Summary
The pathogenic fungus Cryptococcus neoformans must adapt to glucose-limited conditions in the lung and glucose replete conditions upon dissemination to the brain. We report that glucose controls ribosome biogenesis and translation by modulating mRNA decay through a balance of PKA and Hog1 signaling. Glucose signaling through PKA stabilized ribosomal protein (RP) mRNAs whereas glucose starvation destabilized RP transcripts through Hog1. Glucose starvation induced oxidative stress response genes, and treatment of glucose-fed cells with Reactive Oxygen Species (ROS) generating compounds repressed RP transcripts, both of which were dependent on Hog1. Stabilization of RP transcripts led to retention of polysomes in a hog1Δ mutant, whereas stabilization of RP transcripts by cyclic AMP did not affect translation repression, suggesting that Hog1 alone signals translation repression. In sum, this work describes a novel antagonism between PKA and Hog1 controlling ribosome biogenesis via mRNA stability in response to glucose availability in this important human pathogen.
Introduction
Cryptococcus neoformans is a basidiomycetous, yeast-like, ubiquitous fungus that has emerged as an important etiological agent of meningoencephalitis in humans. Of the 19 species that are characterized within the genus Cryptococcus, C. neoformans and C. gatti are pathogenic, causing disease in predominantly immunocompromised and immunocompetent individuals, respectively (Mitchell & Perfect, 1995, Pappas, 2013). C. neoformans var grubii (Serotype A), based on capsular serotyping, is known to cause the majority of opportunistic cryptococcal infections in HIV infected individuals in the US and Africa, as well as in patients with T cell deficiencies, hematologic malignancies and those undergoing immunosuppressive therapies (Mitchell & Perfect, 1995, Pappas, 2013, Evans, 1950). Available epidemiologic data reveal a substantial global burden of cryptococcal meningitis, with an estimated 600,000 deaths in sub-Saharan Africa, where limited availability and high cost of antifungal drugs hinder effective medical treatment (Park et al., 2009).
Spores or desiccated yeast cells are the infectious propagules which, upon inhalation by susceptible hosts, get lodged in the pulmonary spaces to be eventually engulfed by alveolar macrophages. Depending upon the immune competence of the host and virulence of the strain, C. neoformans may be cleared from the lungs or remain latent within macrophages (Chretien et al., 2002, Feldmesser et al., 2000). Initial pulmonary infection may spread by hematogenous dissemination to other parts of the body with a tropism for the central nervous system leading to meningoencephalitis that is fatal if untreated.
C. neoformans possesses a number of virulence factors which include capsule, melanin and mannitol production, and multiple secreted enzymes such as protease, phospholipase C and others (Buchanan & Murphy, 1998). Along with these, another important feature in the pathogenesis of C. neoformans is its ability to withstand stresses and evade host immune system (Voelz & May, 2010, Brown et al., 2007). Being an environmental microbe, C. neoformans encounters and survives exposure to a multitude of stresses during growth in the human host that include oxidative and nitrosative stress, high temperature, hypoxia, and nutrient deprivation (Brown et al., 2007). Stress response pathways sense changes in the environment and signal reprogramming of gene expression on cellular processes promoting stress adaptation. Multiple studies over the years have revealed an involvement of several signaling mediators like Pkc1, Hog1, Pkh2 and calcineurin-calmodulin to regulate responsive mechanisms against diverse stresses encountered in the host (Brown et al., 2007, Bahn et al., 2006, Bloom et al., 2013, Upadhya et al., 2013, Chabrier-Rosello et al., 2013). Although glucose is one of the foremost limiting factors the pathogen faces in the host pulmonary niche, C. neoformans is able to withstand this adverse environment, remain latent and later disseminate to the CNS which is comparatively more nutrient rich (Hu et al., 2008). It is thus important to identify mediators of the adaptive response of C. neoformans to changes in nutrient availability.
Prompt and precise regulation of gene expression is required for cellular survival during adverse growth conditions. Nutrient limitation in S. cerevisiae has been reported to result in a global transcriptional and post-translational reprogramming to modulate ribosome biogenesis (Gasch & Werner-Washburne, 2002, Marion et al., 2004, Lopez et al., 2011). It has been shown that a majority of ribosomal protein genes (RP) undergo more than two-fold reduction in expression during amino acid and glucose starvation mainly as a mechanism to conserve energy (Warner, 1999, Gasch et al., 2000, Natarajan et al., 2001). Similar patterns of RP gene repression have been reported in transcriptional profiles of Candida albicans in infected macrophages and murine infection studies with C. neoformans (Hu et al., 2008, Lorenz et al., 2004). Alterations in ribosomal gene expression and a role for mRNA turnover in response to nutritional changes have been studied in S. cerevisiae, but the importance of mRNA degradation and its downstream effects during starvation stress have not been investigated in the human pathogen, C. neoformans (Munchel et al., 2011). The specific signaling mediators that engage the mRNA decay machinery in response to nutrient availability also remain unknown.
mRNA decay is a key post-transcriptional mechanism of gene regulation. It is a multistep process initiated by deadenylation or trimming of the 3′ poly (A) tail and is catalyzed by Ccr4p, the major deadenylase in C. neoformans (Chen & Shyu, 2011, Parker & Song, 2004). Deadenylation being the rate limiting step for mRNA decay is also instrumental in silencing translation, thereby playing an essential role in modulating gene expression (Wiederhold & Passmore, 2010, Goldstrohm et al., 2006, Benoit et al., 2005). Studies in S. cerevisiae and C. neoformans have revealed that degradation of RP transcripts accounted for a substantial part of adaptive response to temperature stress (Grigull et al., 2004, Bloom et al., 2013). These studies have paved a way for further investigations with the mRNA decay pathway and its involvement in general stress adaptation.
Here we have elucidated a novel regulation of RP gene expression with respect to glucose availability in C. neoformans. We report that glucose starvation is accompanied by oxidative stress which activates Hog1 to trigger the critical cellular response of RP transcript decay. We also show that an absence of RP degradation leads to a defect in translation repression, thereby affecting the adaptive cellular response during starvation stress. Following starvation, when cells encounter glucose rich environment, RP transcripts are induced and stabilized in a PKA dependent manner to resume protein synthesis enabling growth of the pathogen. Overall, our results indicate that the abundance of RP transcripts and in turn ribosome biogenesis is controlled at the level of mRNA stability by the dual and opposing actions of Hog1 and PKA.
