SUMMARY
Borrelia burgdorferi, the causative agent of Lyme disease, is a highly motile spirochete, and motility, which is provided by its periplasmic flagella, is critical for every part of the spirochete’s enzootic life cycle. Unlike externally flagellated bacteria, spirochetes possess a unique periplasmic flagellar structure called the collar. This spirochete-specific novel component is linked to the flagellar basal body; however, nothing is known about the proteins encoding the collar or their function in any spirochete. To identify a collar protein and determine its function, we employed a comprehensive strategy that included genetic, biochemical, and microscopic analyses. We found that BB0286 (FlbB) is a novel flagellar motor protein, which is located around the flagellar basal body. Deletion of bb0286 has a profound effect on collar formation, assembly of other flagellar structures, morphology, and motility of the spirochete. Orientation of the flagella toward the cell body is critical for determination of wild-type spirochete’s wave-like morphology and motility. Here, we provide the first evidence that FlbB is a key determinant of normal orientation of the flagella and collar assembly.
INTRODUCTION
Lyme disease, which is caused by the spirochete, Borrelia burgdorferi, is the most prevalent vector-borne illness in the United States (Mead, 2015). In nature, survival of B. burgdorferi depends on migration by the bacteria to sites of colonization in Ixodes ticks and mammalian hosts (Zhang et al., 2006; Kuehn, 2013). The spirochete is a motile organism and motility is reported to be crucial for every parts of the spirochete’s pathogenic life cycle, e.g., viability of B. burgdorferi in ticks, transmission from ticks to mice, persistent infection and dissemination in the mammalian host (Sultan et al., 2013; Sultan et al., 2015; Motaleb et al., 2015).
The bacterial flagellar motor, such as those examined in Escherichia coli, is a highly efficient nano-machine, with a rotation frequency greater than 100 Hz, even though the diameter of the motor is only ca. 45 nm. The flagellum is a complex structure that is composed of three substructures whose assembly requires at least 25 different proteins. The basal body-motor portion of the flagellum is connected to the filament by the rod-hook assembly. When torque is generated by proton (or sodium in some organisms) flux, the flagellum stator rotates the filament propelling the organism to run or swim. The C-ring or switch complex, which is composed of FliG, FliM, and FliN proteins, is attached to the MS-ring basal body (FliF proteins) and stator (MotA-MotB). The C-ring determines whether a motor rotates clock-wise (CW) or counter clock-wise (CCW) (Sowa; Berry, 2008; Chaban et al., 2015; Minamino; Imada, 2015; Kojima, 2015).
Spirochetes are a group of motile bacteria that are distinct from other externally flagellated bacteria such as those seen in E. coli in many different aspects (Chen et al., 2011; Charon et al., 2012; Zhao et al., 2014). For example, B. burgdorferi is a long organism (10–20 μm) that possesses 7–11 flagella inserted at each pole of the cell (up to 22 flagella per cell). Unlike E. coli, B. burgdorferi flagella are located between the outer membrane and peptidoglycan layer i.e., in the periplasmic space (Kudryashev et al., 2009; Charon et al., 2009; Charon et al., 2012; Wolgemuth, 2015; Motaleb et al., 2015). B. burgdorferi motility and chemotaxis genes are controlled by housekeeping σ70 promoters. This results in the flagella of this spirochete being assembled in a sequential manner. Moreover, the flagellar motors of spirochetes, specifically those of B. burgdorferi, are much larger than E. coli motors (~80 vs. ~45 nm) and thus require more gene products for their assembly (Chevance; Hughes, 2008; Charon et al., 2012; Zhao et al., 2013).
The periplasmic flagella originate near the cell poles and extend toward the other pole of the cell or toward the cell body. In motile cells, the periplasmic flagellar filaments form a ribbon-like structure that wraps around the cell body, resulting in a distinctive flat-wave morphology (Kudryashev et al., 2009; Charon et al., 2009; Sultan et al., 2015; Motaleb et al., 2015). It has been proposed that the spirochete’s flagella rotate asymmetrically during a “run” mode, i.e., flagella at one pole rotate CW whereas the flagella at the other end rotate CCW. When flagella at both poles rotate in the same direction, the spirochete flexes/tumbles (Li et al., 2002; Charon et al., 2012). While flagella in most other bacteria are involved in motility, periplasmic flagella in B. burgdorferi determine the cellular morphology as well as motility. For example, a mutant that lacks FlaB encoding the protein component of the periplasmic flagellar filaments produces a rod-shaped cell in addition to being non-motile (Motaleb et al., 2000; Sultan et al., 2013). Due to their involvement in cellular morphology and the fact that these flagella rotate within the periplasmic space, it is not surprising that spirochetes possess extra or unique flagellar structures that offer flexibility or rigidity that is required to rotate their flagella within the periplasmic space. Recently, cryo-electron tomography (cryo-ET) of spirochete flagellar motors revealed unique features that are absent from all other bacterial motors studied to-date. One of these structures is called the “collar” (Murphy et al., 2006; Liu et al., 2010; Chen et al., 2011; Raddi et al., 2012; Zhao et al., 2014). The collar is appeared to locate adjacent to FliL. FliL homologs are found in several species of bacteria and its function is distinct in those organisms (Jenal et al., 1994; Belas; Suvanasuthi, 2005; Attmannspacher et al., 2008; Suaste-Olmos et al., 2010; Kudryashev et al., 2010; Motaleb et al., 2011; Zhu et al., 2015). In B. burgdorferi, we found that periplasmic flagellar filaments were partially and abnormally tilted toward the cell pole in the ΔfliL mutant (Motaleb et al., 2011).
Importantly, the collar is apparently integrated with the major components of the periplasmic flagella such as the MS-ring. Because of these connections and its central location in the motor, we hypothesize that the collar is critical for flagellar assembly as well as for providing proper rigidity or flexibility of flagella during rotation. However, nothing is known about the proteins encoding the unique collar structure or their function in any spirochete. In this communication, we show that mutations in bb0286 (flbB) has a profound effect on collar formation, flagellar orientation, morphology, motility, and the assembly of FliL as well as the stator. Moreover, using green fluorescent protein (GFP) we determined the location of FlbB in the collar. A mechanism underlying the orientation of the periplasmic flagella is also demonstrated.
RESULTS
Identification of a protein encoding the collar structure
Genomic analysis suggests that over 50 genes or 5–6% of the B. burgdorferi genome are potentially involved in motility and chemotaxis (Fraser et al., 1997; Charon et al., 2012). In order to identify proteins involved in the collar structure, we employed a strategy by subtracting common gene homologs that are present in other bacterial genomes especially those with externally flagellated bacteria whose motors have been determined by cryo-ET (Chen et al., 2011). To ensure that we did not overlook any gene that may share low homology but could encode a collar protein, we systematically mutated almost all genes annotated as “flagellar/motility-related” in the B. burgdorferi genome (Fraser et al., 1997; Charon et al., 2012) (our unpublished observation). Through these analyses, BB0286 (FlbB) was identified as a potential candidate for the collar structure. flbB is a spirochete-specific gene that is located within the flagellar flgB polycistronic operon, increasing the likelihood that this protein may encode for a flagellar gene (Ge et al., 1997; Ge and Charon, 1997; Charon et al., 2012). FlbB is a small protein (205 a.a.) that possess a transmembrane domain at its N-terminal end and shares no significant amino acid sequence identity with proteins from non-spirochetal bacteria (data not shown; see below).
