Abstract
Purpose
The aim of this proof-of-concept work is to propose an unsupervised framework that combines multiple parameters, in “positive-if-all-positive” manner, from different models to localize tumors.
Methods
A voxel-by-voxel analysis of the DW-MRI images of whole prostate was performed to obtain parametric maps for D*, D, f, and K using the IVIM and kurtosis models. Ten patients with moderate or high-risk prostate cancer were included in study. The mean age and serum PSA for these 10 patients were 65 years (range 54 – 78) and 21.9ng/mL (range 4.84 – 44.81), respectively. These patients were scanned using a DW spin-echo sequence with echo-planar readout with 16 equidistantly spaced b-values in the range of 0–2000 s/mm2 (TE=58ms; TR=3990ms; spatial resolution 2.19×2.19×2.73mm3, slices = 26, FOV = 140×140mm, slice gap = 0.27mm, NSA = 2).
Results
The proposed framework detected 24 lesions with 14 were true positive with 58% tumor detection rate on lesion-based analysis with sensitivity of 100%. The mpMRI evaluation (PIRADSv2) identified 12 of 14 true positive lesions with sensitivity of 86%, positive predictive value of mpMRI was 92%. The index lesions were visible on all framework maps and were coded as the most suspicious in 9 of 10 patients.
Conclusion
Preliminary results of the proposed framework indicate high patient-based sensitivity with 100% detection rate for identifying moderate-high risk aggressive index lesions.
Keywords: diffusion-weighted MRI, DWI, prostate cancer, tumor localization
Introduction
In American men, prostate cancer is the most common non-cutaneous cancer type and the second leading cause of cancer death [1]. Currently its diagnosis is based on transrectal ultrasound (TRUS) guided biopsy after an abnormal screening, which includes serum prostate specific antigen (PSA), and digital rectal examination [2]. Multi-parametric MRI (mpMRI), which includes modalities such as T2-weighted (T2W), diffusion-weighted MRI (DW-MRI), and dynamic contrast-enhanced (DCE) imaging [3–5], is an important non-invasive tool for the localization of prostate cancer, enabling targeted biopsies of suspicious lesions in the prostate. Several studies of mpMRI reported improvement in clinically significant prostate cancer detection [6–8].
Diffusion-weighted MRI, a major component of mpMRI, is sensitive to the self-diffusion of water molecules in tissue. DW-MRI is the dominant modality used in interpreting the peripheral zone of the prostate, the site of the majority of prostate cancers. DW-MRI has been credited with the rapid rise of mpMRI as a useful modality for prostate cancer. The DW-MRI portion of the mpMRI consists of an apparent diffusion coefficient (ADC) component based on a series of b-values between 0 and 1000 s/mm2 and a “high b-value” component, generally a single image obtained using b-values between 1400 and 2000 s/mm2 [9–15]. The correlation between ADC values and tissue morphology of prostate cancer has been previously established [9, 16–18].
The main idea behind using multi-parametric MRI is to improve tumor localization using different MRI sequences since the combined detection ability is superior to any single imaging sequence. Our approach has the same underlying idea. However, we are achieving the same goal of improved detection by using unique characteristics estimated from different diffusion models, rather than using different MRI sequences. It is becoming more common to use T2W and ADC maps[16], as it has been shown that ADC on its own is not sufficient. Instead of T2W and ADC maps, in the proposed examples for our framework, we combine ADC, fraction of pseudo-diffusion, and kurtosis [19, 20] obtained from the same diffusion-weighted dataset that consists of only 16 images. Thus, there is no need to perform extra steps such as image registration and resolution reconciliation between the different modalities.
While it is clear that DW-MRI is important for the recognition of prostate cancer, it is unclear whether the current two-component method (fast and slow diffusion) extracts all of the potentially useful information obtained from DW-MRI sequences. Several models of diffusion measurement have been proposed and we utilize two of them as an example to explain multi-characteristic nature of our framework. The intravoxel incoherent motion (IVIM) [19, 21, 22] and kurtosis [20, 23–25] models are widely reported as methods to integrate the various components of DW-MRI. The IVIM model describes the signal decay in prostate tissue using a two-compartment model consisting of both the faster (pseudo-diffusion) and slower (self-diffusion) components. It has been previously shown that diffusion is reduced in prostate cancer using the IVIM model [21, 22]. The kurtosis model attempts to characterize the non-Gaussian part of the displacement distribution. Several groups have shown that the kurtosis was significantly elevated in malignant prostatic tissue compared to normal tissue [23, 25]. While prior studies have shown promising results in separating malignant and normal prostatic tissue, this has only been achieved by manually drawing regions of interest (ROIs) generated by trained observers and not by unsupervised methods [26]. Manual segmentation is not only labor-intensive and time-consuming but is prone to considerable inter-observer variability that may influence the reproducibility and accuracy of DW-MRI analysis.
Recently, automated computer-aided detection (CAD) techniques were introduced to identify prostate cancer automatically by applying classifiers to multi-parametric MRI [27–32]. These classifiers are trained with the information obtained from a combination of T2W MRI, ADC maps, and DCE MRI. This approach has a number of challenges, including the need to spatially register images obtained at different times and to reconcile image resolution differences. Additional difficulties may arise due to peristalsis in the rectum or accumulation of rectal gas due to longer acquisition time required by mpMRI compared to one MRI modality. Furthermore, DCE-MRI requires administration of intravenous contrast agent with added expense, setup time and scan duration. With the exception of a method proposed by Firjani et al. [33], current mpMRI and single modality MRI techniques are either based on classifiers or require segmentation by a trained radiologist, thus they are considered supervised. In contrast, we propose a more simplified unsupervised CAD system that utilizes a combination of multiple characteristics estimated from data obtained during a shorter acquisition with a single imaging modality (DW-MRI) that does not require training sets.
The aim of this proof-of-concept work is to propose a framework that combines multiple parameters from different models to localize tumors using an unsupervised technique. In this work, we utilize both the well-established IVIM and kurtosis models applied to diffusion-weighted images to generate unsupervised multi-characteristic localization maps of intermediate-high risk lesions.
Theory
IVIM Model
Le Bihan et al. [19] proposed the intravoxel incoherent motion (IVIM) model by describing a bi-exponential DW-MRI signal decay. In the biological tissue, these two components represent the fast molecular displacement in the vasculature due to a pressure gradient (pseudo-diffusion) and a slower self-diffusion process (Brownian motion). Eq. 1 represents the IVIM model where D* represents the pseudo-diffusion component and D represents the self-diffusion component:
| (1) |
where S0 represents signal intensity at b=0 s/mm2, f is the fraction of signal dominated by the pseudo-diffusion, α = b·D and β = b·(D* + D), where b is a value derived from the gradients used to acquire DW-MRI scans (b-value).
