Abstract
Cotton (Gossypium spp.) is the single most important spinning fiber that has economic significance worldwide. Cotton is one of the most value-added crops and an excellent model system for the analysis of polyploidization and cell development. Thus, the Cotton Genome Consortium has made rapid and significant progress in whole genome sequencing studies in the last decade. Developments in cotton genome sequencing and assembly provide powerful tools for dissecting the genetic and molecular bases of agronomically important traits and establishing regulatory networks on these processes, which leads to molecular breeding. Here, we briefly review these advances, emphasizing their implications in the genetic improvement of cotton with a particular focus on fiber quality and yield. Moreover, major progresses in chloroplast and mitochondrial genomes have also been summarized.
Keywords: Genomes, Functional genomics, Agronomic traits, Molecular breeding, Cotton
Introduction
Cotton is a globally important natural fiber and oilseed crop of crucial economic significance (Chen et al. 2007). The unique history of domestication, special fiber structure, and abundant genetic resources make cotton an excellent model system to study polyploidization, cell elongation, and cell wall biosynthesis (Ruan et al. 2003; Qin and Zhu 2011; Wang et al. 2004; Shan et al. 2014). In the last 20 years, biologists and cotton breeders have made rapid and impressive progress in breeding insect-resistant and herbicide-resistant genetically modified (GM) cotton varieties (Guo et al. 2015; Yu et al. 2015; Malik et al. 2015). However, slow progress has been made in genetic improvements of fiber yield and quality, seed modification for food and feed, and cultivation of an ideal cotton architecture for mechanical harvesting. The successful implementation of Arabidopsis and rice genome projects has paved the way for consortium-based cotton genome research. The availability of these well-established genome sequences has expedited the progress of cotton functional genomics, improved the understanding of the underlying genetic bases of important agronomic traits, and eventually will apply in cotton molecular breeding. Thus, to sequence cotton genomes, the Cotton Genome Consortium was launched in 2007 (Chen et al. 2007). Recently, many publications have provided draft genome sequences of Gossypium, which have implications for genetic improvements and genomics-based breeding (Zhang et al. 2015; Wang et al. 2012a; Liu et al. 2015; Li et al. 2014, 2015).
Genome sequence of Gossypium provides genomic resources for genetic improvement
The Gossypium genus constitutes six tetraploid ((AtDt)1 to (AtDt)5, where ‘t’ indicates the tetraploid, 2n = 4x = 52) and at least 46 diploid (2n = 2x = 26) species, which are believed to have evolved from a common ancestor approximately 5–10 million years ago (MYA) (Grover et al. 2008; Chen et al. 2007). According to their evolutionary relationships and geographic distribution, diploid cotton species have been classified into eight genomic groups (A-G and K) (Cao 2015; Page et al. 2013). They share the same chromosome number (n = 13) and have significant genomic synteny with each other (Rong et al. 2004; Desai et al. 2006). Cultivated tetraploid cotton originated approximately 1–2 MYA via hybridization and subsequent allopolyploidization between the native D-genome and after transoceanic dispersion of the A-genome ancestor to the New World (Wendel 1989; Paterson and Wendel 2015). Domestication and artificial selection resulted in two major cultivated tetraploid cotton species, G. hirsutum and G. barbadense, that have higher fiber yields and quality than improved diploids (Reinisch et al. 1994; Sunilkumar et al. 2006). The recently released genome sequences of two diploid progenitor species, G. raimondii and G. arboretum, have provided pivotal insight into cotton evolution and the dynamics of genome structures. Additionally, the assembly of two tetraploids, G. hirsutum and G. barbadense, has opened new avenues for revealing the allotetraploid formation and molecular regulatory mechanisms for some important traits, particularly fiber development.
