Abstract
Purpose:
With raw digital mammograms (DMs), which retain the relationship with x-ray attenuation of the breast tissue, not being routinely available, processed DMs are often the only viable means to acquire imaging measures. The authors investigate differences in quantitative measures of breast density and parenchymal texture, shown to have value in breast cancer risk assessment, between the two DM representations.
Methods:
The authors report data from 8458 pairs of bilateral raw (“FOR PROCESSING”) and processed (“FOR PRESENTATION”) DMs acquired from 4278 women undergoing routine screening evaluation, collected with DM units from two different vendors. Breast dense tissue area and percent density (PD), as well as a range of quantitative descriptors of breast parenchymal texture (statistical, co-occurrence, run-length, and structural descriptors), were measured using previously validated, fully automated software. Feature measurements were compared using matched-pairs Wilcoxon signed-ranks test, correlation (r), and linear-mixed-effects (LME) models, where potential interactions with woman- and system-specific factors were also assessed. The authors also compared texture feature correlations with the established risk factors of the Gail lifetime risk score (rG) and breast PD (rPD), and evaluated the within woman intraclass feature correlation (ICC), a measure of bilateral breast-tissue symmetry, in raw versus processed images.
Results:
All density measures and most of the texture features were strongly (r ≥ 0.6) or moderately (0.4 ≤ r < 0.6) correlated between raw and processed images. However, measurements were significantly different between the two imaging formats (Wilcoxon signed-ranks test, pw < 0.05). The association between measurements varied across features and vendors, and was substantially modified by woman- and system-specific image acquisition factors, such as age, BMI, and mAs/kVp, respectively. The strongest correlation, combined with minimal LME-model interactions, was observed for structural texture features. Overall, texture measures from either image representation were weakly associated with Gail lifetime risk (−0.2 ≤ rG ≤ 0.2), weakly to moderately associated with breast PD (−0.6 ≤ rPD ≤ 0.6), and had overall strong bilateral symmetry (ICC ≥ 0.6).
Conclusions:
Differences in measures from processed versus raw DM depend highly on the feature, the DM vendor, and image acquisition settings, where structural features appear to be more robust across the different DM settings. The reported findings may serve as a reference in the design of future large-scale studies on mammographic features and breast cancer risk assessment involving multiple DM representations.
Keywords: digital mammography, breast density, parenchymal texture, image representation
1. INTRODUCTION
With 1.67 × 106 breast cancer cases diagnosed annually worldwide and a sharp increase (20%) in incidence of the disease since 2008,1 which is projected to rise further over the next two decades,2 early breast cancer detection is a major clinical challenge. Mammography remains the cornerstone for early breast cancer detection,3,4 and the transition from screen-film to digital mammograms (DMs) has enhanced the potential for imaging-based measures. In particular, image-derived indices of breast density5,6 and parenchymal texture patterns7 have emerged as promising risk factors for breast cancer in epidemiologic studies, paving the way toward personalized recommendations regarding women’s cancer risk evaluation and screening.8
Breast density, which measures the amount of fibroglandular tissue within the breast, is a key biomarker not only because it is considered a well-established independent risk factor for breast cancer,5 but also because it influences the sensitivity of screening mammography.9 Its critical role has motivated multiple approaches toward its assessment in DMs, from visual categorical assessment10 and interactive quantitative techniques11 to fully automated measurements.12,13 Growing evidence also suggests that localized measures of the parenchymal tissue patterns may reflect changes in modifiable risk factors for breast cancer (i.e., factors, such as hormonal exposure, and BMI, which can be controlled or treated) and have a complementary role to breast density.14,15
Digital mammography acquisition produces two types of images, both of which can be used for breast density and texture analysis.16–25 The raw (i.e., “FOR PROCESSING”) images are proportional to the x-ray attenuation of the breast tissue, while the processed (i.e., “FOR PRESENTATION” or “FOR CLINICAL DISPLAY”) images are generated after vendor-specific postprocessing is applied to the raw images in an attempt to increase lesion conspicuity before clinical evaluation and interpretation.26 Therefore, it would be reasonable to consider that raw images are more appropriate for quantitative analysis than processed images, as they retain the original x-ray attenuation information. However, routine collection of raw images is not widespread due to cost and storage considerations, usually leaving only the processed data available for retrospective examination. As a result, although several algorithms were originally designed to work on raw data,27 many recent studies utilize processed DM images.23–25
This raises the question of potential differences in image-derived measurements from raw and processed mammograms, and subsequent implications in related interpretation. Recent studies investigating associations of digital mammography quantitative descriptors with breast cancer risk28–30 as well as intra- and inter-reader agreement12,28,31 suggest that DM representation may have an effect. However, this topic remains largely unexplored, with breast density measurements in processed versus raw DM compared in few studies,12,28,31 while the literature lacks reports for similar comparisons of parenchymal texture descriptors. To better understand the relationship between such measures from raw and processed DMs, we performed a systematic evaluation of a wide range of fully automated measures on large-population data collected with acquisition systems from two different vendors.
2. METHODS
2.A. Study population and digital mammograms acquisition
We report data from an entire one-year (September 1, 2010 to August 30, 2011) screening cohort at our institution (Hospital of the University of Pennsylvania), which included a total of 10739 consecutive women with no prior history of breast cancer who were screened routinely for breast cancer with digital mammography. The institutional review board waived the requirement to obtain written consent for this retrospective, Health Insurance Portability and Accountability Act (HIPAA)-compliant study. All women underwent bilateral, two-view full-field digital mammography under standard protocol, using either a Senographe Essential (General Electric Healthcare, Little Chalfont, Buckinghamshire, UK, here labeled as “Vendor 1”) or a Selenia Dimensions (Hologic, Inc., Bedford, MA, USA, labeled as “Vendor 2”) unit. The system of Vendor 1 was equipped with anode filter combinations of molybdenum and rhodium and had a pixel size of 0.10 mm/pixel; for the system of Vendor 2, there was a choice of using either a rhodium or a silver filter (determined either manually or automatically by the system) and the pixel size was equal to 0.07 mm/pixel.