Kurtosis Model
The diffusional kurtosis model [20] estimates the non-Gaussianity of the displacement distribution function of water. The kurtosis, K, is a dimensionless parameter that represents the deviation from Gaussian behavior. Eq. 2 describes diffusion patterns given specific kurtosis parameters:
| (2) |
where γ = b·D − b2·D2·K/6, D is the self-diffusion constant and b is a b-value based on the gradient applied during the DW-MRI sequence. This model represents underlying tissue complexity based on the measure of kurtosis, calculated by solving Eq. 2 for K. For example, if K=0, Eq. 2 represents a homogeneous Gaussian displacement distribution, while greater kurtosis (K>0) indicates the presence of hindered and/or restricted diffusion or heterogeneous Gaussian diffusion.
Unsupervised multi-characteristic framework
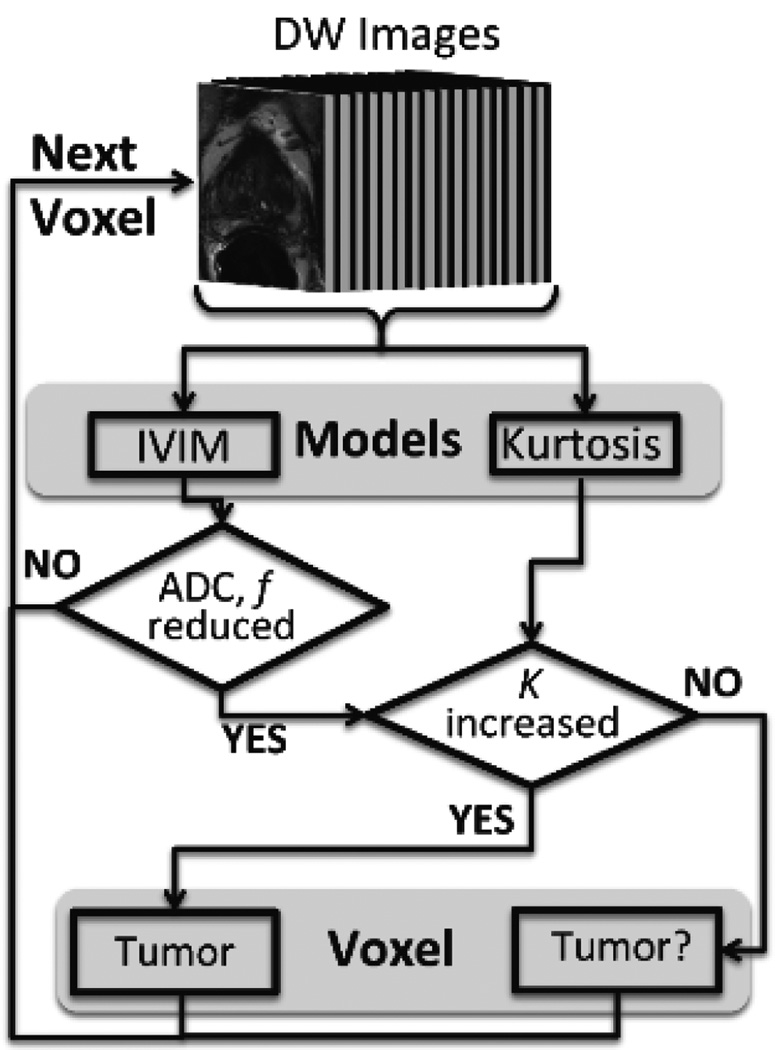
Utilizing previous reports that diffusion parameters are lower in prostate cancer while kurtosis is higher [21–23, 25], we created a framework for multi-characteristic localization of prostate tumors [Figure 1]. This framework combines parameters estimated by independent models (for example, IVIM and kurtosis as were used in this proof-of-concept work) and sets thresholds to distinguish the predicted cancerous and normal regions.
Figure 1.
Flowchart for the unsupervised multi-characteristic framework for prostate cancer localization. This technique utilizes DW-MRI images derived from a 16 b-value DW-MRI as an input to the diffusion models. In this work, two well-established models, intravoxel incoherent motion (IVIM) and kurtosis, were implemented. However, this framework has the capability of being extended to other MRI modalities and corresponding models. Based on previously reported findings for diffusion and kurtosis in prostate cancer tissue, certain criteria were used as shown in this figure to select tumor and tumor suspicious (“Tumor?”) voxels. This process was repeated on a voxel-by-voxel basis.
For each patient, the framework calculates D*, D and f using Eq. 1 and selects voxels with reduced values below a patient-based threshold. Next, kurtosis is taken into account using Eq. 2 and voxels with K greater than the patient-based threshold are selected. Each voxel is marked as “tumor” if all of these conditions are met, with low D*, D and f values and high K values. Voxels with reduced IVIM parameters (pseudo-diffusion fraction and ADC) and kurtosis below the threshold are marked as “tumor suspicious”. In this study, a threshold was set at one standard deviation from the mean value calculated over the entire prostate on a per-patient basis for each of the IVIM and kurtosis parameters.
While the framework requires automated segmentation of the entire prostate using the software iCAD (Nashua, NH), the semi-automated segmentation involves <30–45 seconds of manual correction. This is preferred in order to focus the novel framework on just the prostate tissue. The actual framework for this tumor localization study within the segmented prostate is unsupervised, since the parameter estimation for each model with subsequent threshold calculations are patient-based and do not require training sets.
Methods
Study Design and Patient Population
All patients who underwent 3T prostate MRI with subsequent 12-core systematic and MRI/TRUS fusion-guided biopsy between December 2013 and September 2014 were initially included. Patients with low clinico-pathological risk for prostate cancer or no evidence of prostate cancer were excluded. Thus, a total of ten patients with moderate or high-risk prostate cancer were included in this proof-of-concept study.
These patients were scanned using a DW spin-echo sequence with echo-planar readout with 16 equidistantly spaced b-values in the range of 0–2000 s/mm2 (TE=58ms; TR=3990ms; spatial resolution 2.19×2.19×2.73mm3, slices = 26, FOV = 140×140mm, slice gap = 0.27mm, NSA = 2). A 3T clinical MR scanner (Achieva 3.0T-TX, Philips Healthcare, Best, NL) along with the anterior half of a 32-channel SENSE cardiac coil (In vivo; Gainesville, FL, USA) and an endorectal coil (ERC) filled with fluorinert (BPX-30, Medrad, Pittsburgh, PA, USA) were used. The MRI/TRUS fusion-guided biopsy was based on the results of the mpMRI which included triplanar T2W turbo spin echo (TSE) MRI, ADC maps derived from DW-MRI (obtained from a low b-value DW-MRI using 5 evenly spaced b-values between 0–750 s/mm2 and a high b-value image of 2000 s/mm2), and DCE MRI, which were prospectively evaluated as described previously [22]. No motion correction algorithms were used, though use of the ERC helps to limit patient motion. The scan acquisition time for the framework-specific sequence using 16 equidistantly spaced b-values is 5–6 minutes, and the full standard mpMRI acquisition time is 40–45 minutes.