G. raimondii
For the long-term goal of sequencing cotton genomes, G. raimondii (accession D 5–3, DD), a putative contributor of the D-subgenome, was prioritized by the worldwide cotton community. The integrated genetic-physical map of G. raimondii, which was reported in 2010, provided important information for future assembly and for validating the reference sequence (Lin et al. 2010). Two synchronous but independent studies that reported the draft genomes of G. raimondii were released in 2012 (Wang et al. 2012a; Paterson et al. 2012). Wang and colleagues found that over 73 % of the 78.7-Gb paired-end Illumina reads that covered 103.6-fold of the 775.2-Mb assembled G. raimondii genome were anchored to 13 euchromosomes (Table 1). The genome contained 40,976 predicted protein-coding genes, and 92.2 % of them were confirmed by transcriptome sequencing data, which demonstrated the high accuracy of the gene predictions. A total of 13 pseudomolecules that had 737.8 Mb, 37,505 genes, and 77,267 protein-coding transcripts were annotated (Wang et al. 2012a). Paterson and colleagues also performed a genome assembly of G. raimondii Ulbr with approximately eight longer N50 scaffolds and that were oriented to 98.3 % of the genome (Paterson et al. 2012). Further, the Gossypium genus is the only sequenced plant species that contains an actual CDN1 gene family for gossypol biosynthesis. Importantly, many gene families, such as Sus (sucrose synthase), KCS (3-ketoacyl-CoA synthase), bHLH, and MYB, are expressed predominantly in G. hirsutum ovules, but there are low levels of transcript in the ovules of G. raimondii (Wang et al. 2012a). Additionally, Paterson and colleagues found abundant mutation hotspots in the A-subgenome lineage within the fiber-related quantitative trait locus (QTLs) in tetraploid cotton, which may affect multiple fiber traits. Taken together, these studies imply that some of these genes may act as important regulators that are responsible for fiber cell initiation and elongation, and changes in gene-coordinated expression during domestication is an important contribution to cotton fiber development. Dissecting the G. raimondii genome is a milestone not only because of its templated significance for analyzing largely parallel re-sequencing data from tetraploids but also because of its importance for tracing the origin of genome segments and homologous genes in tetraploid cotton.
Table 1.
A summary annotation of Gossypium genomes, major genes, and agronomically important traits in different cotton species
| G. raimondii | G. arboreum | G. hirsutum | G. barbadense | |
|---|---|---|---|---|
| Variety | D5-3 | Shixiya1 | TM-1 | Xinhai2&3-79 |
| Chromosome number | 26 | 26 | 52 | 52 |
| Chromosomal pattern | DD | AA | (AtDt)1 | (AtDt)2 |
| Genomes size (Mb) | 775.2/737.8 | 1694 | 2173 | 2470/2570 |
| Genes annotated | 40,976/37,505 | 41,330 | 76,943 | 77,526/80,876 |
| LTR size (Mb) | 348/391 | 1145 | 1471 | N/A |
| Number of LTR-retros | 2992/2345 | 8620 | 8624 | 8436 |
| Retrotransposon activity | Stabilization | Very activation | Activation | Activation |
| Fiber length | No | Short (1.3–1.5 cm) | Long (>3 cm) | Extra-long |
| Verticillium resistance | Resistant | Susceptible | Resistant | Resistant |
| Reference | Paterson et al. (2012) and Wang et al. (2012a) | Li et al. (2014) | Li et al. (2015) and Zhang et al. (2015) | Liu et al. (2015) and Yuan et al. (2015) |
t tetraploid
G. arboreum
Both the A- and D-genome diploid species of spinnable fibers are produced by G. arboreum (AA), which are planted on a small scale, whereas G. raimondii hosts some agronomically important traits (Li et al. 2014; Malik et al. 2014). Two years after the publication of the G. raimondii genome, the same researchers sequenced and assembled the genome of the cultivated G. arboreum, Shixiya1 (Li et al. 2014). The quantity of repetitive sequence, including long terminal repeat (LTR)-type retrotransposons, increased from approximately 348 Mb in G. raimondii to 1145 Mb in G. arboreum, which contributed to the formation of the double-sized G. arboreum genome relative to G. raimondii. However, the amount of protein-coding genes in these two species was highly conserved. Furthermore, many syntenic blocks were observed between the two diploid species (Table 1). Strikingly, the ACO (1-aminoyclo-propane-1-carboxylic acid oxidase) gene family, which is involved in the last rate-limiting step in ethylene biosynthesis, exhibited a greater variation between the promoter regions of G. arboreum and G. raimondii. This may be pivotally related to the cotton fiber development. In addition, significant qualitative differences in the TN and TNL subfamilies of NBS-encoding type R gene transcripts were observed in G. arboreum compared with G. raimondii. The rapid expansion and contraction of these R genes in different cotton species may be responsible for difference resistances to Verticillium dahlia. These valuable cotton genes provide a more effective tool for high-yield and disease-resistant genetic engineering.