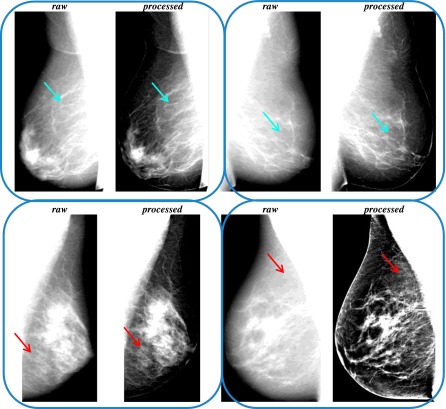
For the purposes of this study, we used the medio-lateral oblique (MLO) view as it offers the opportunity to visualize the maximum amount of breast tissue. Of the entire screening cohort, both raw and processed MLO images were available for 4389 women. From these, 295 pairs of raw and processed DM images were excluded due to woman- or DM-specific image artifacts (i.e., paddles, pacemakers, implants, etc.) in either one or both DM representations, leading to a final dataset of 8458 (Vendor 1: 2903, Vendor 2: 5555) pairs of raw and processed DM images, corresponding to unilateral or bilateral, when available, breast images from 4278 women. Example pairs of the collected raw and processed DMs are shown in Fig. 1.
FIG. 1.
Pairs of raw and processed medio-lateral oblique digital mammograms for different women, showing minor (upper row, blue arrows) or substantial (lower row, red arrows) effects of vendor-specific postprocessing algorithms on the breast parenchymal appearance (left: Vendor 1, right: Vendor 2). (See color online version.)
2.B. Automated estimation of image-derived quantitative descriptors
Raw images were first log-transformed, then inverted, and, finally, intensity-normalized by a z-score transformation specifically within the breast region which was automatically segmented in each image as described below; only z-score normalization was applied to processed images. These steps comprise common preprocessing which helps to alleviate differences between studies via intensity histogram alignment, while also maintaining the overall pattern, spatial relationship, and relative contrast of the pixels, toward appropriate texture analysis.32 Subsequently, 30 quantitative measures (Table I), characterizing breast density and parenchymal texture, were estimated from the two image representations. All measurements were performed using fully automated computational tools, which have been previously developed, extensively optimized, and validated.12,14,15,32,33
TABLE I.
Breast density and parenchymal texture features measured in raw and processed digital mammograms.
| Density features | T12 | 95th | |
| DA | Dense tissue area | T13 | 95th mean |
| PD | Breast percent density | T14 | Entropy |
| Structural texture features | T15 | Kurtosis | |
| T1 | Box-counting fractal dimension | T16 | Max |
| T17 | Mean | ||
| T2 | Local binary pattern | T18 | Min |
| Co-occurrence texture features | T19 | Sigma | |
| T3 | Cluster shade | T20 | Skewness |
| T4 | Correlation | T21 | Sum |
| T5 | Energy | Run-length texture features | |
| T6 | Entropy | T22 | Gray level non-uniformity |
| T7 | Haralick correlation | T23 | High gray level run emphasis |
| T8 | Inertia | T24 | Long run emphasis |
| T9 | Inverse difference moment | T25 | Low gray level run emphasis |
| Gray-level histogram texture features | T26 | Run length non-uniformity | |
| T10 | 5th | T27 | Run percentage |
| T11 | 5th mean | T28 | Short run emphasis |
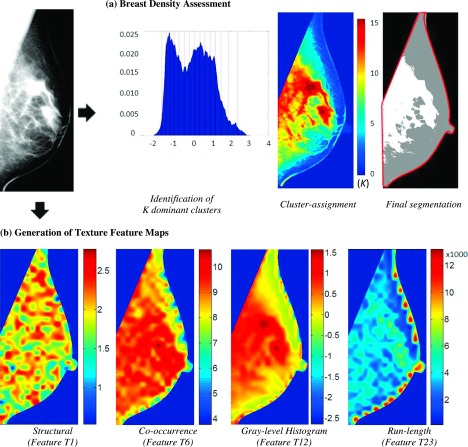
Specifically, the “Laboratory for Individualized Breast Radiodensity Assessment” (libra) software12,33 was used to measure breast density in terms of both total dense tissue area (DA) and breast percent density (PD), defined as the ratio of the segmented DA to the total breast area. The libra algorithm is an adaptive multicluster fuzzy c-means segmentation approach, which has been shown to have good agreement to the widely used semiautomated Cumulus method for both raw and processed DM.12 Briefly, the algorithm first applies an edge-detection algorithm to delineate the boundary of the breast and the pectoral muscle which together define the total breast tissue area. Then, the number of dominant clusters of similar gray-level intensity is determined as the number of local peaks in the gray-level histogram and a fuzzy c-means algorithm assigns each image pixel to the cluster for which that pixel’s intensity value has the highest membership score. In this manner, the breast tissue area is partitioned into clusters of similar gray-level intensity, which are finally aggregated by a trained support-vector machine classifier to the final DA segmentation [Fig. 2(a)]. In addition to the automated density measures, BIRADS density was also available from the clinical evaluation report on record.
FIG. 2.
Automated quantitative assessment of breast density and parenchymal texture. (a) Illustration of breast density assessment using the libra algorithm. The detected boundary of the total breast tissue is marked in red and the segmented dense tissue areas are marked in white in the final segmentation of the digital mammogram. (b) Examples of texture feature maps produced by the lattice-based texture analysis pipeline. The varying color patterns within the breast show that each texture feature reveals different aspects of the mammographic texture. (See color online version.)
An automated image analysis pipeline,14,15 based on a novel lattice-based strategy, was used to extract a wide range of parenchymal texture features from the entire breast region. With this method, texture feature values are calculated within nonoverlapping 6.3 × 6.3 mm2 local square regions surrounding each lattice point of a grid virtually overlaid on the entire segmented breast region, thereby generating a corresponding texture map for each texture descriptor. For breast regions close to the boundary of the breast, where the local square regions may not entirely fall within the breast, the pixels outside the breast were excluded from texture feature calculation. The particular approach of breast sampling has been shown to maximize the robustness of texture features across different systems, as well as their association with breast cancer risk.15,32 In a second step, a feature vector, comprising of the mean of the corresponding texture values over all lattice points within the breast for all descriptors, characterizes the overall breast parenchymal texture. Following this two-step approach, we estimated a total of 28 texture descriptors, including structural,34,35 co-occurrence,36 gray-level histogram,37 and run-length features,38,39 which have all been previously established for mammographic pattern analysis and breast cancer risk assessment.7,14,15,23,35,40,41
In Fig. 2(b), we show representative texture maps for each feature category, while detailed descriptions and mathematical notations of the features are available in Appendix A. Generally, these indices describe different aspects of the mammographic texture pattern. Gray-level histogram features are common first-order statistics which describe the distribution of gray-level intensity values within the breast tissue region. Co-occurrence and run-length features also consider the spatial relationships of pixel gray-level intensities and they reflect the coarseness, contrast, local variation, and spatial dependence of these image intensities in the breast parenchyma. Finally, structural features provide the architectural composition of the breast tissue by capturing the intrinsic complexity and the intensity variations between central and neighboring pixels.