Image Analysis
Whole prostate contour DW-MRI images were analyzed in a blinded fashion, since the construction of the model (by RZ, a computer engineer) was performed without prior knowledge of the risk of prostate cancer. A voxel-by-voxel analysis of the segmented DW-MRI prostate images was performed using the proposed unsupervised multi-characteristic framework that integrated parameters estimated from the IVIM and kurtosis models. Parametric maps for D*, D, f were obtained in two steps. The cutoff b-value was set to 133 s/mm2, within the range of what is typically used in previous literature. The D and f parameters were estimated using a monoexponential model for the diffusion-weighted signal acquired at b-values between 133–800 s/mm2. The D* parameter was estimated according to Eq. 1 using all b-values between 0–800 s/mm2 with the D and f parameters set to values estimated in the first step. The K parameter was estimated over the entire range of b-values (0–2000 s/mm2) according to Eq. 2. In-house post-processing software was developed using Matlab (MathWorks Inc, Natick, Massachusetts).
Evaluation of unsupervised multi-characteristic framework maps
Three readers, including one radiologist (BT with 8 years of cumulative experience in prostate MRI) and 2 radiology trainees (SS and AB with 18 months and 8 months of experience in prostate MRI, respectively), evaluated the unsupervised multi-characteristic framework maps in consensus while blinded to the histopathology and mpMRI results. The lesions suspicious for prostate cancer were recorded on the unsupervised multi-characteristic framework map of each patient and then the readers were asked to determine a possible index lesion as the lesion harboring the most biologically aggressive prostate cancer within a patient’s prostate gland. Finally, each reader separately scored the overall performance of the unsupervised multi-characteristic framework maps to identify index lesions independently on a 5-point scale (1-not helpful at all, 5-very helpful). In a separate session (7 months after the evaluation of the unsupervised multi-characteristic framework maps) the same three readers evaluated the mpMRI data by using the most recent version of Prostate Imaging Reporting and Data System (PIRADSv2) independently and blinded to histopathology and the multi-characteristic framework maps[23]. The median score of the three readers was used to compare mpMRI results with the framework maps and histopathology. The detected lesions were correlated to histopathology from 12-core systematic and MRI/TRUS fusion-guided biopsy results and whole mount histopathology when available. Lesion-based sensitivity and positive predictive values for framework maps and mpMRI were calculated. Specificity and negative predictive values could not be estimated since the readers were only asked to identify tumor suspicious foci but not tumor negative (benign) regions.
Results
The mean age and serum PSA for the 10 patients included in this study were 65 years (range 54 – 78) and 21.9ng/mL (range 4.84 – 44.81), respectively. The highest Gleason scores from prostate biopsy and whole mount histopathology ranged from 7(3+4) to 9(4+5). The full list of Gleason scores is shown in Table 1.
Table 1.
Performance evaluation of the unsupervised multi-characteristic framework in 10 patients by the three independent readers.
| Case # | Age | Serum PSA |
Highest Gleason |
Total detected lesions |
True positive lesions |
Tumor detection rate |
Index lesion detection |
Performance evaluation |
|---|---|---|---|---|---|---|---|---|
| 1 | 73 | 38.6 | 9 (4 + 5) | 2 | 1 | 0.5 | yes | 4.7 |
| 2 | 71 | 7.63 | 9 (4+5) | 5 | 1 | 0.2 | yes* | 3 |
| 3 | 61 | 8.52 | 7 (3+4) | 4 | 2 | 0.5 | yes | 4 |
| 4 | 55 | 5.05 | 7 (3+4) | 3 | 2 | 0.67 | yes | 3.7 |
| 5 | 65 | 14.71 | 9 (4+5) | 1 | 1 | 1 | yes | 4 |
| 6 | 54 | 35.55 | 8 (4+4) | 1 | 1 | 1 | yes | 4.3 |
| 7 | 71 | 16.09 | 9 (4+5) | 3 | 3 | 1 | yes | 4.7 |
| 8 | 78 | 43.18 | 9 (4+5) | 2 | 1 | 0.5 | yes | 5 |
| 9 | 60 | 44.81 | 9 (4 +5) | 2 | 1 | 0.5 | yes | 4 |
| 10 | 58 | 4.84 | 7 (4+3) | 2 | 1 | 0.5 | yes | 4.3 |
| Mean | 65 | 21.9 | 2.5 | 1.4 | 0.64 | 4.2 |
Framework development
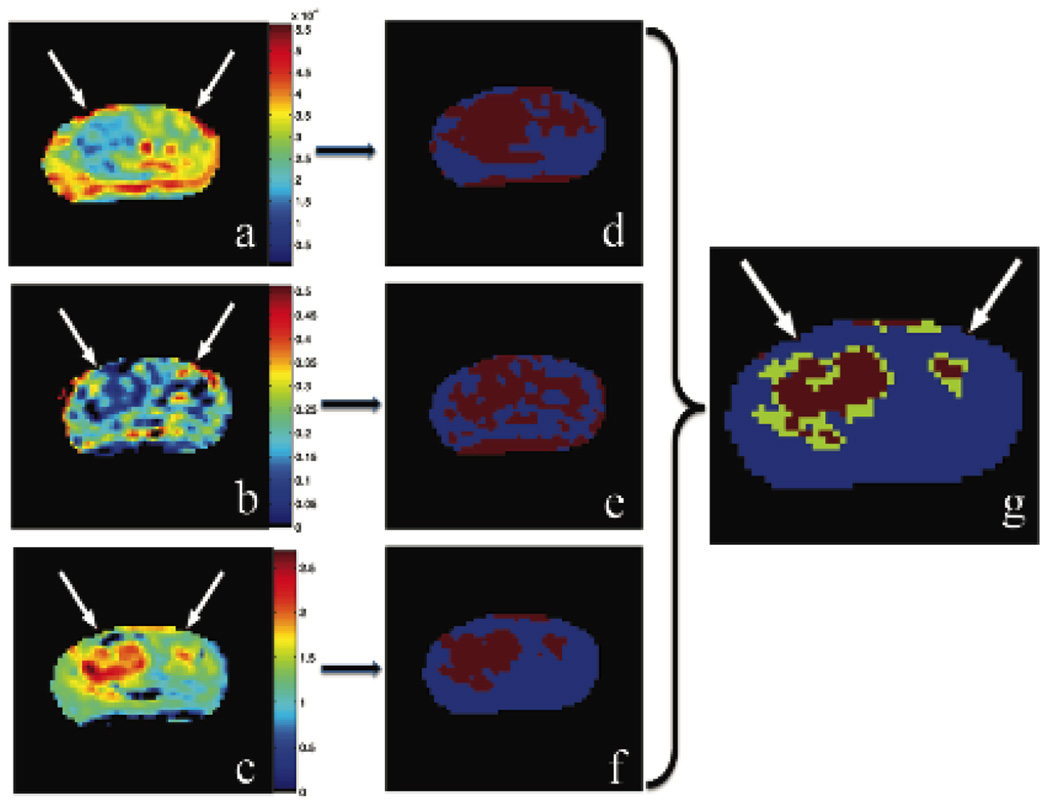
In this study we used both the IVIM and kurtosis models separately as an example of how our framework could be extended to any number of models and/or parameters. The diffusion parameters D*, D, and f and the kurtosis value K were separately calculated and spatially mapped onto the diffusion-weighted b=0 s/mm2 MRI scans. Figure 2 shows an example of the output of the multi-characteristic framework.
