SUMMARY
CENP-A is a centromere-specific histone 3 variant essential for centromere specification. CENP-A partially replaces canonical histone H3 at the centromeres. How the particular CENP-A/H3 ratio at centromeres is precisely maintained is unknown. It also remains unclear how CENP-A is excluded from non-centromeric chromatin. Here we identify Ccp1, an uncharacterized NAP family protein in fission yeast that antagonizes CENP-A loading at both centromeric and non-centromeric regions. Like the CENP-A loading factor HJURP, Ccp1 interacts with CENP-A, and is recruited to centromeres at the end of mitosis in a Mis16-dependent manner. These data indicate that factors with opposing CENP-A loading activities are recruited to centromeres. Furthermore, Ccp1 also cooperates with H2A.Z to evict CENP-A assembled in euchromatin. Structural analyses indicate that Ccp1 forms a homodimer that is required for its anti-CENP-A loading activity. Our study establishes mechanisms for maintenance of CENP-A homeostasis at centromeres and the prevention of ectopic assembly of centromeres.
Graphical abstract
INTRODUCTION
The centromere is the part of a chromosome responsible for kinetochore assembly. It is crucial for the proper segregation of chromosomes (Allshire and Karpen, 2008; Malik and Henikoff, 2009). Most eukaryotes contain large and complex “regional” centromeres, which have multiple microtubule attachment sites per centromere (Fukagawa and Earnshaw, 2014; Westhorpe and Straight, 2015). The underlying DNA sequences in regional centromeres across species are highly divergent across species. Epigenetic mechanisms play a key role in governing the position specification and function of the centromeres (Gomez-Rodriguez and Jansen, 2013; Westhorpe and Straight, 2015). Centromere Protein-A (CENP-A), a conserved centromere-specific histone 3 (H3) variant, is indispensible for centromere function, and has emerged as the best candidate for the epigenetic mark for centromeres (Gomez-Rodriguez and Jansen, 2013; Muller and Almouzni, 2014; Palmer et al., 1991).
The amount of CENP-A at regional centromeres is tightly regulated. CENP-A partially replaces canonical histone H3 at the centromere to serve as the structural and functional foundation for kinetochore assembly (Black and Cleveland, 2011; Bodor et al., 2014; Sullivan and Karpen, 2004). In fact, modifications of histone H3 at centromeres are important for proper assembly of CENP-A chromatin (Bergmann et al., 2012; Bergmann et al., 2011; Stimpson and Sullivan, 2011). How CENP-A is deposited into centromeres has been extensively studied. One key factor known to promote CENP-A loading is the conserved histone chaperone, Scm3/HJURP, which is in turn recruited by the Mis16-Mis18 complex (Dunleavy et al., 2009; Foltz et al., 2009; Pidoux et al., 2009; Williams et al., 2009; Yu et al., 2015). However, how CENP-A and histone H3 levels are precisely maintained at the centromere remains unknown.
In most eukaryotic organisms, each chromosome contains a single centromere. Misincorporation of CENP-A to non-centromeric regions can lead to formation of ectopic centromeres, which has a devastating impact on chromosome segregation (Burrack and Berman, 2012; Gonzalez et al., 2014; Heun et al., 2006; Scott and Sullivan, 2014). Maintaining the H3 nucleosome integrity is important for preventing mis-incorporation of CENP-A. Changes in histone H3 level, or mutations in histone chaperones and the FACT chromatin remodeling complex can lead to promiscuous CENP-A incorporation, likely due to disturbance of H3 chromatin integrity (Au et al., 2008; Choi et al., 2012; Lacoste et al., 2014; Lopes da Rosa et al., 2011). Ubiquitin-mediated proteolysis is another mechanism implicated in protecting euchromatic regions from assembling ectopic CENP-A chromatin (Au et al., 2013; Choi et al., 2012; Collins et al., 2004; Moreno-Moreno et al., 2006). It has recently been shown that the FACT complex in the budding yeast Saccharomyces cerevisiae can prevent mislocalization of the CENP-A homolog Cse4 by directly mediating ubiquitin-dependent proteolysis of Cse4 (Deyter and Biggins, 2014). However, the mechanisms involved in preventing the ectopic assembly of CENP-A remain poorly understood.
Studies on the centromeres of budding yeast have been instrumental in helping us understand centromere structure and function. Budding yeast contains “point” centromeres, which are genetically defined by a 125 bp DNA sequence (Clarke and Carbon, 1980; Cottarel et al., 1989). In contrast, the fission yeast, Schizosaccharomyces pombe, contains large regional centromeres, which are governed by epigenetic mechanisms (Allshire and Karpen, 2008; Carroll and Straight, 2006). In fission yeast, the CENP-A homolog, Cnp1 (CENP-Acnp1), is enriched within 10-12 kb of the central domain region of centromeres, which are flanked with pericentromeric heterochromatin. The loading of CENP-Acnp1 also depends on the CENP-A loading factor, HJURPscm3. Recrutment of HJURPscm3 is cell cycle-regulated, and requires the conserved Mis16-Mis18 complex (Pidoux et al., 2009; Williams et al., 2009). Like in other eukartyotes that harbor regional centromeres, the mislocalization of CENP-Acnp1 at non-centromeric regions can lead to assembly of ectopic CENP-Acnp1 chromatin and consequent chromosome missegregation (Castillo et al., 2013; Gonzalez et al., 2014; Ishii et al., 2008). In addition to ubiquitin-mediated proteolysis and the FACT complex (Choi et al., 2012; Gonzalez et al., 2014), Pht1, the homolog of histone H2A.Z in S. pombe, has also been implicated in preventing ectopic assembly of CENP-Acnp1 (Ogiyama et al., 2013).
In this study, we identify an uncharacterized protein in fission yeast, Ccp1, antagonizing the loading of CENP-Acnp1 at both centromeric and non-centromeric regions. Our data show that Ccp1 physically asosciates with CENP-Acnp1. Like HJURPscm3, Ccp1 is recruited to centromeres by Mis16 at the end of mitosis. The simultaneous recruitment of CENP-A loading and anti-loading factors provides an explanation for how the proper ratio of CENP-A to histone H3 is maintained at centromeres. Ccp1 also functions together with H2A.Z to displace ectopic CENP-A at non-centromeric regions. We report the crystal structure of Ccp1. Our data suggests that Ccp1 functions as a novel CENP-A chaperone specifically for preventing CENP-A loading. Together, these findings provide insights into how the right balance of CENP-A and histone H3 levels is achieved at centromeres, and uncover a mechanism for how cells protect themselves from excessive assembly of CENP-A at non-centromeric regions.