G. hirsutum
Upland cotton, G. hirsutum (AtDt)1, is widely cultivated in over 80 countries in approximately 33 million ha (5 % of the arable land on earth) (Shan et al. 2014) and it provides more than 90 % of the world’s raw cotton fiber. The annual global economic benefits for the textile industry are approximately 630 billion US$ (Cao 2015; Chen et al. 2007). Compared to diploid cotton species, G. hirsutum exhibited significant differences in both plant morphology and economic characteristics, implying that rigorous natural and artificial selection has occurred during evolution. Using the sequence base of two progenitor species, two synchronous but independent cotton genome teams simultaneously completed the genome sequence of the same allotetraploid cultivar, Texas Marker Stock (TM)-1, using second-generation high-throughput sequencing technology that was assisted by traditional paired bacterial artificial chromosome (BAC)-end sequences (Zhang et al. 2015; Li et al. 2015). Despite several differences between the two genomes, such as the number of annotated genes and abundance of retrotransposons and DNA transposons, there are several reasons to convince scientists that the assembly and analyses of the allotetraploid upland cotton genome will begin a new era in functional genomics and marker-assisted breeding (Table 1). First, upland cotton genomes provide not only insights into cotton structural genomics and evolution but also are an important clue for dissecting allopolyploid formation. Second, widespread sharing of G. hirsutum genomes will provide new abilities to screen key target genes that are important for fiber development, seed formation, and cotton plant architecture. This will further unravel the molecular mechanisms that underlie these processes. Third, these genomes provide a valuable template for genotyping, phenotyping, and genome-wide association studies that aim to identify important targets of regulators and accelerate cotton crop improvement. Finally, combined with the increased insight, these genomic data and resources will gradually facilitate genomic-assisted cotton breeding from theory to practice. However, understanding whole cotton genomes will initiate a new way of breeding superior cotton cultivars.
G. barbadense
Sea island cotton, G. barbadense (AtDt)2, contributes to approximately 5 % of the annual world cotton production and is famous for its high-quality extra-long fiber for the production of high-grade textiles (Wang et al. 2012b; Reinisch et al. 1994). The quality of fiber is largely dependent on three key factors: fiber length, strength, and fineness. The recently released genome sequences of G. barbadense, Xinhai21 (Liu et al. 2015) and 3-79 (Yuan et al. 2015), provided important insight into the molecular mechanisms that are required for superior quality fiber development. Liu and colleagues found that among the 77,526 predicted protein-coding genes of G. barbadense, 2483 and 1879 genes are highly and specifically expressed in the fibers and ovules, respectively (Table 1) (Liu et al. 2015). Similarly, Yuan and colleagues identified 708 and 425 homologous gene pairs that are specifically or preferentially expressed in the fiber development stages among the 80,876 predicted protein-coding genes, respectively (Table 1) (Yuan et al. 2015). Interestingly, 58 % of the fiber elongation-related genes and 67 % of the secondary cell wall synthesis-related genes were exclusively either At-biased or Dt-biased (Yuan et al. 2015). These phenomena were also observed in Liu’s studies (Liu et al. 2015). For example, a group of positive cell elongation regulators, Paclobutrazol Resistance (PRE), are significantly A-subgenome tendentious and fiber specific, but the homolog transcripts in the D-subgenome were undetectable, which further supports the origin of the spinnable fiber to be from the A-genome. The expansion, translocation, and subsequent selection of the PRE genes in G. barbadense suggested a genetic factor that is responsible for the rate and duration of fiber elongation-determined. Additionally, two independent studies found that compared with G.hirsutum, several genes of cellulose synthases (CesAs), such as CesA4, CesA7, and CesA8, underlie secondary cell wall biosynthesis and affect fiber strength and fineness and exhibited a laggard but more significant up-regulation in developing fiber cells of G. barbadense. Functional allocation of CesA members conferred extended duration of the elongation stage and highly active secondary wall deposition during the course of extra-long fiber development. Overall, this draft sequence provides valuable information regarding the genes involved in cotton fiber development, and sequencing of the G. barbadense genome will facilitate breeding practices aimed at superior fiber quality.
Ultra-dense genetic linkage maps serve as a valuable tool for cotton genetic research and mark-assisted breeding
Considering the obvious differences in plant characteristics, interspecific crosses between G. hirsutum and G. barbadense have been widely used for constructing genetic linkage maps, gene cloning, and QTLs that correspond to important agronomic traits. Prior to the release of the four draft sequences of Gossypium genome, limited ultra-precision genetic maps were the primary bottleneck that prevented deeper genetic research and breeding of cotton varieties that have a high-yield and extra-long fibers. A comprehensive study was conducted by Wang and colleagues to review the cotton genome structure, and a total of 4,999,048 single nucleotide polymorphisms (SNPs) were detected in 59 interspecific F2 individuals, and two parents, G. hirsutum acc. TM-1 and G. barbadense cv. Hai7124 (Wang et al. 2015a). Genomic analysis revealed that these SNPs covered a total of 4042 cM but were distributed disproportionally in 26 allotetraploid cotton linkage groups. Meanwhile, another high-density genetic map was developed using simple sequence repeat (SSR) and SNP markers that were derived from an interspecific cross between the sequenced cotton varieties, TM-1 and 3-79. Using the genotype 186 recombinant inbred lines (RILs), 2,027 loci were mapped to 26 chromosomes with an average marker interval of 1.63 cM (Yu et al. 2012). In addition, a high-resolution intraspecific linkage genetic map between the two upland cotton cultivars, Acala Prema and 86-1, was constructed using restriction site-associated DNA (RAD) sequencing technology (Wang et al. 2015b). Combined with the two interspecific maps, these studies indicated a high degree of collinearity in the marker order between these two maps. These high-precision genetic and physical maps provide a useful tool to reveal genome rearrangements, to rectify the anomalies in the cotton reference genome assemblies, to merge scaffolds into pseudomolecules that correspond to the chromosomes, and to detect centromeric regions in allotetraploid cottons. These high-precision genetic maps will not only provide a valuable genomic resource to enhance our understanding of polypoid genome structure, evolution, and tagging of the genome-wide linkage disequilibrium but will also open new avenues for marker-assisted selection in cotton and cloning of QTLs for valued traits.