Results
Ccr4 mediates RP repression by accelerated decay of RP transcripts following carbon starvation
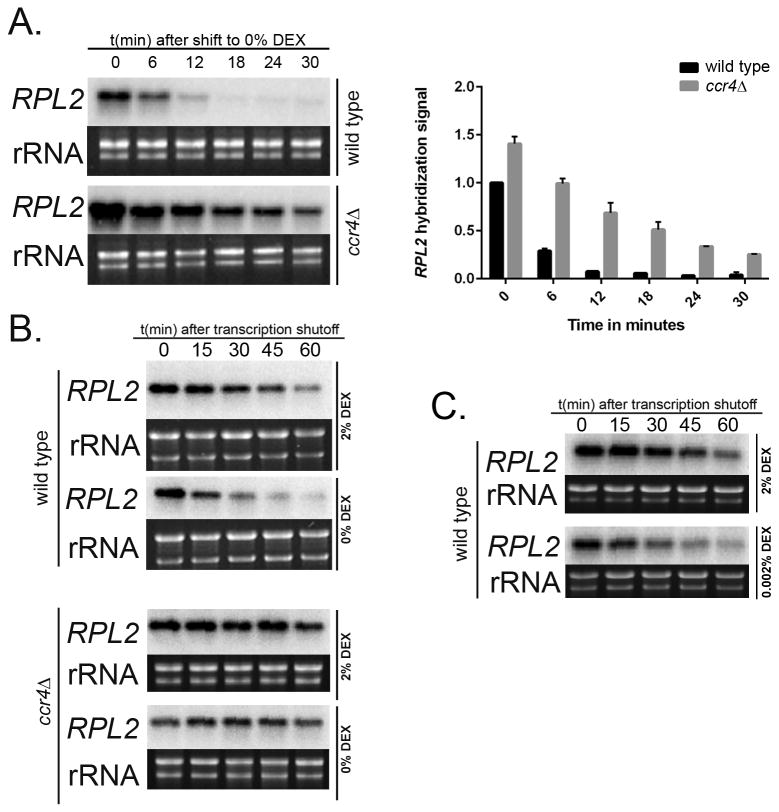
To determine the effect of carbon starvation on overall RP transcript abundance, we performed a Northern Blot time-course assay to analyze the steady state mRNA levels of our representative RP transcript, RPL2 (CNAG_05232), in the wild type every 6 minutes over a 30 minute period following a shift from nutrient rich Yeast Peptone Dextrose (YPD-2% Dextrose) broth to minimal Yeast Nitrogen Base medium (YNB-0% Dextrose). We have previously shown that RPL2 and RPL27e mRNA abundance and decay kinetics show similar profiles during temperature stress in C. neoformans (Bloom et al., 2013). Studies in other systems have also demonstrated a tight co-regulation of RP transcripts in both synthesis and decay patterns in response to environmental stresses (Tavazoie et al., 1999, Grigull et al., 2004, Wang et al., 2002, Mayer & Grummt, 2006). We observed a significant reduction in RPL2 levels by 12 minutes (Fig. 1A, top panel) suggesting that carbon starvation results in a rapid repression of RP genes. Given the role of Ccr4 in repression of RP genes during temperature stress in C. neoformans (Bloom et al., 2013), we investigated the RPL2 transcript steady state levels in a ccr4Δ mutant post shift to YNB media. Our results demonstrate a more gradual and moderate change in RP transcript abundance in the mutant (Fig. 1A, bottom panel) suggesting that deadenylation-dependent mRNA decay mediated by Ccr4 is responsible for a large component of the RP transcript repression seen during starvation.
Figure 1. Carbon starvation in C. neoformans results in RP repression by Ccr4 mediated accelerated decay of RP mRNA.
A) Northern Blot analysis of steady-state RNA extracted from wild type and ccr4Δ mutant cells shifted from YPD (2% Dextrose) to YNB (0% Dextrose) showed reduced levels of RPL2 in wild type in contrast to comparatively higher abundance of the same in the mutant. Graph represents relative abundance of RPL2 in wild type and ccr4Δ mutant cells with SEM values. Northern Blot analyses showed RPL2 decay kinetics when (B, top panel) wild type and (B, bottom panel) ccr4Δ mutant were shifted to minimal media. RPL2 was rapidly destabilized with a shorter high life in wild type when starved whereas a significant stabilization of RPL2 was observed in the ccr4Δ mutant following starvation. Hybridization signal in bands corresponding to indicated transcripts were normalized to rRNA. C) Northern blot analysis showed rapid destabilization of RPL2 transcript with shorter half-life in wild type following a shift from YPD to YNB supplemented with 0.002% dextrose.
To verify that the reduced RPL2 expression seen in the wild type was due to a change in the stability of the RPL2 transcript, we monitored the starvation induced decay kinetics of RP transcripts in both wild type and ccr4Δ mutant in a time course following transcriptional shutoff. Mid-log cells of both strains grown in either YPD or shifted to YNB were treated with 1, 10-phenanthroline to inhibit transcription. RNA was harvested over a one hour time course with intervals at every 15 minutes, and RPL2 abundance measured by northern blot. We observed that wild type RPL2 transcript degraded rapidly and drastically when shifted to YNB with the half life reducing from 33 minutes to 15 minutes (Fig. 1B, top panel) (P<0.0001). In contrast, the same transcripts were highly stabilized in ccr4Δ mutant, both in YPD and YNB, with half lives of >60 minutes in both cases (Fig. 1B, bottom panel) (P<0.0001 compared to wild type RPL2 in YPD, P<0.0001 compared to wild type RPL2 in YNB). These data confirm that carbon starvation induces an accelerated deadenylation-dependent decay of RP transcripts mediated by Ccr4. We also performed a reciprocal stability experiment where mid-log wild type cells were shifted from glucose-rich YPD medium to YNB supplemented with glucose (0.002%), at a concentration that models host nutrient deficient environment of lungs and macrophages (Lorenz et al., 2004, Williams & Del Poeta, 2011, Hu et al., 2008, Kronstad et al., 2012). Our result from Fig. 1C again demonstrated a shorter half-life of RPL2 in low glucose medium, (P<0.0001 compared to wild type RPL2 in YPD) suggesting that carbon deficiency induced an accelerated decay of RP transcripts in C. neoformans.
Reduction in polysome abundance on glucose withdrawal
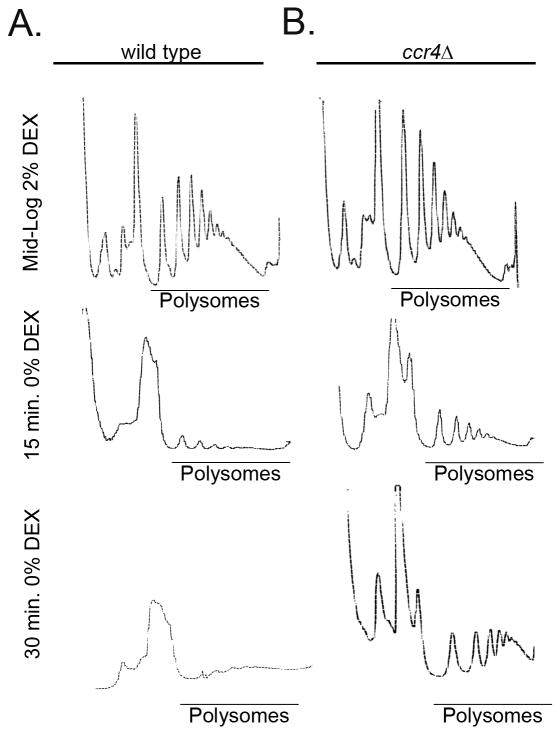
Since deadenylation occurs on mRNAs associated with ribosomes and is a regulatory factor for translation, we investigated the translation state of the wild type and ccr4Δ mutant in both glucose replete and depleted conditions by polysome profile analysis (Chen & Shyu, 2011, Garneau et al., 2007). The profiles of both wild type and the mutant in YPD media revealed a normal distribution of ribosomes with expected peak of 40S ribosome subunit, 60S subunit, an 80S monosome peak followed by multiple polysome peaks (Fig. 2A and B, top panels). Following a shift to glucose deficient media for 15 minutes and 30 minutes, we observed a significant redistribution of ribosomes from polysomes to the ribosomal subunit peaks in the wild type with a marked increase in the 60S peak and a concomitant decrease in the polysome pool leading to an almost absence of polysomes at the 30 minute time point (Fig. 2A middle and bottom panels, respectively). Interestingly, polysome profiles from starved cultures of the ccr4Δ mutant revealed an accumulation of 60S subunit as well, but a comparatively modest change in the polysome abundance compared to the wild type, both at 15 minutes and 30 minutes after starvation (Fig. 2B, middle and bottom panels respectively). Our findings suggest that glucose withdrawal results in translation repression and a significant loss of polysomes. Additionally, a defect in mRNA degradation leads to retention of polysomes during starvation.