2.C. Statistical analyses
Bland-Altman analysis was performed to ensure agreement in the segmented breast tissue area between processed and raw images toward appropriate subsequent comparisons of feature measurements.
To investigate the relationship between feature measurements in the two image representations, feature values were compared via Spearman’s rank correlation coefficient and the matched-pairs Wilcoxon signed-ranks test. A linear-mixed-effects (LME) model42 (unit of analysis: individual woman) was also applied to assess the linear relationship between feature values in processed versus raw images, in which we tested for potential interactions43 with woman- [age, body mass index (BMI), and ethnicity] and system- (kVp and mAs) specific factors known to affect the mammographic parenchymal tissue patterns.32 These factors were handled as categorical variables using the following levels: age (≤49, [50 60), [60 70), [70 80), ≥80 y), BMI (<25, [25 30), ≥30 kg/m2), ethnicity (White, African-American, Other or missing), kVp (Vendor 1: ≤29, >29; Vendor 2: ≤30, >30), and mAs (Vendor 1: ≤52, (52 75], >75; Vendor 2: ≤121, (121 165], >165). The level definitions of kVp and mAs were based on the 25th and 75th percentiles of the corresponding distributions. Given that the processing algorithms and detector technology utilized by the two vendors are different, which, in turn, may impact the appearance of the breast parenchyma, comparisons were stratified by vendor.
Finally, we also assessed texture feature correlations with the established risk factors of the Gail lifetime risk score44 (rG) and breast PD (rPD), using the Spearman’s rank correlation coefficient, and evaluated their bilateral symmetry, using the intraclass correlation (ICC) to assess within-woman texture feature similarity compared to the texture feature variability across all women.
All statistical experiments were performed using the stata (Release 13, StataCorp LP, College Station, TX, USA) and the sas (Release 9.4, SAS Institute, Inc., Cary, NC, USA) statistical software packages.
3. RESULTS
Our study population is summarized in Table II. Bland-Altman limits showed agreement in segmented breast tissue area in processed versus raw images for Vendor 1 (mean difference −0.48 cm2, 95% limits of agreement [−17.62, 16.66] cm2, range of averages [37.86, 516.04] cm2) and Vendor 2 (mean difference −7.75 cm2, 95% limits of agreement [−45.06, 29.56] cm2, range of averages [37.54, 554.79] cm2).
TABLE II.
Study population characteristics, stratified by the two different digital mammography vendor units.
| Vendor 1 | Vendor 2 | |
|---|---|---|
| Number of women | 1474 | 2804 |
| Age (mean ± SD) | 58.6 y ± 11.2 | 56.5 y ± 10.8 |
| Age at first menstrual period (mean ± SD) | 13.0 y ± 3.6 | 12.8 y ± 3.3 |
| Age at first child birth (mean ± SD) | 24.7 y ± 6.7 | 24.5 y ± 6.7 |
| BMI (mean ± SD) | 28.7 kg/mm2 ± 7.0 | 29.8 kg/mm2 ± 7.4 |
| BI-RADS density | ||
| Type A | 162 (11.0%) | 330 (11.8%) |
| Type B | 792 (53.7%) | 1560 (55.7%) |
| Type C | 482 (32.7%) | 852 (30.4%) |
| Type D | 31 (2.1%) | 41 (1.5%) |
| Missing | 7 (0.5%) | 21 (0.8%) |
| Ethnicity | ||
| White | 630 (42.7%) | 1127 (40.2%) |
| African-American | 740 (50.2%) | 1425 (50.8%) |
| Other or missing | 104 (7.1%) | 252 (9.0%) |
| Gail lifetime risk score (mean ± SD) | 8.2% ± 4.4 | 8.6% ± 4.3 |
Note: SD = standard deviation; BMI = body mass index; BIRADS = breast imaging reporting and data system.
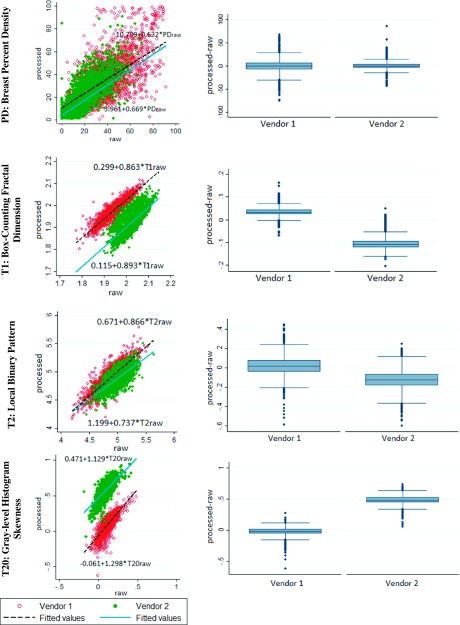
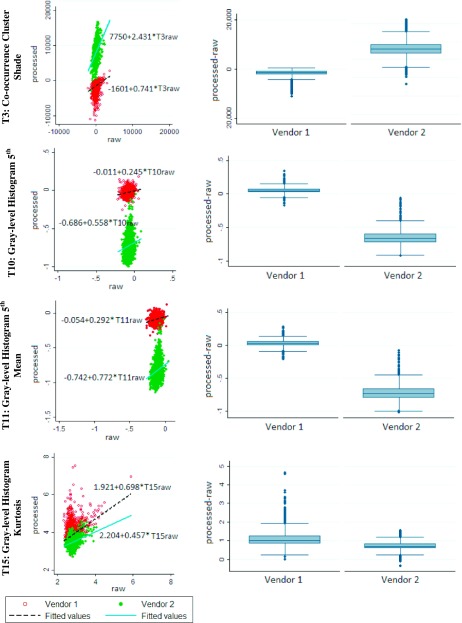
All feature measurements were statistically significantly different between raw and processed images (Wilcoxon signed-ranks test, pw < 0.05), however, most features were strongly (r ≥ 0.6) or moderately (0.4 ≤ r < 0.6) correlated (Table III). For the correlated features, the LME slope was statistically different from zero and generally below unity. The magnitude of the slope varied substantially, not only across different features (e.g., features DA and T25 in Table III), but also between different vendors for the same feature (e.g., T23 in Table III). Strong agreement across the different DM representations, demonstrated as strong correlations and close-to-unity LME slopes in both vendors, was observed for breast PD and for three texture descriptors reflecting intrinsic parenchymal pattern complexity (features T1 and T2) and symmetry (feature T20) (Fig. 3). On the other hand, the lowest correlations in both vendors were seen in features T3, T10, T11, and T15, representing gray-level intensity co-occurrence and histogram statistics, respectively (Fig. 4).