RESULTS
Ccp1 Antagonizes CENP-A Loading
To identify genes required for proper CENP-A distribution, we performed a candidate visual genetic screen using fission yeast cells carrying CENP-Acnp1-GFP under the native promoter at the ade6+ locus. In wild type interphase cells, all centromeres are clustered at the nuclear envelope; thus, a single fluorescent focus was observed in cells expressing CENP-Acnp1-GFP at endogenous level (Figure 1A) (Takahashi et al., 2000). Through the screen, we found that deletion of the gene SPBC36B7.08C results in more than 12% of cells exhibiting multiple foci of CENP-Acnp1-GFP (Figure 1A). We also found that the centromere clustering and the level of CENP-Acnp1-GFP are not affected in the mutant (Figures S1A and S1B). These results indicate that the gene is important for preventing mis-targeting of CENP-Acnp1-GFP to non-centromeric regions. To confirm this, we also examined the distribution pattern of N- or C-terminal GFP-tagged CENP-Acnp1 under the control of its native promoter at endogenous locus in the mutant background. Both fusion proteins display a single fluorescent focus in wild type (Gonzalez et al., 2014; Takayama et al., 2008), but they also mislocalize in the mutant (Figures S1C-S1E), similar to CENP-Acnp1-GFP at ade6+ locus. We have thus named the gene, ccp1+ (counteracter of CENP-A loading protein 1). ccp1+ encodes an uncharacterized NAP (Nucleosome Assembly Protein) domain-containing protein of 244 amino acids, which is 26 and 27% identical to human SET protein and budding yeast Vps75, respectively. NAP family proteins often act as histone chaperones and play crucial roles in both assembly and disassembly of nucleosomes (Lorch et al., 2006; Park and Luger, 2008).
Figure 1. Ccp1 Antagonizes the Loading of CENP-Acnp1 at Both Centromeric and Non-centromeric Regions. (See also Figure S1 and S2).
(A) CENP-Acnp1-GFP is mislocalized to non-centromeric regions in the ccp1Δ mutant. The nuclear envelope is visualized with Ish1-mCherry. The percentage of cells containing multiple CENP-Acnp1-GFP foci (>2) is indicated.
(B) CENP-Acnp1-GFP is enriched at rDNA regions as shown by ChIP assays. ChIP assays were performed using an antibody against GFP and primers specific for an rDNA region and a control gene, act1+ (act). Three independent experiments were performed. The relative fold enrichment is shown in the right panel. The value for WT was normalized to 1.0. WCE, whole-cell extracts. Data represent mean ± S.D.
(C) ChIP assay showed that CENP-A-GFP level at centromere (cen) is largely unchanged. ChIP assays were performed using a GFP antibody and primers specific for a centromeric region. Data represent mean ± S.D.
(D) CENP-Acnp1-GFP signal is enhanced in a subpopulation of ccp1Δ mutant cells displaying a single GFP focus. Quantifications were performed using ImageJ software (v1.50i) (McCloy et al., 2014). The percentage of cells exhibiting a single GFP signal with an intensity twice the average of wild type was plotted.
(E) ccp1Δ mutant cells are sensitive to TBZ. Serially diluted cells were plated on rich media containing 17.5μg/ml TBZ.
(F) Cells overexpressing CENP-Acnp1 in the ccp1Δ mutant show slower growth than wild-type cells overexpressing CENP-Acnp1. Overexpression was induced in minimal medium without thiamine (−thiamine). Scale bars, 2 μm.
To confirm that CENP-Acnp1-GFP is misincorporated into non-centromeric region in the absence of Ccp1, we analyzed ccp1Δ mutant cells using chromatin immunoprecipitation (ChIP) along with primers specific to an rDNA region. Consistent with our cytological observation, we found that CENP-Acnp1-GFP is significantly enriched at the rDNA region, indicating that CENP-Acnp1-GFP associates with ectopic regions (Figure 1B), consistent with our cytological observation.
Intriguingly, our ChIP assay showed that the CENP-Acnp1-GFP level at the centromere in the mutant is largely unchanged (Figure 1C). However, we found that a significant number of ccp1Δ mutant cells displaying a single fluorescence focus has disproportionately brighter CENP-Acnp1-GFP signal (Figure 1D). These results suggest that in ccp1Δ cells exhibiting a single GFP focus, the level of CENP-Acnp1-GFP at this region can substantially increase.
To further verify the mislocalization of CENP-Acnp1 in the ccp1Δ mutant, we analyzed ccp1Δ cells carrying CENP-Acnp1-GFP by ChIP followed by high-throughput sequencing (ChIP-seq). Our ChIP-seq data showed that CENP-Acnp1-GFP is significantly enriched in euchromatin in ccp1Δ mutant relative to wild type (Figure S1F). Together, our results indicate that Ccp1 is required for maintaining proper amounts of CENP-Acnp1 at both centromeric and non-centromeric regions. Consistent with this, we found that the mutant cells exhibited sensitivity to the microtubule-depolymerizing drug, thiabendazole (TBZ) (Figure 1E), which indicates that chromosome segregation in the ccp1 mutant is abnormal. Chromosome missegragation defects were further confirmed by DAPI staining (Figure S1G).
Previous studies have shown that CENP-Acnp1-GFP still largely associates with centromeres when lowly overexpressed in S. pombe (Castillo et al., 2013). To further examine the role of Ccp1 in counteracting the CENP-Acnp1 loading at non-centromeric regions, we induced a low level of overexpression of CENP-Acnp1-GFP using the intermediate-strength, thiamine-repressible nmt42 promoter in the ccp1Δ mutant. After a 32-hour induction, 94% of wild-type cells display one or two fluorescence foci. In contrast, multiple CENP-Acnp1-GFP foci were observed in 46% of the ccp1Δ cells analyzed under the same induction conditions (Figure S2A and B). This indicates that low level overexpression of CENP-Acnp1-GFP in ccp1Δ cells results in overall euchromatic localization.
Overexpression of CENP-Acnp1-GFP in wild-type cells causes slow cell growth (Castillo et al., 2013; Gonzalez et al., 2014). We found that the overexpression of CENP-Acnp1-GFP under the strong nmt1 promoter in the ccp1Δ mutant results in even stronger inhibition of cell growth (Figures 1F and S2C). This is in agreement with the idea that overexpression of CENP-Acnp1 enhances the ectopic distribution of CENP-Acnp1-GFP in ccp1Δ cells. This is reminiscent of the overexpression of N-terminal deleted CENP-Acnp1, which leads to increased level of misincorporation of CENP-Acnp1 and slower growth (Gonzalez et al., 2014).