The complete chloroplast and mitochondrial genomes of Gossypium provide insight into the cytoplasmic–nuclear interaction
Chloroplast (cp) and mitochondria (mt) DNA are maternally inherited. A low rate of nucleotide variation in the cp and mt genomes is a compelling reason for their use in plant evolutionary analysis. Currently available evidence indicates that cytoplasmic-nuclear incompatibility causes cytoplasmic male sterility (CMS) in plants (Wu et al. 2015). Chloroplast genetic engineering provides a new strategy to confer plant herbicide and insect resistance and abiotic tolerance (Jin and Daniell 2015).
The cp genome sequences of G. hirsutum and G. barbadense were published in 2006 (Lee et al. 2006; Ibrahim et al. 2006). Soon afterwards, large-scale cp genome sequence variations among the different cotton species were investigated by re-sequencing two A-genome lineages, two D-genome lineages, two G. hirsutum, three G. barbadense, and three wild allotetraploid cottons accessions (Xu et al. 2012). Combined with the aforementioned two cp genomes, the complete length of 14 cp genomes varied from 159,959 to 160,433 with the same set of 112 unique genes and 19 duplicated genes. A general comparison indicated that there was a high similarity among the different cotton cp genomes regarding genome size, genome structure, gene number, and gene order. However, the boundary junctions between the inverted repeat (IR) and small single copy (SSC) regions were different. The lower proportion of the divergence in the protein-coding regions suggested that natural variations in the major part of the cp genome that are responsible for cotton co-evolution and domestication should be attributed to the intergenic regions. The completion of the Gossypium cp genome sequencing provided vital information for both fundamental theoretical research and practical application. In particular, the available cotton cp sequences provided useful information for chloroplast genetic engineering to confer beneficial agronomic traits and serve as bioreactors for valuable bio-production.
The mitochondrion is the core manufacturer of cellular ATP in eukaryotes. In the practical breeding programs, the detrimental mt-nuclear interaction induces plant CMS, enabling ‘three-line’ hybrid development to increase plant yield. To gain insights into the Gossypium mt genomes and structure, Hua’s group first completed the sequencing of the upland cotton mt genome, and they identified a total of 35 coding-protein genes, 4rRNA genes, and 29 tRNA genes (Liu et al. 2013). Like cp, the cotton mt genome possesses the majority of the same characteristics as other higher plants. The existence and conservation of syntenic gene clusters, intergenic sequences, and genetic contents in the cotton mt genome indicated that the evolution of mt genomes is largely congruent with plant taxonomy. Importantly, the research on the cotton mt genome will not only help us to identify the effect of male-sterile genes on the stamen in the male-sterile line but also provide insights into the mechanism of the nucleo-cytoplasmic interaction in plants.
What is the future for breeding superior cotton varieties based on these draft genome sequences?