TABLE III.
Statistical analysis of paired measurements in processed versus raw digital mammograms. For each of the 30 image-derived features, the linear-mixed-effects model, i.e., intercept, slope (b), 95% confidence interval for b and the coefficient of determination (R2), and the Spearman’s rank correlation coefficient (r) are provided.
| Vendor 1 (N = 2903 image pairs) | Vendor 2 (N = 5555 image pairs) | ||||||
|---|---|---|---|---|---|---|---|
| LME model | LME model | ||||||
| Feature | (Intercept) b (CI) | R2 | r | (Intercept) b (CI) | R2 | r | |
| Density | DA | (13.84) 0.70a [0.67 0.74] | 0.68 | 0.81 | (12.48) 0.41a [0.38 0.43] | 0.50 | 0.53 |
| PD | (10.71) 0.63a [0.60 0.66] | 0.83 | 0.68 | (3.96) 0.67a [0.64 0.69] | 0.68 | 0.73 | |
| Texture | T1 | (0.30) 0.86a [0.85 0.88] | 0.87 | 0.92 | (0.12) 0.89a [0.87 0.91] | 0.56 | 0.76 |
| T2 | (0.67) 0.87a [0.85 0.88] | 0.75 | 0.85 | (1.20) 0.74a [0.72 0.75] | 0.51 | 0.71 | |
| T3 | (−1 601.11) 0.74a [0.62 0.86] | 0.08 | 0.25 | (7 749.71) 2.43a [2.27 2.60] | 0.17 | 0.43 | |
| T4 | (0.00) 0.60a [0.58 0.62] | 0.65 | 0.79 | (0.00) 0.05a [0.04 0.05] | 0.30 | 0.42 | |
| T5 | (0.00) 0.59a [0.55 0.63] | 0.34 | 0.59 | (0.00) 0.09a [0.08 0.10] | 0.07 | 0.27 | |
| T6 | (4.02) 0.53a [0.51 0.55] | 0.56 | 0.72 | (9.92) −0.01a [−0.02 0.00] | 0.01 | −0.24 | |
| T7 | (1 183 546) 0.35a [0.34 0.37] | 0.64 | 0.78 | (13 080 807) 1.64a [1.56 1.71] | 0.42 | 0.65 | |
| T8 | (28.68) 0.68a [0.66 0.69] | 0.79 | 0.86 | (251.56) 1.27a [1.20 1.34] | 0.29 | 0.54 | |
| T9 | (0.08) 0.57a [0.54 0.59] | 0.56 | 0.72 | (0.06) 0.14a [0.13 0.16] | 0.22 | 0.34 | |
| T10 | (−0.01) 0.25a [0.20 0.29] | 0.10 | 0.25 | (−0.69) 0.56a [0.46 0.66] | 0.06 | 0.28 | |
| T11 | (−0.05) 0.29a [0.25 0.34] | 0.15 | 0.29 | (−0.74) 0.77a [0.67 0.87] | 0.10 | 0.34 | |
| T12 | (0.16) 0.51a [0.50 0.52] | 0.82 | 0.88 | (0.59) 0.83a [0.78 0.89] | 0.25 | 0.46 | |
| T13 | (0.18) 0.50a [0.49 0.51] | 0.83 | 0.89 | (0.77) 0.85a [0.79 0.92] | 0.20 | 0.42 | |
| T14 | (1.57) 0.72a [0.69 0.74] | 0.56 | 0.62 | (5.43) 0.14a [0.13 0.15] | 0.20 | 0.47 | |
| T15 | (1.92) 0.70a [0.62 0.78] | 0.08 | 0.15 | (2.20) 0.46a [0.42 0.50] | 0.09 | 0.21 | |
| T16 | (0.24) 0.49a [0.48 0.50] | 0.85 | 0.90 | (1.28) 0.82a [0.74 0.90] | 0.11 | 0.29 | |
| T17 | (0.10) 0.52a [0.49 0.54] | 0.60 | 0.72 | (−0.03) −0.06a [−0.10–0.03] | 0.05 | −0.03 | |
| T18 | (−0.12) 0.49a [0.45 0.53] | 0.35 | 0.47 | (−0.90) 0.87 [0.78 0.96] | 0.11 | 0.32 | |
| T19 | (0.05) 0.54a [0.52 0.56] | 0.68 | 0.77 | (0.31) 1.54a [1.46 1.62] | 0.30 | 0.53 | |
| T20 | (−0.06) 1.30a [1.26 1.34] | 0.75 | 0.81 | (0.47) 1.13a [1.09 1.17] | 0.52 | 0.62 | |
| T21 | (389.85) 0.52a [0.49 0.54] | 0.60 | 0.72 | (−113.81) −0.06a [−0.10 −0.03] | 0.05 | −0.03 | |
| T22 | (29.22) 0.68a [0.65 0.71] | 0.48 | 0.59 | (41.52) 0.24a [0.23 0.26] | 0.36 | 0.57 | |
| T23 | (164.73) 1.07a [1.04 1.10] | 0.73 | 0.79 | (1 248.74) 0.48a [0.46 0.51] | 0.31 | 0.40 | |
| T24 | (0.80) 0.20a [0.19 0.22] | 0.37 | 0.71 | (1.04) −0.04a [−0.08 0.00] | 0.00 | 0.03 | |
| T25 | (0.00) 0.17a [0.15 0.18] | 0.23 | 0.45 | (0.00) 0.22a [0.18 0.25] | 0.01 | 0.11 | |
| T26 | (2 459.26) 0.35a [0.34 0.36] | 0.59 | 0.72 | (2 897.15) 0.24a [0.23 0.25] | 0.19 | 0.40 | |
| T27 | (0.62) 0.35a [0.34 0.36] | 0.58 | 0.72 | (0.73) 0.24a [0.23 0.25] | 0.19 | 0.40 | |
| T28 | (0.81) 0.19a [0.17 0.20] | 0.36 | 0.69 | (1.01) −0.01a [−0.02 0.00] | 0.00 | 0.02 | |
Indicates statistical significance of the deviation of b from unity, assessed with a two-sided Wald test at the α = 0.05 level with the Bonferroni adjustment for 30 comparisons per vendor (i.e., α = 0.05/30 = 0.0017). All correlations were statistically significant at the α = 0.05 level. Boldface indicates strongly correlated features (r ≥ 0.6) in processed versus raw images. LME = linear-mixed-effects; CI = confidence interval.