Figure 2.
Estimated characteristics: apparent diffusion coefficient, D (a); pseudo-diffusion fraction, f, (b); kurtosis, K, (c); corresponding threshold maps (d–e); unsupervised framework map with cancerous voxels in red and tumor suspicious voxels in green (f).
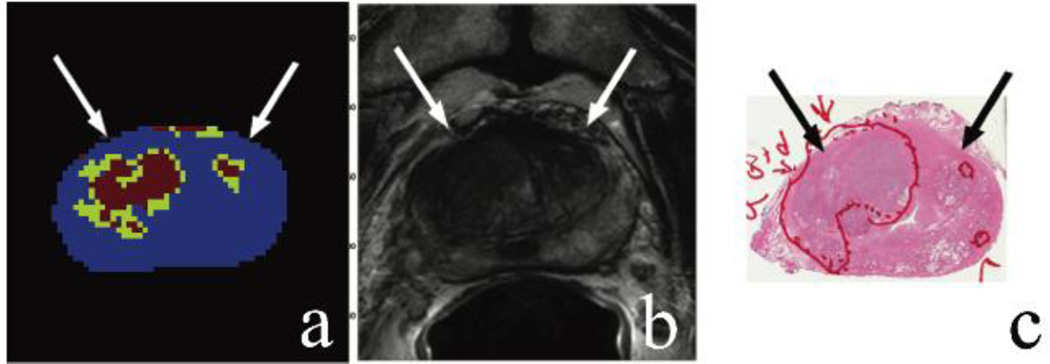
Three parameters, D, f, and K, from the two models were combined in a “positive-if-all-positive” manner into a final composite framework map. The pseudo-diffusion coefficient showed no significant decrease, thus it was not included in our framework [21]. Figure 3a shows an example of this framework map that combines the independent parameters displayed in Figure 2. The “tumor” voxels are shown in red and the “tumor suspicious” voxels are shown in green. These areas correlated with the T2-weighted MRI lesions and histopathology findings as shown in Figure 3b and 3c, respectively. Uncolored regions are considered not suspicious for cancer.
Figure 3.
Unsupervised framework map with cancerous voxels in red and tumor suspicious voxels in green (a); axial T2-weighted MR image confirming large regions high level of risk for prostate carcinoma (b); corresponding histology slide confirms a large focus of Gleason 4+4 cancer (c).
Evaluation of unsupervised multi-characteristic framework maps
The framework maps were evaluated independently by the three readers. The unsupervised multi-characteristic framework detected 24 lesions of which 14 were true positive based on histopathology. This resulted in an overall 58% positive predictive value on lesion-based analysis. Moreover, the sensitivity of framework maps was 100%. The mpMRI evaluation using PIRADSv2 identified 12 of 14 true positive lesions, which resulted in a sensitivity of 86%, whereas the positive predictive value of mpMRI was 92% (Tables 1 and 2). In all patients, the index lesion as determined by mpMRI was visible on the unsupervised framework maps representing a per-patient sensitivity of 100%. The true positive index lesions were coded as the most suspicious lesion on the framework maps in 9 of 10 patients.
Table 2.
Clinical evaluation of prostate lesions on mpMRI using PIRADSv2 and the multi-characteristic framework.
| Case # |
Age | PSA | Lesion | Index lesion |
Median PIRADSv2 score |
Framewor k result |
Histology result |
Overall performance Evaluation |
|---|---|---|---|---|---|---|---|---|
| 1 | 73 | 38.6 | 1 | Y | 5 | Positive | Gleason 4+5 | 4.7 |
| 2 | N | 1 | Positive | NA | ||||
| 2 | 71 | 7.63 | 1 | N | 1 | Positive | BPH | 3 |
| 2 | Y | 5 | Positive* | Gleason 4+5 | ||||
| 3 | N | 1 | Positive | BPH | ||||
| 4 | N | 1 | Positive | BPH | ||||
| 5 | N | 1 | Positive | NA | ||||
| 3 | 61 | 8.52 | 1 | Y | 5 | Positive | Gleason 3+4 | 4 |
| 2 | N | 4 | Positive | Gleason 3+3 | ||||
| 3 | N | 4 | Positive | Benign | ||||
| 4 | 55 | 5.05 | 1 | Y | 5 | Positive | Gleason 3+4 | 3.7 |
| 2 | N | 1 | Positive | Gleason 3+3 | ||||
| 3 | N | 1 | Positive | BPH | ||||
| 5 | 65 | 14.71 | 1 | Y | 5 | Positive | Gleason 4+5 | 4 |
| 6 | 54 | 35.55 | 1 | Y | 5 | Positive | Gleason 4+4 | 4.3 |
| 7 | 71 | 16.09 | 1 | Y | 5 | Positive | Gleason 4+5 | 4.7 |
| 2 | N | 5 | Positive | Gleason 4+5 | ||||
| 3 | N | 1 | Positive | Gleason 4+3 | ||||
| 8 | 78 | 43.18 | 1 | Y | 5 | Positive | Gleason 4+5 | 5 |
| 2 | N | 1 | Positive | NA | ||||
| 9 | 60 | 44.81 | 1 | Y | 5 | Positive | Gleason 4+4 | 4 |
| 2 | N | 1 | Positive | Benign | ||||
| 10 | 58 | 4.84 | 1 | Y | 5 | Positive | Gleason 4+3 | 4.3 |
| 2 | N | 1 | Positive | NA |
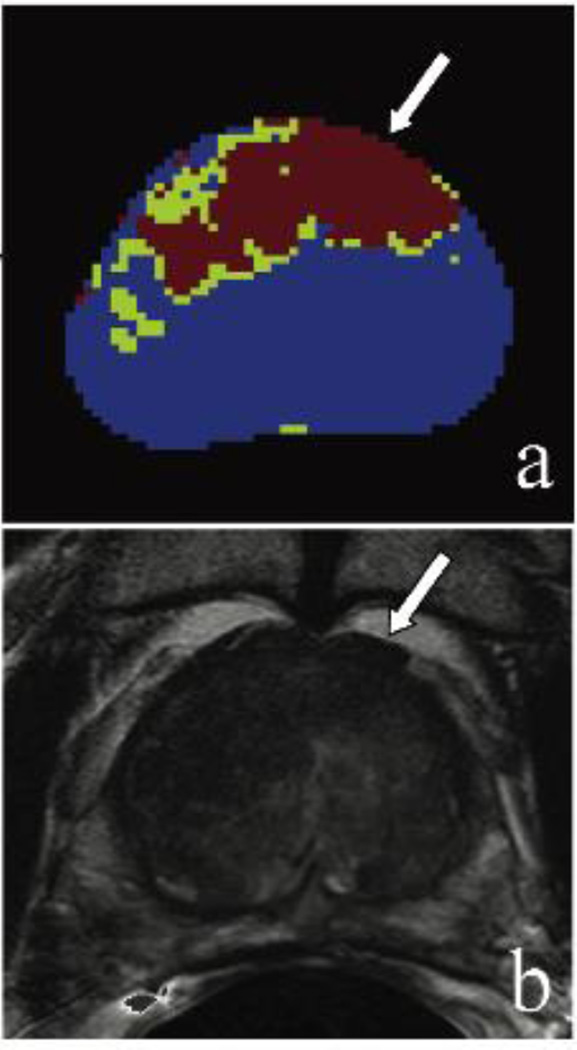
Examples of the performance of the unsupervised framework maps are shown in Figures 4–6. Figures 4 and 6 show results for cases with performance scores of 5 (very helpful) on clinical evaluation, whereas the mean performance scores for the case in Figure 5 were lower.