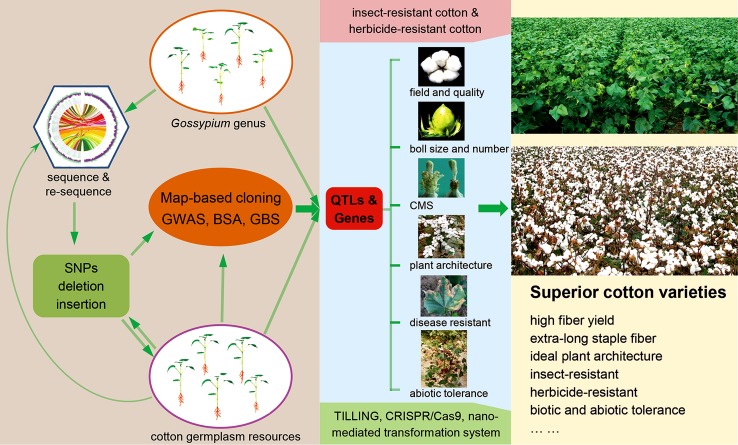
Whole genome sequencing has paved the way for cotton improvement from the bench to molecular breeding. The implementation of genome-assisted molecular breeding must include three coordinated steps (Fig. 1). First, the development of an efficient system to mine major genes and QTLs for agronomical important traits is required. As demonstrated by recent studies in Arabidopsis (Koornneef et al. 2004), rice (Zuo and Li 2014; Huang and Han 2014), and maize (Yang et al. 2013; Jiao et al. 2012), because of the rapid advances in whole genome sequencing, genetic mapping of mutations has been revolutionized by map-based cloning, bulk segregant analysis (BSA), genotyping-by-sequencing (GBS), and genome-wide association studies (GWAS). There is no doubt that sequenced-based high-precision molecular markers of intraspecific and interspecific populations of cotton will play a crucial role in map-based cloning QTLs and major genes. Unlike Arabidopsis and rice, the progress of mutant population generation has not occurred at the same pace because of several factors, including complex and huge genome and the low efficiency of genetic transformation. Therefore, in the coming years, the dissection and cloning of complex QTLs control phenotypic variations, especially agronomically important traits within the germplasm, will be a major focus and great challenge for cotton genetic studies and molecular breeding. The completion of cotton whole genome sequencing will enable rapid identification of SNPs or small indels on a genome-wide and population-wide scale (Hulse-Kemp et al. 2015). Consequently, in light of model plant results, whole genome genotyping of hundreds of cotton varieties with well-characterized phenotypes will be feasible for mapping important agronomic traits. Second, the development of an efficient system to identify genes and QTLs that can be manipulated is required. The forward genetic approach and reverse genetic approach, such as mutants, near-isogenic lines (NILs), and transgenic approach, are essential methods for gene functional analysis. However, in many cases, a complex trait is usually regulated by multiple loci, whereas a single gene is also directly or indirectly involved in controlling different phenotypes. Therefore, experimental studies on the biological functions of genes are facing enormous challenges. Recently, several scientific research groups have been in the process of developing a multi-scale crop system (Long et al. 2015; Chu 2015; Chew et al. 2014), such as the ePlant model (Zhu et al. 2013), which can be used for high-throughput and systematic association studies of multiple loci and complex traits. Finally, the development of an efficient system for molecular breeding by multiple function modules is necessary. The widespread use of recently developed genome editing technologies, such as the clustered regularly interspaced short palindromic repeats associated system (CRISPR-Cas9) (Shan et al. 2013), transcription activator-like effector nucleases (TALENs) (Miller et al. 2011), and target-induced local lesions in genomes (TILLING) (McCallum et al. 2000), as well as the nanocarrier-mediated genetic transformation system, provide excellent alternatives for cotton improvement. The studies of genome-based molecular breeding in rice and maize should be effectively utilized to accelerate cotton breeding. However, unlike rice and maize, transgenic cotton, including Bt-toxin-containing cotton, herbicide tolerance, and the stacked traits of insect-resistant and herbicide-tolerant cotton have been successfully commercialized in the past 20 years (Guo et al. 2015; Meng et al. 2015). Thus, scientifically, fast and fluent communication and feedback channels between molecular biologists and cotton breeders need to be established to better guide and facilitate breeding and engineering to produce ideal agronomic cotton traits in the future.
Fig. 1.
A scheme of the sequential research processes of genome-based breeding from whole genome sequencing to practical engineering in cotton. SNPs single nucleotide polymorphisms, GWAS genome-wide association study, BSA bulked segregant analysis, GBS genotyping-by-sequencing, CMS cytoplasmic male sterility, TILLING targeting induced local lesions in genomes. The diagram of the genomes is reproduced from Li et al. (2014)
Challenges and future perspectives
During the past decade, we have witnessed tremendous progress in sequencing and assembly of whole cotton genomes, including two diploid cotton species, two tetraploid cotton species, and several Gossypium cp and mt genomes. These achievements have made breakthroughs in several important fields, including the identification and characterization of several important genes or gene families. These achievements have included the regulation of fiber development and gossypol biosynthesis, dissection of signal pathway responses to Verticillium wilt resistance and abiotic stresses, and the elucidation of regulatory mechanisms of genome evolution, domestication, and polyploidization. These achievements will serve as a bridge between comparative genomics, functional genomics, and modern cotton breeding. However, compared to Arabidopsis and rice, there are still large knowledge gaps regarding the molecular regulation of the basic biological processes that are related to important agronomic traits. These gaps exist because of the difficulty to identify the targets of key regulators underlying these processes and to establish a quick and simple genetic transformation system. Obviously, cloning and functional characterization of more pivotal genes and QTLs that control complex physiological and agronomy traits will be a major challenge for future studies.
The ample quantitative differences in agronomic traits, especially those involved in fiber quality and yield between the intra- and inter-species of Gossypium, represent variables that can be improved upon in breeding programs. The accumulation of genomic and functional genomic studies will be multiplied upon in the next generation of cotton cultivars. These data will ideally result in extra-long fibers, high fiber yields, and improved plant architectures that are suitable for mechanical harvesting. In addition, knowledge from systems biology approaches, especially from different omics studies, such as transcriptomics, proteomics, and metabolomics, will allow us to dissect the related traits in more detail and explore the regulation pathways and networks governing these traits. Therefore, there is no doubt that with the continuous development of cotton genome sequencing and genome editing technologies, as well as improved transgenic technology and use of marker-assisted selection, we can innovate germplasms and cotton breeding in effective ways.