FIG. 3.
Graphs for strongly correlated feature measurements. (Left column) Scatterplots and LME models for measurements in processed (y-axis) versus raw (x-axis) digital mammograms, and (right column) box plots of the differences between the corresponding measurements (processed minus raw).
FIG. 4.
Graphs for weakly correlated feature measurements. (Left column) Scatterplots and LME models for measurements in processed (y-axis) versus raw (x-axis) digital mammograms, and (right column) box plots of the differences between the corresponding measurements (processed minus raw).
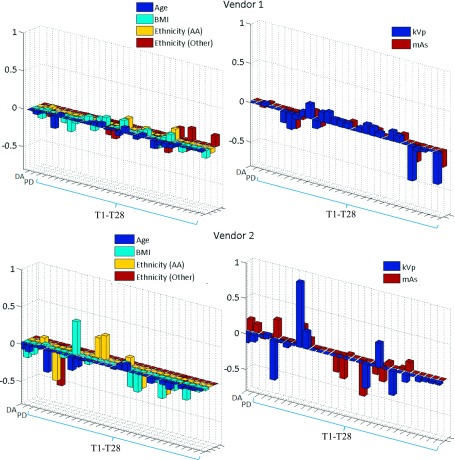
The linear relationship between measures in processed versus raw images varied considerably across different acquisition settings and woman-specific characteristics (Fig. 5). The LME slope was mostly modified by system-specific image acquisition factors, such as mAs and kVp, followed by age and BMI, while ethnicity had limited impact (Tables IV and V, Appendix B). Interestingly, the LME slopes for features T1 and T2 remained substantially stable for both vendors.
FIG. 5.
Modification of the LME-model slope by woman- and system-specific factors. Amount of change in the slope of the line fitted between feature values in processed versus raw digital mammograms, when the woman-specific factors of age, BMI, and ethnicity or the system-specific factors of kVp and mAs increase by one level. In case of the non-ordinal variable of ethnicity, this value represents the amount of change in the slope for the “African-American” (AA) and “Other or missing” (Other) subgroups with respect to the “White” subgroup.
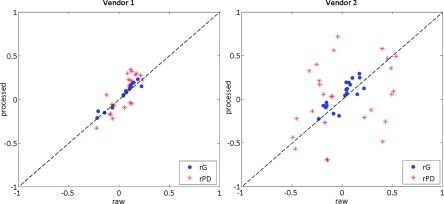
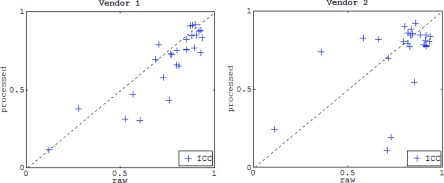
Overall, texture features from both image representations showed weak correlations with the Gail lifetime risk score and weak to moderate correlations with breast PD (Fig. 6). In terms of their association with breast PD, large differences between raw and processed DM texture measures were observed, especially when comparing different vendors, with several measures having inverted associations with breast PD depending on whether raw or processed DMs were used for texture analysis (e.g., for feature T6 in vendor 2, rPD was equal to 0.41 and −0.49 in raw and processed images, Table VI, Appendix C). Most features demonstrated strong bilateral symmetry in both image representations, while intrawoman similarity was statistically significantly stronger in raw images especially within separate vendors (Fig. 7).
FIG. 6.
Feature correlations with the Gail lifetime risk score (blue circles) and breast percent density (red asterisks) for parenchymal texture feature measures in processed versus raw digital mammograms. Each mark denotes a texture descriptor. The dashed identity line is provided for reference. (See color online version.).
FIG. 7.
Per-woman bilateral texture feature similarity for feature measurements in processed versus raw digital mammograms. Each mark denotes a texture descriptor. The dashed identity line is provided for reference.
4. DISCUSSION
Our study represents an extensive evaluation of key quantitative descriptors obtained from processed versus raw DMs, and could have implications for designing large-scale studies where DM images from different vendors and presentation formats are utilized. Overall, our results suggest that the extent of differences in breast density and parenchymal texture measures from processed versus raw DMs depends highly on the specific feature as well as the machine vendor and physics settings of the image acquisition.
First, the consistently correlated breast PD confirmed the previously reported associations in density measurements across DM representations;12,28,31,45 however, the linear relationship between breast PD measured in processed and raw images seems to interact highly with woman- and system-specific factors, such as age, BMI, and mAs/kVp, respectively. Our results are generally in line with previous similar analyses in smaller datasets,28,31 with any significant differences likely occurring due to the larger size of our sample, spanning an entire one-year screening cohort.
Texture features, which provide more subtle characterization of the breast parenchyma, seem to be overall more affected by vendor-specific postprocessing than the breast density measures. Moreover, the slope values of the LME models showed that per-unit feature changes were generally lower in processed images, suggesting that raw images may be more sensitive to texture differences (Table III). However, texture features were also significantly correlated and associations between measurements from the two image representations could be sufficiently explained by linear relationships. It should be emphasized, though, that both the strength of the correlation and the linear model relating the feature measurements in processed versus raw DM depend highly on the specific feature and the DM unit.