Overexpression of Ccp1 Evicts CENP-A
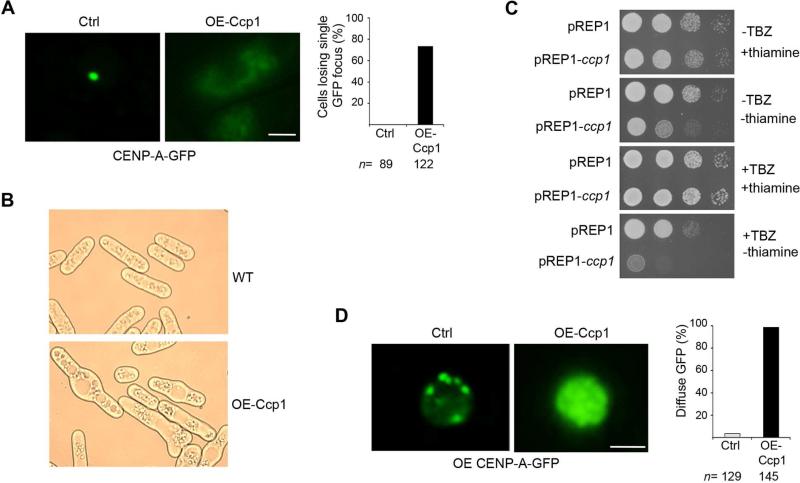
To further investigate the role of Ccp1 in CENP-Acnp1 positioning at centromeres, we overexpressed Ccp1 from the strong nmt1 promoter in wild type cells expressing CENP-Acnp1-GFP at endogenous level. We found that the single CENP-Acnp1 focus is lost from centromeres in 76% of the cells analyzed (Figure 2A), even though the level of CENP-Acnp1-GFP in these cells remained similar to that of wild-type (Figure S3A). These results demonstrate that overexpresssion of Ccp1 causes significant eviction of CENP-Acnp1-GFP from native centromeres. Consistent with this, cells overexpressing Ccp1 exhibit severe growth defects and are highly sensitive to TBZ (Figures 2B and 2C).
Figure 2. Overexpression of Ccp1 Results in Eviction of CENP-Acnp1 at Centromeric and Noncentromeric Regions. (See also Figure S3).
(A) CENP-Acnp1-GFP signal at centromeres is diminished in cells overexpressing Ccp1. The percentage of cells showing loss of the CENP-Acnp1-GFP focus at centromeres is indicated.
(B) Abnormal morphology in cells overexpressing Ccp1. Wild-type cells carrying nmt1-Ccp1 were plated on minimal media without thiamine for 24 hrs.
(C) Cells overexpressing Ccp1 are highly sensitive to TBZ. Serially diluted cells were plated on media containing 17.5μg/ml TBZ. –TBZ, media without TBZ.
(D) Overexpression of Ccp1 causes eviction of ectopically-assembled CENP-A from noncentromeric loci. Cells carrying nmt1-CENP-Acnp1-GFP and nmt1-Ccp1 were used (right panel). Cells overexpressing CENP-Acnp1-GFP alone were used as a control (left panel). The percentage of cells showing diffuse GFP pattern in nucleoplasm is indicated. Scale bars, 2 μm.
The overexpression of CENP-Acnp1 can induce the assembly of stable ectopic CENP-A chromatin at non-centromeric regions (Castillo et al., 2013; Gonzalez et al., 2014). To investigate whether Ccp1 can promote the exclusion of CENP-Acnp1 from ectopic loci, we overexpressed Ccp1 in wild-type cells carrying ectopic CENP-Acnp1 chromatin induced by massive overexpression of CENP-Acnp1-GFP. Consistent with previous studies, multiple distinct, stable fluorescent foci were observed in control cells overexpressing CENP-Acnp1-GFP alone, after 24 hrs induction (Figure 2D). However, when we overexpressed Ccp1 in cells also overexpressing CENP-Acnp1-GFP, we found that almost all cells exhibited diffusion of the CENP-Acnp1-GFP signal, indicating that the ectopically assembled CENP-Acnp1-GFP was evicted from non-centromeric chromatin (Figures 2D, and S3B). Together, our results show that overexpression of Ccp1 results in the eviction of CENP-Acnp1 from both centromeric and ectopic loci, consistent with its role as an anti-CENP-A loading factor.
Ccp1 Interacts With CENP-A
To further investigate the relationship between Ccp1 and CENP-Acnp1, ccp1Δ was crossed into a CENP-Acnp1 mutant, cnp1-1, to create a ccp1Δ cnp1-1 double mutant. The double mutant displays a severe growth impairment at 32°C. The synthetic growth defects are even more apparent at 36°C (Figure 3A). Our data predict that Ccp1 physically associates with CENP-Acnp1. To test the idea, we created a TAP-tag version of Ccp1 under the control of its native promoter at its endogenous locus. This strain shows no sensitivity to TBZ (Figure S4A), indicating that Ccp1-TAP is functional. In the Ccp1-TAP strain massively overexpressing CENP-Acnp1-GFP, we found that Ccp1-TAP co-immunoprecipitated with CENP-Acnp1-GFP (Figure 3B), supporting the idea that Ccp1 physically associates with CENP-Acnp1. We also conducted Co-IP experiments using cells carrying Ccp1-S tag and GFP-tagged histone H3. Ccp1-S tag was not able to co-immunoprecipitate H3-GFP from cell lysates (Figure S4B). To further confirm the interaction between Ccp1 and CENP-Acnp1, we purified chromatin-bound CENP-Acnp1 using the TAP-tag purification method, and followed with mass spectrometry (mass spec) analysis to identify its interacting proteins. As expected, in addition to CENP-Acnp1, histones were identified by our mass spec analysis of purified chromatin-bound CENP-Acnp1 (Figure 3C). We also found that the chromatin-remodeling complex FACT, including Pof3 and Spt16, interacts with CENP-Acnp1 (Figure 3C). Consistent with these findings, CENP-A has been shown to be associated with the FACT complex in Drosophila and human cells (Boltengagen et al., 2015; Chen et al., 2015; Foltz et al., 2006; Okada et al., 2009). In addition, a conserved centromere protein, CENP-T (Cnp20 in fission yeast) copurified with CENP-Acnp1. CENP-T also copurified with CENP-A nucleosome in human cells (Foltz et al., 2006). Importantly, we identified Ccp1 in the TAP-tag purified product (Figure 3C). We next performed an in vitro binding assay to examine whether Ccp1 directly interacts with CENP-A. We found that GST-tagged Ccp1, but not GST alone, was able to pull down CENP-Acnp1/H4 dimer (Figure 3D), indicating a direct interaction between Ccp1 and CENP-Acnp1. Notably, we did not observe direct interaction between Ccp1 and H3/H4 complex in the parallel pull-down experiments (3D). Together, these data indicate that Ccp1 physically interacts with CENP-Acnp1.