ΔflbB mutant cells are rod-shaped and non-motile
We deleted the flbB gene by using a promoter-less kanamycin resistance cassette that results in nonpolar mutations (Fig. 1A) (Sultan et al., 2010; Pitzer et al., 2011; Sultan et al., 2011). PCR analysis of the kanamycin-resistant B. burgdorferi clones confirmed the deletion of the flbB (data not shown). Immunoblotting with anti-FlbB antisera indicated that FlbB protein synthesis was inhibited in the ΔflbB mutant cells; yet, expression of proteins encoded by genes downstream of flbB, i.e., flgE, motB, and fliL, were not altered in the mutant cells compared to the wild-type B. burgdorferi (Fig. 1B). This suggests that the mutant phenotype is due solely to loss of flbB function (see below). Although the mutant did not exhibit a polar effect on downstream genes expression, we attempted to complement the ΔflbB mutant both in cis (genetic recombination) and in trans (using a shuttle vector). While multiple attempts to genetically complement the ΔflbB mutant have failed, we report our findings with the mutant as others have done in the past (Stewart et al., 2008; Rogers et al., 2009; Dresser et al., 2009; Hyde et al., 2009; Pappas et al., 2011; Motaleb et al., 2011; Brisson et al., 2012; Miller et al., 2013).
Figure 1. Construction of ΔflbB mutant and determination of polar effect on downstream genes expression.
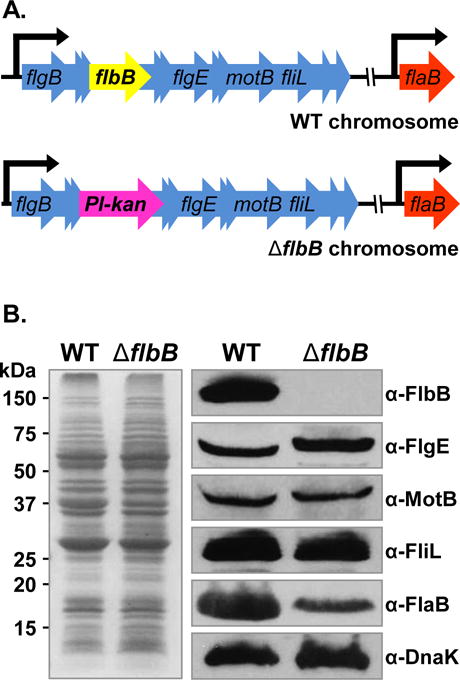
(A) Schematic diagrams of wild-type and ΔflbB mutant genomes. flaB gene (bb0147) is separated from the targeted flbB gene (bb0286) by approximately 100 kb. WT B. burgdorferi with the flgB polycistronic operon containing the targeted flbB is shown in top panel. The Pl-kan cassette replacing the flbB gene by allelic exchange is shown in bottom panel. The model lists only a few of the 26 genes of the flgB operon, and other genes are indicated by multiple arrowheads. (B) Confirmation of flbB gene-deletion and determination of polar effect by western blotting. WT and ΔflbB mutant cell lysates were subjected to SDS-PAGE (left) followed by immunoblotting (right). Immunoblotting was performed with B. burgdorferi FlbB, FlgE, MotB, FliL, or FlaB-specific antibodies. DnaK was used as a loading control. FlbB antiserum reacted with a 14 kDa protein in the wild-type lysate that is absent in the ΔflbB lysates indicate that this protein is the FlbB (FlbB blot).
Dark-field microscopy and swarm plate assays were used to assess cell morphology and motility of ΔflbB cells (Motaleb et al., 2007; Moon et al., 2016). These measurements indicated that the mutant cells are non-motile and display a rod-shaped morphology (Fig. 2), despite the synthesis of periplasmic flagella, FlaB, albeit at a reduced level compared to the wild-type (Fig. 1B). Furthermore, swarm plate motility assays indicate that the mutant cells produced colony diameters that are significantly smaller than the wild-type cells (~1 mm vs. ~6 mm swarm produced by the wild-type; Fig. S1). The morphology and motility phenotypes of the ΔflbB mutant cells are similar to the non-motile, rod-shaped ΔflaB mutants that lack flagellar filaments (Motaleb et al., 2000; Sultan et al., 2013). Taken together, our results indicate that FlbB is important for morphology and motility of B. burgdorferi.
Figure 2. Morphology phenotype of the ΔflbB mutant cells.
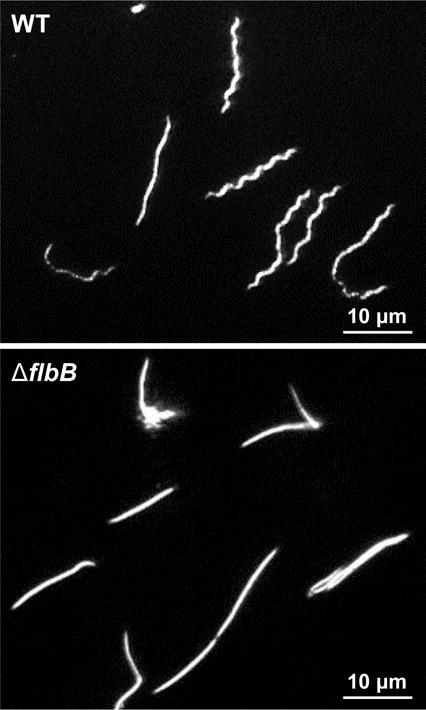
Dark-field microscopic images showing the distinct rod-shaped morphology of ΔflbB spirochetes whereas the wild-type (WT) cells exhibit a flat-wave morphology. Growing B. burgdorferi cells were visualized using a dark-field microscope (40×) and images were captured using a digital camera. The mutant cells were also non-motile (see Fig. S1).
ΔflbB mutant displays abnormal periplasmic flagellar orientation
Previous studies showed that periplasmic flagella are crucial not only for motility but also for the cellular flat-wave morphology of B. burgdorferi (Motaleb et al., 2000; Sultan et al., 2013). Because the ΔflbB mutant cells synthesize periplasmic flagella but are rod-shaped, we investigated the basis of those defects using cryo-ET (Fig. 3). Our reconstructions of the native cellular structures shown in Figs. 3C, D indicate that the wild-type periplasmic flagella form ribbon-like structures that are oriented inwards toward the center of the cell. In contrast, the mutant’s periplasmic flagella are short and the flagellar ribbon is distorted. Most striking and opposite to wild-type, ΔflbB flagella are oriented abnormally toward the cell pole (compare Fig. 3A–B with 3C–D or Movie 1 with Movie 2). In fact, the majority of periplasmic flagella (82% vs. 1–2% in the wild-type) are found to be abnormally oriented toward the cell pole in the mutant cells (Table 1; Movies 1–2). These results indicate that FlbB is essential for normal orientation of periplasmic flagella (toward the cell body).
Figure 3. Periplasmic flagellar orientation in wild-type and ΔflbB mutant.