Figure 2. Glucose withdrawal leads to loss of polysomes following RP decay.
Polysome profile analysis of wild type (A) showed reduced abundance of polysomes following a 15 minutes and 30 minutes shift to glucose deficient YNB medium whereas (B) a significant retention of polysomes was observed in the ccr4Δ mutant at both time points following starvation.
RP transcript degradation is specific to the absence of glucose signaling
C. neoformans has a functional gluconeogenetic pathway and can utilize glycerol and acetate among others as an alternative carbon source for growth (Price et al., 2011, Hu et al., 2008). To investigate if the RP transcript degradation was a response to the absence of glucose specifically or to lack of a carbon source in general, we performed a stability time-course experiment where mid-log wild type cells were shifted from YNB+2% dextrose to YNB supplemented with 2% glycerol as an alternative carbon source. The half-life of RPL2 was calculated to be 35 minutes in YNB+2% dextrose (Fig. 3A), similar to half-life of RPL2 in another dextrose containing media YPD (P value=0.5262). The RPL2 transcript underwent an accelerated degradation when cells were shifted to the glycerol containing media with a half life of 17 minutes (Fig. 3A), (P<0.0002 compared to RPL2 half-life in YNB-2% dextrose), suggesting that the RP transcript decay response was specific to glucose depletion, and that an alternative carbon source was unable to suppress this response. There are two major ways in which glucose can signal to the mRNA decay machinery. Glucose can be sensed directly and signal through the cAMP-dependent PKA pathway, or the cells can sense a change in metabolic state through ATP/AMP ratios. To address if RP transcript degradation was specific to the absence of glucose as a metabolite, we performed a stability time course assay to look at RPL2 decay kinetics in wild type cells shifted from YNB+2% dextrose to YNB supplemented with a non-metabolizable glucose analogue, 2-Deoxy-D-glucose (2-DG) that would provide the glucose signal, but not result in ATP production (Panepinto & Williamson, 2006). In the presence of 2-DG, RPL2 transcripts degraded with a normal half-life of 33 minutes (Fig. 3B) (P value=0.4577 compared to RPL2 half life in YNB+2% dextrose), which suggested that activation of glucose signaling even in the absence of glucose itself had a stabilizing effect on RP transcripts.
Figure 3. Activation of cAMP/PKA signaling stabilizes RP transcripts.
A) Northern Blot analysis of wild type cells when shifted from YNB+2% dextrose to YNB supplemented with 2% glycerol as alternate carbon source showed a rapid decay of RPL2 mRNA with a shorter half-life. Activation of glucose signaling by incubation of wild type cells in the presence of (B) exogenous 2-Deoxy-D-glucose (C) and cyclic AMP led to a stabilization of RPL2 even in the absence of glucose. Decay kinetics was plotted and nonlinear regression analysis was used to determine half-lives. D) Polysome profile analysis of mid-log wild type cells incubated in YNB supplemented with cAMP showed reduced levels of polysomes similar to the profile obtained during glucose starvation. E) Inactivation of cAMP/PKA pathway did not influence RPL2 decay as exhibited in Northern Blot analyses in gpa1Δ and pka1Δ mutants, both in YPD (2% Dextrose) and YNB media (0% Dextrose).
Activation of cAMP/PKA pathway stabilizes RP transcripts
Once we established that activation of glucose signaling stabilized RP transcripts, we next investigated the role of the cAMP/PKA pathway, which is a reputed key player in the cell's ability to sense and adapt to glucose availability (Choi et al., 2015, Santangelo, 2006). cAMP/PKA is known to regulate RP gene expression subject to nutrient availability, but the contribution of this pathway to post-transcriptional regulation of RP gene expression in C. neoformans has not been investigated (Zurita-Martinez & Cardenas, 2005, Klein & Struhl, 1994, Martin et al., 2004). Studies in mammalian cells revealed that cAMP is instrumental in stabilizing selective mRNAs which prompted us to investigate whether cAMP/PKA exerted any effect on RP transcript stability in C. neoformans (Chen et al., 1993, Hod & Hanson, 1988). To answer this, we performed a stability time course experiment where mid-log wild type cells were shifted from YPD to YNB supplemented with 10 mM cAMP to activate the pathway even in the absence of glucose. Addition of cAMP stabilized RPL2 transcript, with a half life of >60 minutes, (Fig. 3C) (P<0.0001, compared to RPL2 half-life in YNB), confirming a role for PKA as a positive regulator of mRNA stability. We further wanted to examine whether this increase in RP transcript abundance due to cAMP activation had any effect on the ribosome population. Polysome analysis of mid-log wild type cells that were shifted from YPD to cAMP-supplemented minimal media revealed that there was a marked reduction in the polysome pool with a concurrent increase in 60S subunit (Fig. 3D, bottom panel) a profile similar to wild type cells during starvation (Fig. 3D, top panel). Data from above experiments convey that exogenously added cAMP activated the PKA pathway rendering a stabilizing effect on RP transcripts, but that stabilization does not relieve translation repression or lead to retention of polysomes. We know that eukaryotic mRNA decay is a multi-step process that is initiated by deadenylation of poly-A tail followed by the sequential process of decapping and exonucleolytic cleavage at 5′-end in the P-bodies or by the exosome at the 3′-end after the transcript is dissociated from ribosome. Any defect in these later steps of mRNA decay would hence result in stabilized deadenylated transcripts. Our data from experiments with cAMP led us to hypothesize that stabilization of RPL2 transcripts occurs in the mRNA degradation pathway after the point of deadenylation because the mRNAs remain stable, but the ribosomes have dissociated.
We further hypothesized that if RP transcripts were stabilized by activation of PKA, then inactivating the same, even in the presence of glucose, would lead to a starvation response resulting in rapid decay of RPL2 mRNA. To address this question, we performed a stability time course assay and Northern blot analyses to measure RPL2 decay kinetics in gpa1Δ mutant lacking the Gα subunit that is an activator of PKA and a pka1Δ mutant that lacks a PKA catalytic subunit (Alspaugh et al., 1997, D'Souza et al., 2001). We did not observe any difference in the rate of RPL2 degradation in either of the mutants as compared to wild type, the half-lives of RPL2 being 31 minutes in both gpa1Δ and pka1Δ mutants during their growth in YPD (Fig. 3E middle and right panels respectively), (P value=.5183, compared to wild type RPL2 half-life in YPD). This suggested that although activating PKA exhibited a stabilizing effect on RP transcripts, inactivation of the pathway did not elicit an opposing effect, suggesting that another signaling pathway is providing a destabilizing signal in the absence of glucose.