Figure 3. Ccp1 Interacts with CENP-A. (See also Figure S4).
(A) Synthetic genetic interactions of ccp1Δ with a CENP-Acnp1 mutant, cnp1-1. Serial dilutions of cells on YES medium were incubated at 25°C, 32°C, or 36°C for 3 days.
(B) Cell lysates from cells expressing Ccp1-TAP and CENP-Acnp1-GFP were subjected to immunoprecipitation with an antibody specific for the TAP tag. CENP-Acnp1-GFP was intermediately overexpressed under the nmt42 promoter. Precipitated proteins were analyzed by Western blotting using indicated antibodies. Cells expressing CENP-Acnp1-GFP or Ccp1-TAP only were used as control. The percentage of immunoprecipitated CENP-Acnp1-GFP relative to input was indicated.
(C) Summary of mass spectrometry analysis of purified chromatin-bound CENP-Acnp1-TAP. The number of peptides identified and sequence coverage are shown.
(D) Coomassie blue staining showing binding between GST-Ccp1 and the refolded heterodimeric complexes: CENP-A/H4 (lane 5), and H3/H4 complexes (lane7). GST protein alone was served as control (Lane 6 and Lane 8). Glutathione sepharose beads were used to pull down the GST-Ccp1 or GST proteins in the pull-down assays. Input: Lane 1, 2, 3 and 4 mark the size of CENP-A/H4 dimer, H3/H4 dimer, GST-Ccp1 and GST alone, respectively. Individual bands of these proteins on the gel were indicated by lower-case letters: a, CENP-A-TFH; b, H4-TFH; c, H3-TFH. Note: CENP-A-TFH and H4-TFH have similar molecular weights and comigrated in one band. Right panel: quantification of CENP-A/histones pulled down by the in vitro binding assays. Data represent mean ± S.D.
Ccp1 Associates with Centromeres in a Cell Cycle-dependent Manner
To investigate how Ccp1 is distributed in the cells, we replaced the endogenous Ccp1 with GFP-tagged Ccp1 at its C-terminus. Cells expressing Ccp1-GFP from its endogenous locus are not sensitive to TBZ, indicating that Ccp1-GFP is functional (Figure S4A). Ccp1-GFP is faintly distributed throughout the nucleus, but also forms a distinct single fluorescence focus (Figure 4A). We also found that Ccp1 with N-terminal tagged GFP has the same distribution pattern (Data not shown). Ccp1-GFP and CENP-Acnp1-mCherry colocalize, indicating that the single Ccp1-GFP focus associates with centromeres (Figure 4A). The centromere association of Ccp1 was further confirmed by ChIP assays (Figure S5A).
Figure 4. Centromere Localization of Ccp1 Depends on Mis16. (See also Figure S4 and S5).
(A) Ccp1-GFP is localized in the nucleus, preferentially enriched at centromeres. CENP-Acnp1-mCherry expressed at endogenous level was used to visualize centromeres.
(B) Ccp1 is recruited to centromeres at the end of mitosis. Wild-type cells carrying Ccp1-GFP were used. The nucleus can be visualized by Ccp1-GFP signal, which is also faintly diffuse throughout the nucleus. Cells were assigned to cell cycle stages based on nuclear morphology.
(C) The centromere localization of Ccp1-GFP was abolished in mis16-53 mutant. The percentage of interphase cells showing diffuse GFP pattern within the nucleus is indicated.
(D) The centromere localization of Mis16-GFP is not affected in the ccp1Δ mutant. The percentage of interphase cells showing single GFP focus is indicated.
(E) Yeast two-hybrid assay showing that Ccp1 interacts with Mis16. BD, GAL4 DNA-binding domain fusion; AD, GAL4 activation domain fusion. Scale bars, 2 μm.
Interestingly, our time-lapse microscopy demonstrated that the centromeric focus of Ccp1-GFP is present throughout interphase. However, the signal vanishes at the onset of mitosis and reappears in telophase, indicating that Ccp1 delocalizes from centromeres during mitosis (Figure 4B). Therefore, the association of Ccp1 with centromeres is cell cycle-regulated. Intriguingly, this temporal pattern is similar to that observed for the CENP-A loading factors HJURPscm3 and the Mis16- Mis18 complex (Pidoux et al., 2009; Williams et al., 2009).
Mis16 Recruits Ccp1 to Centromeres
It is known that Mis16 recruits the CENP-A loading factor, HJURPscm3 at the end of mitosis, which in turn mediates the loading of CENP-A at centromeres (Pidoux et al., 2009; Williams et al., 2009). We investigated whether the centromere association of Ccp1 also depends on Mis16 using a temperature-sensitive mis16-53 mutant (Hayashi et al., 2004). We observed that the centromeric localization of Ccp1-GFP in mis16-53 cells was completely abolished after incubation at 36°C for four hours (Figure 4C). However, in the null mutant of ams2+, which produces another key CENP-A loading factor (Chen et al., 2003), Ccp1-GFP remains at centromeres (Figure S5B). Also, in the ccp1 mutant, Mis16-GFP still associates with centromeres (Figure 4D). These results indicate that Mis16 is required for the localization of Ccp1 at centromeres. Furthermore, our yeast two-hybrid assays indicate that Ccp1 physically interacts with Mis16 (Figure 4E). Together, our data suggest that the Mis16-Mis18 complex is responsible for recruiting factors with opposing activities for CENP-A loading at the end of mitosis to centromeres to mediate the CENP-A level at these loci.
Ccp1 Cooperates with H2A.Z to Prevent Mistargeting of CENP-A
Pht1, a homolog of H2A.Z, has been suggested to play a role in preventing ectopic incorporation of CENP-A (Ogiyama et al., 2013). Consistent with this study, we found that approximately 14% of pht1Δ cells exhibited multiple foci of CENPcnp1-A-GFP (Figure 5A), a percentage similar to that observed in ccp1Δ cells (Figure 1A). Our yeast two-hybrid analysis also showed that Ccp1 associates with Pht1 (Figure 5B). Furthermore, double mutant cells of ccp1Δ and pht1Δ displayed a higher percentage of multiple CENPcnp1-A-GFP foci than either single mutant strain (Figure 5A). Consistent with this, the ccp1Δ pht1Δ double mutant is more sensitive to TBZ than the single mutants (Figure 5C). These data suggest that Ccp1 functions in parallel with Pht1 to prevent ectopic formation of CENP-A chromatin.