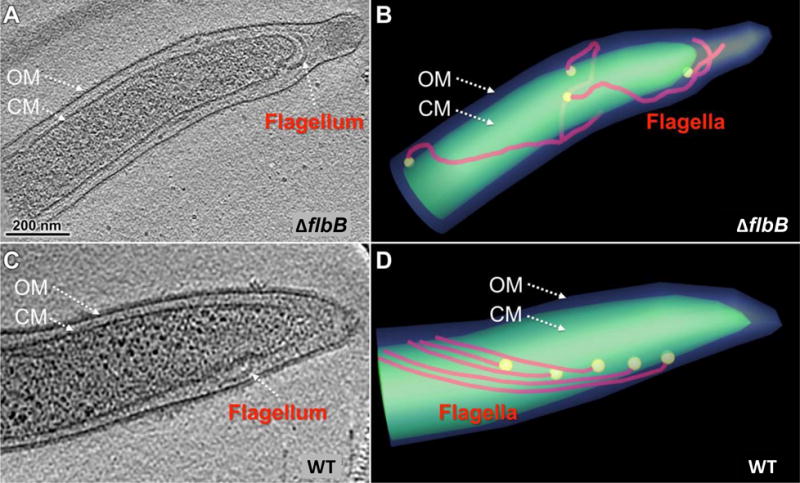
(A) A representative tomographic slice of a ΔflbB cell showing that the periplasmic flagella are abnormally oriented toward the cell pole. (B) A cartoon model of the ΔflbB mutant shown in (A) clearly illustrated the abnormal tilting of the flagella. (C) A representative tomographic slice of a WT cell showing the periplasmic flagella that are extended toward the cell body but not the cell pole. (D) A cartoon model of the WT cell showing the normal orientation of the flagella toward the cell body.
Table 1.
Periplasmic flagella of ΔflbB mutant cells are oriented abnormally toward the cell pole.
| Strain | No. of cells analyzed | No. of irregular periplasmic flagellaa | No. of normal periplasmic flagellab | % irregular periplasmic flagella |
|---|---|---|---|---|
| Wild-type | 43 | 5 | 288 | 1.7 |
| ΔfliL | 41 | 55 | 208 | 21 |
| ΔflbB | 44 | 144 | 32 | 82 |
Irregular periplasmic flagella were tilted toward the cell pole.
Normal periplasmic flagella were tilted toward the cell body. ΔfliL mutant was used as a reference strain (Motaleb et al., 2011).
The collar structure is absent in the ΔflbB mutant cells
We compared the motor structures in wild-type and the ΔflbB mutant cells by cryo-ET and found that the collar structure is absent from the mutant’s periplasmic flagella (Figs. 4A, B). To reveal the motor structure in detail, we used subtomogram averaging to analyze approximately 1000 motor structures extracted from tomographic reconstructions. The averaged structure reveals major features of the flagellar motor, such as the export apparatus, the C-ring, the MS-ring, the rod, and the P-ring (Fig. 4C). These major features of the wild-type motor are also detected in the ΔflbB motor (Fig. 4D). However, a large portion of densities surrounding the central rod and the P-ring are absent in the ΔflbB motor (Fig. 4D). Specifically, the ΔflbB motor lacks the collar structure detected in wild-type cells (compare Figs. 4C, E with 4D, F).
Figure 4. Comparative analysis of in situ flagellar motors from wild-type and ΔflbB reveals the 3D collar structure for the first time.
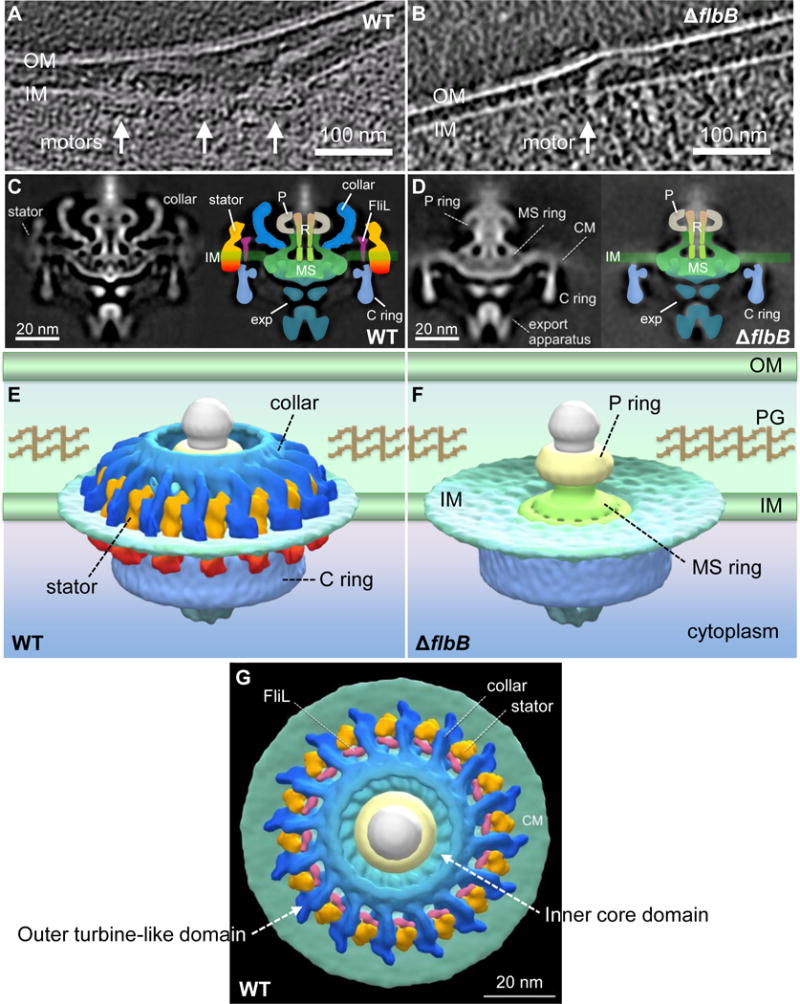
(A) A tomographic section from a WT cell shows the motors that are embedded in the cytoplasmic inner membrane (IM/CM). (B) A tomographic section from a ΔflbB cell shows a motor that is embedded in the cytoplasmic membrane. (C) The central section (left) and schematic diagram (right) of the WT flagellar motor. (D) The central section (left) and schematic diagram (right) of the ΔflbB flagellar motor. (E) The surface rendering of the 3D averaged WT and (F) ΔflbB motor structures are shown in side view. (G) The surface rendering of the 3D averaged WT motor structure is shown in tilted top view (90°). Compared to the motor structures from WT (C, E, G), the ΔflbB motor lacks the entire collar (blue), the stator (orange-red), and FliL (pink) structures. Noticeably, the collar is a large and complex structure comparing to FliL and the stator. OM, outer membrane; PG, peptidoglycan; P, P-ring; R, central rod; exp, export apparatus.
By comparing structures from a deletion mutation in motB (J. Liu and M. Motaleb-unpublished), fliL (Motaleb et al., 2011), and the current ΔflbB strain, we were able to define the 3D structure of the collar and the stator (Figs. 4E, G). The overall dimension of the collar is ~71 nm in diameter and ~24 nm in height. This unique structure consists of two major layers along the radial direction—for clarity, labeled here as the inner core domain and the outer turbine-like domain (Fig. 4G). The inner domain of the intact collar appears to consist of 16 truss-like subassemblies joined together to form a chamber-like structure that surrounds the rod and the P-ring (Figs. 4E, G). FliL is attached to each subassembly at the membrane region (Figs. 4C, G). The outer domain—the sixteen extended “turbine blades”—is the most distinct feature appearing in the spirochetal flagellar motor. Sixteen stator units are inserted between two adjacent “turbine blades”, forming a stator ring that packs around the C-ring in the cytoplasm (Figs. 4E, G). Furthermore, the stator (MotA-MotB) and FliL structures are also disappeared in the mutant even though MotB and FliL proteins are stably expressed at wild-type levels, suggesting that FlbB/collar is important for the assembly of those flagellar structures (compare Figs. 4C, E with 4D, F). Together, our cryo-ET data indicate that FlbB is essential for the formation of the collar structure and the assembly or stability of FliL and the stator.
