systematic molecular phenotyping of patients in the emergency department. The challenges for this model are further discussed below in “Translation to the ED”.
In the following two sections, we provide specific clinical examples of how systematic molecular phenotyping approaches have already been applied to acute care conditions and how they may be applied in the future. Unlike our opening case, the case presentations that follow are hypothetical.
Improving Diagnosis and Prognosis
Case Presentation
A 55-year-old man presents to the emergency department with chest pain. He is unable to reliably characterize the pain. He has a past medical history of diabetes but has never had prior risk stratification for acute coronary syndrome. His initial work up, including troponin and electrocardiogram, is non-diagnostic. He is placed in an emergency department observation unit but the next day fails to attain target heart rate on an exercise stress test. A comprehensive metabolomic profile performed from a blood sample collected during his initial ED workup shows low levels of dicarboxylacylcarnitines, suggesting his clinical risk for death or AMI in the next year is low17. As a result, he is discharged with reassurance and without further cardiac testing in the ED.
In this hypothetical example, the clinician overlays objective measures, such as troponin, EKG, and stress testing onto the patient’s clinical history and risk factors for coronary artery disease. While many patients are adequately diagnosed and managed using this model, a significant number have indeterminate tests, and risk persists. Additional noninvasive tests could play a substantial role in such scenarios.
Systematic molecular phenotyping research is one way to develop such noninvasive tests. Under the current research translation paradigm, the development of a candidate biomarker assay typically begins with assumption about what the relevant analyte should be. For example, troponin-I was identified as an intracellular, cardiac muscle-specific marker that increases in the blood among patients with myocardial injury. It was subsequently validated in a series of studies to determine its performance as a diagnostic test.
However, developing a new assay that measures a different molecule requires another series of studies. This process is laborious, costly, and requires time. The use of systematic molecular phenotyping techniques may be a more efficient means of discovering such biomarkers. A well-designed and adequately powered study has the potential for multiple, parallel avenues for discovery:
genetic factors (e.g. gene for factor V Leiden mutation) that identify patients at high risk of disease;
genetic risk scores that accommodate existing clinical characteristics or diagnostic schemes associated with the disease;
single, previously unknown biomarkers to use as diagnostic tests;
complex patterns or panels of biomarkers that identify disease; and/or
factors that predict prognosis or likely side effects of treatment.
Any of these may be used to assist in diagnosis of a condition individually, or in combination to more accurately diagnose patients or predict prognosis.
For example, one group of investigators identified a pattern of gene expression that occurred in healthy individuals who had been inoculated with respiratory viruses18. They then used this gene expression pattern to create and validate a set of biomarkers to differentiate respiratory viral infections from bacterial infections19. Although not yet available in clinical practice, this novel biomarker could be used to differentiate clinically similar but etiologically different diseases, a hallmark of the precision medicine effort. It also demonstrates the ability to quickly identify which molecules are important to measure out of several possible targets.
Additionally, more complex patterns of multiple biomarkers can be used to better differentiate phenotypes of interest. These patterns can be used as a panel for diagnosis or prognosis. For example, a proteomic analysis of patients with severe burns not only discovered novel associations between certain proteins and burn injuries, but also found 43 proteins with different concentrations in survivors versus nonsurvivors20. These protein biomarkers were then combined into a model that predicts survival. One could envision a test that produces an aggregate report that incorporates all of these proteins into a single risk-stratification measure for the clinician. Such complex relationships would have been extremely difficult to establish using a single biomarker approach. Similar approaches have also suggested methods which may predict mortality and other complications from trauma21, sepsis13,22, and intracerebral hemorrhage23.
Incorporation of these basic science approaches may augment existing clinical risk stratification models in several acute disease states. As an example, two recent studies used transcriptomic analysis24 and metabolomic profiling25 to improve prediction of death in acute myocardial infarction (AMI). In the first study, researchers identified mRNA that were more prevalent in the blood of patients with coronary artery disease. They then created and validated a “gene expression score” incorporating these mRNA changes with traditional cardiac risk factors. In the latter study, investigators identified the fatty acids, ketones, and other metabolites associated with risk for AMI or death. They then combined these factors with a traditional risk stratification model.
