Abstract
Sporulating Bacillus subtilis cells assemble a multimeric membrane complex connecting the mother cell and developing spore that is required to maintain forespore differentiation. An early step in the assembly of this transenvelope complex (called the A-Q complex) is an interaction between the extracellular domains of the forespore membrane protein SpoIIQ and the mother cell membrane protein SpoIIIAH. This interaction provides a platform onto which the remaining components of the complex assemble and also functions as an anchor for cell-cell signaling and morphogenetic proteins involved in spore development. SpoIIQ is required to recruit SpoIIIAH to the sporulation septum on the mother-cell side, however the mechanism by which SpoIIQ specifically localizes to the septal membranes on the forespore side has remained enigmatic. Here, we identify GerM, a lipoprotein previously implicated in spore germination, as the missing factor required for SpoIIQ localization. Our data indicate that GerM and SpoIIIAH, derived from the mother cell, and SpoIIQ, from the forespore, have reciprocal localization dependencies suggesting they constitute a tripartite platform for the assembly of the A-Q complex and a hub for the localization of mother cell and forespore proteins.
Keywords: protein localization, sporulation, sigmaG, specialized secretion systems
Introduction
Bacteria possess highly organized internal architectures that are intimately linked to essential biological processes (Lenz & Sogaard-Andersen, 2011, Rudner & Losick, 2010, Shapiro et al., 2009). The mechanisms by which proteins and protein complexes localize to specific subcellular sites remain incompletely understood (Rudner & Losick, 2010). In many cases, the localization of one protein, or a set of proteins, to a specific subcellular position depends on others and this dependency underlies the ordered assembly of large macromolecular complexes (Goehring et al., 2006, Gamba et al., 2009, Diepold et al., 2010, Li & Sourjik, 2011). An example of this ordered assembly can be found in specialized secretion systems (Diepold et al., 2010, Lybarger et al., 2009, Chandran, 2013). These multicomponent nano-machines are involved in the transport of proteins (and sometimes DNA) between bacterial cells or between bacteria and host cells (Chandran, 2013, Buttner, 2012, Portaliou et al., 2016, Dalbey & Kuhn, 2012). Their assembly typically involves the formation of a basal platform followed by the ordered association of the remaining components (Diepold et al., 2010, Lybarger et al., 2009, Morimoto et al., 2014, Diepold et al., 2011). Here, we define the molecular basis for the localization of the basal platform of a novel transenvelope complex that resembles a specialized secretion system connecting two daughter cells during the developmental process of sporulation.
In response to starvation, B. subtilis enters a developmental pathway that culminates in the formation of a stress-resistant spore (Errington, 2003, Higgins & Dworkin, 2012, Tan & Ramamurthi, 2014). The first morphological event in this process is the formation of an asymmetric septum, generating two cells of unequal size and distinct developmental fates. The smaller cell (called the forespore) differentiates into the dormant spore while the larger cell (referred to as the mother cell) nurtures the forespore and prepares it for dormancy. During this developmental process, the mother cell and forespore follow cell-type-specific programs of gene expression that are linked to each other by cell-cell signaling pathways. Polar division triggers the activation of the σF transcription factor in the forespore, which, in turn, leads to the activation of σE in the mother cell. At a later stage, transcription under σG control in the forespore triggers σK activation in the mother cell. Initially, the forespore and mother cell lie side-by-side separated by a double membrane septum. However, shortly after polar division, cell wall hydrolases produced in the mother cell degrade the septal peptidoglycan and aid in the migration of the mother-cell membranes around the forespore in a phagocytic-like process called engulfment. Upon completion of engulfment, the forespore resides as a free protoplast in the mother cell. At this late stage, the mother cell packages the spore in protective layers while the spore prepares for dormancy. Finally, mother cell lysis releases the mature spore into the environment.
During the morphological process of engulfment the mother cell and forespore assemble a multimeric complex that spans the double membrane between them (Doan et al., 2005, Doan et al., 2009, Blaylock et al., 2004). This transenvelope complex (called the A-Q complex) is composed of eight mother cell proteins (SpoIIIAA-SpoIIIAH, referred to as AA-AH, for simplicity) encoded in the spoIIIA operon (Illing & Errington, 1991) and one forespore protein SpoIIQ (Q) (Londono-Vallejo et al., 1997). Sporulating cells lacking any of these factors produce forespores that are smaller in size, develop membrane invaginations, and in some instances lose their integrity (Doan et al., 2009, Rodrigues et al., 2013). In addition, these forespores are unable to maintain transcriptional potential including gene expression under σG control (Doan et al., 2009, Camp & Losick, 2009, Sun et al., 2000). Thus, this complex is essential to maintain forespore development. Several of the SpoIIIA proteins share remote homology to components of specialized secretion systems found in Gram-negative bacteria, suggesting that the A-Q complex functions as a novel secretion apparatus (Doan et al., 2009, Camp & Losick, 2008, Meisner et al., 2008). In support of this idea, the extracellular domain of SpoIIIAH (AH) has been shown to share structural homology with the PrgK/EscJ ring-forming proteins found in Type III secretion systems (Yip et al., 2005, Meisner et al., 2012, Levdikov et al., 2012). It remains unclear what the A-Q complex transports; however, the mother cell protein SpoIIIAA (AA) resembles a secretion ATPase suggesting that if this complex is a secretion system then transport likely occurs from mother cell to forespore (Doan et al., 2009). In the context of this model, the secreted factor(s) would be necessary to maintain the metabolic and/or transcriptional potential of the forespore (Camp & Losick, 2009).
One of the earliest steps in the assembly of the A-Q complex is the interaction between the forespore membrane protein Q and the mother cell membrane protein AH. The extracellular domains of these two proteins associate in the space between the double membrane septum (Doan et al., 2005, Blaylock et al., 2004). This transenvelope interaction is required for the assembly of the rest of the complex and is thought to function as a basal platform (Camp & Losick, 2008, Doan et al., 2009). However, protein localization studies indicate that the assembly of this platform requires additional factors (Rodrigues et al., 2013, Rubio & Pogliano, 2004, Fredlund et al., 2013). In particular, although the specific localization of AH to the septal membrane on the mother-cell side depends on Q, the septal localization of Q on the forespore side is largely unaffected by the absence of AH (Rodrigues et al., 2013, Fredlund et al., 2013). We have previously shown that, in addition to AH, proper localization of Q requires degradation of the septal peptidoglycan and an additional unidentified protein produced in the mother cell under σE control (Rodrigues et al., 2013).
Here, we report that GerM, a lipoprotein previously implicated in spore germination (Sammons et al., 1987, Slynn et al., 1994), is the missing mother cell protein required for Q localization. We show that GerM and AH are required to anchor Q at the septum and in their absence Q becomes uniformly distributed in the forespore membranes. Furthermore, forespores in the double mutant fail to thrive, do not maintain σG activity, and exhibit a synergistic sporulation defect. Consistent with the idea that GerM is Q's elusive partner, we show that GerM is sufficient to localize Q to the septal membrane in the absence of all other σE-dependent proteins, provided the septal peptidoglycan is thinned. Furthermore, protein localization studies reveal that GerM is surface-exposed and, like AH (Doan et al., 2005, Blaylock et al., 2004), localizes to the septal membrane in a manner that depends on Q. Finally, we show that GerM is required for the proper localization of SpoIIIAG (AG), an essential component of the A-Q complex. Collectively, our results suggest that AH, Q and GerM form a tripartite basal platform in the assembly of the A-Q transenvelope complex.