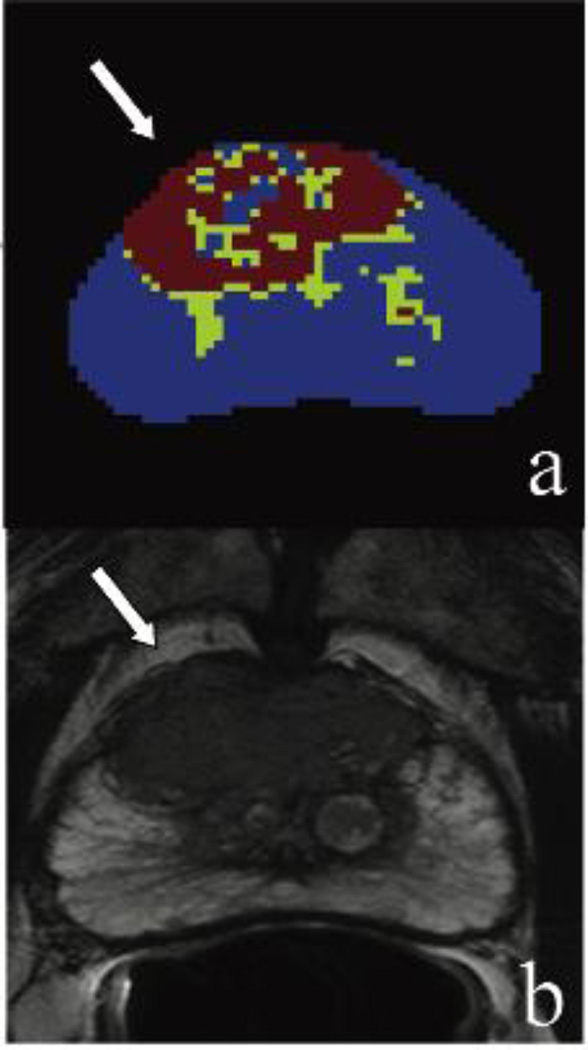
Figure 4.
(Case #1) A 73 year old patient with a PSA of 38.6 ng/mL with multiple prior negative systematic biopsies. Framework map identifying lesion voxels marked in red and green (a); axial T2-weighted MR image confirming the large anterior lesion with broad capsular base, border irregularity, and capsular bulge – highly suspicious for extracapsular extension (b). Lesion is displayed with the white arrows. MRI/TRUS fusion-guided biopsy confirmed Gleason 4+5 cancer.
Figure 6.
(Case #9) A 60 year old patient with a PSA of 44.81 ng/mL with prior negative systematic biopsy. Framework map identifying lesion voxels marked in red and green (a); axial T2-weighted MR image confirming the midline apical-mid transition zone lesion with broad capsular base (b). Lesion is displayed with the white arrows. MRI/TRUS fusion-guided biopsy targeting this lesion confirmed Gleason 4+4 cancer.
Figure 5.
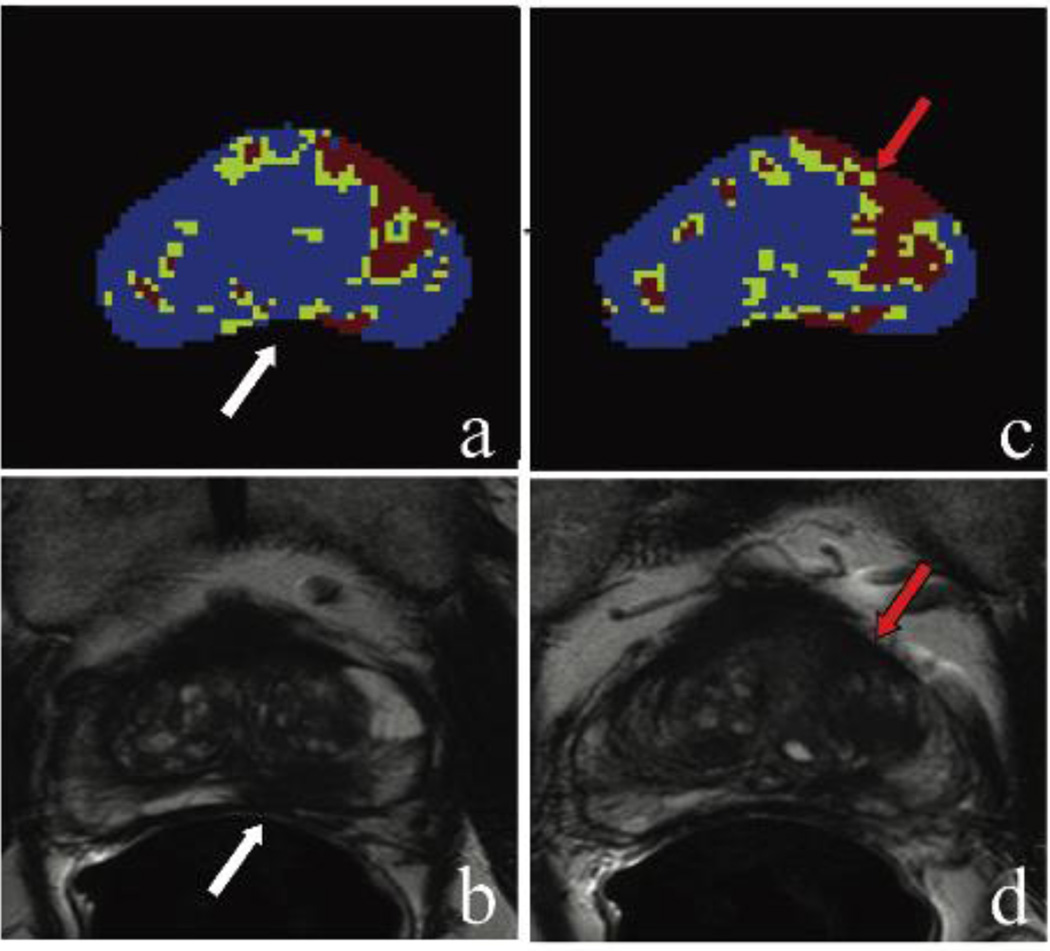
(Case #2) A 71 year-old patient with a PSA of 7.63 ng/mL with no prior biopsy. Framework map identifying lesion voxels marked in red and green (a); axial T2-weighted MR image confirming the left apical to base peripheral zone lesion (b) which included Gleason 4+5 cancer detected on the MRI/TRUS fusion-guided biopsy. Adjacent slice in the same patient is shown in (c)–(d) with the framework map identifying lesion voxels marked in red and green (c); however, axial T2-weighted MR image does not identify a lesion at this level (d). True positive lesion is displayed with the white arrows. The framework map identified a lesion in the left anterior transition zone which is most likely a false-positive finding (red arrows).