FlbB—FliL interactions
As shown above, majority of ΔflbB mutant’s periplasmic flagella are abnormally tilted toward the cell pole. Interestingly, we observed a similar phenotype with our ΔfliL mutant cells (Table 1) (Motaleb et al., 2011). These results led us to predict that (a) FlbB and FliL proteins interact and direct the periplasmic flagella to orient toward the cell body but not the cell pole; (b) FlbB and FliL are located in close proximity to each other; and (c) FlbB/collar is assembled before FliL and the stator because the FliL and stator structures are not assembled in the ΔflbB. To determine if FlbB interacts with FliL, we performed co-immunoprecipitation (co-IP) assays with wild-type and ΔflbB cell extracts. Our co-IP data indicate that FlbB specifically interacts with FliL (Fig. 5). The FlbB-FliL binding is verified further by using an alternative bacterial two-hybrid assay (BACTH). Our two-hybrid assays also confirmed that FlbB interacts with FliL (Fig. S2). These FliL-FlbB interaction results suggest that FlbB is located adjacent to the FliL, near the base of the collar structure (see below).
Figure 5. FlbB directly interacts with FliL.
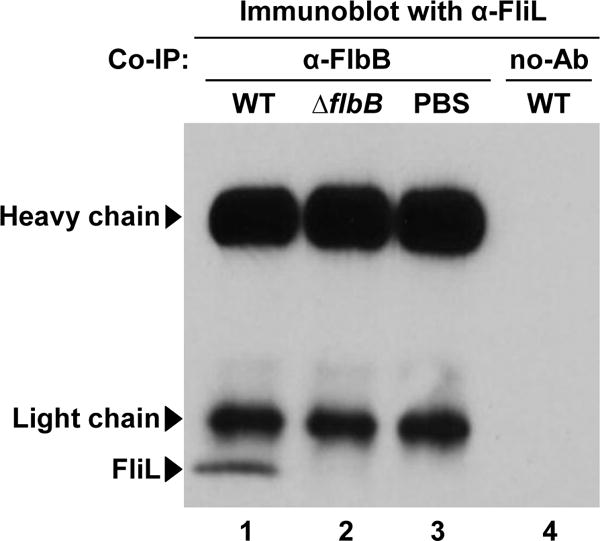
A co-IP assay showing the interaction between FlbB and FliL. FlbB-specific antibody conjugated with Dynabeads was incubated with wild-type or ΔflbB cell extracts. The proteins that were immunoprecipitated with the FlbB-antibody were separated using a gel, transferred to a PVDF-membrane, and subsequently, the membrane was blotted with anti-FliL (Motaleb et al., 2011). FliL proteins are co-precipitated with B. burgdorferi wild-type cell extracts (lane 1), but not with ΔflbB mutant extracts (lane 2). PBS buffer was used as a negative control (lane 3). To check for the non-specific protein binding on Dynabeads, we used empty Dynabeads (no Ab/antisera) in the co-IP with wild-type extracts (lane 4). Arrowheads indicate the positions of antibody heavy chain (~55 kDa), light chain (~25 kDa), or FliL (~20 kDa). Lane numbers are shown at the bottom of the figure. See also Figure S2.
Our motor structures show that the periplasmic domain of the stator is adjacent to the collar and that the stator structure is missing in the ΔflbB (Fig. 4). To test if there is any interaction between the stator and FlbB, and this interaction is important for the assembly of the stator, we performed co-IP and BACTH assays, as described above. Using these assays, we failed to detect any FlbB-MotA or FlbB-MotB interaction (data not shown), indicating that FlbB is not directly interacting with the stator and supporting our proposal that FlbB is just a small part of the collar that is located at its base (below).
Localization of FlbB by GFP fusion
To determine the location of FlbB in the periplasmic flagella, the gene encoding GFP was fused at the 3′-end of flbB (flbB-gfp) and then ligated such that flgB promoter drives the expression of flbB-gfp (PflgB-flbB-gfp) from the shuttle vector pBSV2G. The placement of gfp at the 3′-end of flbB is suitable for the expression of FlbB-GFP since the N-terminal region (7–29 amino acid residues) of FlbB is found to possess a transmembrane domain using TMHMM Server, ver. 2.0 (Fig. S3) (Sonnhammer et al., 1998; Krogh et al., 2001). Subsequent introduction of pBSV2G::PflgB-flbB-gfp into ΔflbB mutant cells resulted in expression of FlbB-GFP, as confirmed by immunoblotting using anti-GFP and anti-FlbB (not shown). Confocal microscopy shows the FlbB-GFP clusters at ~73% of the cell tips of the ΔflbB/pBSV2G::PflgB-flbB-gfp cells where motors are typically located (ΔflbB/flbB-GFP cells; Fig. 6, right). As expected, this pattern (FlbB-GFP clusters) was not observed in the wild-type cells expressing only GFP using pBSV2G::PflgB-gfp plasmid (wild-type GFP; Fig. 6, left) or in the ΔflbB mutant negative control cells that does not express GFP (not shown).
Figure 6. Expression and location of FlbB-GFP in B. burgdorferi.
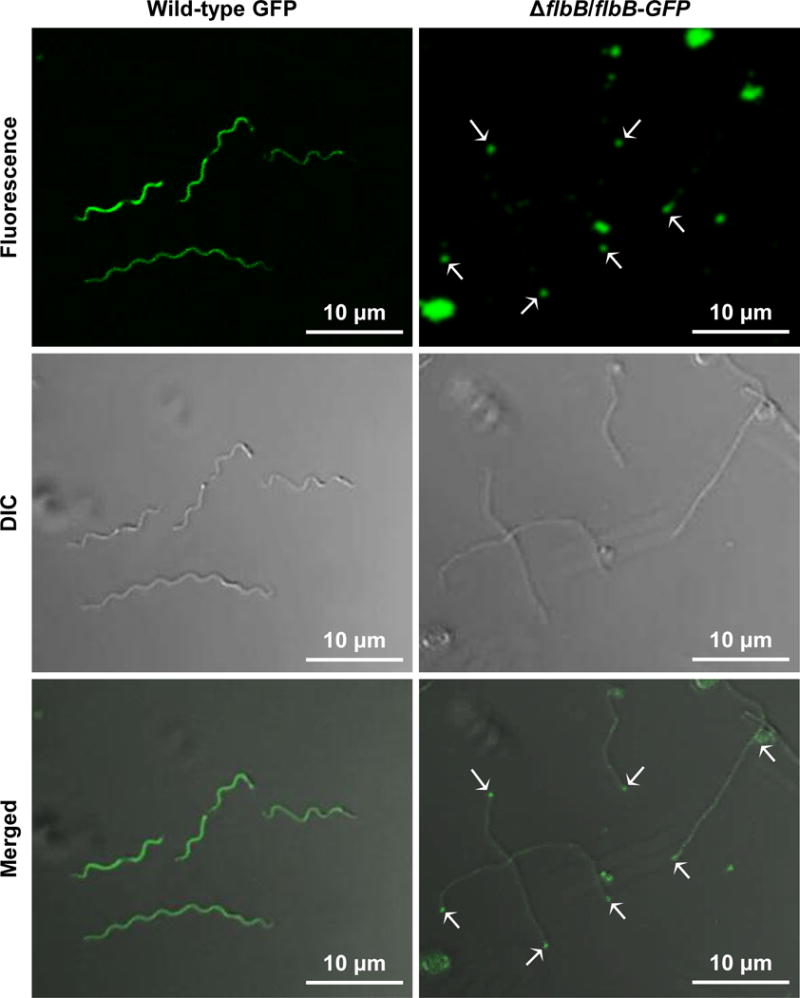
Confocal microscopy showing the fluorescence (top), differential interference contrast (DIC; middle), and merged (bottom) micrographs of the wild-type cells expressing GFP (wild-type GFP) and of ΔflbB cells expressing FlbB-GFP (ΔflbB/flbB-GFP) at 64×. The white arrows indicate the location of FlbB-GFP in the ΔflbB/flbB-GFP cell tips (FlbB-GFP clusters were detected in approximately 73% cells tips). Even distribution of the GFP signal was observed throughout the wild-type GFP cells, as expected.