Results
GerM is required for σG activity and forespore development
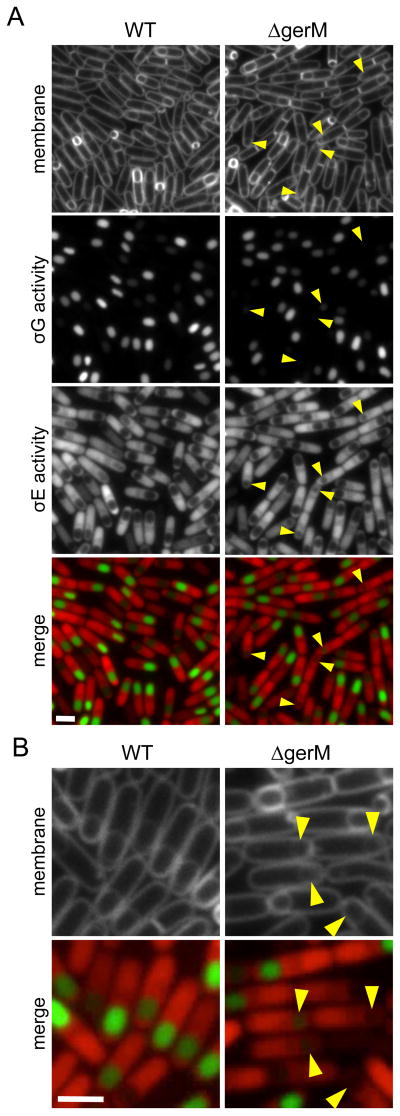
In the course of our analysis of new sporulation genes identified by Transposon-Sequencing (Meeske et al., 2016), we characterized a subset of previously identified sporulation mutants that had not been examined cytologically. One of these was gerM, a gene identified almost 30 years ago in a screen for germination mutants (Sammons et al., 1987). Cells lacking gerM were reported to have pleiotropic defects in sporulation and impaired germination (Sammons et al., 1987, Slynn et al., 1994). We introduced the gerM null mutant into a strain that harbored fluorescent reporters for all four sporulation-specific sigma factors (Meeske et al., 2016) and analyzed the cells during a sporulation time course (Fig. 1 and Fig. S1). No obvious defects were observed at the early stages of sporulation (Fig. S1); however, at 3.5 hours (T3.5) after the onset of sporulation, a subset of sporulating cells lacking GerM had reduced gene expression under the control of the late-acting forespore transcription factor σG (Fig. 1A and quantified in Fig. 2). Furthermore, in many cases these forespores appeared smaller in size (Fig.1 A and B). Introduction of gerM at an ectopic locus restored σG activity, forespore morphology, and wild-type levels of sporulation (Fig. S2). The mutant phenotypes associated with the absence of GerM were similar to those observed in cells lacking proteins in the A-Q transenvelope complex (Doan et al., 2009, Rodrigues et al., 2013), suggesting that gerM might function in the same genetic pathway.
Figure 1. GerM is required for σG activity and forespore morphology.
A. Representative images of wild-type (WT, BCR1071) and ΔgerM (BAM833) sporulating cells at hour 3.5 (T3.5) of sporulation. Images (from top to bottom) are membrane staining with TMA-DPH, σG activity (PsspB-cfp), σE activity (PspoIID-mCherry) and merge of σG activity (green) and σE activity (red). Small and/or collapsed forespores with reduced σG activity are highlighted (yellow carets). Scale bar indicates 2 μm. A complete sporulation time-course comparing wild-type and ΔgerM can be found in Figure S1. B. Larger images highlighting the defects in σG activity and forespore morphology in the ΔgerM mutant.
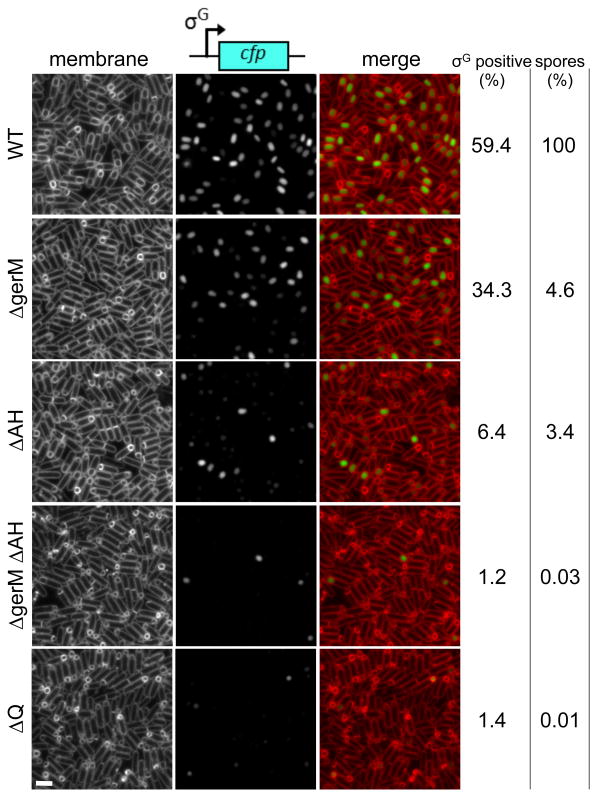
Figure 2. Synergistic defects in the gerM AH double mutant.
Representative images of sporulating cells harboring the σG-dependent reporter PsspB-cfp at hour 4 after the onset of sporulation. Images are wild-type (WT, BTD1609), ΔgerM (BCR1190), ΔAH (BCR1233), ΔgerM ΔAH double mutant (BCR1200), and ΔQ (BCR151). TMA-DPH-stained membranes (left), σG activity (middle) and a merged image (right) are shown. Scale bar represents 2 μm. The percentage of σG positive cells (n>600) at hour 4 are shown (see Materials and Methods for details). Spore titers relative to wild-type at hour 30 are indicated on the right. The data are representative of two biological replicates.
gerM is in the A-Q pathway
To investigate whether gerM is in the A-Q pathway, we took advantage of the partially penetrant phenotypes of the AH mutant. Unlike all other components of the A-Q complex, cells lacking AH have a relatively mild sporulation defect (1-10% sporulation efficiency compared to 0.01- 0.001% for mutants in any other component) and ∼95% of the forespores are smaller in size and have reduced σG activity (Fig. 2) (Doan et al., 2009). The ΔgerM mutant had a similar reduction in sporulation efficiency but a less penetrant defect in σG activity and forespore size (Fig. 2). If the role of GerM in maintaining forespore morphology and σG activity is related to that of the A-Q complex, then we would expect a ΔgerM ΔAH double mutant to display synergistic phenotypes and this is what we observed (Fig. 2). The ΔgerM ΔAH double mutant had a sporulation efficiency of 0.03%, similar to the ΔQ mutant (Fig. 2). Furthermore, almost all of the sporulating cells lacking GerM and AH had smaller forespores and reduced σG activity (Fig. 2). Introduction of gerM at an ectopic locus in the ΔgerM ΔAH double mutant restored sporulation efficiency and σG activity to levels observed in the ΔAH null (Fig. S2). These results are consistent with the idea the GerM functions in the A-Q pathway and raised the possibility it is part of this transenvelope complex.