In both studies, combining –omics data with existing clinical prediction scores led to a higher proportion of patients being classified appropriately. The gene expression score allowed the safe exclusion of some patients from the need for cardiac angiography while increasing the diagnostic yield of cardiac angiograms performed from 36.2% to 48.2%24. Likewise, metabolomic profiling increased the proportion of patients correctly risk-stratified for AMI or death by 11%25. Should subsequent studies validate these findings, such approaches may provide more accurate diagnosis and prognosis while using fewer healthcare resources.
Targeting Treatment
Case Presentation
A 62-year-old man is brought in by his family because of fever and lethargy. He is found to be in septic shock. His electronic health record supports the integration of genetic information. He is flagged as having the AA genotype, rs1042717, in the beta(2)-adrenergic receptor gene (ADRB2), which makes him susceptible to deleterious effects of adrenergic vasopressors along with an increased mortality compared to other genotypes. Moreover, the decision support tool associated with this notation suggests the increased mortality can be mitigated by the administration of steroids26. He is given intravenous steroids and his pressure is supported with vasopressin.
This hypothetical case demonstrates how systematic molecular phenotyping approaches may allow greater therapeutic success by more accurately targeting treatments. Within existing disease classifications, it may be possible to identify subcohorts that preferentially benefit from a particular treatment. This area of research has the potential to “rescue” previously discarded therapies by more accurately targeting them toward patients who might benefit. Alternatively, more accurate classification of patients may allow prevention of serious adverse drug effects.
For example, pharmacometabolomics and proteomics have been used to predict drug responsiveness in conditions like sepsis8,27 and pain control28. One specific example of this is a post hoc study of patients in septic shock randomized to l-carnitine or placebo29. Pretreatment blood samples were analyzed for a comprehensive panel of fatty acids and other metabolites. This analysis showed that patients with a low concentration of 3-hydroxybutyrate who were treated with l-carnitine had decreased vasopressor requirements and reduced 1-year mortality. This demonstrates how an –omics analysis can identify an appropriate subgroup for a particular treatment within a common phenotype to improve clinical outcomes.
Similar approaches might also identify patients at risk for pharmacologic adverse events. For example, Del Río-Espínola et al.30 performed genetic analyses on 1,172 acute ischemic stroke patients who were treated with recombinant tissue plasminogen activator (rtPA), of whom 20.9% developed hemorrhagic transformation and 10.6% died. They studied 140 single nucleotide variants (SNVs) in 97 candidate genes and identified one SNV that predicted hemorrhagic transformation and death. This was subsequently validated in a separate cohort. This example highlights how in the near future, the decision to administer rtPA will can be informed by a systematic approach using biologic data to determine a patient’s risk of bleeding.
The preceding examples are illustrative of the broad ways in which systematic molecular phenotyping research lead to findings that can directly impact acute care. Several others are presented in Table 2. While not all of these techniques can be applied at the bedside, it is readily apparent how they might be incorporated in the future once certain hurdles are overcome. In the following section, we discuss obstacles to this process of translation and some potential solutions.
Table 2.