To conclusively determine FlbB location, cryo-ET and subtomogram averaging were utilized to visualize the motor structure of the ΔflbB/flbB-GFP cells. Compared to the cellular density of ΔflbB motor, the ΔflbB/flbB-GFP motor shows extra densities near the basal body MS-ring structure as shown in Fig. 7, suggesting the location of FlbB-GFP. Together, these data imply that FlbB proteins are located at the base of the collar and they are anchored to the cytoplasmic membrane to form the base for the assembly of the collar complex.
Figure 7. Location of FlbB-GFP as determined by cryo-ET.
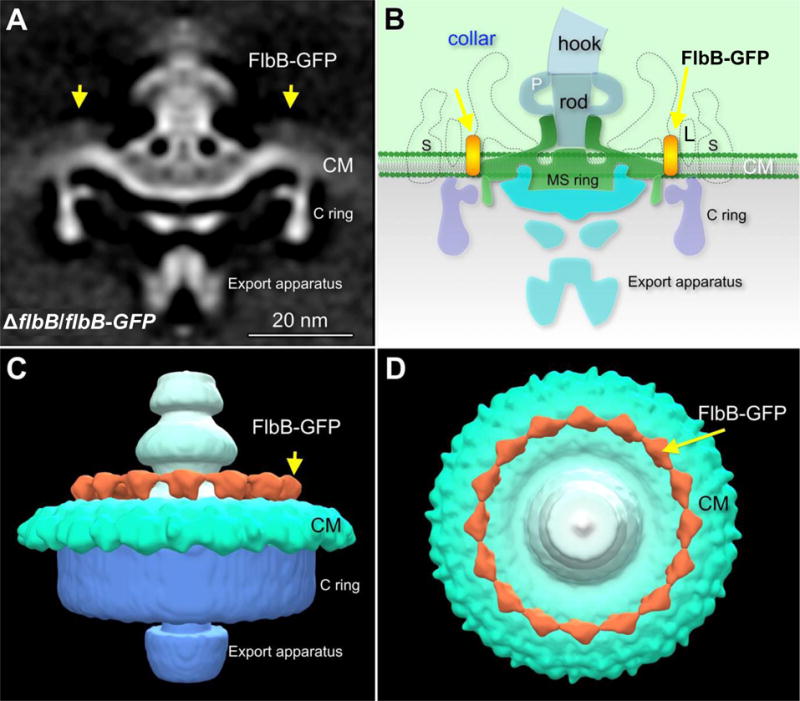
(A) The averaged 3D motor structures of the ΔflbB cells expressing FlbB-GFP (ΔflbB/flbB-GFP), and (B) schematic diagram of the ΔflbB/flbB-GFP flagellar motor illustrating the location of FlbB at the base of the collar. (C) Three dimensional isosurface rendering of the ΔflbB/flbB-GFP flagellar motor is shown in side view. (D) Three dimensional isosurface rendering of the ΔflbB/flbB-GFP flagellar motor is shown in tilted (top) view. The yellow arrows indicate the location of FlbB-GFP. S, stator; L, FliL; P, P-ring.
DISCUSSION
Despite the fact that periplasmic flagellar motility is crucial for host colonization or disease production by the spirochetes including B. burgdorferi, there is still very limited knowledge about what genes encode for the spirochete-specific flagellar components. FlbB identified as a collar protein in this communication has profound effects in motility, morphology, orientation of periplasmic flagella, and assembly of motor proteins. The ΔflbB mutant cells are rod-shaped and non-motile despite the possession of periplasmic flagella (Figs. 1, 2, and 3), however, those flagella are inactive due to their missing stators (Figs. 4D, F). Stator proteins use proton flux to produce torque in order for the flagella to rotate, which in turn enables the organism to translocate. Because the periplasmic flagella are oriented abnormally and their stators are missing, it was obvious that those B. burgdorferi mutant cells exhibited rod-shaped and non-motile phenotypes. However, the number of periplasmic flagella or level of flagellar filament FlaB protein is reduced in the mutant compared to the wild-type cells (Fig. 1B). The flaB gene is not genetically linked with the targeted flbB or other genes in the flgB operon. However, we observed this reduced FlaB protein synthesis or fewer flagellar filaments not only in ΔflbB but also in other non-motile mutants such as ΔmotB (Sultan et al., 2015). These observations suggest that the stator or collar-stator is important for the wild-type level of periplasmic flagellar filament synthesis in B. burgdorferi.
One of the most remarkable findings here is the abnormal orientation of flagella in the ΔflbB mutant (Fig. 3; Table 1; Movie 2). Normal orientation of the flagella toward the cell body and not the cell pole is critical in producing the wild-type spirochete’s wave-like morphology and smooth swimming (Figs. 2 and 3) (Motaleb et al., 2011). We have previously reported that FliL is partially responsible for determination of flagellar orientation (Motaleb et al., 2011). As shown in Fig. 3 and Table 1, 82% of the flagella in the ΔflbB are abnormally oriented. Thus, based on ΔflbB and ΔfliL flagellar orientation phenotypes as well as protein-protein interaction data shown in Figs. 5 and S2, we propose that FlbB/collar—FliL structures enforce the periplasmic flagella to orient toward the cell body—an observation that has never been demonstrated in any spirochete. We, however, postulate that this irregular periplasmic flagellar phenotype associated with the mutant is a combined effect of collar-stator-FliL rather than just the FlbB/collar since the stator and FliL structures were diminished along with the collar (in the ΔflbB mutant). Moreover, it is important to note that the ΔflbB mutant was not complemented. Thus, the phenotypes observed with the mutant could be due to a secondary mutation elsewhere in the genome rather than just because of the deletion of flbB even though the mutant is non-polar (Fig. 1B).
The stator and FliL structures were disappeared in the mutants despite the synthesis of MotB and FliL proteins at the wild-type levels (Figs. 1 and 4D, F), indicating that those structures were not assembled due to the lack of the collar structure. It is noteworthy to mention that the collar structure is intact in our ΔmotB (J. Liu and M. Motaleb-unpublished) or ΔfliL mutants (Motaleb et al., 2011). Furthermore, the stator is intact in the ΔfliL mutant—a very good indication that FliL and stator structures were diminished not because of a secondary alteration or polar effect (Motaleb et al., 2011). These results also suggest that the collar is assembled before FliL or the stator. Moreover, FliL and stator structures were not assembled in the ΔflbB likely because the collar provides the stability/foundation for those two motor structures similar to what was observed with the flagellar filament proteins, FlaA and FlaB. FlaA and FlaB proteins interact and we found that unless the filament FlaB is synthesized and assembled, FlaA protein is not assembled in the ΔflaB (Motaleb et al., 2000; Sultan et al., 2013).