GerM and AH are required for Q localization
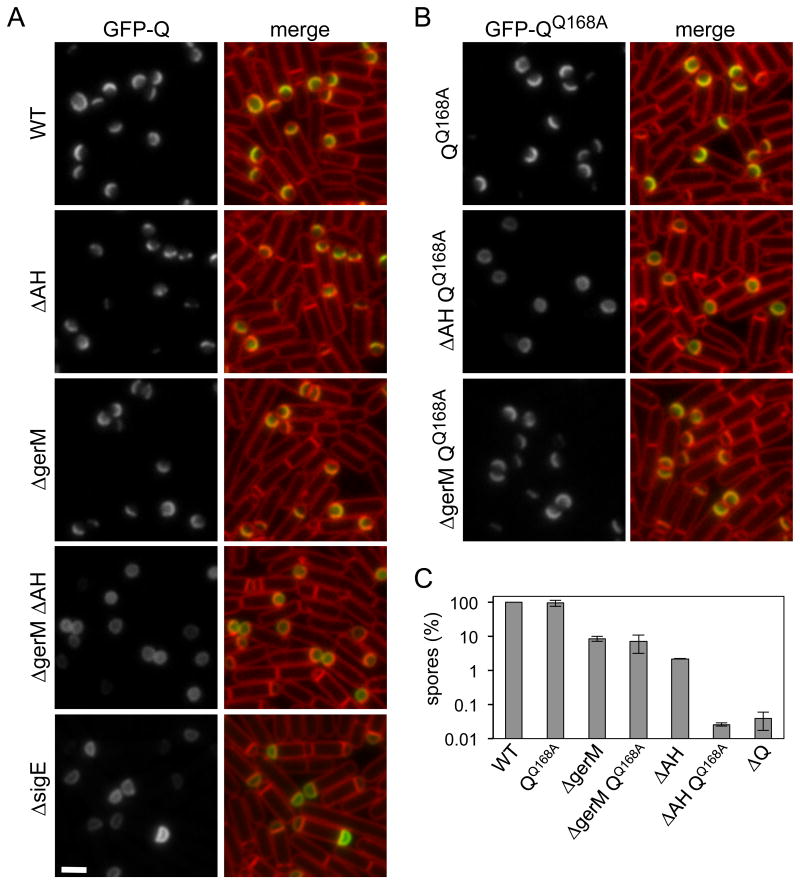
Our previous work indicated that Q localization requires AH and a second unidentified protein synthesized in the mother cell under σE control (Rodrigues et al., 2013). gerM is predicted to encode a secreted lipoprotein (PRED-LIPO; reliability score of 0.995, (Bagos et al., 2008)) and expression profiling, 5′ end mapping (Eichenberger et al., 2004, Feucht et al., 2003, Steil et al., 2005) and our fluorescence microscopy indicate that gerM is part of the σE regulon (Fig. S3). Accordingly, we investigated whether GerM was the missing mother-cell protein required to localize Q to the septal membranes on the forespore side. In the absence of AH or GerM, a functional GFP-Q fusion retained much of its localization in the septal membranes with weaker and heterogenous signal in the peripheral membranes (Fig. 3A). Strikingly, in cells lacking both GerM and AH, GFP-Q was evenly distributed in all forespore membranes (Fig. 3A). The extent of mislocalization was indistinguishable from that observed in cells lacking the mother cell transcription factor σE (Fig. 3A) (Rubio & Pogliano, 2004). Finally and as expected, introduction of gerM at an ectopic locus in the ΔgerM ΔAH double mutant restored proper Q localization (Fig. S4).
Figure 3. GFP-Q localization requires GerM and AH.
A. Representative images of GFP-Q localization in sporulating cells at hour 2 of sporulation. Images are from wild-type (BCR46), ΔAH (BCR56), ΔgerM (BCR1211), the ΔAH ΔgerM double mutant (BCR1197), and ΔsigE (BKM1930). Scale bar represents 2 μm. B. Representative images of GFP-QQ168A localization in sporulating cells at hour 2 of sporulation. Images show GFP-QQ168A in an otherwise wild-type background (BCR87), ΔAH (BCR80), or ΔgerM (BCR1313). C. Bar graph showing sporulation efficiencies of wild-type (BCR163), QQ168A (BCR152), ΔgerM (BCR1314), the ΔgerM QQ168A double mutant (BCR1313), ΔAH (BCR1335), the ΔAH QQ168A double mutant (BCR1334) and ΔQ (BTD1541). Error bars represent standard deviations (n=2).
The extracellular domain of Q contains a degenerate LytM domain (Meisner & Moran, 2011). Functional LytM domains cleave peptide crossbridges that link the glycan strands in peptidoglycan (Odintsov et al., 2004). Although Q's LytM domain lacks the catalytic residues required for endopeptidase activity, the domain adopts a similar fold (Meisner et al., 2012, Levdikov et al., 2012). We have previously shown that the substrate-binding groove in the degenerate LytM domain of Q is likely to function as the interaction surface for its second anchoring protein (Rodrigues et al., 2013). Specifically, we showed that a point mutation in this groove (Q168A) had no discernable impact on the localization of Q or on sporulation efficiency (Fig 3B and C). However, in combination with an AH null mutant, GFP-QQ168A was almost completely mislocalized (Fig. 3B) (Rodrigues et al., 2013). Moreover, the QQ168A ΔAH double mutant had a synergistic sporulation defect (Fig. 3C). Accordingly, if GerM is the second anchoring protein for Q, our data predict it would act through the LytM groove. Consistent with this idea, GFP-QQ168A retained most of its septal localization in the absence of GerM and the sporulation efficiency of the ΔgerM QQ168A double mutant was no worse than the ΔgerM single mutant (Fig. 3B and C).
GerM localizes to the outer forespore membrane in a manner that depends on the Q's LytM groove and AH
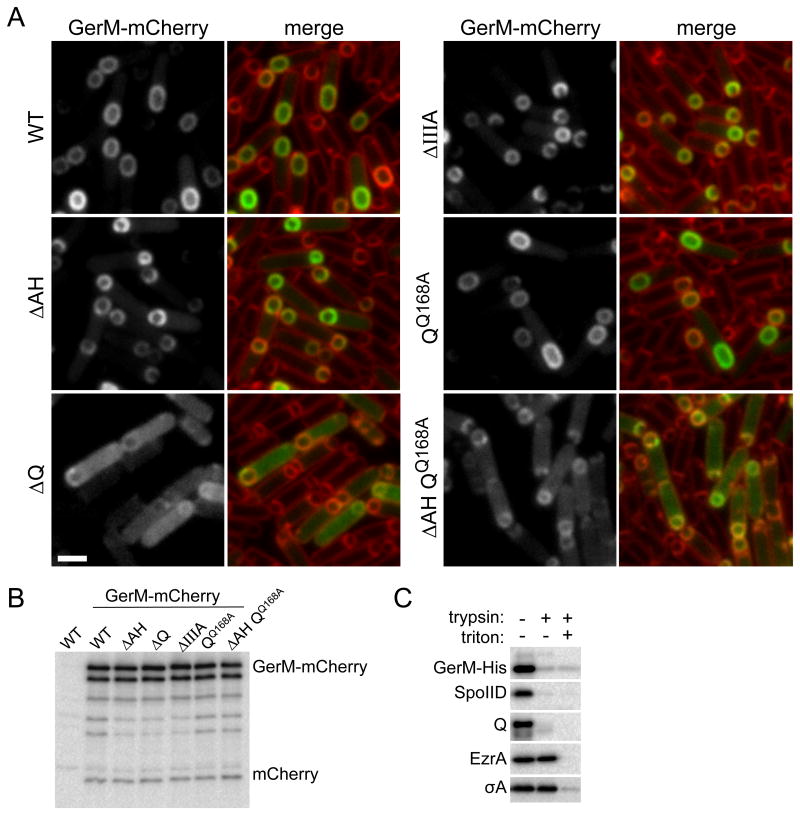
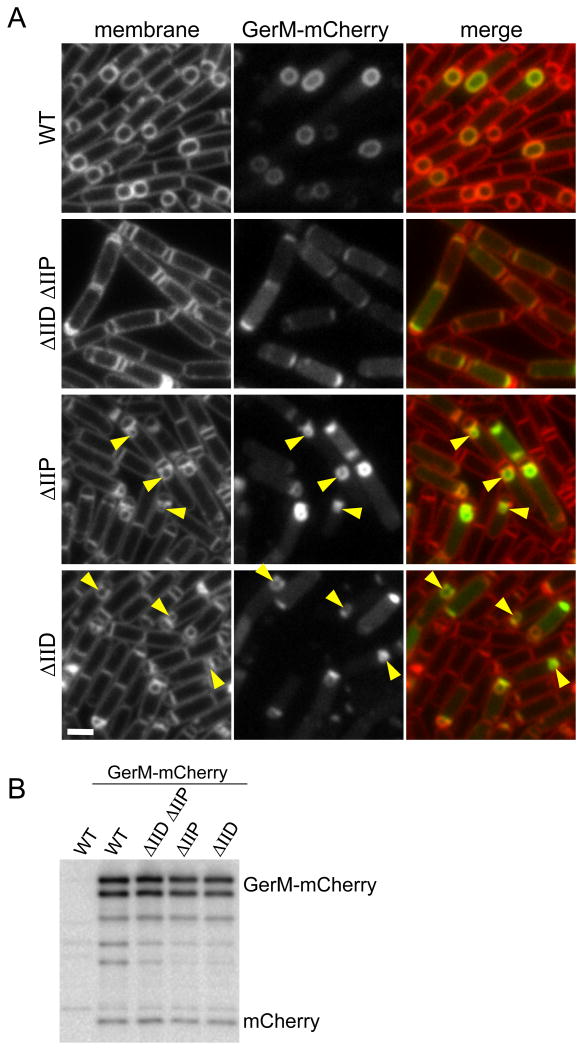
GerM is predicted to be a lipoprotein that is anchored in the outer leaflet of the mother cell membrane. In support of this idea, we found that a functional GerM-His6 fusion (Fig. S2 and S4) was membrane-associated and susceptible to trypsin cleavage in a protease accessibility assay (Fig. 4C). To determine the subcellular localization of GerM in the mother cell membranes, we generated a functional GerM-mCherry fusion (Fig. S5) and monitored its localization during a sporulation time course. Because of the slow maturation of mCherry (Shaner et al., 2005, Merzlyak et al., 2007), the earliest time point at which we could detect the fluorescent fusion was hour 2.5. However, consistent with the idea that GerM is part of the A-Q complex, GerM-mCherry localized to the outer forespores membrane during and after the completion of engulfment (Fig. 4A). Moreover, GerM localization to the forespore membrane was significantly reduced in the absence of Q (Fig. 4A). The mislocalized GerM-mCherry appeared as a diffuse cytoplasmic haze as if the fusion was cleaved releasing soluble mCherry. However, immunoblot analysis revealed that most of the fusion remained full-length and that the small degree of proteolysis was similar in all backgrounds examined (Fig. 4B). A similar diffuse localization has been observed for the polytopic membrane protein SpoIIE (King et al., 1999) and CFP-AH in the absence of Q (Doan et al., 2005). The molecular basis of this localization pattern is currently unknown.