Figure 5. Ccp1 Interacts with H2A.Z.
(A) Distribution of CENP-A-GFP in the pht1Δ mutant, and the ccp1Δ pht1Δ double mutant. Scale bars, 2 μm.
(B) Yeast two-hybrid assay showing that Ccp1 interacts with Pht1. BD, GAL4 DNA-binding domain fusion; AD, GAL4 activation domain fusion.
(C) Serial dilutions of the ccp1Δ pht1Δ double mutant were spotted on the rich media containing 17.5μg/ml TBZ. ccp1Δ and pht1Δ mutants were used as a control.
Structure Determination of the Ccp1 Protein
To learn more about the molecular function of Ccp1, we determined its structure at 2.1 Å by single-wavelength anomalous diffraction (Se-SAD). The final model contains two Ccp1 molecules (residues 2-207) and 258 water molecules per asymmetric unit, and it was refined to an Rwork/Rfree of 0.193/0.236. Another region (residues 208-244), which contains a stretch of acidic Asp/Glu amino acids, is not included owing to disorder in the crystals. All the structural models have excellent geometrical parameters (Table 1). Our crystal structure indicates that Ccp1 is a homodimer (Figures 6C and 6D). This is confirmed by Co-IP using a strain expressing both TAP-tagged and mCherry-tagged Ccp1 (Figure S6A).
Table 1.
Data collection and refinement statistics.
| Parameter* | Ccp1_mu2 | Ccp1_wt |
|---|---|---|
| Data collection statistics | ||
| Wavelength (Å) | 0.97923 | 0.97892 |
| Resolution range (Å) | 29. 29-2. 10(2. 18-2. 10) | 38. 76 -2. 10 (2. 18-2. 10) |
| Space group | P 41 2 2 | P 41 2 2 |
| Cell dimensions | ||
| a,b,c (Å) | 86.81 86.81 158.78 | 86.66 86.66 158.77 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Unique reflections | 36000 (3501) | 35996 (3534) |
| Multiplicity | 13.6 (13.9) | 14. 4 (14. 7) |
| Completeness (%) | 99.74 (98.93) | 99.69 (99.33) |
| Mean I/sigma(I) | 37.5 (11.1) | 23.8 (5.6) |
| Wilson B-factor | 23.86 | 23.87 |
| R-merge (%)† | 9.1(60.6) | 8. 0 (69. 8) |
| Structure refinement | ||
| Rwork/Rfree (%) | 17.9/22.1 | 19.3/23.6 |
| Number of non-hydrogen atoms | 3917 | 3774 |
| macromolecules | 3451 | 3488 |
| ligands | 28 | |
| waters | 466 | 258 |
| Protein residues | 414 | 414 |
| RMSD | ||
| Bond length (Å) | 0.006 | 0.003 |
| Bond angles (°) | 1.07 | 0.73 |
| Average B-factor | 37.3 | 34.6 |
| macromolecules | 35.6 | 34.0 |
| ligands | 53.5 | |
| solvent | 49.9 | 41.0 |
| Ramachandran plot (%) | ||
| favored region | 99 | 97 |
| allowed region | 0.76 | 2.76 |
| Outliers region | 0.24 | 0.24 |
Values in parentheses are for highest-resolution shell.
Rmerge= Σhkl |I-<I>|/ΣhklI where I is the intensity of unique relfection hkl and <I> is the average over symmetry-related observations of unique reflection hkl.
Figure 6. Crystal Structure of Ccp1 Dimer. (See also Figure S6).
(A) Ribbon drawing of the Ccp1 Monomer, colored spectrally from dark blue at its N-terminal segment to red at its C-terminal segment.
(B) Topology diagram of the Ccp1 monomer.
(C and D) Ribbon diagram of the Ccp1 dimer.
(E and F) Electrostatic potential at the molecular surface viewed as in C and D, respectively. The contour level is at ±5 kT/e; red for negative potential and blue for positive potential.
The structures of the Ccp1 molecules in the non-crystallographic dimer are essentially identical, with a root mean squared deviation (r.m.s.d.) of 0.62 Å for the superimposition of all ~200 Cα atoms. Ccp1 has highest structural similarity to NAP family proteins, which include budding yeast Nap1, Vps75 and the human SET/ TAF-Ib/INHAT(Muto et al., 2007; Park and Luger, 2006; Park et al., 2008; Tang et al., 2008) (Figure S6B). The closest structural homologue was budding yeast Vps75 (PDB 2ZD7; Z-score of 19.3, 27% sequence identity) (Park et al., 2008; Tang et al., 2008), with a r.m.s.d. of 2.5 Å between 212 structurally equivalent α-carbon atomic pairs.
Similar to other NAP family proteins, the Ccp1 protein has a “headphone” shaped topology with three major domains: the structurally defined dimerization domain, earmuff domain and a disordered acidic C-terminal domain (Figures 6A and 6B). In brief, the dimerization domain is comprised of a long helix (α1, residues 1-44) that is arranged in an antiparallel fashion with the helix from the opposing subunit. The globular shaped “earmuff” domain contains a four-stranded β-sheet (β1–β4) and five helices (the N-terminal α2, α3, α4 and the C-terminal α5, α6) that together form a hydrophobic core (Figures 6A and 6B). Interestingly, unlike the Nap1 and Vps75 proteins, we found that Ccp1 contains relatively short β1 and β2 strands but long β3 and β4 strands. The C-terminal domain is enriched with acidic residues of Asp/Glu, probably devoid of stable secondary and tertiary structure, and therefore is disordered in the structure, suggesting that the domain is highly mobile in aqueous solution.
The earmuff domain of the Ccp1 dimer is attached to the concave side of the dimerization domain helices, therefore forming a pronounced cleft near the center of the homodimer (Figures 6C and 6D; Figure S6C). The electrostatic potential surface is largely negative, especially near the edge of the cleft (Figures 6E and 6F). The dimensions of the cleft are ~ 35 Å height with a width of ~ 12 Å at the narrowest point near the dimerization domain, and ~ 55 Å width between the tips of the turn between β3 and β4. The NAP1 and Vps75 homodimers also form a prominent central cleft by the earmuff domains, which has been shown to accommodate the histone H3-H4 tetramer (Bowman et al., 2011; Park and Luger, 2006; Tang et al., 2008).