A Sample of Genomic, Transcriptomic, and Metabolomic Studies Relevant to Emergency Medicine
| Study | Disease Area | Approach | Primary Finding | Emergency Care Import |
|---|---|---|---|---|
| Biffi et al.23 | Cerebral hemorrhage | Genetic association study | Vasculopathic changes associated with the APOE epsilon2 allele might have a role in the severity and clinical course of lobar intracerebral hemorrhage. | This information could be used early in the course of a patient with intracerebral hemorrhage to guide treatment and even goals-of-care discussions. |
| Warren et al.21 | Trauma | Genomic | Assessment of genome-wide gene expression provides useful clinical information different from that provided by currently utilized anatomic or physiologic scores. | Consideration of the patient’s underlying physiology is as important as the traumatic injury event characteristics. This information could be used to guide leveling criteria or operative treatment decisions. |
| Linnstaedt et al.37 | Pain | Genetic association study | Patients with an AG or GG genotype at the OPRM1 A118G allele have less severe pain at six weeks after trauma. | This information could be used to rationally guide which patients would need specialized pain service follow-up after traumatic injury. |
| DeCoux et al.27 | Sepsis | Proteomic | Distinct proteins in common pathways predicted sepsis outcomes. | This information could be used to guide disposition decisions or to identify patients with occult sepsis. |
| Subudhi et al.38 | Altitude-Related Illness | Metabolomics | Identified metabolites associated with hemoglobin adaptation to high altitude sickness. | This information could be used to guide the decision of whether to transport patients out of high altitude settings, a resource-intensive treatment. |
| Fernández-Cadenas et al.39 | Stroke | Genomic | Three single nucleotide polymorphisms were associated with recanalization in rTPA- treated stroke patients | These data could be used to guide interventional or TPA administration treatment decisions. |
| Voellenkle et al.40 | Congestive heart failure | Transcriptomic | Identified miRNAs differentially modulated in non- ischemic vs. ischemic cardiomyopathy patients. | This information could guide the decision whether to pursue coronary imaging and transplant options. |
Direct-to-Consumer Tests
As demonstrated in our opening clinical case, the prospect of patients directly obtaining comprehensive genomic analyses could have a profound impact on emergency care. Such direct-to-consumer tests bypass the physician as gatekeeper between patients and their genomic information, but also leave patients without professional medical guidance on how to interpret and act on the information they receive. It remains to be seen how patients will respond to such information, particularly given the probabilistic nature of such data. As can be seen from our real-life case example, patients may turn to emergency physicians for guidance, particularly when they are concerned about time-sensitive conditions such as venous thrombosis. Compounding the difficulty in interpretation is the fact that these tests are generally not certified by the Clinical Laboratory Implementation Amendments (CLIA). Thus, even hospitals that are able to do these analyses and that have systems to integrate results into clinical decision support would require re-testing in their own clinical laboratories.
Beyond the logistics of where and how patients obtain guidance on interpreting the results, regulation of direct-to-consumer genomic tests represents a major challenge for translating these findings to the bedside5,31. While the technology currently exists to provide patients direct access to their genome, it remains unclear whether the recommendations and risk assessments provided by these direct-to-consumer commercial firms have been properly vetted5. Firms often fail to disclose which genetic variants inform their predictions, let alone reveal the underlying evidence base32. A further challenge is that tests can identify an essentially unlimited number of variants based on the over 3 billion base pairs that compose the human genome. Evaluating whether each data point is accurately measured would take years and thus delay the public’s access to the benefits of this technology.
The FDA has acknowledged the competing interests of evidentiary mandate and public access to this technology through some of their novel regulatory approaches to this area. Rather than requiring a company to provide evidence that a proposed test can accurately measure every possible gene variant in the genome, the FDA asks for evidence for only a representative subset of possible gene variants. They also leverage existing federally funded databases of genetic correlations with disease to validate clinical predictions5. Many such databases already exist, such as the publically funded Database of Genotypes and Phenotypes (DbGAP), developed to “archive and distribute the results of studies that have investigated the interaction of genotype and phenotype.”33
Direct-to-consumer whole genome sequencing represents a challenge to clinicians. ED providers know how to act on existing user-performed testing, such as home pregnancy tests. However, unlike a pregnancy test, whole genome sequencing cannot currently be rapidly repeated or verified in the ED with present technology. As such, emergency physicians will have to decide whether, and how, to incorporate these results into their clinical decision making.
Translation to the ED
For systematic molecular phenotyping approaches to reach their full clinical potential for ED patients, other barriers must be surmounted as well.
Time
Current systematic molecular phenotyping techniques cannot provide turnaround times in an ED timeframe. Recently a group reported a 26-hour turnaround time for a whole genome sequence6, which is rapid, but still not useful for emergency physicians. Thus, it is currently more likely that emergency physicians will encounter previously-run analyses that are available via electronic medical records or directly from patients16. As of 2014, almost 100,000 people have had their genome sequenced and this number is expected to increase exponentially2. A more likely scenario is that the results of comprehensive measurements will be done in the research setting. Once candidate markers have been identified, tests can then be developed to rapidly identify only the relevant targets. Such targeted assays could conceivably provide results in a time frame suitable for ED use.