Figure 4. GerM-mCherry localizes to septal membrane in a manner that depends on Q.
A. Representative images of GerM-mCherry localization at 2.5 hours after the onset of sporulation. Images are from wild-type (BCR1332), ΔAH (BCR1344), ΔQ (BCR1345), ΔspoIIIA (BCR1346), QQ168A (BCR1348) and the ΔAH QQ168A double mutant (BCR1353). Scale bar represents 2 μm. B. GerM-mCherry levels and proteolytic products are similar in all mutant backgrounds tested. Immunoblot analysis using anti-mCherry antibodies of sporulating cells from the same strains described above and with a true wild-type control (PY79, no mCherry) harvested at hour 2.5 of sporulation. The nature of the GerM-mCherry doublet is currently unknown. C. GerM-His6 is surface exposed and thus accessible to trypsin digestion. Immunoblot analysis using anti-His antibodies of protoplasted sporulating cells (strain BCR1306) treated with Trypsin in the presence and absence of TritonX-100. Consistent with the idea that GerM is a lipoprotein, it remained cell-associated after the generation of protoplasts. As controls, the immunoblot was probed for two membrane proteins with extracellular domains (SpoIID and Q), a membrane-anchored cytoplasmic protein (EzrA), and a cytoplasmic protein (SigA).
To investigate whether Q's LytM groove was required for GerM localization, we monitored GerM-mCherry in the QQ168A mutant. The localization of the mCherry fusion in this background was similar to wild-type (Fig. 4A). One possible explanation for the absence of a localization defect is that GerM also interacts with AH, which associates with a distinct interface on Q (Meisner et al., 2012, Levdikov et al., 2012). To test this idea, we examined GerM-mCherry in a ΔAH, QQ168A double mutant. Under these conditions, GerM-mCherry had a pronounced mislocalization phenotype (Fig.4A) although it was not as strong as in the Q null suggesting that the Q168A substitution is not sufficient to completely disrupt Q-dependent localization of GerM. Altogether, our results are consistent with the idea that GerM, Q, and AH form the basement layer of the A-Q transenvelope complex.
GerM localization requires septal peptidoglycan hydrolysis
Our previous studies revealed that the unidentified mother cell protein that helps anchor Q in the forespore membrane requires degradation of septal peptidoglycan (PG), presumably to bring the two membranes into close apposition to allow for efficient interaction with Q (Rodrigues et al., 2013). If GerM is indeed this anchor, its interaction with Q and by extension its localization, should depend on thinning of the septal PG. To test this, we monitored GerM-mCherry localization in a strain lacking the two cell wall hydrolases SpoIID and SpoIIP that degrade the septal PG after polar division (Abanes-De Mello et al., 2002, Chastanet & Losick, 2007, Morlot et al., 2010). In the absence of both enzymes, GerM-mCherry had a diffuse membrane localization phenotype similar to what was observed in the absence of Q (Fig. 5A). Furthermore, consistent with the idea that septal PG hydrolysis allows GerM to interact with Q, in sporulating cells lacking either SpoIID or SpoIIP, GerM-mCherry was enriched at sites where the PG was thinned and the septal membranes bulged into the mother cell cytoplasm (Fig 5A and S6). Finally, immunoblot analysis indicates that GerM-mCherry was predominantly full-length with similar amounts of smaller proteolytic products in all backgrounds examined (Fig. 5B).
Figure 5. GerM-mCherry localization to the septal membrane requires thinning of the septal peptidoglycan.
A. Representative images of GerM-mCherry localization at hour 2.5 of sporulation. Images are from wild-type (BCR1332), the ΔspoIID ΔspoIIP double mutant (BCR1381), ΔspoIIP (BCR1347), and ΔspoIID (BCR1414). Enrichment of GerM-mCherry at septal bulges is highlighted (yellow carets). Larger fields of cells can be found in Figure S6. Scale bar represents 2 μm. B. GerM-mCherry levels and proteolytic products are similar in all mutant backgrounds tested. Immunoblot analysis using anti-mCherry antibodies of sporulating cells from the same strains described above and with a true wild-type control (PY79, no mCherry) harvested at hour 2.5 of sporulation.