Acknowledgments
We would like to apologize to all the investigators whose research could not be cited appropriately owing to space limitations. We are grateful to Dr. Yiqin Wang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for her critical comments on the manuscript.
Compliance with ethical standards
Funding
This work was supported by grants from the Ministry of Agriculture of China (Grant No. 2013ZX08005004, 2016ZX08005004, 2016ZX08009003-003-004).
Conflict of interest
The authors declare no competing financial interests.
Footnotes
Rong Yan and Chengzhen Liang contributed equally to this work.
Contributor Information
Xuefeng Zong, Email: zxfeng@swu.edu.cn.
Sandui Guo, Phone: +86-10-82106128, Email: guosandui@caas.cn.
Rui Zhang, Phone: +86-10-82106127, Email: zhangrui@caas.cn.
References
- Cao X. Whole genome sequencing of cotton—a new chapter in cotton genomics. Sci China Life Sci. 2015;58(5):515–516. doi: 10.1007/s11427-015-4862-z. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD, Arioli T, Brubaker C, Cantrell RG, Lacape JM, Ulloa M, Chee P, Gingle AR, Haigler CH, Percy R, Saha S, Wilkins T, Wright RJ, Van Deynze A, Zhu Y, Yu S, Abdurakhmonov I, Katageri I, Kumar PA, Mehboob Ur R, Zafar Y, Yu JZ, Kohel RJ, Wendel JF, Paterson AH. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007;145(4):1303–1310. doi: 10.1104/pp.107.107672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew YH, Wenden B, Flis A, Mengin V, Taylor J, Davey CL, Tindal C, Thomas H, Ougham HJ, de Reffye P, Stitt M, Williams M, Muetzelfeldt R, Halliday KJ, Millar AJ. Multiscale digital Arabidopsis predicts individual organ and whole-organism growth. Proc Natl Acad Sci USA. 2014;111(39):E4127–E4136. doi: 10.1073/pnas.1410238111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C. A new era for crop improvement: from model-guided rationale design to practical engineering. Mol Plant. 2015;8(9):1299–1301. doi: 10.1016/j.molp.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Desai A, Chee PW, Rong J, May OL, Paterson AH. Chromosome structural changes in diploid and tetraploid A genomes of Gossypium. Genome. 2006;49(4):336–345. doi: 10.1139/G05-116. [DOI] [PubMed] [Google Scholar]
- Grover CE, Yu Y, Wing RA, Paterson AH, Wendel JF. A phylogenetic analysis of indel dynamics in the cotton genus. Mol Biol Evol. 2008;25(7):1415–1428. doi: 10.1093/molbev/msn085. [DOI] [PubMed] [Google Scholar]
- Guo S, Wang Y, Sun G, Jin S, Zhou T, Meng Z, Zhang R. Twenty years of research and application of transgenic cotton in China. Sci Agi Sin. 2015;48(17):3372–3387. [Google Scholar]
- Huang X, Han B. Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol. 2014;65:531–551. doi: 10.1146/annurev-arplant-050213-035715. [DOI] [PubMed] [Google Scholar]
- Hulse-Kemp AM, Lemm J, Plieske J, Ashrafi H, Buyyarapu R, Fang DD, Frelichowski J, Giband M, Hague S, Hinze LL, Kochan KJ, Riggs PK, Scheffler JA, Udall JA, Ulloa M, Wang SS, Zhu QH, Bag SK, Bhardwaj A, Burke JJ, Byers RL, Claverie M, Gore MA, Harker DB, Islam, Jenkins JN, Jones DC, Lacape JM, Llewellyn DJ, Percy RG, Pepper AE, Poland JA, Mohan Rai K, Sawant SV, Singh SK, Spriggs A, Taylor JM, Wang F, Yourstone SM, Zheng X, Zheng X, Lawley CT, Ganal MW, Van Deynze A, Wilson IW, Stelly DM. Development of a 63 K SNP array for cotton and high-density mapping of intraspecific and interspecific populations of Gossypium spp. G3 Bethesda. 2015;5(6):1187–1209. doi: 10.1534/g3.115.018416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim RI, Azuma J, Sakamoto M. Complete nucleotide sequence of the cotton (Gossypium barbadense L.) chloroplast genome with a comparative analysis of sequences among 9 dicot plants. Genes Genet Syst. 2006;81(5):311–321. doi: 10.1266/ggs.81.311. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Zhao H, Ren L, Song W, Zeng B, Guo J, Wang B, Liu Z, Chen J, Li W, Zhang M, Xie S, Lai J. Genome-wide genetic changes during modern breeding of maize. Nat Genet. 2012;44(7):812–815. doi: 10.1038/ng.2312. [DOI] [PubMed] [Google Scholar]