Positive regulation of mRNA stability via PKA is important during refeeding
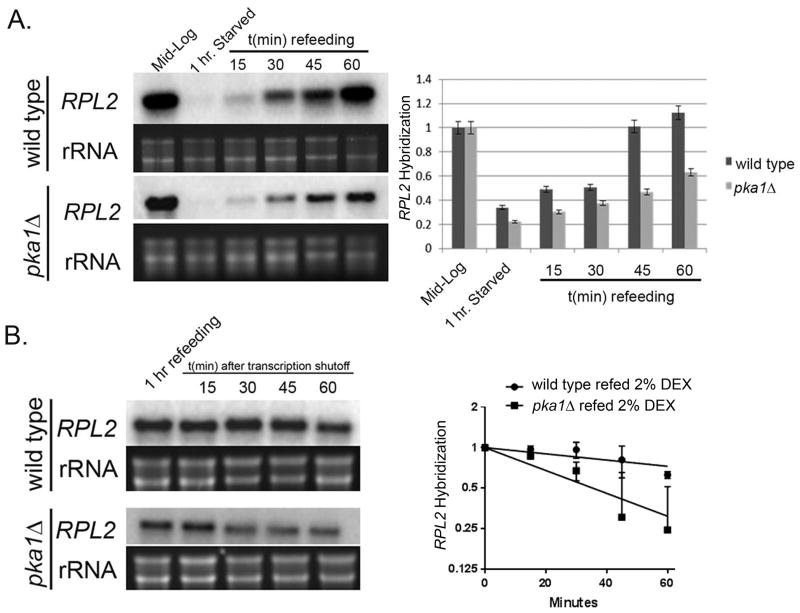
Given the role of cAMP/PKA in induction of RP genes during nutrient replete condition in S. cerevisiae, we next wanted to determine if similar phenomenon occurred in C. neoformans following starvation (Zurita-Martinez & Cardenas, 2005, Neumansilberberg et al., 1995, Kraakman et al., 1999). Mid-log wild type and pka1Δ mutant cells were starved for 2 hours in YNB, followed by re-addition of dextrose for an hour. Cells were harvested from mid-log, starved and refed cultures (every 15 minutes) and RNA was extracted to perform Northern Blot and probed for RPL2 steady state levels. We observed a significant decrease in RPL2 levels following starvation in the wild type, which gradually increased upon refeeding with glucose and reached almost pre-starved levels by the end of 1 hour (Fig. 4A). In contrast, transcript abundance did not increase as efficiently in pka1Δ mutant with RPL2 reaching only about 60% of wild type level, (Fig. 4A) suggesting that this increase in RPL2 mRNA is dependent on the functionally active PKA pathway. Given our earlier observation that cAMP activation of PKA led to a stabilizing effect on RPL2 transcripts (Fig. 3D), we next wanted to investigate whether the increase in RPL2 abundance during refeeding phase was due to a pro-stabilizing effect by this pathway on RP transcripts. To address this, mid-log wild type and pka1Δ cells were shifted from YPD to YNB for 2 hours and supplemented with dextrose for 30 minutes to induce RP transcripts. Once RP genes were induced, 1, 10-phenanthroline was added to halt further transcription and cells were harvested every 15 minutes for an hour to analyze RP decay kinetics during the refeeding stage. During refeeding, RPL2 mRNA was highly stabilized in the wild type with a half life of >60 minutes in contrast to the significantly less stable RPL2 transcript in the pka1Δ mutant (Fig. 4B) (P<0.001), thereby confirming a role for PKA1 in positively regulating RP mRNA stability during glucose refeeding.
Figure 4. Stabilization of RP transcripts by PKA is important during refeeding of glucose.
A) Northern Blot steady state analysis of starved wild type cells showed a progressive induction of RPL2 following refeeding with glucose over a one hour period. This increase in RPL2 abundance was defective in a pka1Δ mutant. Mean expression of RPL2 from 3 biological replicates are demonstrated in the histogram with error bars depicting SEM. B) Northern Blot analysis in starved wild type cells showed a greater stabilization of RPL2 following refeeding with 2% dextrose. This stabilization was comparatively deficient in a pka1Δ mutant with a shorter half life as depicted in the regression plot.
Common nutrient and stress signaling cascades do not mediate the RP decay response to glucose starvation
Once we established a pro-stabilizing effect of PKA on RP transcripts during glucose availability, we focused our interest on other well characterized nutrient sensing and stress responsive pathways for a possible role in accelerating RP mRNA decay during glucose starvation. We hypothesize that if RP decay during glucose depletion was mediated by any of these cascades, then inactivating the pathway would lead to a stabilization of RP transcripts under starvation conditions. Earlier studies with S. cerevisiae have revealed a role for TOR signaling in regulating RP gene expression and ribosome biogenesis in response to nutrients (Marion et al., 2004, Lopez et al., 2011, Powers & Walter, 1999). To determine if TOR signaling mediated RP decay in response to glucose starvation, mid-log wild type cells growing in YPD were pre-treated with rapamycin (inhibitor of TOR kinases) for twenty minutes before shifting them to minimal media containing rapamycin and a stability time course was performed for an hour. Northern blot analyses revealed that inhibiting TOR did not stabilize RPL2 and its half life was 15 minutes (Fig. 5A, left panel). This acceleration was similar to that observed in wild type during starvation, (P=0.9813). To exclude the role of TOR more convincingly, we performed a reciprocal experiment where mid-log wild type cells were treated with rapamycin for twenty minutes during its growth in glucose-rich YPD before shutting off-transcription by adding 1,10-phenanthroline. (Fig. 5A, right panel) demonstrates that simply inactivating TOR did not have any effect on the RPL2 degradation and transcript half-lives were the same both in rapamycin treated and control cells (P=0.6731), thereby excluding a role for TOR in regulating RPL2 stability in C. neoformans. Glucose depletion and an altered AMP:ATP ratio is known to activate the AMP activated kinase, SNF1, that regulates a multitude of processes including stress response and mRNA metabolism (Hu et al., 2008, Braun et al., 2014). We therefore created a snf1Δ knockout mutant and assayed RPL2 stability as above. The snf1Δ mutant also exhibited a wild type response to starvation with the half life of RPL2 calculated as 15 minutes (Fig. 5C, P=0.7832), ruling out this regulator in starvation induced RP transcript degradation in C. neoformans. Given the role for Pkh2 signaling in RP mRNA decay during host-temperature adaptation in C. neoformans (Bloom et al., 2013), we performed similar experiment on pkh2Δ-02 mutant to investigate for a possible interaction between Pkh2 signaling and RP transcript decay during glucose deficiency. RPL2 degraded rapidly with a half life of 16 minutes in the pkh2Δ-02 mutant, (Fig. 5D) (P=0.6642, compared to RPL2 half-life of wild type in YNB) ruling out Pkh2 as a mediator of RP transcript decay.
Figure 5. Common stress signaling pathways do not mediate starvation induced RP decay.
Northern Blot analyses of (A, left panel) rapamycin treated wild type cells in YNB (A, right panel) rapamycin treated wild type cells in YPD (B) snf1Δ mutant and (C) pkh2Δ-02 mutant did not exhibit any stabilization of RPL2 mRNA during glucose unavailability, excluding their role as a signaling regulator for RP decay.