Cost
Although the calculated cost of performing genomic analyses has decreased exponentially (Figure 3)34, these costs may not be representative of all fixed costs required to perform such analyses. Other systematic molecular phenotyping costs likely will follow suit in seeing a trend toward lower costs. While direct-to-consumer testing has demonstrated a willingness of some patients to pay for this technology, it is unlikely that all patients would be willing to do so, and unclear whether third-party payors would cover these costs. It is likely that insurers will require some demonstration of economic value and improved clinical outcomes prior to agreeing to pay for general systematic molecular phenotype screening.
Figure 3.
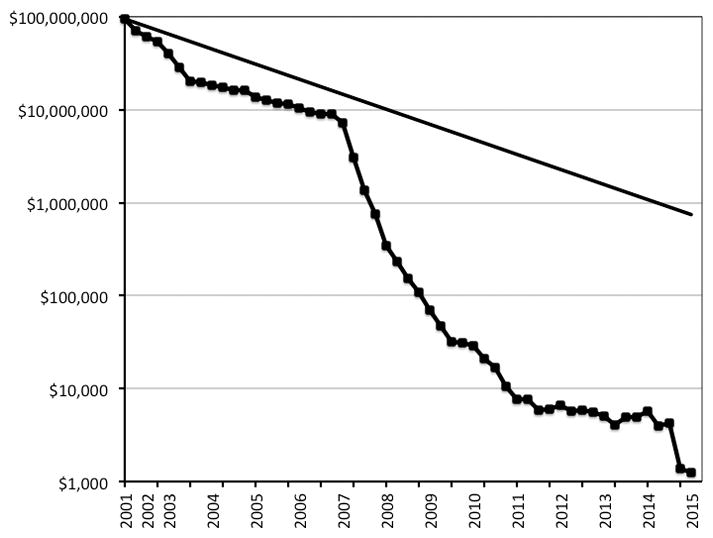
Reproduced from www.genome.gov/sequencingcosts. Direct costs associated with DNA sequencing as reported by the National Human Genome Research Institute’s Genome Sequencing Program. To illustrate the nature of the reductions in costs, this graph shows hypothetical data reflecting Moore's Law, which describes a long-term trend in the computer hardware industry that involves the doubling of 'compute power' every two years. If the costs per genome were to follow Moore’s Law going forward, the cost per genome in the year 2022 would be approximately $100.
Capacity
Indeed, even if such costs are overcome, it is not clear that each hospital or health system would have the necessary data infrastructure and technical expertise to carry out such analyses on a widespread scale35. Hospitals that conduct such analyses currently do so primarily for research purposes. Increased investments in analyzers, data storage, and personnel would be required to scale up to mass testing.
Validation
Some genomic studies may require thousands of patients and multiple centers to perform. Therefore replication and validation of findings can be difficult35. Furthermore, the relationship between genetic associations and disease mechanisms often requires further study. For most SNVs discovered through modern genotyping techniques, the actual mechanism relating the SNV to the disease is unknown. More commonly, the causes of disease are polygenic11. Therefore, it may be hard to discern which associations are valid and which are false positive discoveries.
Adoption
Furthermore, developing a scheme for integrating clinically actionable results into ED decision-making will be another challenge. Assuming technical barriers of analysis can be overcome, physicians will still have to learn what to do with the vast amount of information generated. Given the large number of molecule concentrations that may be measured in any given sample, practicing clinicians will not be able to realistically incorporate the numerous relationships into their knowledge base. Indeed, even at a leading academic medical institution, a survey of providers indicated a lack of knowledge of the scope of existing indications for pharmacogenetic-guided therapeutics36.
Implementation
Given the overwhelming breadth and depth of such data, even the most supportive clinicians will likely need sophisticated technological assistance to interpret the results. Decision support systems will need to accommodate not only large amounts of molecular data but also the patient’s clinical context. Challenges already exist in making decision support user-friendly when the inputs for decision rules are relatively simple. It remains to be seen whether such systems can tolerate large volumes of data and numerous potential inputs. Plus, these systems must have the flexibility to evolve with the growing understanding of associations between molecular phenotypes and correlated diseases.
Overtesting
Like all diagnostic tests, it is possible that the availability of such testing may not improve diagnostic accuracy but only increase resource utilization. Ensuring that the availability of molecular phenotyping does not drive unnecessary testing will be particularly difficult given the time constraints in managing acute illness. Such information would either need to be already available or be available using technologies that provide results in clinically relevant time frames suitable for ED care. Future research will need to examine the impact on overall resource utilization and outcomes that both the test context and the test results themselves would have.