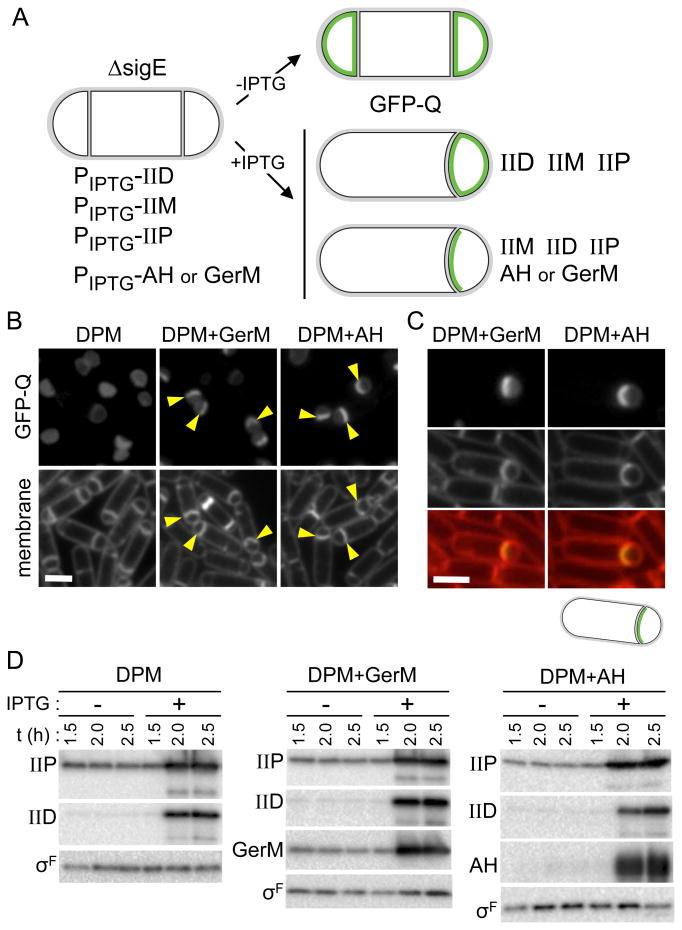
GerM is sufficient to localize Q in the absence of all other σE-dependent proteins
All of our data thus far are consistent with the idea that GerM is the missing σE-dependent protein that, together with AH, anchors Q in the septal membrane on the forespore side. However, despite extensive effort using in vitro and in vivo protein-protein interaction assays, we were unable to detect a direct interaction between GerM and the extracellular domains of Q, AH or both. We reasoned that if GerM is in fact Q's missing σE-dependent anchor then expression of GerM in the absence of all other σE-dependent proteins should be sufficient to localize Q. To test this, we took advantage of a strain we used previously to investigate whether the second Q anchor is a protein under σE control (Fig. 6A) (Rodrigues et al., 2013). This strain lacks sigE but contains IPTG-inducible alleles of spoIID, spoIIP and spoIIM encoding the cell wall degrading machinery to ensure thinning of the septal PG. GFP-Q fails to localize to the septal membranes in this background (Fig. 6A and B) (Rodrigues et al., 2013). To test if GerM is sufficient to localize Q, we introduced an IPTG-inducible allele of gerM-his6 into this strain (Fig. 6A). As a control for this experiment, we constructed a complementary strain containing an IPTG-inducible allele of AH. When GerM was expressed in addition to SpoIID, SpoIIP and SpoIIM, GFP-Q was enriched at the septum (Fig. 6B, C and D) (Fig. S7A, B and C). As expected, a similar enrichment of GFP-Q was observed when AH was expressed (Fig. 6B, C and D) (Fig. S7A, B and C). These results indicate that GerM or AH alone is sufficient to localize Q. Collectively, the data presented here and our previous analysis of Q localization (Rodrigues et al., 2013) indicate that GerM and AH are the two σE-controlled proteins required to anchor Q in the septal membrane.
Figure 6. GerM is sufficient to localize GFP-Q at the sporulation septum.
A. Experimental rationale and schematic outcomes for GFP-Q localization (in green) when SpoIID, SpoIIP and SpoIIM, alone (BCR1444) or together with either GerM (BCR1447) or AH (BCR1446) are artificially produced in a ΔsigE mutant. B. Representative images of cells at hour 2.5 of sporulation in which IPTG (1 mM final) was added 1.5 hours after the onset of sporulation. Examples of engulfing septal membranes with enrichment of GFP-Q in the forespore membranes are indicated (yellow carets). Scale bar represents 2 μm. As described previously (Rodrigues et al., 2013), in a subset of sporulating cells (in strain BCR1444 and derivatives) in which IPTG was added GFP-Q lost compartmentalization (not shown). Images of the same strains from the same time point in the absence of IPTG can be found in Figure S8. C. Larger images highlighting GFP-Q enrichment in the engulfing membrane when either GerM or AH was induced. D. Immunoblot analysis monitoring SpoIID, SpoIIP, GerM-His6 and AH accumulation upon the addition of IPTG. σF was used to control for loading.
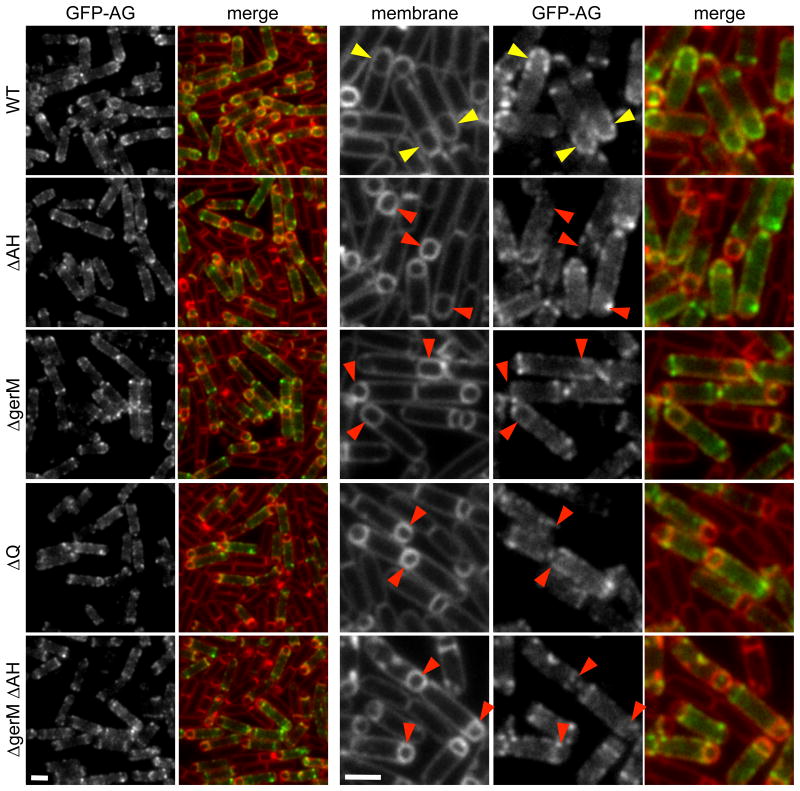
GerM and AH are necessary for SpoIIIAG localization
In previous work, we showed by co-immunoprecipitation that at least five of the SpoIIIA proteins (SpoIIIAB, SpoIIIAD, SpoIIIAE, SpoIIIAF, SpoIIIAG) reside in a multimeric membrane complex (Doan et al., 2009). Furthermore, we found that the localization of a partially functional CFP-SpoIIIAG (CFP-AG) fusion in the outer forespore membrane was substantially reduced in cells lacking Q and was impaired in the absence of AH (Doan et al., 2009). Since our data suggest that GerM functions as part of the basement layer of the A-Q complex, we investigated whether GerM was required for the localization of SpoIIIAG (AG). We monitored CFP-AH (Doan et al., 2005) and separately a partially functional GFP-AG fusion during a sporulation time course in the presence and absence of GerM. Consistent with our finding that GerM-mCherry retains its proper localization in the absence of AH (Fig. 4A), in the gerM mutant, CFP-AH localized to the outer forespore membrane in a manner indistinguishable from wild-type (Fig. S9). In a wild-type background, GFP-AG localized as bright puncta predominantly in the membranes surrounding the forespore as previously reported (Fig. 7) (Doan et al., 2009). In the gerM mutant, we observed a reduction in the number of forespores with bright GFP-AG puncta; instead, the GFP-AG signal was more dispersed in the peripheral membranes (Fig. 7). This mislocalization phenotype was enhanced by the loss of AH and qualitatively resembled the Q mutant (Fig. 7). These results suggest that GerM, AH, and Q function as a scaffold for AG and likely the entire SpoIIIA complex.
Figure 7. GerM is required for efficient localization of GFP-AG.
Representative images of GFP-AG localization at hour 2.5 of sporulation. Images are from wild-type (BCR1193), ΔAH (BCR1228), ΔgerM (BCR1328), ΔQ (BCR1340), and the ΔAH ΔgerM double mutant. (BCR1343). Larger images are shown on the right. Wild-type forespores with GFP-AG puncta are highlighted with yellow carets. Mutant forespores with reduced accumulation of GFP-AG puncta around the forespore are highlighted with red carets. Scale bar represents 2 μm.