Hog1 signals accelerated RP transcript degradation during starvation stress
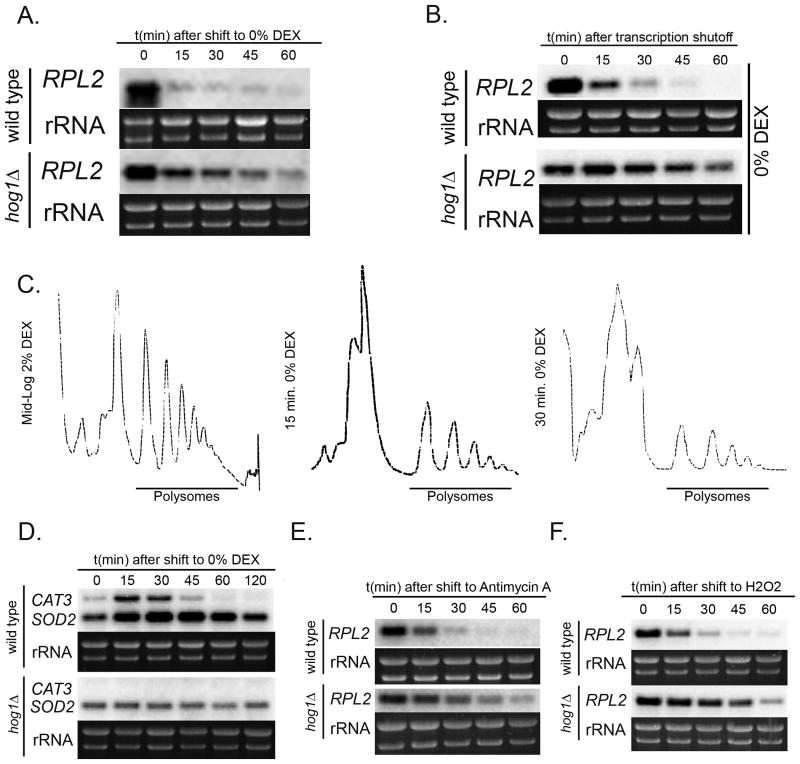
We know Hog1 plays a pivotal role in conferring cellular adaptive responses during multiple environmental stresses (Bahn et al., 2006, Giles et al., 2006). To determine a possible role of Hog1 signaling in RP gene expression in C. neoformans during glucose deprivation, we performed steady state time course experiments where both wild type and hog1Δ mutant mid-log cells were shifted from YPD to YNB (Bahn et al., 2005). Northern blot analyses of RPL2 expression revealed relatively higher levels of steady state mRNA in the mutant compared to wild type (Fig. 6A). In accordance with this result, stability time course assay with hog1Δ mutant cells demonstrated a relative stabilization of the RPL2 transcript in the hog1Δ mutant with a longer half life (>60 minutes) (Fig. 6B), (P<.001 compared to wild type). This suggested that Hog1 provides the signal for accelerated RP degradation during glucose starvation in C. neoformans, and RP transcripts were stabilized in its absence. To determine the effect of RP transcript stabilization on ribosome abundance, we performed polysome profile analysis on mid-log hog1Δ mutant cells in YPD and 15 minutes and 30 minutes post shift to YNB. Results showed that in normal growth conditions, hog1Δ mutant had a typical profile with high levels of polysomes (Fig. 6C left panel). Interestingly, polysomes were retained when cells were incubated in YNB media at both time points (Fig. 6C, middle panel and right panel), with comparatively higher abundance of polysomes in hog1Δ mutant compared to wild type. Taken together, our data indicate that Hog1 positively regulates RP mRNA decay in the absence of glucose and is also involved in modulating ribosome biogenesis and translation repression during starvation stress.
Figure 6. Hog1 signals accelerated RP transcript degradation during starvation stress.
(A) Northern blot steady state analysis in hog1Δ mutant cells showed higher abundance of RPL2 following a shift from YPD (2% Dextrose) to YNB (0% Dextrose) compared to wild type. (B) This increased steady state RPL2 levels in the hog1Δ mutant was due to a greater stabilization of the transcript compared to wild type as depicted by stability time course data. (C) Polysome profile analysis of hog1Δ mutant showed retention of polysomes when cells were shifted to minimal media for 15 or 30 minutes. (D) Induction of oxidative stress during glucose starvation was demonstrated in wild type by a transient increase in steady state levels of CAT3 and SOD2 during growth in YNB. This induction was absent in hog1Δ mutant. Treatment of wild type cells with ROS generating compounds (E) Hydrogen peroxide and (F) antimycin exhibited a rapid and drastic repression in RPL2 that was absent in hog1Δ mutant.
Studies in yeast and mammalian cells have shown that nutrient starvation can additionally lead to oxidative stress in cells (Varela et al., 1995, Suzuki et al., 2011). To determine whether glucose starvation was accompanied by oxidative stress in C. neoformans, we measured steady state mRNA levels of oxidative stress induced genes CAT3 (CNAG_00575) and SOD2 (CNAG_04388) which encode for catalase and superoxide dismutase, respectively (Giles et al., 2006, Cox et al., 2003). Wild type and hog1Δ mid-log cells were shifted from YPD to YNB and harvested at designated time points over two hours. RNA was extracted and northern blot performed to reveal a rapid but transient induction of both CAT3 and SOD2 transcripts by 15 minutes which then reduced to minimal levels by the end of 1 hour in the wild type (Fig. 6D, top panel). On the contrary, we observed only basal levels SOD2 which did not elevate during starvation, along with a total absence of CAT3 induction in hog1Δ mutant (Fig. 6D, bottom panel). We hypothesized that if our earlier observation of RP transcript repression and decay was an effect of starvation induced oxidative stress, then treatment with Reactive Oxygen Species (ROS) generating agents would also show similar response. In our study, treatment of wild type cells with two ROS generating compounds antimycin and hydrogen peroxide also caused a Hog1 dependent repression of the RPL2 mRNA (Fig. 6E and 6F), consistent with the hypothesis that glucose starvation is accompanied by oxidative stress that leads to Hog1 mediated regulation of RP genes. In order to verify our hypothesis, we next wanted to determine if adding anti-oxidants in glucose depleted medium prevented RP repression and so we performed reciprocal experiments using multiple anti-oxidants to determine any compensatory mechanism that prevented accelerated degradation of RPL2 transcripts. Stability experiments were performed with wild type cells that were pre-treated with tocopherol (Raspor et al., 2005), EUK-134 (Batinic-Haberle et al., 2010) and mito-Tempo (He et al., 2014) for 30 minutes before shifting them to glucose deficient conditions to investigate for RPL2 decay kinetics. Results demonstrated rapid decay of RPL2 with significantly shorter half-lives in all the cases suggesting that pre-treatment with anti-oxidants singly and in combination failed to abrogate the oxidative stress induced upon starvation. Result demonstrated drastically reduced levels of RPL2 transcript (Supplementary Fig. S1, bottom panel) which suggested that pre-treatment of cells with a mixture of anti-oxidants failed to prime them against the effects of oxidative stress. We also probed for CAT3 and SOD2 transcripts in the same samples to reveal transient induction of both the genes verifying that pre-treatment of cells with anti-oxidants was insufficient to negate the effect of starvation induced oxidative stress and thus failed to stabilize RPL2 transcript (Supplementary Fig. S1, top panel). We hypothesize that failure to prevent the induction of oxidative stress despite using the anti-oxidants was presumably due to the inability of the anti-oxidants to permeate the cells or reach an effective intracellular concentration. Alternatively, carbon starvation may activate Hog1 independent of ROS production. The specific mechanism by which oxidative stress activates Hog1 has not yet been defined.