In one patient (case #2 shown in Figure 5) the actual true positive index lesion was evaluated as the second most suspicious lesion on the unsupervised framework map. For this patient, another lesion in the left mid-base transition zone of the prostate was read as the most suspicious (index) lesion, and this area was in fact a benign prostatic hyperplasia (BPH) nodule. The most common causes of false positive lesions were BPH nodules (n=7) and anterior fibrous stroma (n=3) on histology.
Performance evaluation revealed a mean score of 4 or above in eight patients, whereas in one patient the mean score was 3.7 and for case #2 with the incorrectly identified index lesion, it was 3 [Table 1].
Discussion
In this work, we evaluated the feasibility of a novel unsupervised method of identifying prostate cancers based on a multi b-value DW-MRI modality analyzed with a multi-characteristic framework. The proposed technique enabled the identification of suspicious areas on the framework maps corresponding to the index lesion on mpMRI. There were, however, several false positives corresponding to BPH and fibrous stromal tissue, which have similar diffusivity properties as malignant tumors [5, 11, 16]. Additional refinements of the framework may be needed to exclude these types of benign lesions in the future. The overall performance of the framework maps was evaluated as helpful (mean score ≥ 4 out of 5) in 8 of 10 patients, whereas in the remaining two patients the framework map delineated lesions that were also benign. Ultimately, in all patients the dominant lesion was captured with the proposed technique.
Our approach consists of combining a number of unique parameters that characterize underlying tissue structure estimated from the DW-MRI signal attenuation. To demonstrate the main principle of our framework, we combined three unique parameters derived from the well-established IVIM and kurtosis models. The models were chosen based on the uniqueness of the estimated parameters rather than for their biologic significance. Although, we do not know the exact biophysical processes that drive estimation of pseudo-diffusion and kurtosis parameters, nevertheless these parameters no doubt reflect behavior of the tissue, i.e., diffusion and fraction of pseudo-diffusion decrease and kurtosis increases in the abnormal tissue compared to normal. These three unique parameters included the apparent diffusion coefficient (D), fraction of pseudo-diffusion (f), and kurtosis (K) values. The final framework map with identified “tumor” and “tumor suspicious” regions was generated by applying a threshold to the individual parametric maps and masking voxels that satisfied all conditions. Regions that demonstrated a reduction in diffusivity and increase in kurtosis were labeled as tumor-containing regions. The process of combining unique parameters using multiple models allows for the reduction of false positive and false negative voxels compared to using a single parameter, such as only ADC or kurtosis. This is due to the fact that theoretically a “positive-if-all-positive” selection process, as was applied for our framework, favors higher specificity. However, further work is needed to test the proposed framework such that each independent parameter is evaluated separately to ensure that the multi-characteristic framework indeed yields higher specificity. Additionally, threshold optimization for each parameter will increase robustness of the framework.
Our results show that the diffusion kurtosis parameter correlates best with the final lesion outline based on visual analysis. However, kurtosis may define the lesion size too specifically, while other IVIM parameters might characterize more fully microstructural information in the lesion periphery. Taking into account all of the lesion features, both the core of the lesion and the periphery of the lesion, is a helpful practice for better delineating the lesions for biopsies… The ADC and fraction parameters are almost identical at a higher signal-to-noise ratio (SNR) when the endorectal coil is used. These observations were made through visual examination of the algorithm parameters but were not formally evaluated and scored by the readers. In general, because pathologists following standard clinical procedures do not outline the extent of each lesion, there is no “gold” standard to assess each parametric map in regards to proper lesion(s) size and absolute location. Overall, without comparison with “gold standard, kurtosis outperforms two other parameters.
Existing CAD techniques identify prostate cancer by applying classifiers to multi-parametric MRI [27–32]. These classifiers are trained with information obtained from a combination of different MRI modalities. Besides registering and matching resolution among images obtained from different MRI sequences at different time points, such CAD techniques have a difficult task of training classifiers due to the large variety of cancer types and locations within the patient population. Thus, these CAD techniques remain supervised and biased depending on the specific patient population chosen for training classifiers.
The current patchwork of DW-MRI consisting of lower b-value images obtained to construct ADC maps and a “high b-value” image may not extract as much information as is possible if all the b-values were acquired in one sequence. In this way, the data could be more easily analyzed according to established models [34]. Although DW-MRI itself may be subject to motion, in most cases misregistration between consecutive DW-MRI scans is not as severe as between different classes of imaging such as T2W and DCE MRI.
Several investigators have previously demonstrated the value of DW-MRI for localizing prostate cancer. Firjani et al. [33] used a 3D Generalized Gauss-Markov Random Field model with a voxel neighborhood to smooth the distance between signal intensity in the absence of a diffusion sensitizing gradient (b=0 s/mm2) and the attenuated signal at b=800 s/mm2. This group subsequently identified tumor boundaries with a level set-based deformable model. Such an approach is well suited to large tumors, but it is not clear how it will perform for smaller lesions. It is important to note that our approach is based on voxel-by-voxel post-processing in which each voxel is evaluated independently and stringently. Thus, our model is not limited by the size of a lesion. Another challenge with the approach used by Firjani et al. is the close proximity of benign and malignant tissue, which may become merged due to 3D smoothing. Although Firjani et al. applied K-Nearest Neighbor (KNN) classifier analysis to delineate between benign and malignant tumors, Oto et al. [5] showed that there was no statistical difference in diffusivity between glandular and stromal BPH and tumors in the transition zone. Thus, the smoothing approach used by Firjani et al. will further exaggerate the size of the segmented lesion. Furthermore, the lack of DW-MRI data acquisition standardization reduces the reliability of training sets in the clinical setting, which is a problem unique to supervised approaches requiring training and testing sets. In comparison, our unsupervised approach does not depend on the relative similarity of an initial set of data since there are no training sets involved.