Discussion
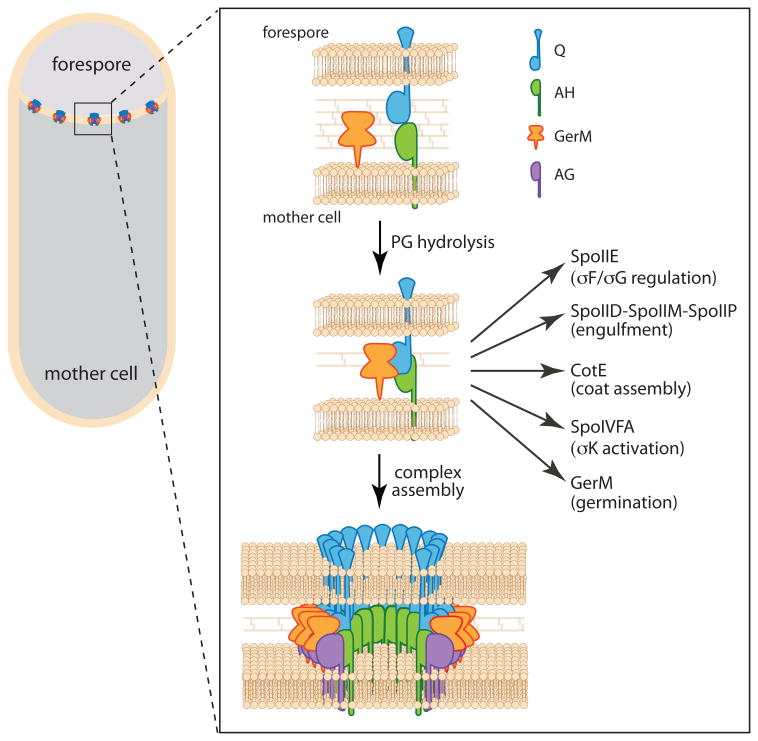
Here, we have identified GerM as the missing σE-controlled protein required to localize Q in the septal membrane. Our data suggest that Q, AH, and GerM constitute a basal platform upon which the A-Q complex is assembled (Fig. 8). In addition to its role in the A-Q complex, this platform has been found to function as an interaction hub, contributing to the localization of a diverse set of sporulation proteins on both sides of the septal membrane (Campo et al., 2008, Doan et al., 2005, Aung et al., 2007, Chastanet & Losick, 2007, McKenney & Eichenberger, 2012, Flanagan et al., 2016, Dworkin, 2014) (Fig. 8). Thus, by defining how this founder complex is localized to the septal membrane, our findings reveal how cell-cell signaling complexes and morphogenetic proteins achieve their proper localization.
Figure 8. Schematic model showing the assembly of the basal platform of the A-Q complex and its localization network.
Upon production of Q in the forespore and AH in the mother cell the two proteins associate across the sporulation septum. Degradation of the septal peptidoglycan by SpoIID and SpoIIP brings the two septal membranes closer together (Tocheva et al., 2013) allowing for stable association with GerM either through direct interaction as shown here or mediated by a unidentified bridging protein. This transenvelope complex directly or indirectly anchors morphogenetic and signaling proteins on both sides of the sporulation septum (see Discussion). The complex also serves as the basal platform for the assembly of AG and the rest of the proteins in the spoIIIA operon (not shown). The complex is shown as a ring-shaped conduit based on the structural similarity between AH and the EcsJ/PrgK family. By analogy to specialized secretion systems, it is hypothesized that one or several membrane proteins encoded in the spoIIIA operon assemble a pore in the mother cell membrane (not shown). Evidence suggests the existence of a pore in the forespore membrane (Meisner et al., 2008) (not shown), however the identity of this protein is unknown.
Evidence for a tripartite complex
In previous work we proposed the existence of an additional σE-dependent protein that anchors Q in the septal membranes (Rodrigues et al., 2013). While we were unable to detect a direct interaction between GerM and the extracellular domains of Q (or the AH-Q complex), by all other measures GerM is the missing σE-dependent protein and fits the requirements for Q's elusive partner: (i) GerM is specifically expressed under the control of σE in the mother cell; (ii) the mutant has a synergistic sporulation defect with ΔAH; (iii) cells lacking GerM and AH fail to localize Q; (iv) GerM's localization requires septal PG hydrolysis and Q; and finally (v) GerM is sufficient to localize Q in the absence of all other σE-dependent genes (provided the cell wall has been sufficiently thinned). Our inability to detect a direct interaction between Q and GerM leaves open the possibility that a protein produced earlier during sporulation or during vegetative growth functions as an additional component of the basal platform bridging GerM and Q. It is also formally possible that GerM indirectly affects assembly of the complex, for example by acting as a chaperone. However, we favor a simpler model in which GerM directly contacts Q and does so, in part, through its LytM groove. In the context of this model, the interaction between GerM and Q is weak and might require a specific conformation that is favored in the context of full-length Q and its transmembrane segment and/or when GerM is anchored in the membrane. Alternatively, some feature of the intermembrane space, like partially degraded peptidoglycan, might be necessary to stabilize the interaction between these proteins.
The data presented here and our previous findings and those of the Pogliano, Moran, Henriques and Losick groups (Blaylock et al., 2004, Fredlund et al., 2013, Meisner et al., 2008, Camp & Losick, 2008, Camp & Losick, 2009, Doan et al., 2005, Doan et al., 2009, Rodrigues et al., 2013) lead us to propose a working model for the assembly of the A-Q complex (Fig. 8). Assembly initiates with the activation of σF in the forespore. σF controls the production of Q (Londono-Vallejo et al., 1997) and is also responsible for triggering the activation of σE in the mother cell (Londono-Vallejo & Stragier, 1995). σE, in turn, directs the synthesis of the cell wall degrading machine (SpoIID/SpoIIM/SpoIIP) (Morlot et al., 2010, Chastanet & Losick, 2007, Abanes-De Mello et al., 2002), AH and the rest of the proteins in the spoIIIA operon, and GerM. Thinning of the septal peptidoglycan allows GerM to contact Q, reinforcing the association between AH and Q (Fig. 8). This tripartite complex then serves as the basal platform for the assembly of the remaining components of the A-Q complex. Based on remote homologies and the structural similarity between AH and EcsJ/PrgK family (Meisner et al., 2012, Levdikov et al., 2012), this complex is thought to assemble into a ring-shaped conduit that connects mother cell and forespore (Fig. 8). The complex could function as a specialized secretion system or feeding tube allowing the mother cell to nurture the forespore (Doan et al., 2009, Camp & Losick, 2009). Defining the structure of this complex and its role in maintaining forespore differentiation are exciting challenges for the future.
GerM is a novel component of the A-Q complex
That GerM's role in the assembly of the A-Q complex was missed by us, and others, for so many years, highlights the power of a name. The gerM mutant was originally defined as having pleiotropic defects during sporulation and a delay in spore germination (Sammons et al., 1987). Since the gene was identified in a germination screen it was given a “ger” designation rather than a “spo” name. Had it been called spoIIT or spoIIIL, its role in the A-Q pathway would likely have been discovered over a decade ago. In the original characterization of GerM (Sammons et al., 1987, Slynn et al., 1994), it was reported that a subset of sporulating cells lacking gerM arrest after polar division and the mutant had a reduction in glucose dehydrogenase activity, an activity associated with a late stage of sporulation. Our cytological analysis failed to detect a stage II block or a defect in engulfment. However, since the time of the original publication on gerM, the gene encoding glucose dehydrogenase (gdh) was found to be under σG control (Nakatani et al., 1989). Accordingly, the reduced activity in the mutant is fully consistent with our finding that a subset of sporulating cells lacking GerM have weak or undetectable σG activity. GerM's role in germination is currently unclear but our data are fully consistent with the idea that GerM has a second function in sporulation and/or germination beyond its role in the A-Q complex. Specifically, the defects in σG activity and forespore morphology were weaker in cells lacking GerM compared to the AH mutant (Fig. 2). Yet, the sporulation efficiencies in the two mutants were similar. Furthermore, unlike AH and Q that are degraded shortly after engulfment is complete (Chiba et al., 2007), GerM persists through late stages of sporulation (Fig. S10). GerM's second function could be related to the accumulation of dipicolinic acid in the spore and/or cortex hydrolysis upon germination (Slynn et al., 1994). Alternatively, GerM could influence germination indirectly by promoting proper coat assembly (McKenney & Eichenberger, 2012).