In E. coli or Salmonella typhimurium, FliL was reported to interact with the stator (Partridge et al., 2015). However, we could not detect any interactions between FliL-MotA, FliL-MotB, FlbB-MotA, or FlbB-MotB. These results suggest that FliL or FlbB may not interact with the stator directly. Alternatively, our BACTH vectors (pKT25 and pUT18C) could not express MotA or MotB properly or our co-IP reaction conditions were not optimized. However, in Rhodobacter sphaeroides, FliL is able to interact with itself but not with the MotB leading to the proposal that FliL may participate in coupling with the flagellar stator in an indirect manner (Suaste-Olmos et al., 2010). Moreover, in Vibrio alginolyticus, FliL was suggested to interact with the stator directly or indirectly (Zhu et al., 2015). Subsequently, we propose that B. burgdorferi FliL (or FlbB/collar) interacts with the stator indirectly using a yet to be identified protein(s) which is important for the assembly of the stator.
It is noteworthy that the full collar structure was not assembled in the ΔflbB cells expressing FlbB-GFP (Fig. 7), and morphology and motility phenotype were also not restored in those cells (not shown). This result is not surprising because in order for the collar structure or function to be restored in the ΔflbB, the FlbB-GFP protein’s stoichiometry should be the same as that of other collar proteins since most motor complexes maintain a ratio (such as the FliG:FliM:FliN protein copies in a switch complex are 34:34:100, and MotA:MotB ratio is 4:2 in a stator complex) (Blair, 2003; Kojima; Blair, 2004; Leake et al., 2006). When GFP or mCherry was fused with flagellar motor MotA or MotB or their homologs, assembly and/or function was reported to be abolished in other bacteria (Fukuoka et al., 2009; Paulick et al., 2009).
The collar is a colossal structural component of the periplasmic flagella. It is noticeably larger than the C-ring or stator (Fig. 4). Considering that the C-ring is composed of three proteins (FliG, FliM, and FliN), the collar is likely comprised of multiple proteins. FlbB is a small protein (205 amino acids) that is comparable to its binding partner FliL (178 amino acids) or MotB (260 amino acids). FliL appears to form a small and elongated structure right next to the edge of the collar (Figs. 4C, G) (Motaleb et al., 2011). Therefore, we propose that FlbB is arranged in a small structure at the base of the collar by embedding in the cytoplasmic membrane using its transmembrane domain (Figs. 7 and S3). Other (unidentified) collar proteins are expected to assemble onto the FlbB base. As such, deletion of flbB had a dramatic effect on the entire collar, and thus, its associated structures are not assembled in the ΔflbB.
Altogether, our data demonstrate that the collar is a highly complex structure that has profound impacts in B. burgdorferi. Importantly, we show for the first time that FlbB assembles around the flagellar basal body and plays critical roles in collar formation. Furthermore, we provided the first 3D structure of the collar and revealed its unprecedented complexity. Moreover, we show that FlbB and FliL are crucial for normal orientation of periplasmic flagella.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
High-passage, avirulent B. burgdorferi strain B31-A was used as a wild-type clone throughout the study (Bono et al., 2000; Elias et al., 2002). Construction of a flbB (bb0286) deletion mutation was achieved as described below. B. burgdorferi cells were cultured in liquid Barbour-Stoenner-Kelly (BSK-II) medium, and plating BSK was prepared using 0.4% agarose (Motaleb et al., 2007; Sultan et al., 2013). Cells were grown at 35°C in a 2.5% CO2 incubator as described previously (Motaleb et al., 2007). E. coli cells were grown at 30°C or 37°C in Luria-Bertani (LB) broth or LB agar (Bertani, 1951). Antibiotics, indicators, and inducers, when required, were included in the bacterial culture medium with the following concentrations: 100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, 100 μg ml−1 spectinomycin, 40 μg ml−1 gentamicin, 80 μg ml−1 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal), 0.5 mM isopropyl-β-D-thiogalactoside (IPTG).
Construction of the flbB-deletion mutant
Construction of the flbB-deletion plasmids, electroporation, and plating conditions were described previously (Motaleb et al., 2007; Sultan et al., 2010; Pitzer et al., 2011; Sultan et al., 2013). Briefly, the 5′-(1198 bp), and 3′-flaking (432 bp) DNA of flbB gene were amplified by PCR from chromosomal DNA of B. burgdorferi strain B31-A using primers FlbB.KO.P1F (GACGATTAGAACCTACTTTCG) and FlbB.KO.P1R (TAAAATTGCTTTTAACTATTATTCACTTTCATTCC), and FlbB.KO.P2F (CGATGAGTTTTTCTAATCATTGGAGTAGTGTG) and FlbB.KO.P2R (TTGGTCCTTAGAGTCATCT), respectively. Promoter-less kanamycin resistance cassette [Pl-kan, 846 bp] was similarly PCR amplified from PflgB-aph1 using primers FlbB.KO.KanF (GGAATGAAAGTGAATAATAGTTAAAAGCAATTTTA) and FlbB.KO.KanR (CACACTACTCCAATGATTAGAAAAACTCATCG) (Sultan et al., 2010). These three pieces of DNA fragments were linked by overlapping PCR, yielding bb0285-Pl-kan-bb0287 (flbB_KO_Pl-kan), then cloned into the pGEM-T Easy (Promega Inc.), yielding plasmid Teasy::flbB_KO_Pl-kan. Competent B31-A cells were electroporated with flbB_KO_Pl-kan PCR amplified linear DNA. The transformants were selected with 200 μg ml−1 kanamycin. The kanamycin-resistant transformants were isolated and confirmed the replacement of flbB gene with the Pl-kan by PCR, and lack of FlbB protein expression was confirmed by immunoblotting as described below.
Construction of a plasmid expressing FlbB-GFP
To construct a B. burgdorferi strain that expresses GFP coupled with FlbB, the flgB promoter (PflgB), and flbB gene were PCR amplified from chromosomal DNA of strain B31-A using primers PflgB-BamHI.F (GGATCCCGAGCTTCAAGGAAGATTTCC) and PflgB-flbB.R (ATAAAAAATTATTCACATGGAAACCTCCCTCATTTAAAA), and PflgB-flbB.F (TGAGGGAGGTTTCCATGTGAATAATTTTTTATCGTTC) and flbB-gfp.R (TCTTCTCCTTTACTCTCCAATGAACTAACAG), respectively (BamHI restriction site is underlined). gfp was PCR amplified from pMC2498 plasmid using primers flbB-gfp.R (CTGTTAGTTCATTGGAGAGTAAAGGAGAAGA) and gfp-HindIII.R (AAGCTTCTATTTGTATAGTTCATCCATGCCATG) (HindIII restriction site is underlined) (Caimano et al., 2015; Iyer et al., 2015). These three pieces of DNA fragments were linked by overlapping PCR and cloned into the pGEM-T Easy, yielding plasmid Teasy::PflgB-flbB-gfp. This and the B. burgdorferi shuttle vector pBSV2G were digested with BamHI and HindIII, and ligated to yield pBSV2G::PflgB-flbB-gfp (Elias et al., 2003). Approximately 50 μg of pBSV2G::PflgB-flbB-gfp plasmid DNA was electroporated into the ΔflbB mutant cells. Transformants were selected with kanamycin and gentamicin. Resistant transformants were analyzed by PCR to confirm the presence of the pBSV2G::PflgB-flbB-gfp plasmids in the transformants (ΔflbB/flbB-GFP cells). Furthermore, the expression of GFP and FlbB proteins in the ΔflbB/flbB-GFP cells were confirmed by immunoblotting with B. burgdorferi FlbB-specific polyclonal and Anti-GFP monoclonal (Roche Life Science) antibodies, respectively. Anti-FlbB was raised in rabbits which immunized with purified recombinant His6-FlbB, as described (Alpha Diagnostic International) (Motaleb et al., 2011).