Strongest agreement in measures between raw and processed DM for both vendors was demonstrated by three texture descriptors (T1, T2, and T20). Moreover, when tested for interactions with woman- and system-specific factors, the LME models of two of these features (T1 and T2) were minimally affected. Interestingly, these two features represent structural texture features, which were recently associated with enhanced robustness across different DM units.32 These findings suggest that features capturing intrinsic characteristics of the breast parenchyma, by considering the overall topology and structure of the pattern, may be more robust and invariant to image intensity transformations by postprocessing.
Regardless of the strength of correlation between the different image types, all texture features, when measured either in processed or raw mammograms, were weakly to moderately correlated with breast PD, and were also weakly correlated with Gail lifetime risk. Given that earlier studies have associated the same texture descriptors with potential value in breast cancer prediction,7,14,23,35,40,41 our results also support a potential independent role of mammographic texture analysis in breast cancer risk assessment. However, it is important to mention that we observed substantial differences between raw and processed DM texture measures in their associations with breast PD. Similar variations have also been reported previously by related studies,23,24,38,46 which together suggest an important effect of DM representation on the associations of parenchymal texture descriptors with density. Furthermore, both image representations had strong bilateral feature similarity, suggesting an inherently strong agreement in bilateral parenchymal symmetry, regardless of the imaging format.
Finally, our findings revealed possible different effects of the vendor-specific postprocessing algorithms, given that feature correlations, their linear relationships, and their interactions with woman- and system-specific factors varied substantially across vendors. A more extensive comparison between DM units is warranted and generalized conclusions should, therefore, be limited. However, our findings also emphasize the need for standardization frameworks,32 which can alleviate differences and ensure comparable measurements across different digital mammography units, acquisition settings, and representations.
Overall, our analysis showed that the DM representation, specific imaging settings (DM unit and mAs/kVp), and the woman’s individual profile (age and BMI), can affect both the feature measurements in DM and potentially their associations with established risk factors for breast cancer. This could also imply potential effects on the robustness of the density and texture measures in assessing a woman’s breast cancer risk across varying screening conditions. A complementary case-control study on this open question would therefore be important future work toward evaluating DM quantitative measures from different imaging representations for inclusion in breast cancer risk assessment models.
The conclusions of this work may serve as a reference in the design of future studies aiming to develop more robust mammographic measures of breast density and parenchymal texture, as well as help guide statistical approaches for calibrating and/or standardizing parenchymal features in processed versus raw DMs. As such, our study could guide the design of future large-scale studies, involving multiple digital mammography representations, and the integration of computerized texture analysis for breast cancer risk assessment in clinical settings, where only processed mammograms are routinely available.
Certain limitations should also be acknowledged for our study. First, we included a set of breast density and texture features estimated using specific algorithms, while additional features have been used in previous studies.16–21,25,28–30 Moreover, our dataset consisted of images which were collected with digital mammography units from two different vendors, although there are generally more vendors with FDA-approved units for use in mammography facilities.47 Furthermore, additional system-specific parameters (e.g., anode filter) might have an effect on the mammographic parenchymal tissue patterns. Our aim with this study was to perform comparisons between key mammographic features. Therefore, we chose to focus on (a) commonly used features that have been previously associated with breast cancer risk, (b) a single, yet systematically optimized and validated, implementation of breast density and texture assessment, (c) two leading DM vendors, and (d) image acquisition settings known to substantially affect mammographic images. Larger datasets, with broader feature implementations, and more complex interactions with system-specific factors could be explored in future studies, aiming at further elucidating the differences between measurements in raw versus processed DM and how these could be calibrated in different scenarios of image acquisition and subsequent image analysis.
5. CONCLUSIONS
In summary, breast density and parenchymal texture features are generally correlated in processed versus raw DMs. The extent of differences in these measures across different imaging representations depends highly on the specific feature and the image acquisition settings. Among the features examined, structural texture features demonstrated stronger agreement between raw and processed images and were minimally affected by woman- and system-specific factors. These results may serve as a reference in the design of future studies on mammographic measures of density and parenchymal texture. They may also contribute to the design of large-scale studies, involving multiple digital mammography representations, and to the integration of computerized texture analysis in breast cancer risk assessment for clinical settings, where only processed mammograms are routinely available.
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute at the National Institutes of Health: Population-based Research Optimizing Screening through Personalized Regimens (PROSPR) Network (Grant No. U54CA163313), an R01 Research Project (No. 5R01CA161749-04), and a Resource-Related Research Project–Cooperative Agreement (No. 1U24CA189523).
APPENDIX A: TEXTURE FEATURE GENERATION
The structural features include (a) the local binary pattern (LBP), which captures intensity variations between central and neighboring pixels34 and (b) the fractal dimension (FD), which reflects the degree of complexity and was estimated using the box-counting method.35
The LBP at pixel (xc, yc) is estimated as
| (A1) |
where Ic and Ip are the gray level values for pixels and , respectively, and q is a function which attributes values zero and one for negative and non-negative inputs, respectively. The neighborhood of each pixel is defined in terms of size and number of neighborhood pixels by the parameters Q and P, respectively, which were set equal to Q = 1 and P = 8 following previous optimization experiments.32
The FD estimation relies on the concept of self-similarity. The fractal dimension of a bounded set S in Euclidean n-space is defined as
| (A2) |
where Nr is the least number of distinct copies of S in the scale r. In case of an image, where fractals are not deterministic, the FD is approximated by the box-counting dimension as follows. We consider a 3D spatial surface, with (x, y) denoting pixel position on the image plane and the third coordinate, z, denoting the pixel gray level. The image plane is partitioned into nonoverlapping blocks of size s × s, with s being an integer corresponding to the scale of the block. On each block, there is a column of boxes of size s × s × s′, where s′ is the height of each box and it is determined by s, the size of the image and the total number of gray levels. If we assign numbers to the boxes of a block and the minimum and maximum gray levels fall into the mth and lth boxes, respectively, then the boxes covering the block are counted in the number as nB = l − m + 1. By considering the contributions of all blocks, Nr is counted as the sum of nr over all blocks. Following this process for different values of r, FD can, then, be estimated from the least-squares linear fit of versus.