There are extensive H-bonds and van der Waals interactions among the anti-parallel pairing of the α1-helices along their entire length of 40 amino acids (Figure 7A). The two helical axes are aligned in an antiparallel “tram-track” fashion, rather than coiling around each other. The kink in the long α1-helices may contribute to the formation of this unique arrangement, which is commonly observed among the NAP family proteins. A conserved Pro-34 is located at a distance of about 2/3 of the α1-helix length from the N-terminus, which generates a ~40° kink. A second kink (~25°) is observed at Leu13 of the helices. As a consequence of the antiparallel pairing of the two helices, Pro34 juxtaposes Leu13 of the second Ccp1 molecule within the dimer (Figure 7A). It is likely that the bend at Leu13 is caused by the need to maintain packing by keeping the two helical axes parallel. The residues contributing to dimer formation are Ala6, Phe7, Leu10, Leu13, Phe17, Ala20, Ile24, Leu31, Phe32, Leu35, Phe36, Ile42, Leu43, and Ile46 (Figure 7A). The dimer is further stabilized by the β3-β4 loop and the α5-helix.
Figure 7. Dimerization of Ccp1 is Required for its Anti-CENP-A Loading Activity. (See also Figure S5 and S7).
(A) The dimerization domain, top and side views. Residues involved in the interactions at the dimerization interface are shown. Residues (Leu13 and Pro34) that generate the kink are indicated. Residues selected for mutagenesis to disrupt dimer interaction are shown.
(B) Upper panel: Gel-filtration chromatography of the wild type protein (red) and a quadruple mutant (L10A/F17A/I24A/F32A, blue). Lower panel: Protein samples of the indicated fractions were subjected to SDS-PAGE and Coomassie blue staining. The retention volume for the quadruple mutant shifted backward ~1.4ml on the gel filtration column Superdex 200, indicating the dissociation of dimer into monomer.
(C) Sensitivity of ccp1-4A mutant to TBZ. Indicated cells were spotted on the rich media containing 17.5μg/ml TBZ.
(D) CENP-Acnp1-GFP distribution in ccp1-4A mutant. Scale bars, 2 μm.
(E) Yeast two-hybrid assay showing that interaction with Ccp1-4A with Mis16 is compromised. BD, GAL4 DNA-binding domain fusion; AD, GAL4 activation domain fusion.
(F) The association of Ccp1-4A-GFP with centromeres is lost. Scale bars, 2 μm.
(G) Yeast two-hybrid assay showing that interaction with Ccp1-4A with Pht1 is abolished. BD, GAL4 DNA-binding domain fusion; AD, GAL4 activation domain fusion.
(H) Model: Mis16 recruits Ccp1 and HJURPScm3 to centromeres to balance the level of CENP-A (red) and histone H3 (green) nucleosomes. In non-centromeric regions, Ccp1 acts together with H2A.Z to evict mis-incorporated CENP-A.
Dimerization of Ccp1 is Required for its Anti-CENP-A Loading Activity
To disrupt the formation of the Ccp1 dimer, a series of residues at the dimer interface, including Leu10, Phe17, Ile24 and Phe32, were selected for mutagenesis (Figures 7A and S7A). Our result indicated that a quadruple mutant, ccp1-4A (L10A/F17A/I24A/F32A) was unable to dimerize, as evidenced from the analysis on the gel-filtration chromatography (Figure 7B). Consistent with this, we found that Ccp1-4A-TAP was unable to co-immunoprecipitate Ccp1-4A-mCherry (Figure S7B and C). To determine whether dimerization of Ccp1 is essential for its function as an anti-CENP-A loading factor in vivo, we replaced the endogenous Ccp1 with ccp1-4A. The ccp1-4A mutant is sensitive to TBZ (Figure 7C). Also similar to the ccp1Δ mutant, 12% of ccp1-4A cells showed multiple CENP-Acnp1-GFP foci (Figure 7D), indicating that dimerization of Ccp1 is required for its capability to counteract CENP-A loading. In addition, we found that the mutation significantly disrupts the interaction between Ccp1 and Mis16 (Figure 7E). Consistent with this, the association of Ccp1-4A-GFP with centromeres is lost (Figures 7F and S5A). Our yeast two-hybrid assay also showed that the dimerization mutant of Ccp1 results in loss of interaction between Ccp1 and Pht1 (Figure 7G).
DISCUSSION
Here using a visual genetic screen, we identified Ccp1, a novel NAP family protein that counteracts the loading of CENP-A at centromeres. Our results further reveal that both loading and anti-loading factors of CENP-A are needed at centromeres to mediate the proper level of CENP-A and histone H3, and provide a plausible explanation for how CENP-A and histone H3 are balanced at centromeres.
Ccp1 belongs to the conserved NAP family proteins. NAP family proteins often function as histone chaperones, and play diverse roles, including nucleosome assembly and disassembly, chromatin remodeling and transcription (Park and Luger, 2006, 2008). NAP domain is known to be required and sufficient for histone binding (Fujii-Nakata et al., 1992). It has been shown that NAP1 and Vps75, two NAP family histone chaperones, directly bind the histone H3-H4 tetramer (Bowman et al., 2011; Selth and Svejstrup, 2007). Our results suggest that Ccp1 may function as a CENP-A chaperone specifically used for dissembling the CENP-A nucleosome. Consistent with this, our structural analysis of Ccp1 reveals that, similar to NAP1 and Vps75, Ccp1 forms a homodimer, which contains a pronounced central cleft between the highly acidic earmuff domains. The central cleft in Nap1 and Vps75 has been shown to interact with the histone H3-H4 tetramer (Bowman et al., 2011). We suggest that Ccp1 may directly interact with CENP-A through the central cleft to disassemble CENP-A nucleosomes. In agreement with this idea, we show that the disruption of Ccp1 dimerization results in loss of its ability to antagonize CENP-A loading. We also noticed that, although the overall structure of Ccp1 is similar to the structures of other NAP family proteins, the relative length of the β strands and the dimension of the central cleft differ. These structural differences may account for its role as a CENP-A chaperone in regulating CENP-A homeostasis.
HJURPscm3 is a well-characterized CENP-A chaperone (Dunleavy et al., 2009; Foltz et al., 2009; Pidoux et al., 2009; Williams et al., 2009). Crystal structures of HJURPScm3 in complex with CENP-A/H4 have been reported (Cho and Harrison, 2011; Zhou et al., 2011). These structural studies indicated that the recognition by HJURP is conferred by CENP-A targeting domain (CATD) in CENP-A (Cho and Harrison, 2011; Zhou et al., 2011), consistent with a previously proposed model (Foltz et al., 2009). It will be interesting to investigate in the future whether Ccp1 specificity depends on the CENP-A/H4 structure conferred by the CATD domain.