Bioinformatics analyses of B. subtilis GerM indicate that GerM homologs are present in virtually all the endospore formers of the Bacillaceae family but are absent in the Clostridiacaea (Fig. S11). Accordingly, if Q and the proteins in the spoIIIA operon assemble a similar transenvelope complex in the Clostridiacaea, the basal platform must differ in protein composition. Consistent with this idea, studies on the A-Q complex in Clostridium difficile point to the possibility that AH may be the sole anchor for Q. This hypothesis is a based on the observation that an AH mutant in C. difficile has a sporulation defect comparable to mutations in the other spoIIIA genes (Fimlaid et al., 2015, Serrano et al., 2015).
GerM contains two tandem copies of a novel domain designated GERMN (Rigden & Galperin, 2008). While this domain organization is restricted to a subset of endospore formers (Fig. S11), the GERMN domain is also present in isolation or fused to other protein domains in a diverse collection of bacterial phyla, including Actinobacteria, Cyanobacteria, Proteobacteria and in Deinococcus-Thermus group (Rigden & Galperin, 2008). Intriguingly, in the bacterium Halothermothrix orenii the GERMN domain is fused to an amidase domain involved in cell wall remodeling, suggesting GERMN could be a PG binding domain (Rigden & Galperin, 2008). If it is, GerM could stabilize the A-Q complex through an interaction with the remodeled cell wall in the intermembrane space. Future biochemical and structural analysis will be required to define the extent of GerM's role in the A-Q complex.
Materials and Methods
General methods
All B. subtilis strains were derived from the prototrophic strain PY79 (Youngman et al., 1983). Sporulation was induced by resuspension at 37°C according to the method of Sterlini-Mandelstam (Harwood & Cutting, 1990) or by exhaustion in supplemented DS medium (Schaeffer et al., 1965). Sporulation efficiency was determined in 24-30 hour cultures as the total number of heat-resistant (80°C for 20 min) colony forming units (CFUs) compared with wild-type heat-resistant CFUs. Deletion mutants were generated by isothermal assembly (Gibson, 2011) and direct transformation into B. subtilis. Tables of strains, plasmids and oligonucleotide primers and descriptions of plasmid construction and isothermal assembly deletion mutants can be found online as supplementary material.
Immunoblot analysis
Whole-cell lysates from sporulating cells (induced by resuspension) were prepared as described previously (Doan et al., 2009). Samples were heated for 10 min at 50°C prior to loading. Equivalent loading was based on OD600 at the time of harvest. Proteins were separated by SDS-PAGE on 12.5% polyacrylamide gels, electroblotted onto Immobilon-P membranes (Millipore) and blocked in 5% nonfat milk in phosphate-buffered saline (PBS)-0.5% Tween-20. The blocked membranes were probed with anti-SpoIID (1:10,000) (Doan & Rudner, 2007), anti-SpoIIQ (1:10,000) (Doan et al., 2005), anti-SpoIIIAH (1:10,000) (Doan et al., 2005), anti-SpoIIP (1:10,000) (Morlot et al., 2010), anti-σA (1:10,000) (Fujita, 2000), anti-EzrA (1:10,000) (Levin et al., 1999), anti-His (Genscript) (1:4000), anti-mCherry (1:10,000), diluted into 3% BSA in 1X PBS-0.05% Tween-20. Primary antibodies were detected using horseradish peroxidase-conjugated goat, anti-rabbit IgG (1:20,000, BioRad) and the Western Lightning reagent kit as described by the manufacturer (PerkinElmer).
Fluorescence microscopy
Fluorescence microscopy was performed with an Olympus BX61 microscope as previously described (Doan et al., 2009). Cells were mounted on a 2% agarose pad containing resupsension medium using a gene frame (BioRad). Fluorescent signals were visualized with a phase contrast objective UplanF1 100x and captured with a monochrome CoolSnapHQ digital camera (Photometrics) using Metamorph software version 6.1 (Universal Imaging). The membrane dye TMA-DPH (Molecular Probes) was used at a final concentration of 0.01 mM and exposure times were typically 200 ms. Images were analyzed, adjusted and cropped using Metamorph software.
Protease Susceptibility
Protease susceptibility assays were preformed as described previously (Doan & Rudner, 2007) in a spoIIIAH mutant (strain BCR1306) to ensure that membrane proteins present in the inner and outer forespore membrane would not be artificially inaccessible due to protoplast engulfment (Broder & Pogliano, 2006). 25 ml of sporulating cells (induced by resuspension) were harvested by centrifugation at 2 hours after the onset of sporulation, washed and resuspended in 2 ml 1X SMM buffer (0.5 M sucrose, 20 mM MgCl2, 20 mM maleic acid pH 6.5) (Harwood & Cutting, 1990). The cells were then protoplasted by lysozyme (5 mg/ml final) for 10 min. The protoplasts were harvested by centrifugation and resuspended in 1 ml of 1X SMM. 100 μl protoplasts were incubated with Trypsin (30 ug/ml) (Worthington), Trypsin and Triton X-100 (2%), or 1XSMM for 15 min. Reactions were terminated by the addition of 100 μl of 2X SDS-sample buffer and boiling for 5 min at 95°C. 5 μl from each reactions was analyzed by immunoblot.
Quantification of σG positive cells
σG activity was assessed in single cells as described previously (Rodrigues et al., 2013) at hour 4 after the onset of sporulation using the fluorescent reporter PsspB-cfp (Doan et al., 2009). A forespore was considered σG positive if it contained forespore fluorescence, it displayed normal forespore membrane morphology, and was of normal size. The second and third criteria were included in the analysis to ensure that those forespores that had just activated σG and therefore had faint forespore fluorescence were scored appropriately. Faint forespore fluorescence in normal-sized forespores with unperturbed membranes was scored as σG positive. Faint forespore fluorescence in small forespores with aberrant membrane morphologies indicative of arrested development (Doan et al., 2009) was scored as σG negative. The percentage of σG positive cells was calculated based on the total number of cells.
Supplementary Material
Acknowledgments
We thank members of the Rudner and Bernhardt labs for advice and encouragement, Olive Tang for anti-mCherry antisera, Lok-To Sham and Padraig Deighan for assistance with protein-protein interaction studies and Mary-Jane Tsang, Ting Pang, Jeffrey Meisner, and Cécile Morlot for stimulating discussions. Support for this work comes from the National Institute of Health Grant GM073831 (D.Z.R.) and RC2 GM092616 (D.Z.R.), F.H.R-G. is a recipient of a Conacyt postdoctoral fellowship (México).