Discussion
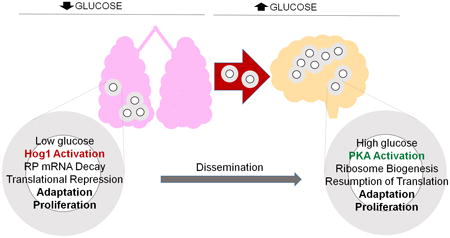
The concentration of glucose is nearly 3-20 times lower in the airway surface liquid (ASL) and lower respiratory tract airways, than the plasma glucose content which is normally maintained at 5 mM L-1 (Garnett et al., 2012). This is also in contrast to the comparatively elevated levels of glucose (2.5-4 mML-1 or 0.18% glucose) in the human cerebrospinal fluid (CSF) which is 50-70% of blood glucose levels (Leen et al., 2012, Pastuszczak et al., 2013). A key factor that differentiates glucose content between lungs and the brain compartments is the presence of multiple glucose transporters in the latter and a substantial decrease of the same in lungs (Wood & Trayhurn, 2003, Garnett et al., 2012). C. neoformans pathogenesis initiates within the human lung and culminates in the brain; both of the compartments are varied in glucose availability. HIV infected individuals have been reported to show a very high burden of C. neoformans in their CSF (Bicanic et al., 2009) suggesting that the glucose rich environment favors proliferation of cryptococcal cells in the brain tissue and subarachnoid spaces. Thus C. neoformans in the course of its pathogenesis transitions from a glucose-deprived niche in the lungs and macrophages and moves to a glucose rich environment in the CNS. Our work is a novel approach to decipher the adaptive mechanisms required by the pathogen to mediate this transition and promote its survival when it encounters environments with variable glucose availability. Our data has led to a hypothetical model of how the antagonism of Hog1 and PKA could promote host adaptation in the glucose-deficient environment of the lung, and promote proliferation in the CNS where glucose is available (Fig. 7).
Figure 7. Hypothetical model for how the antagonism between Hog1 and PKA may promote host adaptation during C. neoformans infection.

Glucose deprivation in the lungs results in Hog1 activation in C. neoformans. Hog1 signal mediates RPL2 decay and translation repression that promotes adaptation to stress. The glucose-rich environment of the brain leads to PKA activation in C. neoformans that result in induction and stabilization of RP genes, and resumption of translation to promote adaptation and proliferation.
We demonstrate that on exposure to glucose-limiting conditions, (Fig. 1C), that models the glucose concentration of lungs and the macrophage intracellular environment, C. neoformans undergoes an acute and prompt response where RP transcripts are rapidly degraded. We believe that this is a cellular response to rid the cell of one of the most abundant classes of transcripts to conserve energy. This is supported by our findings from polysome profiles of starved cells where polysome abundance was reduced in wild type in contrast to a ccr4Δ mutant. Ribosome biogenesis and protein synthesis are high energy driven processes, and inhibiting them in starved cells is an efficient way of conserving energy to utilize it for other stress adaptive mechanisms (Warner, 1999, Warner et al., 2001, Causton et al., 2001). We also determined that this regulation of RP mRNA was mediated by Hog1 signaling. Hog1 has been well documented as a global response mediator to a number of environmental cues along with its importance in virulence; a hog1Δ mutant was reported to be attenuated in virulence in an animal model of cryptococcal infection (Bahn et al., 2006). Transcriptome profiling in hydrogen peroxide treated C. neoformans cells have also shown a down-regulation of RP genes (Upadhya et al., 2013). Our findings now suggest that a defect in Hog1 further leads to impaired RP mRNA decay, and stabilization of the RP transcripts results in retention of polysomes, thereby disrupting stress adaptive mechanisms. We also provide the first evidence of the role of Hog1 in translation repression in C. neoformans through modulation of RP transcript stability during a nutritional down-shift.
Once C. neoformans faces glucose-rich conditions, as it does upon entry into the CNS, it is required to adapt to the increase in the nutrient levels and elicit appropriate responses to utilize these resources and proliferate. In order to resume and sustain protein synthesis, induction of RP genes are of prime importance and previous findings from gene expression studies supports this hypothesis. Transcriptional profiles of C. neoformans cells obtained from CNS of infected rabbits have shown a significant upregulation of ribosomal protein genes, transporters including hexose transporter, and numerous genes involved in carbohydrate metabolism (Steen et al., 2003). We also know that transcription of RP genes and other ribosome biogenesis factors are regulated primarily by PKA and TOR pathways (Zurita-Martinez & Cardenas, 2005, Klein & Struhl, 1994, Martin et al., 2004). The significance of a PKA mediated response during growth of C. neoformans in CNS can be reinforced from studies which demonstrate a virulence defect in a pka1Δ mutant and hypervirulence in a pkr1Δ strain (constitutive activation of PKA) (D'Souza et al., 2001, Hicks et al., 2004). In our study we show that in addition with induction, PKA also exerts a stabilizing effect on newly synthesized RP mRNAs that enable the transcripts to be readily available to resume protein synthesis and ribosome biogenesis. Moreover, stabilization of RP transcripts by exogenous cAMP is witnessed even in the absence of glucose, thereby suggesting a novel role for PKA in mRNA metabolism. Activation of PKA that occurs in the absence of glucose did not relieve translational repression, suggesting that the starvation signal via Hog1 is sufficient for translation repression. Hog1 is known to be activated by ROS, and the response to starvation included the induction of ROS-sensitve genes including SOD2 and CAT3 that was Hog1-depedent, suggesting that ROS production is a component of the starvation response. It is possible, however, that Hog1 is activated by another, novel signal during starvation that leads to the ROS-responsive program downstream of Hog1 activation.
The mechanism by which PKA and Hog1 communicate with the mRNA decay machinery remains unknown. Recent quantitative proteomic analysis of C. neoformans strains overexpressing Pka1 demonstrated regulation of the abundance of ribosomal components, consistent with a role for Pka1 in direct modulation of translation (Geddes et al., 2016). The experimental conditions used in this study, namely galactose induction, may not directly correlate with our in vitro conditions or in vivo conditions encountered in the human host. Nevertheless, findings from both the studies pave the way for further research to determine the targets and mechanisms involved in Pka1 mediated regulation of ribosome biogenesis and translation with respect to glucose availability. No direct targets of Hog1 phosphorylation have been described, and so further investigation is needed to determine the direct targets of Hog1 that regulate translation and mRNA decay.
Our study describes an elegant regulatory control between two signaling pathways to modulate gene expression in C. neoformans in response to glucose availability. This mechanism of translation repression and resumption of protein synthesis by modulating RP mRNA stability via an antagonistic action of Hog1 and PKA signaling is both interesting and novel. Deciphering the biology of these processes will help in the better understanding of pathogenesis and stress response in this important human pathogen.
Experimental Procedures
Strains, media and chemicals
C. neoformans strain H99 (serotype A), ccr4Δ mutant (Bloom et al., 2013, Panepinto et al., 2007), pka1Δ mutant (D'Souza et al., 2001) gpa1Δ mutant and (Alspaugh et al., 1997) hog1Δ mutant (Bahn et al., 2005) were gifts of Joe Heitman (Duke University), pkh2-02Δ mutant and H99 against which the mutant was created are a part of the Madhani collection (Liu et al., 2008) and obtained from the Fungal Genetics Stock Center at Kansas State University.
Production of snf1Δ deletion strain: A C. neoformans snf1Δ strain was constructed as described previously (Panepinto et al., 2005). The knockout construct was verified by sequencing and the construct was introduced into wild type H99 strain using biolistic gene transfer method (Toffaletti et al., 1993). NAT-resistant colonies were screened by PCR with 5′ SNF1 and 3′SNF1 screening primers (S1.Table 1). PCR-positive clones were further verified by Northern and Southern blot.
All strains were maintained in YPD broth (BD Difco) with 80% glycerol (Fisher Chemicals) and streaked fresh on YPD agar plates before conducting experiments. Nutrient rich, mid-log cells were grown in YPD broth whereas glucose starvation experiments were performed either in YNB medium alone (YNB without amino acids, BD Difco) or YNB supplemented with 2% glycerol, 2% 2-Deoxy-D-glucose (Sigma Aldrich), 10 mM cyclic AMP (Sigma Aldrich), or 1 μg ml-1 Rapamycin (Sigma Aldrich). For generating oxidative stress, YPD medium was supplemented with 1 mM Hydrogen Peroxide solution (J.T Baker) and 1 μM Antimycin (Sigma Aldrich). All the anti-oxidants used in this study, α-Tocopherol, EUK-134 and Mito-Tempo was obtained from Sigma Aldrich. 1,10-phenanthroline (J.T Baker) was used as a transcriptional inhibitor and cycloheximide (Sigma Aldrich) was used as a translational inhibitor in our studies.