SDS-PAGE and immunoblot analyses
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with an enhanced chemiluminescent detection method (GE Health Inc.) were carried out as reported previously (Motaleb et al., 2000; Sultan et al., 2013). The concentration of protein in cell lysates was determined by a Bio-Rad protein assay kit. Unless otherwise noted, 10 μg of lysate protein was subjected to SDS-PAGE and immunoblotting using proper antibodies.
Dark-field microscopy and measurement of colony size
Growing B. burgdorferi cells (2–4 ×107 cells ml−1) were imaged using a Zeiss Imager M1 dark-field microscope connected to a Zeiss AxioCam MRc digital camera to determine morphology. For measurement of B. burgdorferi colony swarm diameter, approximately 20 to 50 cells were plated on semi-solid BSK-II medium containing 0.4% agarose. Four weeks after inoculation, we measured the diameter of 20 representative colonies from each clone (Moon et al., 2016).
Confocal laser scanning microscopy
B. burgdorferi cells were examined with a confocal microscope (LSM 510; Carl Zeiss, Inc., Thornwood, NY, USA) using 488 nm Argon ion laser excitation and a 505–550 bandpass filter to collect GFP fluorescence emission, with simultaneous collection of a transmitted light image using differential interference contrast (DIC) optics. Images were acquired and analyzed using Zen 2009 software (Zeiss Inc.).
Cryo-electron tomography (cryo-ET) and subtomogram averaging
Frozen-hydrated specimens were prepared as described previously (Zhao et al., 2013; Sultan et al., 2015). Briefly, growing B. burgdorferi wild-type, ΔflbB mutant, and flbB-GFP cells were harvested at low 1,500×g speed, and suspended in 40 μl phosphate buffered saline (PBS, pH 7.4) at a final concentration of ~2 × 109 cells ml−1. Resuspended cells were mixed with 10 nm gold clusters, then 5 μl was deposited onto freshly glow-discharged holey carbon grids for 1 min. Grids were blotted with filter paper to remove excess fluid, followed by rapid freezing in liquid ethane maintained at −180°C using a gravity-driven plunger apparatus (Liu et al., 2009; Zhao et al., 2013). The resulting frozen-hydrated specimens were imaged at −170°C using a Polara G2 electron microscope (FEI Company) equipped with a field emission gun and a K2 direct electron detector (Gatan). The microscope was operated at 300 kV with a magnification of ×9,400, resulting in an effective pixel size of 4.6 Å. Using the FEI batch tomography program, low-dose single-axis tilt series were collected from each bacterium at a −6 μm defocus with a cumulative dose of ~60 e−/Å2 distributed over 60 images. Tilt angles were in the range of −60° and +60° with an angular increment of 2°. Tilt series were aligned and reconstructed using IMOD software and tomoauto (Kremer et al., 1996; Hu et al., 2015).
In total, 285 and 190 reconstructions were generated from ΔflbB mutant and flbB-GFP cells, respectively. A total of 1,742 motor sub-tomograms (256×256×256 voxels) were visually identified, then extracted from the reconstructions. The initial orientation of each particle was estimated by the C-ring and the hook, thereby providing two of the three Euler angles. To accelerate image analysis, 4×4×4 binned sub-tomograms (64×64×64 voxels) were used for initial alignment. The original data was then used for the refinement and averaging as described previously (Liu et al., 2009; Zhao et al., 2013).
3D visualization
Reconstructions of B. burgdorferi cells were visualized and segmented manually using IMOD (Kremer et al., 1996). UCSF Chimera, a visualization system for exploratory research and analysis, was utilized for 3-D surface rendering of sub-tomogram averages (Pettersen et al., 2004).
Co-immunoprecipitation (co-IP)
FlbB and FliL protein-protein interactions were determined using a Dynabeads® Protein A Immunoprecipitation kit, according to the manufacture’s protocol (Novex Inc.). Briefly, 36 μg of rabbit polyclonal FlbB antibodies were diluted in 600 μl of PBS with 0.01% Tween 20, and then coupled the antibody with 1.5 mg of Dynabeads. To prepare cell extracts, wild-type or ΔflbB mutant B. burgdorferi cells were harvested. Cell pellets were resuspended in PBS with 0.01% Tween 20 and then lysed by sonication. Sonicated cell extracts were centrifuged at 16,000 × g for 15 min at 4°C to remove bacterial debris. Approximately 750 μg cell extracts were incubated with FlbB antibody-conjugated Dynabeads with gentle shaking for 10 min at room temperature, and washed with 1 ml of washing buffer for four times. 50 μl of SDS loading dye was added directly to the FlbB antibody-conjugated Dynabeads after the washes, and then heated for 10 minutes in boiling water bath. The boiled samples were subjected to SDS-PAGE and immunoblotting using B. burgdorferi FliL-specific antibodies (Motaleb et al., 2011). FlbB-MotB and FliL-MotB co-IP assays were similarly performed using B. burgdorferi FlbB, FliL, or MotB-specific polyclonal antisera (Sultan et al., 2015).
Bacterial two-hybrid system
Protein-protein interactions between FlbB and FliL were measured with the bacterial adenylate cyclase two hybrid system, according to the manufacture’s protocol (BACTH; Euromedex Inc.). Briefly, flbB and fliL genes were amplified by PCR from chromosomal DNA of B. burgdorferi strain B31-A using primers FlbB.BamHI.F (GGATCCCAATAATTTTTTATCG) and FlbB.KpnI.R (GGTACCCTCCAATGAACTAAC), and FliL.BamHI.F (GGATCCCCCTAATAAAGACG) and FliL.KpnI.R (GGTACCCATATCAAAAATATCAATT), respectively (restriction enzyme sites are shown in bold). These DNA fragments were cloned into the pUT18C and pKT25. Both pUT18C::flbB (or fliL) and pKT25::fliL (or flbB) were co-transformed into the BTH101 E. coli host cell. Transformants were grown on LB plates containing X-gal, ampicillin, and kanamycin. Appearance of blue colored colonies in those plates is an indication of a positive protein-protein interaction. FliL-MotA, FliL-MotB, FlbB-MotA, and FlbB-MotB interactions were performed as described for FliL-FlbB.
Statistical analysis
A paired Student’s t-test was used to determine statistical significance. A P-value of ≤0.05 between samples was considered significant.
Supplementary Material
Movie 1. A typical 3D reconstruction of a wild-type B. burgdorferi cell tip shows well-organized periplasmic flagella that are oriented toward the other pole of the cell.
Movie 2. A typical 3D reconstruction of a ΔflbB cell tip shows the shortened periplasmic flagella—majority of them are abnormally tilted toward the cell pole.
Acknowledgments
We thank Dr. Robert Belas for critical reading of this manuscript and Dr. Douglas Weidner for help in confocal microscopy. This work was supported by grants from National Institute of Allergy and Infectious Diseases (NIAID) (R01AI087946, R21AI113014), National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) (R01AR060834), and Welch Foundation (AU-1714).