- Jin S, Daniell H. The engineered chloroplast genome just got smarter. Trends Plant Sci. 2015;20(10):622–640. doi: 10.1016/j.tplants.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genom. 2006;7:61. doi: 10.1186/1471-2164-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, Chen W, Liang X, Shang H, Liu W, Shi C, Xiao G, Gou C, Ye W, Xu X, Zhang X, Wei H, Li Z, Zhang G, Wang J, Liu K, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, Liang X, Huang G, Percy RG, Liu K, Yang W, Chen W, Du X, Shi C, Yuan Y, Ye W, Liu X, Zhang X, Liu W, Wei H, Wei S, Huang G, Zhang X, Zhu S, Zhang H, Sun F, Wang X, Liang J, Wang J, He Q, Huang L, Wang J, Cui J, Song G, Wang K, Xu X, Yu JZ, Zhu Y, Yu S. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33(5):524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- Lin L, Pierce GJ, Bowers JE, Estill JC, Compton RO, Rainville LK, Kim C, Lemke C, Rong J, Tang H, Wang X, Braidotti M, Chen AH, Chicola K, Collura K, Epps E, Golser W, Grover C, Ingles J, Karunakaran S, Kudrna D, Olive J, Tabassum N, Um E, Wissotski M, Yu Y, Zuccolo A, Rahman M, Peterson DG, Wing RA, Wendel JF, Paterson AH. A draft physical map of a D-genome cotton species (Gossypium raimondii) BMC Genom. 2010;11:395. doi: 10.1186/1471-2164-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Cao D, Li S, Su A, Geng J, Grover CE, Hu S, Hua J. The complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PLoS ONE. 2013;8(8):e69476. doi: 10.1371/journal.pone.0069476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhao B, Zheng HJ, Hu Y, Lu G, Yang CQ, Chen JD, Chen JJ, Chen DY, Zhang L, Zhou Y, Wang LJ, Guo WZ, Bai YL, Ruan JX, Shangguan XX, Mao YB, Shan CM, Jiang JP, Zhu YQ, Jin L, Kang H, Chen ST, He XL, Wang R, Wang YZ, Chen J, Wang LJ, Yu ST, Wang BY, Wei J, Song SC, Lu XY, Gao ZC, Gu WY, Deng X, Ma D, Wang S, Liang WH, Fang L, Cai CP, Zhu XF, Zhou BL, Jeffrey Chen Z, Xu SH, Zhang YG, Wang SY, Zhang TZ, Zhao GP, Chen XY. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci Rep. 2015;5:14139. doi: 10.1038/srep14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Marshall-Colon A, Zhu XG. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell. 2015;161(1):56–66. doi: 10.1016/j.cell.2015.03.019. [DOI] [PubMed] [Google Scholar]
- Malik W, Ashraf J, Iqbal MZ, Khan AA, Qayyum A, Ali Abid M, Noor E, Ahmad MQ, Abbasi GH. Molecular markers and cotton genetic improvement: current status and future prospects. Sci World J. 2014;2014:607091. doi: 10.1155/2014/607091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik W, Abid MA, Cheema HMN, Khan AA, Iqbal MZ, Qayyum A, Hanif M, Bibi N, Yuan SN, Yasmeen A, Mahmood A, Ashraf J. From Qutn to Bt cotton: development, adoption and prospects. A review. Cytol Genet. 2015;49(6):408–419. doi: 10.3103/S0095452715060055. [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 2000;123(2):439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Meng Z, Zhang R, Liang C, Wan J, Wang Y, Zhai H, Guo S. Expression of the rice arginase gene OsARG in cotton influences the morphology and nitrogen transition of seedlings. PLoS ONE. 2015;10(11):e0141530. doi: 10.1371/journal.pone.0141530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Page JT, Huynh MD, Liechty ZS, Grupp K, Stelly D, Hulse AM, Ashrafi H, Van Deynze A, Wendel JF, Udall JA. Insights into the evolution of cotton diploids and polyploids from whole-genome re-sequencing. G3 (Bethesda) 2013;3(10):1809–1818. doi: 10.1534/g3.113.007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Wendel JF. Unraveling the fabric of polyploidy. Nat Biotechnol. 2015;33(5):491–493. doi: 10.1038/nbt.3217. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- Qin YM, Zhu YX. How cotton fibers elongate: a tale of linear cell-growth mode. Curr Opin Plant Biol. 2011;14(1):106–111. doi: 10.1016/j.pbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Reinisch AJ, Dong JM, Brubaker CL, Stelly DM, Wendel JF, Paterson AH. A detailed RFLP map of cotton, Gossypium hirsutum x Gossypium barbadense: chromosome organization and evolution in a disomic polyploid genome. Genetics. 1994;138(3):829–847. doi: 10.1093/genetics/138.