How is Ccp1 recruited to centromeres? We demonstrated that the temporal localization pattern of Ccp1 to centromeres is the same as the CENP-A chaperone, HJURPscm3. Incorporation of HJURPscm3 at the end of mitosis depends on Mis16-Mis18 complex (Pidoux et al., 2009; Williams et al., 2009; Yu et al., 2015). We found that centromeric incorporation of Ccp1 also requires the Mis16-Mis18 complex. Our results suggest that the Mis16-Mis18 complex promotes targeting of both CENP-A loading and anti-loading factors to centromeres at the end of mitosis (Figure 7H). Coordinated recruitment of factors with opposite activities for CENP-A loading at this stage may ensure that the proper amount of CENP-A is incorporated into centromeres during later stage of cell cycles.
How non-centromeric regions are protected from mistakenly assembling CENP-A is another key question in chromatin regulation. So far, only a few of factors have been shown to be involved in this process. Ubiquitin-mediated proteolysis appears to be a conserved mechanism to inhibit the assembly of CENP-A at ectopic loci by controlling CENP-A protein level (Collins et al., 2004; Gonzalez et al., 2014; Moreno-Moreno et al., 2006). CENP-A also can mis-incorporate to euchromatic region if histone H3 chromatin integrity is perturbed (Au et al., 2008; Choi et al., 2012; Lopes da Rosa et al., 2011). Recently, the H2A.Z homolog Pht1 in fission yeast has been implicated in the exclusion of CENP-A from non-centromeric chromatin (Ogiyama et al., 2013). In this study, we show that, in agreement with Ccp1 as a CENP-A chaperone for dissembling CENP-A nucleosome, Ccp1 is also important for excluding CENP-A from non-centromeric chromatin. Our results further suggest that Ccp1 may cooperate with Pht1 to remove mis-incorporated CENP-A at non-centromeric regions (Figure 7H), and provide new insight into the mechanism underlying prevention of ectopic centromere formation. Redundant pathways involved in excluding the formation of ectopic centromere likely explain the relative small percentage of cells showing CENP-A mislocalization in ccp1Δ. Importantly, we demonstrate that Ccp1 as an anti-CENP-A loading factor helps maintain the integrity of both centromere and non-centromeric regions. Our results suggest that the duality of its function is achieved through its ability to interact with different proteins at different loci (Figure 7H).
One caveat associated with this study is that our results largely rely on GFP-fused CENP-Acnp1. C-terminally tagged CENP-A in Drosophila, mouse and budding yeast is functionally impaired (Kalitsis et al., 2003; Schuh et al., 2007; Wisniewski et al., 2014). We and others found that C-terminal GFP-tagged CENP-Acnp1 in fission yeast is also impaired, but largely functional (Gonzalez et al., 2014; Takayama et al., 2008). Tagging CENP-Acnp1-GFP at N-terminus causes no detectable defects (Takayama et al., 2008), but we cannot rule out the possibility that the N-terminus might have subtle effect on the function of CENP-Acnp1.
Taken together, our findings uncover a critical missing link for how CENP-A homeostasis is maintained at centromeres, and provides insight into how the cells protect themselves from erroneously assembling CENP-A at ectopic regions. Since major features of centromere regulation in S. pombe are conserved, we expect that similar mechanisms are used by other eukaryotes, including humans, for balancing specific CENP-A/H3 ratios at centromeric and non-centromeric loci.
EXPERIMENTAL PROCEDURES
Strains, Media and Genetic Analysis
Fission yeast strains used in this study are listed in Table S1. Standard media and genetic analysis for fission yeast were used (Moreno et al., 1991).
TAP-tag Purification
TAP-tag purification was performed as described (Cheeseman et al., 2001). Briefly, cell lysates in 1× lysis buffer (50 mM bis-Tris propane, pH 7.0, 0.1 M KCl, 5 mM EDTA, 5 mM EGTA and 10% glycerol) were cleared by ultra-centrifugation. After the salt concentration was adjusted to 0.4 M KCl, the extract was incubated with IgG sepharose (GE Healthcare) for 2 hr. Immediately following washing, protein was eluted from the beads and incubated overnight with TEV protease (Invitrogen). S protein agarose beads (Novagen) was then added and incubated for 3 hr. The eluted fractions from S protein agarose were subjected to mass spectrometry analysis at Proteomics Resource Center at NYU School of Medicine (New York, NY).
Immunoprecipitation
Mild over-expression of CENP-Acnp1-GFP was conducted by supplement with 0.05 μM thiamine for 24 hr. Cells were collected and lysed in lysis buffer (20 mM HEPES at pH 7.5, 5 mM EDTA, 100 mM NaCl, 0.1% Nonidet P-40) with 1 mM PMSF and proteinase inhibitor (P8215; Sigma). Lysates were incubated with S-protein agarose (Novagen) or lgG sepharose (GE Healthcare) at 4 °C for 2 h. After washing with lysis buffer three times, proteins were eluted in SDS loading buffer. Eluates were analyzed by Western blotting using commercial anti-S-peptide (MA1-981; Thermo Scientific), Peroxidase Anti-Peroxidase (P1291; Sigma), anti-GFP (11814460001; Roche) antibodies.
Yeast Two-hybrid Assays
Yeast two-hybrid analyses were performed using the ProQuest two-hybrid system according to the manufacturer's instructions (Invitrogen). Details are described in the Supplemental Information.
in vitro pull-down assays
Details on in vitro pull-down assays are described in the Supplemental Information.
Microscopy
Cells were imaged using the Delta Vision System (Applied Precision, Issaquah, WA). Images were taken as z-stacks of 0.2 μm increments with an oil immersion objective (×100), and deconvolved using SoftWoRX2.50 software (Applied Precision). For the candidate gene screen, candidate gene deletion strains carrying CENP-Acnp1-GFP at endogenous level were visually examined by epi-fluorescence microscopy (Olympus BX53).
Crystallization and Structure Determination
Ccp1 was solved by single-wavelength anomalous dispersion. Details for protein purification, crystallization, data collection and refinement are described in the Supplemental Information.
Mutational Studies on the Dimer Interface
The construct for the Ccp1 quadruple mutant (L10A/F17A/I24A/F32A) was generated by PCR with pairs of primers harboring the corresponding mutated nucleotides, and was confirmed by sequencing. The expression and purification for the quadruple mutant protein were conducted in the same way as wild type Ccp1. To confirm the disruption of the dimer formation of the quadruple mutant, ~2 mg proteins from the wild type and quadruple mutant were analyzed by Superdex 200 gel filtration chromatography.