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S, Pogliano K. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol Microbiol. 2007;65:1534–1546. doi: 10.1111/j.1365-2958.2007.05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ. Prediction of lipoprotein signal peptides in Gram-positive bacteria with a Hidden Markov Model. J Proteome Res. 2008;7:5082–5093. doi: 10.1021/pr800162c. [DOI] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Jr, Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008;69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N, Marquis KA, Rudner DZ. SpoIIQ anchors membrane proteins on both sides of the sporulation septum in Bacillus subtilis. J Biol Chem. 2008;283:4975–4982. doi: 10.1074/jbc.M708024200. [DOI] [PubMed] [Google Scholar]
- Chandran V. Type IV secretion machinery: molecular architecture and function. Biochem Soc Trans. 2013;41:17–28. doi: 10.1042/BST20120332. [DOI] [PubMed] [Google Scholar]
- Chastanet A, Losick R. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol Microbiol. 2007;64:139–152. doi: 10.1111/j.1365-2958.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- Chiba S, Coleman K, Pogliano K. Impact of membrane fusion and proteolysis on SpoIIQ dynamics and interaction with SpoIIIAH. J Biol Chem. 2007;282:2576–2586. doi: 10.1074/jbc.M606056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey RE, Kuhn A. Protein traffic in Gram-negative bacteria--how exported and secreted proteins find their way. FEMS Microbiol Rev. 2012;36:1023–1045. doi: 10.1111/j.1574-6976.2012.00327.x. [DOI] [PubMed] [Google Scholar]
- Diepold A, Amstutz M, Abel S, Sorg I, Jenal U, Cornelis GR. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 2010;29:1928–1940. doi: 10.1038/emboj.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Wiesand U, Cornelis GR. The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol Microbiol. 2011;82:502–514. doi: 10.1111/j.1365-2958.2011.07830.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr, Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Rudner DZ. Perturbations to engulfment trigger a degradative response that prevents cell-cell signalling during sporulation in Bacillus subtilis. Mol Microbiol. 2007;64:500–511. doi: 10.1111/j.1365-2958.2007.05677.x. [DOI] [PubMed] [Google Scholar]
- Dworkin J. Protein Targeting during Bacillus subtilis Sporulation. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.TBS-0006-2012. TBS-0006-2012. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Feucht A, Evans L, Errington J. Identification of sporulation genes by genome-wide analysis of the sigmaE regulon of Bacillus subtilis. Microbiology. 2003;149:3023–3034. doi: 10.1099/mic.0.26413-0. [DOI] [PubMed] [Google Scholar]
- Fimlaid KA, Jensen O, Donnelly ML, Siegrist MS, Shen A. Regulation of Clostridium difficile Spore Formation by the SpoIIQ and SpoIIIA Proteins. PLoS Genet. 2015;11:e1005562. doi: 10.1371/journal.pgen.1005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan KA, Comber JD, Mearls E, Fenton C, Wang Erickson AF, Camp AH. A membrane-embedded amino acid couples the SpoIIQ channel protein to anti-sigma factor transcriptional repression during Bacillus subtilis sporulation. J Bacteriol. 2016 doi: 10.1128/JB.00958-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund J, Broder D, Fleming T, Claussin C, Pogliano K. The SpoIIQ landmark protein has different requirements for septal localization and immobilization. Mol Microbiol. 2013;89:1053–1068. doi: 10.1111/mmi.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells. 2000;5:79–88. doi: 10.1046/j.1365-2443.2000.00307.x. [DOI] [PubMed] [Google Scholar]
- Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Gonzalez MD, Beckwith J. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol Microbiol. 2006;61:33–45. doi: 10.1111/j.1365-2958.2006.05206.x. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Wiley, New York: 1990. [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N, Errington J. The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the sigma E form of RNA polymerase. Mol Microbiol. 1991;5:1927–1940. doi: 10.1111/j.1365-2958.1991.tb00816.x. [DOI] [PubMed] [Google Scholar]
- King N, Dreesen O, Stragier P, Pogliano K, Losick R. Septation, dephosphorylation, and the activation of sigmaF during sporulation in Bacillus subtilis. Genes Dev. 1999;13:1156–1167. doi: 10.1101/gad.13.9.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz P, Sogaard-Andersen L. Temporal and spatial oscillations in bacteria. Nat Rev Microbiol. 2011;9:565–577. doi: 10.1038/nrmicro2612. [DOI] [PubMed] [Google Scholar]
- Levdikov VM, Blagova EV, McFeat A, Fogg MJ, Wilson KS, Wilkinson AJ. Structure of components of an intercellular channel complex in sporulating Bacillus subtilis. Proc Natl Acad Sci U S A. 2012;109:5441–5445. doi: 10.1073/pnas.1120087109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sourjik V. Assembly and stability of flagellar motor in Escherichia coli. Mol Microbiol. 2011;80:886–899. doi: 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Frehel C, Stragier P. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- Lybarger SR, Johnson TL, Gray MD, Sikora AE, Sandkvist M. Docking and assembly of the type II secretion complex of Vibrio cholerae. J Bacteriol. 2009;191:3149–3161. doi: 10.1128/JB.01701-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Eichenberger P. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol. 2012;83:245–260. doi: 10.1111/j.1365-2958.2011.07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Rodrigues CD, Brady J, Lim HC, Bernhardt TG, Rudner DZ. High-Throughput Genetic Screens Identify a Large and Diverse Collection of New Sporulation Genes in Bacillus subtilis. PLoS Biol. 2016;14:e1002341. doi: 10.1371/journal.pbio.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Maehigashi T, Andre I, Dunham CM, Moran CP., Jr Structure of the basal components of a bacterial transporter. Proc Natl Acad Sci U S A. 2012;109:5446–5451. doi: 10.1073/pnas.1120113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Moran CP., Jr A LytM domain dictates the localization of proteins to the mother cell-forespore interface during bacterial endospore formation. J Bacteriol. 2011;193:591–598. doi: 10.1128/JB.01270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr A channel connecting the mother cell and forespore during bacterial endospore formation. Proc Natl Acad Sci U S A. 2008;105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- Morimoto YV, Ito M, Hiraoka KD, Che YS, Bai F, Kami-Ike N, Namba K, Minamino T. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol Microbiol. 2014;91:1214–1226. doi: 10.1111/mmi.12529. [DOI] [PubMed] [Google Scholar]
- Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Nicholson WL, Neitzke KD, Setlow P, Freese E. Sigma-G RNA polymerase controls forespore-specific expression of the glucose dehydrogenase operon in Bacillus subtilis. Nucleic Acids Res. 1989;17:999–1017. doi: 10.1093/nar/17.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odintsov SG, Sabala I, Marcyjaniak M, Bochtler M. Latent LytM at 1.3A resolution. J Mol Biol. 2004;335:775–785. doi: 10.1016/j.jmb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Portaliou AG, Tsolis KC, Loos MS, Zorzini V, Economou A. Type III Secretion: Building and Operating a Remarkable Nanomachine. Trends Biochem Sci. 2016;41:175–189. doi: 10.1016/j.tibs.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Rigden DJ, Galperin MY. Sequence analysis of GerM and SpoVS, uncharacterized bacterial ‘sporulation’ proteins with widespread phylogenetic distribution. Bioinformatics. 2008;24:1793–1797. doi: 10.1093/bioinformatics/btn314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CD, Marquis KA, Meisner J, Rudner DZ. Peptidoglycan hydrolysis is required for assembly and activity of the transenvelope secretion complex during sporulation in Bacillus subtilis. Mol Microbiol. 2013;89:1039–1052. doi: 10.1111/mmi.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A, Pogliano K. Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. EMBO J. 2004;23:1636–1646. doi: 10.1038/sj.emboj.7600171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2:a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons RL, Slynn GM, Smith DA. Genetical and molecular studies on gerM, a new developmental locus of Bacillus subtilis. J Gen Microbiol. 1987;133:3299–3312. doi: 10.1099/00221287-133-12-3299. [DOI] [PubMed] [Google Scholar]
- Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Crawshaw AD, Dembek M, Monteiro JM, Pereira FC, de Pinho MG, Fairweather NF, Salgado PS, Henriques AO. The SpoIIQ-SpoIIIAH complex of Clostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis. Mol Microbiol. 2015 doi: 10.1111/mmi.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slynn GM, Sammons RL, Smith DA, Moir A, Corfe BM. Molecular genetical and phenotypical analysis of the gerM spore germination gene of Bacillus subtilis 168. FEMS Microbiol Lett. 1994;121:315–320. doi: 10.1111/j.1574-6968.1994.tb07119.x. [DOI] [PubMed] [Google Scholar]
- Steil L, Serrano M, Henriques AO, Volker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology. 2005;151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- Sun YL, Sharp MD, Pogliano K. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J Bacteriol. 2000;182:2919–2927. doi: 10.1128/jb.182.10.2919-2927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva EI, Lopez-Garrido J, Hughes HV, Fredlund J, Kuru E, Vannieuwenhze MS, Brun YV, Pogliano K, Jensen GJ. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip CK, Kimbrough TG, Felise HB, Vuckovic M, Thomas NA, Pfuetzner RA, Frey EA, Finlay BB, Miller SI, Strynadka NC. Structural characterization of the molecular platform for type III secretion system assembly. Nature. 2005;435:702–707. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.