The proposed unsupervised multi-characteristic framework helped readers identify the index lesions in all patients, resulting in a sensitivity of 100% for index lesion detection. However, the lesion-based positive predictive value was relatively low at 58% and eight of the framework detected lesions were not regarded as suspicious for cancer on mpMRI. This was mainly due to false positive lesions as a result of BPH and anterior stromal tissue with diffusion properties similar to malignancy. The tissue composition of BPH can have dense cellularity and reduced extracellular fluid components [5]. Several groups have reported overlapping findings of prostate cancer and BPH on ADC maps of DW-MRI [5, 11, 16]. A potential solution to overcome the challenge of separating prostate cancer from BPH is to further refine the current algorithm with the incorporation of histologically validated BPH samples.
While comparing the results of our framework directly with other studies using diffusion MRI to identify prostate cancer may provide some insights, in clinical practice radiologists rely on multiple MRI parameters to detect prostate cancer. Multiple reports have demonstrated that the combination of MRI sequences rather than a single sequence alone provides the best results for lesion detection [26, 35]. Thus, a more fruitful approach for comparing our framework to standard clinical practice involved evaluation of the same patient cohort on mpMRI.
We aimed to compare the current multi-characteristic framework maps with mpMRI by using PIRADSv2 methodology. The lesion-based comparison revealed that the framework maps are slightly more sensitive than mpMRI; however, the positive predictive value of the framework maps was lower than that of mpMRI. Although specificity analysis could not be done in the current study design, mpMRI evaluation using PIRADSv2 methodology can be more specific for index lesions. Joint use of the framework maps and mpMRI can improve prostate cancer detection yield and potentially reduce the false positive rate. However, this idea requires further research and validation in a larger scale study.
During the clinical evaluation of the framework maps, we observed the presence of “tumor suspicious” voxels with reduced diffusion coefficients and normal kurtosis. From a biological standpoint, our best explanation is that these areas may represent the tumor boundary, which harbors both healthy and cancerous tissue [36]. These voxels may represent the safety margin around the tumor core that needs to be included in targeted focal therapeutic strategies. However, further exploration is needed to confirm the nature of these “tumor suspicious” regions, which typically abutted the “tumor voxels.”
There are a number of limitations to our framework. First, our patient population included intermediate-high risk patients with highest Gleason scores ranging from 3+4=7 to 4+5=9 and thus, the tumors detected tended to be larger. We evaluated higher risk lesions to determine the initial feasibility of our approach. Moving forward with this work, we fully intend to conduct this study on a larger cohort of patients with a broader range of risk for prostate cancer. Our approach also used images obtained using an ERC, which optimizes the signal to noise ratio (SNR). There is a growing trend away from the use of ERC and we do not yet have experience in this setting. Another limitation is that our DW-MRI images were obtained with 16 equally spaced b-values, which is time-consuming to acquire although comparable in time to separately acquired low b and high b-value images. Minimizing the number of b-values used for image acquisition is highly desirable for clinical applications of our model. A further concern is the small sample size examined in our proof-of-concept study, since only 10 patients with moderate to high-risk prostate cancer who underwent biopsy were included. The main intent in choosing such a patient population was to test this new method in detecting clinically relevant cancer lesions which need definitive therapy. There is a need to evaluate the performance of our approach on a larger patient population with different levels of risk for prostate cancer to overcome this bias towards more aggressive tumors.
In conclusion, in this work we present a novel unsupervised multi-characteristic framework for prostate cancer localization based on a 16 b-value DW-MRI sequence analyzed concurrently with two models of diffusion measurement, IVIM and kurtosis. Our preliminary results have shown high sensitivity with a 100% detection rate for identifying the moderate-high risk aggressive index lesions. This approach has comparable results with mpMRI and it utilizes parameters obtained using a single MRI technique with much less scanning time required, thus potentially can be an adjunct to mpMRI. Further analysis in a larger patient population with less aggressive disease is warranted to expand this technique from the proof-of-concept stage to a clinically useful tool.
Acknowledgments
We are grateful to Peter Basser, Ph.D., Elizabeth Hutchinson, Ph.D. and Alexandru Avram for providing helpful suggestions.
This research was made possible through the National Institutes of Health (NIH) Medical Research Scholars Program, a public-private partnership supported jointly by the NIH and generous contributions to the Foundation for the NIH from Pfizer Inc, The Doris Duke Charitable Foundation, The Newport Foundation, The American Association for Dental Research, The Howard Hughes Medical Institute, and the Colgate-Palmolive Company, as well as other private donors. For a complete list, please visit the Foundation website at: http://fnih.org/work/education-training-0/medical-research-scholars-program
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, et al. Cancer Statistics, 2014. CA: A Cancer Journal for Clinicians. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Rothwax JT, et al. Multiparametric MRI in biopsy guidance for prostate cancer: fusion-guided. Biomed Res Int. 2014;2014:439171. doi: 10.1155/2014/439171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noworolski SM, et al. Dynamic contrast-enhanced MRI and MR diffusion imaging to distinguish between glandular and stromal prostatic tissues. Magn Reson Imaging. 2008;26(8):1071–1080. doi: 10.1016/j.mri.2008.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ocak I, et al. Dynamic contrast-enhanced MRI of prostate cancer at 3 T: a study of pharmacokinetic parameters. AJR Am J Roentgenol. 2007;189(4):849. doi: 10.2214/AJR.06.1329. [DOI] [PubMed] [Google Scholar]
- 5.Oto A, et al. Prostate cancer: differentiation of central gland cancer from benign prostatic hyperplasia by using diffusion-weighted and dynamic contrast-enhanced MR imaging. Radiology. 2010;257(3):715–723. doi: 10.1148/radiol.10100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salami SS, et al. Multiparametric magnetic resonance imaging outperforms the Prostate Cancer Prevention Trial risk calculator in predicting clinically significant prostate cancer. Cancer. 2014;120(18):2876–2882. doi: 10.1002/cncr.28790. [DOI] [PubMed] [Google Scholar]