The co-occurrence features reflect the spatial relationship between pixels and they are based on the assumption that this information is adequately encoded by the so-called gray-level co-occurrence matrix (GLCM).36 GLCM is an M × M matrix, where each element, f(i, j), corresponds to the relative frequency with which two neighboring pixels, one with gray level i and the other with gray level j, occur in the image. Such matrices of gray-tone spatial dependence frequencies are a function of the distance (d) and the orientation between the neighboring pixels.
In this study, the GLCM matrices were estimated using M = 128 gray levels to balance computational precision withefficiency and d = 11 pixels, and the following features were computed by averaging over four orientations (0°, 45°, 90° and 135°), based on the premise that these features are orientation invariant and one single direction might not give sufficient information.14,15,32
| Feature | Mathematical notation | Qualitative description |
| Cluster shade | Asymmetry in gray-level values | |
| Correlation | Linear gray-level dependence | |
| Haralick correlation | ||
| Energy | Certainty of gray-level co-occurrence | |
| Entropy | Uncertainty of gray-level co-occurrence | |
| Inertia | Local variation of gray-level intensity | |
| Inverse difference moment | Local homogeneity in gray-level values |
where μi = ∑ji*f(i, j), μj = ∑ij*f(i, j), , and .
The gray-level histogram features are 12 well-known first-order statistics,37 which were calculated from the gray-level intensity histogram of the image using 128 histogram bins.14,32
| Feature | Mathematical notation | Qualitative description |
| Mean | ∑k(k*g(k))/(K) | Mean gray-level value |
| Min | Min(k) | Minimum gray-level value |
| Max | max(k) | Maximum gray-level value |
| Fifth percentile | k : 5% of values ≤k | The histogram bin that 5% of gray-level values are less than or equal to |
| Fifth mean | for k ≤ fifth percentile | Mean value of the gray-level values which less than or equal to the 5th percentile |
| 95th percentile | k : 95% of values ≥k | The histogram bin that 95% of gray-level values are less than or equal to |
| 95th mean | for k ≥ 95th percentile | Mean value of the gray-level values which larger than or equal to the 95th percentile |
| Sum | ∑kk*g(k) | Sum of gray-level values |
| Sigma | Variation of gray-level values around the mean | |
| Entropy | −∑kg(k)*log(g(k)) | Measure of histogram uniformity |
| Kurtosis | Sigma−4∑k(k − Mean)4*g(k) − 3 | Measure of histogram flatness |
| Skewness | Sigma−3∑k(k − Mean)3*g(k) | Measure of histogram symmetry |
where k is the histogram bin, g is the frequency of the histogram bin, and K = ∑kg(k).
Run-length features capture the coarseness of a texture in specified directions.38,39 A run is defined as a string of consecutive pixels which have the same gray-level intensity along a specific linear orientation. Fine textures tend to contain more short runs with similar gray-level intensities, while coarse textures have more long runs with significantly different gray-level intensities. Similarly to the GLCM, a run-length matrix R is defined, with each element, R(i, j), representing the number of runs with pixels of gray-level intensity equal to i and length of run equal to j along a specific orientation. The size of the matrix R is M × N, where N is equal to the possible maximum run length.
In correspondence with the co-occurrence features, the following run-length statistics were estimated for M = 128 gray levels, N = 11 pixels, and averaged values over four orientations (0°, 45°, 90° and 135°).14,15,32
| Feature | Mathematical notation | Qualitative description |
| Gray level nonuniformity | Dissimilarity in runs across gray-level values | |
| High gray level run emphasis | Runs of high-gray-level values | |
| Long run emphasis | Emphasis on the long runs | |
| Low gray level run emphasis | Runs of low-gray-level values | |
| Run length nonuniformity | Dissimilarity in runs across lengths | |
| Run percentage | nr/#pixels | Homogeneity and distribution of runs |
| Short run emphasis | Emphasis on the short runs |
where is the total number of runs.
APPENDIX B: TESTS FOR INTERACTIONS WITH WOMAN- AND SYSTEM-SPECIFIC FACTORS
TABLE IV.
Tests for interactions for Vendor 1. Slope of the line fitted between feature values in processed versus raw images for the first (b1) and last (bL) levela of each interaction factor in case of Vendor 1. Features are classified into three groups, corresponding to G1: nonstatistically significant interaction (95% confidence intervals including zero value), G2: limited interaction (|b1 − bL| ≤ 0.15), and G3: high interaction (|b1 − bL| > 0.15) with the corresponding factor.