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J, Abbey C, Bowers JE, Brubaker CL, Chang C, Chee PW, Delmonte TA, Ding X, Garza JJ, Marler BS, Park CH, Pierce GJ, Rainey KM, Rastogi VK, Schulze SR, Trolinder NL, Wendel JF, Wilkins TA, Williams-Coplin TD, Wing RA, Wright RJ, Zhao X, Zhu L, Paterson AH. A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium) Genetics. 2004;166(1):389–417. doi: 10.1534/genetics.166.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT. Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell. 2003;15(4):952–964. doi: 10.1105/tpc.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- Shan CM, Shangguan XX, Zhao B, Zhang XF, Chao LM, Yang CQ, Wang LJ, Zhu HY, Zeng YD, Guo WZ, Zhou BL, Hu GJ, Guan XY, Chen ZJ, Wendel JF, Zhang TZ, Chen XY. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat Commun. 2014;5:5519. doi: 10.1038/ncomms6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci U S A. 2006;103(48):18054–18059. doi: 10.1073/pnas.0605389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell. 2004;16(9):2323–2334. doi: 10.1105/tpc.104.024844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44(10):1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhu Y, Song X, Cao Z, Ding Y, Liu B, Zhu X, Wang S, Guo W, Zhang T. Inheritance of long staple fiber quality traits of Gossypium barbadense in G. hirsutum background using CSILs. Theor Appl Genet. 2012;124(8):1415–1428. doi: 10.1007/s00122-012-1797-7. [DOI] [PubMed] [Google Scholar]
- Wang S, Chen J, Zhang W, Hu Y, Chang L, Fang L, Wang Q, Lv F, Wu H, Si Z, Chen S, Cai C, Zhu X, Zhou B, Guo W, Zhang T. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 2015;16:108. doi: 10.1186/s13059-015-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ning Z, Hu Y, Chen J, Zhao R, Chen H, Ai N, Guo W, Zhang T. Molecular mapping of restriction-site associated DNA markers in allotetraploid upland cotton. PLoS ONE. 2015;10(4):e0124781. doi: 10.1371/journal.pone.0124781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. New world tetraploid cottons contain old world cytoplasm. Proc Natl Acad Sci USA. 1989;86(11):4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun Y, Zhao Y, Zhang J, Luo L, Li M, Wang J, Yu H, Liu G, Yang L, Xiong G, Zhou JM, Zuo J, Wang Y, Li J. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Res. 2015;25(5):621–633. doi: 10.1038/cr.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Xiong G, Li P, He F, Huang Y, Wang K, Li Z, Hua J. Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: origin and evolution of allotetraploids. PLoS ONE. 2012;7(8):e37128. doi: 10.1371/journal.pone.0037128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Li Z, Li W, Ku L, Wang C, Ye J, Li K, Yang N, Li Y, Zhong T, Li J, Chen Y, Yan J, Yang X, Xu M. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci USA. 2013;110(42):16969–16974. doi: 10.1073/pnas.1310949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JZ, Kohel RJ, Fang DD, Cho J, Van Deynze A, Ulloa M, Hoffman SM, Pepper AE, Stelly DM, Jenkins JN, Saha S, Kumpatla SP, Shah MR, Hugie WV, Percy RG. A high-density simple sequence repeat and single nucleotide polymorphism genetic map of the tetraploid cotton genome. G3 (Bethesda) 2012;2(1):43–58. doi: 10.1534/g3.111.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LH, Wu SJ, Peng YS, Liu RN, Chen X, Zhao P, Xu P, Zhu JB, Jiao GL, Pei Y, Xiang CB. Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol J. 2015 doi: 10.1111/pbi.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D, Tang Z, Wang M, Gao W, Tu L, Jin X, Chen L, He Y, Zhang L, Zhu L, Li Y, Liang Q, Lin Z, Yang X, Liu N, Jin S, Lei Y, Ding Y, Li G, Ruan X, Ruan Y, Zhang X. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep. 2015;5:17662. doi: 10.1038/srep17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- Zhu XG, Wang Y, Ort DR, Long SP. e-Photosynthesis: a comprehensive dynamic mechanistic model of C3 photosynthesis: from light capture to sucrose synthesis. Plant, Cell Environ. 2013;36(9):1711–1727. doi: 10.1111/pce.12025. [DOI] [PubMed] [Google Scholar]
- Zuo J, Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet. 2014;48:99–118. doi: 10.1146/annurev-genet-120213-092138. [DOI] [PubMed] [Google Scholar]