Steady-state and stability time-courses
Overnight cultures of respective strains were inoculated in YPD at an OD600=0.2 and grown to mid-log phase (OD600=0.6) at 30°C. Cells were harvested by centrifugation at 4000 X g for 5 minutes. For steady state analyses of transcripts, cells were resuspended in 30 ml YNB media and incubated for 30 minutes or one hour as required for each experiment. 5 ml aliquots were pelleted by centrifugation at specific time intervals depending upon the experiments and further processed for RNA extraction. For transcript stability analyses, mid-log cells were re-suspended in 30 ml YNB alone or supplemented with reagents as mentioned for each experiment. A transcriptional inhibitor, 1,10-phenanthroline (250 μg ml-1) was added to the medium and cells were incubated for an hour. Aliquots of 5 ml were harvested every 15 minutes over a one hour time period and processed for RNA extraction.
RNA extraction and Northern blot analyses
RNA was extracted using Qiagen RNAeasy Kit following manufacturer's instructions. RNA was quantified and 2 μg RNA was loaded on formaldehyde-agarose gel. Upon electrophoresis, RNA was transferred to nitrocellulose membrane, cross-linked, and probed with P32-labelled DNA probes that hybridized to the transcripts being tested, as described previously (Bloom et al., 2013). After washing, blots were exposed to phosphor screen, and imaged on a Typhoon 9400 Variable Mode Imager. Hybridization signal was quantified using Quantity One software and normalized to the signal of the ribosomal bands. Statistical analysis for steady state experiments were performed by Two-way ANOVA grouped analyses method to detect standard errors (SEM) between replicate data sets using Graphpad Prism software. Statistical analyses to calculate stability data were performed by determining the least squares fit of one-phase exponential decay non-linear regression with Graphpad Prism software. Significance between curves was detected by sum-of-squares F-test, with P<0.05 as significant P-value, which determined that the data from two comparable experiments fall on different regression lines thereby confirming different rates of decay and ultimately different half-lives of the transcripts.
Polysome profile analyses
Sample was prepared by harvesting mid-log cells growing in YPD or cells shifted to YNB medium for 15 or 30 minutes. Following the induction of starvation stress, translating polyribosomes were halted on the mRNAs by the addition of cycloheximide to 0.1 mg/ml and chilled on ice for 5 minutes prior to centrifugation. Cells were washed twice with 10 ml water followed by a final wash with 15 ml of lysis buffer (20 mM Tris-HCl [pH 8], 140 mM KCl, 5 mM MgCl2, 0.5mM DTT, 0.1 mg ml-1 cycloheximide, and 0.5 mg ml-1 heparin, 1% Triton X-100), flash frozen in liquid nitrogen and stored at -80°C. Cells were thawed, 200 μl of lysis buffer was added to resuspend the cells, and an equal volume of pre-chilled glass beads were added. Cells were mechanically crushed by vortexing every 20 seconds followed by incubation on ice for 40 seconds for a total of 5 minutes. The lysate was cleared by two subsequent microcentrifugation steps (15,000 x g, for 5 min at 4°C) and volume was brought to an equal level by adding buffer. Equal volumes of the samples were loaded onto a 10 ml linear sucrose gradients (10%-50%) prepared in gradient buffer (20mM Tris-HCl, 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.5 mg ml-1 heparin, and 0.1 mg ml-1 cycloheximide). The gradients were centrifuged at 39,000 × rpm for 2 hours at 4°C, using an SW41 Ti rotor. Following centrifugation, sucrose gradients were pushed through a flow cell and RNA was recorded using UV-Vis detector at 254 nm.
All data represented in the work are representatives of 3 biological replicates. Supplementary material includes list of primers used in the study (S1.Table 1)
Supplementary Material
Acknowledgments
We are grateful to Joseph Heitman, Hiten Madhani and the Fungal Genetics Stock Center for providing the strains used in this study.
Funding: This work was supported by the NIH grant 1R01 AI089920-01A1 to J.C.P.
Footnotes
Author contributions: D.B. was responsible for designing and conducting the experiments, interpreting the results and writing the manuscript. A.L.M.B. developed the method for polysome analysis and contributed to the writing of the manuscript. J.C.P. contributed to the design and analyses of experiments, writing and final revision of the manuscript.
References
- Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Gene Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell. 2005;16:2285–2300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Kojima K, Cox GM, Heitman J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell. 2006;17:3122–3135. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batinic-Haberle I, Reboucas JS, Spasojevic I. Superoxide dismutase mimics: chemistry, pharmacology, and therapeutic potential. Antioxid Redox Signal. 2010;13:877–918. doi: 10.1089/ars.2009.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tall length control and early development in Drosophila. Dev Cell. 2005;9:511–522. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Bicanic T, Muzoora C, Brouwer AE, Meintjes G, Longley N, Taseera K, Rebe K, Loyse A, Jarvis J, Bekker LG, Wood R, Limmathurotsakul D, Chierakul W, Stepniewska K, White NJ, Jaffar S, Harrison TS. Independent Association between Rate of Clearance of Infection and Clinical Outcome of HIV-Associated Cryptococcal Meningitis: Analysis of a Combined Cohort of 262 Patients. Clin Infect Dis. 2009;49:702–709. doi: 10.1086/604716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom ALM, Solomons JTG, Havel VE, Panepinto JC. Uncoupling of mRNA synthesis and degradation impairs adaptation to host temperature in Cryptococcus neoformans. Mol Microbiol. 2013;89:65–83. doi: 10.1111/mmi.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun KA, Vaga S, Dombek KM, Fang F, Palmisano S, Aebersold R, Young ET. Phosphoproteomic analysis identifies proteins involved in transcription-coupled mRNA decay as targets of Snf1 signaling. Sci Signal. 2014;7 doi: 10.1126/scisignal.2005000. [DOI] [PubMed] [Google Scholar]
- Brown SM, Campbell LT, Lodge JK. Cryptococcus neoformans, a fungus under stress. Curr Opin Microbiol. 2007;10:320–325. doi: 10.1016/j.mib.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchananl KL, Murphy JW. What makes Cryptococcus neoformans a pathogen? Emerg Infect Dis. 1998;4:71–83. doi: 10.3201/eid0401.980109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabrier-Rosello Y, Gerik KJ, Koselny K, DiDone L, Lodge JK, Krysan DJ. Cryptococcus neoformans Phosphoinositide-Dependent Kinase 1 (PDK1) Ortholog Is Required for Stress Tolerance and Survival in Murine Phagocytes. Eukaryot Cell. 2013;12:12–22. doi: 10.1128/EC.00235-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CYA, Shyu AB. Mechanisms of deadenylation-dependent decay. Wires Rna. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Schnermann J, Smart AM, Brosius FC, Killen PD, Briggs JP. Cyclic-Amp Selectively Increases Renin Messenger-Rna Stability in Cultured Juxtaglomerular Granular Cells. J Biol Chem. 1993;268:24138–24144. [PubMed] [Google Scholar]
- Choi J, Jung WH, Kronstad JW. The cAMP/protein kinase A signaling pathway in pathogenic basidiomycete fungi: Connections with iron homeostasis. J Microbiol. 2015;53:579–587. doi: 10.1007/s12275-015-5247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien F, Lortholary O, Kansau I, Neuville S, Gray F, Dromer F. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J Infect Dis. 2002;186:522–530. doi: 10.1086/341564. [DOI] [PubMed] [Google Scholar]
- Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, Casadevall A, Perfect JR. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun. 2003;71:173–180. doi: 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza CA, Alspaugh JA, Yue CL, Harashima T, Cox GM, Perfect JR, Heitman J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EE. The antigenic composition of Cryptococcus neoformans. I. A serologic classification by means of the capsular and agglutination reactions. J Immunol. 1950;64:423–430. [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Bio. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Garnett JP, Baker EH, Baines DL. Sweet talk: insights into the nature and importance of glucose transport in lung epithelium. Eur Respir J. 2012;40:1269–1276. doi: 10.1183/09031936.00052612. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschl AP, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- Geddes JM, Caza M, Croll D, Stoynov N, Foster LJ, Kronstad JW. Analysis of the Protein Kinase A-Regulated Proteome of Cryptococcus neoformans Identifies a Role for the Ubiquitin-Proteasome Pathway in Capsule Formation. MBio. 2016;7:e01862–01815. doi: 10.1128/mBio.01862-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, Stajich JE, Nichols C, Gerrald QD, Alspaugh JA, Dietrich F, Perfect JR. The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot Cell. 2006;5:1447–1459. doi: 10.1128/EC.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Harris N, Ren J, Han X. Mitochondria-targeted antioxidant prevents cardiac dysfunction induced by tafazzin gene knockdown in cardiac myocytes. Oxid Med Cell Longev. 2014;2014:654198. doi: 10.1155/2014/654198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, D'Souza CA, Cox GM, Heitman J. Cyclic AMP-dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2004;3:14–26. doi: 10.1128/EC.3.1.14-26.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl Y, Hanson RW. Cyclic-Amp Stabilizes the Messenger-Rna for Phosphoenolpyruvate Carboxykinase (Gtp) against Degradation. J Biol Chem. 1988;263:7747–7752. [PubMed] [Google Scholar]
- Hu GG, Cheng PY, Sham A, Perfect JR, Kronstad JW. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol. 2008;69:1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinl C, Struhl K. Protein-Kinase-a Mediates Growth-Regulated Expression of Yeast Ribosomal-Protein Genes by Modulating Rap1 Transcriptional Activity. Mol Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman L, Lemaire K, Ma PS, Teunissen AWRH, Donaton MCV, Van Dijck P, Winderickx J, de Winde JH, Thevelein JM. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- Kronstad J, Saikia S, Nielson ED, Kretschmer M, Jung W, Hu G, Geddes JM, Griffiths EJ, Choi J, Cadieux B, Caza M, Attarian R. Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot Cell. 2012;11:109–118. doi: 10.1128/EC.05273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leen WG, Willemsen MA, Wevers RA, Verbeek MM. Cerebrospinal Fluid Glucose and Lactate: Age-Specific Reference Values and Implications for Clinical Practice. Plos One. 2012;7 doi: 10.1371/journal.pone.0042745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen CB, Madhani HD, Noble SM. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AD, Tar K, Krugel U, Dange T, Ros IG, Schmidt M. Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10. Mol Biol Cell. 2011;22:528–540. doi: 10.1091/mbc.E10-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. P Natl Acad Sci USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Mayerl C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- Mitchelll TG, Perfect JR. Cryptococcosis in the Era of Aids - 100 Years after the Discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol Biol Cell. 2011;22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumansilberberg FS, Bhattacharya S, Broach JR. Nutrient Availability and the Ras/Cyclic Amp Pathway Both Induce Expression of Ribosomal-Protein Genes in Saccharomyces cerevisiae but by Different Mechanisms. Mol Cell Biol. 1995;15:3187–3196. doi: 10.1128/mcb.15.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto J, Liu LD, Ramos J, Zhu XD, Valyi-Nagy T, Eksi S, Fu JM, Jaffe HA, Wickes B, Williamson PR. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J Clin Invest. 2005;115:632–641. doi: 10.1172/JCI200523048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto JC, Komperda KW, Hacham M, Shin S, Liu XG, Williamson PR. Binding of serum mannan binding lectin to a cell integrity-defective Cryptococcus neoformans ccr4Δ mutant. Infect Immun. 2007;75:4769–4779. doi: 10.1128/IAI.00536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepintol JC, Williamson PR. Intersection of fungal fitness and virulence in Cryptococcus neoformans. Fems Yeast Res. 2006;6:489–498. doi: 10.1111/j.1567-1364.2006.00078.x. [DOI] [PubMed] [Google Scholar]
- Pappas PG. Cryptococcal infections in non-HIV-infected patients. Trans Am Clin Climatol Assoc. 2013;124:61–79. [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TA. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. Aids. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Parkerl R, Song HW. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pastuszczak M, Wojas-Pelc A, Jaworek AK. Association of CSF glucose concentration with neurosyphilis diagnosis. Cent Eur J Med. 2013;8:48–51. [Google Scholar]
- Powersl T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, Kronstad JW, Perfect JR. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. MBio. 2011;2:e00103–00111. doi: 10.1128/mBio.00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspor P, Plesnicar S, Gazdag Z, Pesti M, Miklavcic M, Lah B, Logar-Marinsek R, Poljsak B. Prevention of intracellular oxidation in yeast: the role of vitamin E analogue, Trolox (6-hydroxy-2,5,7,8-tetramethylkroman-2-carboxyl acid) Cell Biol Int. 2005;29:57–63. doi: 10.1016/j.cellbi.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen BR, Zuyderduyn S, Toffaletti DL, Marra M, Jones SJM, Perfect JR, Kronstad J. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot Cell. 2003;2:1336–1349. doi: 10.1128/EC.2.6.1336-1349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One. 2011;6:e17412. doi: 10.1371/journal.pone.0017412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene-Transfer in Cryptococcus neoformans by Use of Biolistic Delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Campbell LT, Donlin MJ, Aurora R, Lodge JK. Global Transcriptome Profile of Cryptococcus neoformans during Exposure to Hydrogen Peroxide Induced Oxidative Stress. Plos One. 2013;8 doi: 10.1371/journal.pone.0055110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela JCS, Praekelt UM, Meacock PA, Planta RJ, Mager WH. The Saccharomyces cerevisiae Hsp12 Gene Is Activated by the High-Osmolarity Glycerol Pathway and Negatively Regulated by Protein-Kinase-A. Mol Cell Biol. 1995;15:6232–6245. doi: 10.1128/mcb.15.11.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelzl K, May RC. Cryptococcal Interactions with the Host Immune System. Eukaryot Cell. 2010;9:835–846. doi: 10.1128/EC.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. P Natl Acad Sci USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harb Symp Quant Biol. 2001;66:567–574. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- Wiederholdl K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem Soc T. 2010;38:1531–1536. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamsl V, Del Poeta M. Role of glucose in the expression of Cryptococcus neoformans antiphagocytic protein 1, App1. Eukaryot Cell. 2011;10:293–301. doi: 10.1128/EC.00252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodl IS, Trayhurn P. Glucose transporters (GLUT and SGLT): expanded families of sugar transport proteins. Br J Nutr. 2003;89:3–9. doi: 10.1079/BJN2002763. [DOI] [PubMed] [Google Scholar]
- Zurita-Martinezl SA, Cardenas ME. Tor and cyclic AMP-protein kinase A: Two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell. 2005;4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.