- 7.Delongchamps NB, et al. Multiparametric magnetic resonance imaging for the detection and localization of prostate cancer: combination of T2-weighted, dynamic contrast-enhanced and diffusion-weighted imaging. BJU Int. 2011;107(9):1411–1418. doi: 10.1111/j.1464-410X.2010.09808.x. [DOI] [PubMed] [Google Scholar]
- 8.Turkbey B, et al. Multiparametric 3T prostate magnetic resonance imaging to detect cancer: histopathological correlation using prostatectomy specimens processed in customized magnetic resonance imaging based molds. J Urol. 2011;186(5):1818–1824. doi: 10.1016/j.juro.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issa B. In vivo measurement of the apparent diffusion coefficient in normal and malignant prostatic tissues using echo-planar imaging. J Magn Reson Imaging. 2002;16(2):196–200. doi: 10.1002/jmri.10139. [DOI] [PubMed] [Google Scholar]
- 10.Sato C, et al. Differentiation of noncancerous tissue and cancer lesions by apparent diffusion coefficient values in transition and peripheral zones of the prostate. J Magn Reson Imaging. 2005;21(3):258–262. doi: 10.1002/jmri.20251. [DOI] [PubMed] [Google Scholar]
- 11.Hosseinzadeh K, Schwarz SD. Endorectal diffusion-weighted imaging in prostate cancer to differentiate malignant and benign peripheral zone tissue. J Magn Reson Imaging. 2004;20(4):654–661. doi: 10.1002/jmri.20159. [DOI] [PubMed] [Google Scholar]
- 12.Costouros NG, et al. Diagnosis of prostate cancer in patients with an elevated prostate-specific antigen level: role of endorectal MRI and MR spectroscopic imaging. AJR Am J Roentgenol. 2007;188(3):812–816. doi: 10.2214/AJR.06.0165. [DOI] [PubMed] [Google Scholar]
- 13.deSouza NM, et al. Magnetic resonance imaging in prostate cancer: the value of apparent diffusion coefficients for identifying malignant nodules. Br J Radiol. 2007;80(950):90–95. doi: 10.1259/bjr/24232319. [DOI] [PubMed] [Google Scholar]
- 14.Turkbey B, et al. Imaging localized prostate cancer: current approaches and new developments. AJR Am J Roentgenol. 2009;192(6):1471–1480. doi: 10.2214/AJR.09.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma S, Rajesh A. A clinically relevant approach to imaging prostate cancer: review. AJR Am J Roentgenol. 2011;196(3 Suppl):S1–S10. doi: 10.2214/AJR.09.7196. Quiz S11-4. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs P, et al. Comparison of quantitative T2 mapping and diffusion-weighted imaging in the normal and pathologic prostate. Magn Reson Med. 2001;46(6):1054–1058. doi: 10.1002/mrm.1298. [DOI] [PubMed] [Google Scholar]
- 17.Tamada T, et al. Apparent diffusion coefficient values in peripheral and transition zones of the prostate: comparison between normal and malignant prostatic tissues and correlation with histologic grade. J Magn Reson Imaging. 2008;28(3):720–726. doi: 10.1002/jmri.21503. [DOI] [PubMed] [Google Scholar]
- 18.Hambrock T, et al. Relationship between apparent diffusion coefficients at 3.0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology. 2011;259(2):453–461. doi: 10.1148/radiol.11091409. [DOI] [PubMed] [Google Scholar]
- 19.Le Bihan D, et al. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161(2):401–407. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- 20.Jensen JH, et al. Diffusional kurtosis imaging: the quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magn Reson Med. 2005;53(6):1432–1440. doi: 10.1002/mrm.20508. [DOI] [PubMed] [Google Scholar]
- 21.Dopfert J, et al. Investigation of prostate cancer using diffusion-weighted intravoxel incoherent motion imaging. Magn Reson Imaging. 2011;29(8):1053–1058. doi: 10.1016/j.mri.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Riches SF, et al. Diffusion-weighted imaging of the prostate and rectal wall: comparison of biexponential and monoexponential modelled diffusion and associated perfusion coefficients. NMR Biomed. 2009;22(3):318–325. doi: 10.1002/nbm.1328. [DOI] [PubMed] [Google Scholar]
- 23.Rosenkrantz AB, et al. Prostate cancer: feasibility and preliminary experience of a diffusional kurtosis model for detection and assessment of aggressiveness of peripheral zone cancer. Radiology. 2012;264(1):126–135. doi: 10.1148/radiol.12112290. [DOI] [PubMed] [Google Scholar]
- 24.Prostate Imaging Reporting and Data System version 2.0. 2015 [Google Scholar]
- 25.Suo S, et al. Non-Gaussian water diffusion kurtosis imaging of prostate cancer. Magn Reson Imaging. 2014;32(5):421–427. doi: 10.1016/j.mri.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Turkbey B, et al. Prostate cancer: value of multiparametric MR imaging at 3 T for detection--histopathologic correlation. Radiology. 2010;255(1):89–99. doi: 10.1148/radiol.09090475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan I, et al. Detection of prostate cancer by integration of line-scan diffusion, T2-mapping and T2-weighted magnetic resonance imaging; a multichannel statistical classifier. Med Phys. 2003;30(9):2390–2398. doi: 10.1118/1.1593633. [DOI] [PubMed] [Google Scholar]
- 28.Langer DL, et al. Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2009;30(2):327–334. doi: 10.1002/jmri.21824. [DOI] [PubMed] [Google Scholar]
- 29.Artan Y, et al. Prostate Cancer Localization With Multispectral MRI Using Cost-Sensitive Support Vector Machines and Conditional Random Fields. Ieee Transactions on Image Processing. 2010;19(9):2444–2455. doi: 10.1109/TIP.2010.2048612. [DOI] [PubMed] [Google Scholar]
- 30.Niaf E, et al. Computer-aided diagnosis of prostate cancer in the peripheral zone using multiparametric MRI. Physics in Medicine and Biology. 2012;57(12):3833–3851. doi: 10.1088/0031-9155/57/12/3833. [DOI] [PubMed] [Google Scholar]
- 31.Vos PC, et al. Automatic computer-aided detection of prostate cancer based on multiparametric magnetic resonance image analysis. Physics in Medicine and Biology. 2012;57(6):1527–1542. doi: 10.1088/0031-9155/57/6/1527. [DOI] [PubMed] [Google Scholar]
- 32.Hambrock T, et al. Prostate Cancer: Computer-aided Diagnosis with Multiparametric 3-T MR Imaging-Effect on Observer Performance. Radiology. 2013;266(2):521–530. doi: 10.1148/radiol.12111634. [DOI] [PubMed] [Google Scholar]
- 33.Firjani A, et al. A diffusion-weighted imaging based diagnostic system for early detection of prostate cancer. J of Biomedical Science and Engineering. 2013;6:346–356. [Google Scholar]
- 34.Loch R, et al. Prostate magnetic resonance imaging: challenges of implementation. Curr Probl Diagn Radiol. 2015;44(1):26–37. doi: 10.1067/j.cpradiol.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Tanimoto A, et al. Prostate cancer screening: The clinical value of diffusion-weighted imaging and dynamic MR imaging in combination with T2-weighted imaging. Journal of Magnetic Resonance Imaging. 2007;25(1):146–152. doi: 10.1002/jmri.20793. [DOI] [PubMed] [Google Scholar]
- 36.Gibson E, et al. Multiparametric MR imaging of prostate cancer foci: assessing the detectability and localizability of Gleason 7 peripheral zone cancers based on image contrasts. Medical Imaging 2014: Digital Pathology. 2014;9041 [Google Scholar]