Future Directions
In the future, it may become possible to test a wide array genes, proteins, or metabolites, in the ED on a routine basis. These tests may report not just individual biomarkers but also patterns of biomarkers. These might be augmented by technological support for automated interpretation. Treatments will not be chosen based not broad diagnostic categories, but rather on individualized recommendations based on systematic molecular analyses. In the more near term, integration of previously-run systematic molecular analyses of patients may inform decision support tools embedded in electronic health records. Additionally, real-time –omic testing might serve as a basis for public health screening or long-term health interventions from the ED.
However, for this future to be realized, emergency physicians must play a key role in producing and consuming this literature. Our specialty’s holistic view of patients that extends beyond single organ systems may advance this field of research by creating new collaborations and synergies. Furthermore, a strategy of blind acceptance of handed-down research findings and implementation strategies from other specialties is likely to lead to burdensome systems that don’t match our unique work environment. Therefore, it behooves us as a specialty to educate ourselves about this body of research and to get involved where we can. At the individual level, this will require engaging local experts in research and education in this area and collaborating on career development or other grants with such experts. Involvement in pharmacy and therapeutics committees is an opportunity for interested emergency physicians to engage in discussions on how EDs should interact with the dozens of pharmaceuticals with FDA-approved pharmacogenomic indications. At the specialty-organization level, this might involve sponsoring conferences and didactics, advocating for or creating new research grant opportunities, and partnering with other specialty organizations to create new opportunities for collaboration and dialogue.
Conclusion
Currently, systematic molecular phenotyping research is making an impact on emergency care in many traditional ways as a tool for researchers. By improving our understanding of disease, systematic molecular phenotyping research will allow us to develop better diagnostics and help us understand which treatments are effective. More accurate application of therapies may improve care while providing cost and time savings for patients in the ED by reducing the need for observation or hospital admission.
Furthermore, systematic molecular phenotyping research is introducing new concepts that may alter how we provide care in the ED. Emergency physicians may increasingly see patients bringing direct-to-consumer genomic reports to their ED visits, and ED physicians will be called upon to help patients interpret the results. Thus, systematic molecular phenotyping research will fundamentally alter the format and even the scope of ED diagnostics and therapeutics. However, before this can become a reality, there are many unique challenges that must be overcome. Technical, financial, and cultural challenges may impede or prevent adoption. The dangers of such research include premature adoption of findings and overtesting. In the future, emergency physicians must play a key role in producing such research and in helping patients to interpret the complex results.
Acknowledgments
Funding Sources: We would like to acknowledge the following grant support: Dr. Limkakeng’s work was supported by an internal grant from the Duke University PDC. Dr. Kabrhel’s work was supported by National Institutes of Health Grant HL 116854. Dr. Puskarich’s work was supported by National Institutes of Health Grant K23 GM113041-01. Dr. Monte’s work was supported by National Institutes of Health Grants K23 GM110516 and UL1 TR000154 (A.A.M).
We would like to acknowledge the proofreading and editing assistance of Ms. Ashley Morgan, MA.
Footnotes
Prior Presentations: This work has not previously been presented.
Disclosures: There are no relevant commercial conflicts of interest.
References
- 1.Collins FS, Varmus H. A New Initiative on Precision Medicine. New England Journal of Medicine. 2015;372:793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topol Eric J. Individualized Medicine from Prewomb to Tomb. Cell. 157:241–53. doi: 10.1016/j.cell.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. [Accessed 6/2/15];International Consortium Completes Human Genome Project. 2003 doi: 10.1517/phgs.4.3.241.22688. at http://www.genome.gov/11006929. [DOI] [PubMed]
- 4.Guinto J. Why is this $99 Home DNA Kit Causing Such an Uproar? Genome. 2014 [Google Scholar]
- 5. [Accessed 6/2/15];Optimizing FDA’s Regulatory Oversight of Next Generation Sequencing Diagnostic Tests—Preliminary Discussion Paper. 2015 at http://www.fda.gov/downloads/MedicalDevices/NewsEvents/WorkshopsConferences/UCM427869.pdf.