References
- Attmannspacher U, Scharf BE, Harshey RM. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol Microbiol. 2008;68:328–341. doi: 10.1111/j.1365-2958.2008.06170.x. [DOI] [PubMed] [Google Scholar]
- Belas R, Suvanasuthi R. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J Bacteriol. 2005;187:6789–6803. doi: 10.1128/JB.187.19.6789-6803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair DF. Flagellar movement driven by proton translocation. FEBS Lett. 2003;545:86–95. doi: 10.1016/s0014-5793(03)00397-1. [DOI] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko JJ, 3rd, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Drecktrah D, Eggers CH, Samuels DS. Genetics of Borrelia burgdorferi. Annu Rev Genet. 2012;46:515–536. doi: 10.1146/annurev-genet-011112-112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Dunham-Ems S, Allard AM, Cassera MB, Kenedy M, Radolf JD. Cyclic di-GMP modulates gene expression in Lyme disease spirochetes at the tick-mammal interface to promote spirochete survival during the blood meal and tick-to-mammal transmission. Infect Immun. 2015;83:3043–3060. doi: 10.1128/IAI.00315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol. 2015;46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, et al. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol. 2012;66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, et al. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol. 2009;191:600–607. doi: 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser AR, Hardy P, Chaconas G. Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathog. 2009;5:e1000680. doi: 10.1371/journal.ppat.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Bono JL, Kupko JJ, 3rd, Stewart PE, Krum JG, Rosa PA. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol. 2003;6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol Microbiol. 2009;71:825–835. doi: 10.1111/j.1365-2958.2008.06569.x. [DOI] [PubMed] [Google Scholar]
- Ge Y, Charon NW. Identification of a large motility operon in Borrelia burgdorferi by semi-random PCR chromosome walking. Gene. 1997;189:195–201. doi: 10.1016/s0378-1119(96)00848-7. [DOI] [PubMed] [Google Scholar]
- Ge Y, Old IG, Saint Girons I, Charon NW. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus sigma70 promoter. J Bacteriol. 1997;179:2289–2299. doi: 10.1128/jb.179.7.2289-2299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, Picking WL, et al. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci U S A. 2015;112:1047–1052. doi: 10.1073/pnas.1411610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JA, Shaw DK, Smith R, III, Trzeciakowski JP, Skare JT. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol Microbiol. 2009;74:1344–1355. doi: 10.1111/j.1365-2958.2009.06951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Caimano MJ, Luthra A, Axline D, Corona A, Iacobas DA, et al. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol Microbiol. 2015;95:509–538. doi: 10.1111/mmi.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, White J, Shapiro L. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J Mol Biol. 1994;243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- Kojima S. Dynamism and regulation of the stator, the energy conversion complex of the bacterial flagellar motor. Curr Opin Microbiol. 2015;28:66–71. doi: 10.1016/j.mib.2015.07.015. [DOI] [PubMed] [Google Scholar]
- Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry (N Y) 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kudryashev M, Cyrklaff M, Baumeister W, Simon MM, Wallich R, Frischknecht F. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol Microbiol. 2009;71:1415–1434. doi: 10.1111/j.1365-2958.2009.06613.x. [DOI] [PubMed] [Google Scholar]
- Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F. Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol. 2010;169:54–61. doi: 10.1016/j.jsb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Kuehn BM. CDC estimates 300,000 US cases of Lyme disease annually. JAMA. 2013;310:1110. doi: 10.1001/jama.2013.278331. [DOI] [PubMed] [Google Scholar]
- Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- Li C, Bakker RG, Motaleb MA, Sartakova ML, Cabello FC, Charon NW. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc Natl Acad Sci U S A. 2002;99:6169–6174. doi: 10.1073/pnas.092010499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol. 2010;403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS. Epidemiology of Lyme Disease. Infect Dis Clin North Am. 2015;29:187–210. doi: 10.1016/j.idc.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Miller CL, Karna S, Seshu J. Borrelia host adaptation Regulator (BadR) regulates rpoS to modulate host adaptation and virulence factors in Borrelia burgdorferi. Mol Microbiol. 2013;88:105–124. doi: 10.1111/mmi.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Imada K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015;23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Moon K, Hobbs G, Motaleb MA. Borrelia burgdorferi CheD promotes various functions in chemotaxis and pathogenic life cycle of the spirochete. Infec Immun. 2016;84:1743–1752. doi: 10.1128/IAI.01347-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Miller MR, Bakker RG, Li C, Charon NW. Isolation and Characterization of Chemotaxis Mutants of the Lyme Disease Spirochete Borrelia burgdorferi Using Allelic Exchange Mutagenesis, Flow Cytometry, and Cell Tracking. Meth Enzymol. 2007;422:419–437. doi: 10.1016/S0076-6879(06)22021-4. [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci U S A. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Pitzer JE, Sultan SZ, Liu J. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol. 2011;193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb M, Liu J, Wooten RM. Spirochetal motility and chemotaxis in the natural enzootic cycle and development of Lyme disease. Curr Opin Microbiol. 2015;28:106–113. doi: 10.1016/j.mib.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- Pappas CJ, Iyer R, Petzke MM, Caimano MJ, Radolf JD, Schwartz I. Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. PLoS Pathog. 2011;7:e1002102. doi: 10.1371/journal.ppat.1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Nieto V, Harshey RM. A new player at the flagellar motor: FliL controls both motor output and bias. MBio. 2015;6:e02367–14. doi: 10.1128/mBio.02367-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulick A, Koerdt A, Lassak J, Huntley S, Wilms I, Narberhaus F, Thormann KM. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol Microbiol. 2009;71:836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, Motaleb MA. Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect Immun. 2011;79:1815–1825. doi: 10.1128/IAI.00075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raddi G, Morado DR, Yan J, Haake DA, Yang XF, Liu J. Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J Bacteriol. 2012;194:1299–1306. doi: 10.1128/JB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Terekhova D, Zhang H, Hovis KM, Schwartz I, Marconi RT. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol. 2009;71:1551–1573. doi: 10.1111/j.1365-2958.2009.06621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008;41:103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Bestor A, Cullen JN, Rosa PA. A tightly regulated surface protein of Borrelia burgdorferi is not essential to the mouse-tick infectious cycle. Infect Immun. 2008;76:1970–1978. doi: 10.1128/IAI.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suaste-Olmos F, Domenzain C, Mireles-Rodriguez JC, Poggio S, Osorio A, Dreyfus G, Camarena L. The flagellar protein FliL is essential for swimming in Rhodobacter sphaeroides. J Bacteriol. 2010;192:6230–6239. doi: 10.1128/JB.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Miller MR, Motaleb MA. Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol Microbiol. 2010;77:128–142. doi: 10.1111/j.1365-2958.2010.07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Manne A, Stewart PE, Bestor A, Rosa PA, Charon NW, Motaleb MA. Motility is crucial for the infectious life cycle of Borrelia burgdorferi. Infect Immun. 2013;81:2012–2021. doi: 10.1128/IAI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR, Motaleb MA. Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect Immun. 2011;79:3273–3283. doi: 10.1128/IAI.05153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Sekar P, Zhao X, Manne A, Liu J, Wooten RM, Motaleb MA. Motor rotation is essential for the formation of the periplasmic flagellar ribbon, cellular morphology, and Borrelia burgdorferi persistence within Ixodes scapularis tick and murine hosts. Infect Immun. 2015;83:1765–1777. doi: 10.1128/IAI.03097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth CW. Flagellar motility of the pathogenic spirochetes. Semin Cell Dev Biol. 2015;46:104–112. doi: 10.1016/j.semcdb.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Meltzer MI, Pena CA, Hopkins AB, Wroth L, Fix AD. Economic impact of Lyme disease. Emerg Infect Dis. 2006;12:653–660. doi: 10.3201/eid1204.050602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Norris SJ, Liu J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry. 2014;53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, et al. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2013;110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Kumar A, Kojima S, Homma M. FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Mol Microbiol. 2015;98:101–110. doi: 10.1111/mmi.13103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. A typical 3D reconstruction of a wild-type B. burgdorferi cell tip shows well-organized periplasmic flagella that are oriented toward the other pole of the cell.
Movie 2. A typical 3D reconstruction of a ΔflbB cell tip shows the shortened periplasmic flagella—majority of them are abnormally tilted toward the cell pole.