ChIP and ChIP-seq
Details on ChIP and ChIP-seq analysis are described in the Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to M. Yanagida and the Japan Yeast Genetic Resource Center for kindly providing the strains used in this study. We thank B. Ueberheide and Proteomics Resource Center at NYU School of Medicine for the mass spectrometry analysis, and J. Qi for data collection at the Shanghai Synchrotron Radiation Facility (BL17U1). We thank S. Broyde, D.A. Corrales, S. Ercan, C. Hicks, and A. Hochwagen for comments on the manuscript. F. L. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. This work was supported by NIH grant R01GM106037 (to F.L.), NSF grant MCB-1330557 (to F.L.), the CAS Strategic Priority Research Program XDB08020301 (to Y.C.), the National Science Foundation of China grant 31322005 (to Y.C.), the Thousand Young Talents and IGDB Start-up Funds (to Y.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Y.C. and F.L. designed the experiments and wrote the manuscript; Q.D. performed most of the functional experiments in fission yeast with assistance from Y.S., M.G., H.H., J.Y., and S.Z.; Y.F. performed crystallography; G.F., F.Z., L.Y. and M.S. helped to solve the Ccp1 crystal structure and data analysis.
ACCESSION NUMBERS
The atomic coordinates have been deposited in the Protein Data Bank (PDB; accession code: 5GPL and 5GPK). The accession number for the ChIP-seq data reported in this paper is GEO: GSE85166.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, and two tables and can be found with this article online.
REFERENCES
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008;179:263–275. doi: 10.1534/genetics.108.088518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Dawson AR, Rawson DW, Taylor SB, Baker RE, Basrai MA. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics. 2013;194:513–518. doi: 10.1534/genetics.113.149898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, et al. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci. 2012;125:411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. The quantitative architecture of centromeric chromatin. eLife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltengagen M, Huang A, Boltengagen A, Trixl L, Lindner H, Kremser L, Offterdinger M, Lusser A. A novel role for the histone acetyltransferase Hat1 in the CENP-A/CID assembly pathway in Drosophila melanogaster. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A, Ward R, Wiechens N, Singh V, El-Mkami H, Norman DG, Owen-Hughes T. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Berman J. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res. 2012;20:607–619. doi: 10.1007/s10577-012-9296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Straight AF. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Castillo AG, Pidoux AL, Catania S, Durand-Dubief M, Choi ES, Hamilton G, Ekwall K, Allshire RC. Telomeric repeats facilitate CENP-A(Cnp1) incorporation via telomere binding proteins. PLoS One. 2013;8:e69673. doi: 10.1371/journal.pone.0069673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Bowers S, Lipinszki Z, Palladino J, Trusiak S, Bettini E, Rosin L, Przewloka MR, Glover DM, O'Neill RJ, et al. Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription. Dev Cell. 2015;34:73–84. doi: 10.1016/j.devcel.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell. 2003;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci U S A. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ES, Stralfors A, Catania S, Castillo AG, Svensson JP, Pidoux AL, Ekwall K, Allshire RC. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLoS Genet. 2012;8:e1002985. doi: 10.1371/journal.pgen.1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Cottarel G, Shero JH, Hieter P, Hegemann JH. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3342–3349. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter GM, Biggins S. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 2014;28:1815–1826. doi: 10.1101/gad.243113.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Fujii-Nakata T, Ishimi Y, Okuda A, Kikuchi A. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. The Journal of biological chemistry. 1992;267:20980–20986. [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rodriguez M, Jansen LE. Basic properties of epigenetic systems: lessons from the centromere. Curr Opin Genet Dev. 2013;23:219–227. doi: 10.1016/j.gde.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Gonzalez M, He H, Dong Q, Sun S, Li F. Ectopic Centromere Nucleation by CENP-A in Fission Yeast. Genetics. 2014;198:1433–1446. doi: 10.1534/genetics.114.171173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Earle E, Griffiths B, Howman E, Newson AJ, Choo KH. Partially functional Cenpa-GFP fusion protein causes increased chromosome missegregation and apoptosis during mouse embryogenesis. Chromosome Res. 2003;11:345–357. doi: 10.1023/a:1024044008009. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, Almouzni G. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Lopes da Rosa J, Holik J, Green EM, Rando OJ, Kaufman PD. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics. 2011;187:9–19. doi: 10.1534/genetics.110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O, Torras-Llort M, Azorin F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Almouzni G. A network of players in H3 histone variant deposition and maintenance at centromeres. Biochim Biophys Acta. 2014;1839:241–250. doi: 10.1016/j.bbagrm.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R, Senda T, Horikoshi M. Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc Natl Acad Sci U S A. 2007;104:4285–4290. doi: 10.1073/pnas.0603762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiyama Y, Ohno Y, Kubota Y, Ishii K. Epigenetically induced paucity of histone H2A.Z stabilizes fission-yeast ectopic centromeres. Nat Struct Mol Biol. 2013;20:1397–1406. doi: 10.1038/nsmb.2697. [DOI] [PubMed] [Google Scholar]
- Okada M, Okawa K, Isobe T, Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell. 2009;20:3986–3995. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci U S A. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Current opinion in structural biology. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol. 2008;15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Scott KC, Sullivan BA. Neocentromeres: a place for everything and everything in its place. Trends Genet. 2014;30:66–74. doi: 10.1016/j.tig.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L, Svejstrup JQ. Vps75, a new yeast member of the NAP histone chaperone family. The Journal of biological chemistry. 2007;282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan BA. Histone H3K4 methylation keeps centromeres open for business. EMBO J. 2011;30:233–234. doi: 10.1038/emboj.2010.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell. 2008;19:682–690. doi: 10.1091/mbc.E07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Meeth K, Jiang E, Luo C, Marmorstein R. Structure of Vps75 and implications for histone chaperone function. Proc Natl Acad Sci U S A. 2008;105:12206–12211. doi: 10.1073/pnas.0802393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF. The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb Perspect Biol. 2015;7:a015818. doi: 10.1101/cshperspect.a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J, Hajj B, Chen J, Mizuguchi G, Xiao H, Wei D, Dahan M, Wu C. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. eLife. 2014;3:e02203. doi: 10.7554/eLife.02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, et al. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell. 2015;32:68–81. doi: 10.1016/j.devcel.2014.11.030. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.