- 6.Miller NA, Farrow EG, Gibson M, Willig LK, Twist G, Yoo B, et al. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome medicine. 2015;7:100. doi: 10.1186/s13073-015-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draghia-Akli R. Enabling personalized medicine in Europe: a look at the European Commission’s funding activities in the field of personalized medicine research. Personalized Medicine. 2012;9:151–5. doi: 10.2217/pme.11.91. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J, Vincent JL, Adhikari NK, Machado FR, Angus DC, Calandra T, et al. Sepsis: a roadmap for future research. Lancet Infect Dis. 2015;15:581–614. doi: 10.1016/S1473-3099(15)70112-X. [DOI] [PubMed] [Google Scholar]
- 9.Jameson JL, Longo DL. Precision Medicine - Personalized, Problematic, and Promising. N Engl J Med. 2015 doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 10.Skibsted S, Bhasin MK, Aird WC, Shapiro NI. Bench-to-bedside review: future novel diagnostics for sepsis - a systems biology approach. Critical care (London, England) 2013;17:231. doi: 10.1186/cc12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyner MJ, Paneth N. Seven Questions for Personalized Medicine. JAMA. 2015 doi: 10.1001/jama.2015.7725. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez JM, Rucavado A, Escalante T, Lomonte B, Angulo Y, Fox JW. Tissue pathology induced by snake venoms: how to understand a complex pattern of alterations from a systems biology perspective? Toxicon. 2010;55:166–70. doi: 10.1016/j.toxicon.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Langley RJ, Tsalik EL, van Velkinburgh JC, Glickman SW, Rice BJ, Wang C, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. 2013;5:195ra95. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janke AT, Overbeek DL, Kocher KE, Levy PD. Exploring the Potential of Predictive Analytics and Big Data in Emergency Care. Annals of emergency medicine. 2016;67:227–36. doi: 10.1016/j.annemergmed.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Peterson JF, Bowton E, Field JR, Beller M, Mitchell J, Schildcrout J, et al. Electronic Health Record Design and Implementation for Pharmacogenomics: a 1 Local Perspective. Genetics in medicine : official journal of the American College of Medical Genetics. 2013;15:833–41. doi: 10.1038/gim.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a Large-Scale De-Identified DNA Biobank to Enable Personalized Medicine. Clinical pharmacology and therapeutics. 2008;84:362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–20. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaas AK, Chen M, Varkey J, Veldman T, Hero AO, 3rd, Lucas J, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–17. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaas AK, Burke T, Chen M, McClain M, Nicholson B, Veldman T, et al. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci Transl Med. 2013;5:203ra126. doi: 10.1126/scitranslmed.3006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finnerty CC, Jeschke MG, Qian WJ, Kaushal A, Xiao W, Liu T, et al. Determination of burn patient outcome by large-scale quantitative discovery proteomics. Crit Care Med. 2013;41:1421–34. doi: 10.1097/CCM.0b013e31827c072e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren HS, Elson CM, Hayden DL, Schoenfeld DA, Cobb JP, Maier RV, et al. A genomic score prognostic of outcome in trauma patients. Mol Med. 2009;15:220–7. doi: 10.2119/molmed.2009.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong HR, Salisbury S, Xiao Q, Cvijanovich NZ, Hall M, Allen GL, et al. The pediatric sepsis biomarker risk model. Critical care (London, England) 2012;16:R174. doi: 10.1186/cc11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biffi A, Anderson CD, Jagiella JM, Schmidt H, Kissela B, Hansen BM, et al. APOE genotype and extent of bleeding and outcome in lobar intracerebral haemorrhage: a genetic association study. Lancet Neurol. 2011;10:702–9. doi: 10.1016/S1474-4422(11)70148-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg S, Elashoff MR, Beineke P, Daniels SE, Wingrove JA, Tingley WG, et al. Multicenter validation of the diagnostic accuracy of a blood-based gene expression test for assessing obstructive coronary artery disease in nondiabetic patients. Ann Intern Med. 2010;153:425–34. doi: 10.7326/0003-4819-153-7-201010050-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah SH, Sun JL, Stevens RD, Bain JR, Muehlbauer MJ, Pieper KS, et al. Baseline metabolomic profiles predict cardiovascular events in patients at risk for coronary artery disease. Am Heart J. 2012;163:844–50. e1. doi: 10.1016/j.ahj.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Nakada T-a, Russell JA, Boyd JH, Aguirre-Hernandez R, Thain KR, Thair SA, et al. β2-Adrenergic Receptor Gene Polymorphism Is Associated with Mortality in Septic Shock. American Journal of Respiratory and Critical Care Medicine. 2010;181:143–9. doi: 10.1164/rccm.200903-0332OC. [DOI] [PubMed] [Google Scholar]
- 27.Decoux A, Tian Y, Deleon-Penelle KY, Nguyen NT, deCarstro LE, Flynn ER, et al. Plasma Glycoproteomics Reveals Sepsis Outcomes Linked to Distinct Proteins in Common Pathways. Critical Care Medicine. 2015 doi: 10.1097/CCM.0000000000001134. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monte AA, Heard KJ, Campbell J, Hamamura D, Weinshilboum RM, Vasiliou V. The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Academic emergency medicine : official journal of the Society for Academic Emergency Medicine. 2014;21:879–85. doi: 10.1111/acem.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puskarich MA, Finkel MA, Karnovsky A, Jones AE, Trexel J, Harris BN, et al. Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Ann Am Thorac Soc. 2015;12:46–56. doi: 10.1513/AnnalsATS.201409-415OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.del Rio-Espinola A, Fernandez-Cadenas I, Giralt D, Quiroga A, Gutierrez-Agullo M, Quintana M, et al. A predictive clinical-genetic model of tissue plasminogen activator response in acute ischemic stroke. Ann Neurol. 2012;72:716–29. doi: 10.1002/ana.23664. [DOI] [PubMed] [Google Scholar]
- 31.Evans BJ, Burke W, Jarvik GP. The FDA and Genomic Tests — Getting Regulation Right. New England Journal of Medicine. doi: 10.1056/NEJMsr1501194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lander ES. Cutting the Gordian Helix — Regulating Genomic Testing in the Era of Precision Medicine. New England Journal of Medicine. 2015;372:1185–6. doi: 10.1056/NEJMp1501964. [DOI] [PubMed] [Google Scholar]
- 33.dbGaP Overview. National Center for Biotechnology Information; 2015. at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/about.html. [Google Scholar]
- 34. [Accessed Accessed [1/18/16]];DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) 2016 at http://www.genome.gov/sequencingcosts.
- 35.Pare G. Genome-wide association studies--data generation, storage, interpretation, and bioinformatics. J Cardiovasc Transl Res. 2010;3:183–8. doi: 10.1007/s12265-010-9181-y. [DOI] [PubMed] [Google Scholar]
- 36.Katsanis SH, Minear MA, Vorderstrasse A, Yang N, Reeves JW, Rakhra-Burris T, et al. Perspectives on genetic and genomic technologies in an academic medical center: the duke experience. J Pers Med. 2015;5:67–82. doi: 10.3390/jpm5020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linnstaedt SD, Hu J, Bortsov AV, Soward AC, Swor R, Jones J, et al. mu-Opioid Receptor Gene A118G Variants and Persistent Pain Symptoms among Men and Women Experiencing Motor Vehicle Collision. J Pain. 2015 doi: 10.1016/j.jpain.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subudhi AW, Bourdillon N, Bucher J, Davis C, Elliott JE, Eutermoster M, et al. AltitudeOmics: the integrative physiology of human acclimatization to hypobaric hypoxia and its retention upon reascent. PLoS One. 2014;9:e92191. doi: 10.1371/journal.pone.0092191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez-Cadenas I, Del Rio-Espinola A, Giralt D, Domingues-Montanari S, Quiroga A, Mendioroz M, et al. IL1B and VWF variants are associated with fibrinolytic early recanalization in patients with ischemic stroke. Stroke. 2012;43:2659–65. doi: 10.1161/STROKEAHA.112.657007. [DOI] [PubMed] [Google Scholar]
- 40.Voellenkle C, van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2010;42:420–6. doi: 10.1152/physiolgenomics.00211.2009. [DOI] [PubMed] [Google Scholar]