| Age | G1 | DA, T1, T2, T5, T6, T8, T13, T16, T19, T25, T26, T27 | |||||||||||||||||||
| T7 | T9 | T12 | T24 | T28 | |||||||||||||||||
| G2 | b1 | 0.33 | 0.53 | 0.53 | 0.17 | 0.16 | |||||||||||||||
| bL | 0.39 | 0.65 | 0.48 | 0.27 | 0.25 | ||||||||||||||||
| PD | T3 | T4 | T10 | T11 | T14 | T15 | T17 | T18 | T20 | T21 | T22 | T23 | |||||||||
| G3 | b1 | 0.67 | 0.96 | 0.54 | 0.18 | 0.20 | 0.78 | 0.54 | 0.57 | 0.43 | 1.43 | 0.57 | 0.74 | 1.10 | |||||||
| bL | 0.52 | 0.32 | 0.70 | 0.36 | 0.45 | 0.59 | 0.85 | 0.41 | 0.62 | 1.02 | 0.41 | 0.51 | 0.93 | ||||||||
| BMI | G1 | T2, T3, T12, T15, T16, T19, T25, T27 | |||||||||||||||||||
| DA | T1 | T6 | T7 | T8 | T13 | T17 | T21 | T23 | T26 | ||||||||||||
| G2 | b1 | 0.74 | 0.84 | 0.67 | 0.35 | 0.66 | 0.44 | 0.37 | 0.37 | 1.04 | 0.39 | ||||||||||
| bL | 0.64 | 0.88 | 0.53 | 0.40 | 0.80 | 0.48 | 0.44 | 0.44 | 0.95 | 0.36 | |||||||||||
| PD | T4 | T5 | T9 | T10 | T11 | T14 | T18 | T20 | T22 | T24 | T28 | ||||||||||
| G3 | b1 | 0.75 | 0.71 | 0.95 | 0.75 | 0.20 | 0.20 | 0.55 | 0.37 | 1.33 | 0.59 | 0.34 | 0.32 | ||||||||
| bL | 0.52 | 0.52 | 0.57 | 0.55 | 0.37 | 0.42 | 0.81 | 0.52 | 1.09 | 0.78 | 0.14 | 0.12 | |||||||||
| Ethnicity | AA | G1 | DA, T2, T3, T6, T7, T8, T10, T11, T12, T13, T15, T16, T18, T19, T23, T25, T26, T27 | ||||||||||||||||||
| PD | T1 | T4 | T5 | T9 | T14 | T17 | T20 | T21 | T22 | T24 | T28 | ||||||||||
| G2 | b1 | 0.69 | 0.84 | 0.65 | 0.68 | 0.63 | 0.65 | 0.47 | 1.35 | 0.47 | 0.61 | 0.23 | 0.22 | ||||||||
| bL | 0.60 | 0.88 | 0.56 | 0.59 | 0.56 | 0.78 | 0.52 | 1.27 | 0.52 | 0.75 | 0.17 | 0.16 | |||||||||
| G3 | b1 | ||||||||||||||||||||
| bL | |||||||||||||||||||||
| Other | G1 | DA, T1, T2, T3, T4, T5, T6, T7, T8, T9, T12, T13, T14, T15, T16, T17, T19, T21, T25, T26, T27 | |||||||||||||||||||
| PD | T22 | T28 | |||||||||||||||||||
| G2 | b1 | 0.69 | 0.61 | 0.22 | |||||||||||||||||
| bL | 0.55 | 0.74 | 0.36 | ||||||||||||||||||
| T10 | T11 | T18 | T20 | T23 | T24 | ||||||||||||||||
| G3 | b1 | 0.23 | 0.28 | 0.49 | 1.35 | 1.06 | 0.23 | ||||||||||||||
| bL | 0.04 | 0.06 | 0.27 | 1.12 | 0.90 | 0.40 | |||||||||||||||
| kVp | G1 | DA, PD, T2, T3, T15, T20, T23, T27 | |||||||||||||||||||
| T1 | T4 | T6 | T7 | T9 | T10 | T11 | T12 | T13 | T14 | T16 | T17 | T18 | T19 | T21 | T22 | T25 | T26 | ||||
| G2 | b1 | 0.85 | 0.67 | 0.62 | 0.36 | 0.67 | 0.23 | 0.25 | 0.49 | 0.48 | 0.68 | 0.48 | 0.44 | 0.42 | 0.57 | 0.44 | 0.65 | 0.19 | 0.38 | ||
| bL | 0.89 | 0.53 | 0.56 | 0.43 | 0.56 | 0.36 | 0.40 | 0.57 | 0.56 | 0.75 | 0.54 | 0.57 | 0.53 | 0.63 | 0.57 | 0.74 | 0.14 | 0.35 | |||
| T5 | T8 | T24 | T28 | ||||||||||||||||||
| G3 | b1 | 0.78 | 0.68 | 0.57 | 0.51 | ||||||||||||||||
| bL | 0.57 | 0.87 | 0.13 | 0.12 | |||||||||||||||||
| mAs | G1 | DA, T2, T3, T10, T11, T15, T19, T20, T23, T25, T26, T27 | |||||||||||||||||||
| PD | T1 | T7 | T8 | T12 | T13 | T14 | T16 | T17 | T18 | T21 | |||||||||||
| G2 | b1 | 0.69 | 0.84 | 0.38 | 0.70 | 0.49 | 0.49 | 0.64 | 0.49 | 0.42 | 0.43 | 0.42 | |||||||||
| bL | 0.56 | 0.89 | 0.42 | 0.85 | 0.56 | 0.55 | 0.77 | 0.54 | 0.53 | 0.54 | 0.53 | ||||||||||
| T4 | T5 | T6 | T9 | T22 | T24 | T28 | |||||||||||||||
| G3 | b1 | 0.73 | 0.94 | 0.69 | 0.75 | 0.60 | 0.58 | 0.53 | |||||||||||||
| bL | 0.51 | 0.50 | 0.53 | 0.54 | 0.76 | 0.13 | 0.12 | ||||||||||||||
For age, BMI, kVp, and mAs, b1 and bL correspond to the lowest and highest level, respectively. For ethnicity, b1 represents the White subgroup and bL corresponds to the African-American (AA) or to the Other or missing (Other) subgroups.
TABLE V.
Tests for interactions for Vendor 2. Slope of the line fitted between feature values in processed versus raw images for the first (b1) and last (bL) levela of each interaction factor in case of Vendor 2. Features are classified into three groups, corresponding to G1: nonstatistically significant interaction (95% confidence intervals including zero value), G2: limited interaction (|b1 − bL| ≤ 0.15), and G3: high interaction (|b1 − bL| > 0.15) with the corresponding factor.
| Age | G1 | T1, T4, T5, T6, T10, T11, T12, T13, T17, T18, T19, T21, T24, T28 | ||||||||||||
| T2 | T9 | T14 | T26 | T27 | ||||||||||
| G2 | b1 | 0.71 | 0.15 | 0.16 | 0.25 | 0.25 | ||||||||
| bL | 0.81 | 0.11 | 0.09 | 0.17 | 0.17 | |||||||||
| DA | PD | T3 | T7 | T8 | T15 | T16 | T20 | T22 | T23 | T25 | ||||
| G3 | b1 | 0.48 | 0.75 | 2.85 | 1.85 | 1.40 | 0.33 | 0.66 | 1.24 | 0.31 | 0.57 | 0.22 | ||
| bL | 0.22 | 0.42 | 1.70 | 1.14 | 0.96 | 0.61 | 1.06 | 0.94 | 0.11 | 0.20 | 0.06 | |||
| BMI | G1 | T1, T3, T8, T10, T11, T12, T13, T14, T18, T19, T24 | ||||||||||||
| T2 | T4 | T6 | T15 | T22 | T26 | T27 | T28 | |||||||
| G2 | b1 | 0.70 | 0.09 | 0.09 | 0.39 | 0.26 | 0.32 | 0.33 | 0.03 | |||||
| bL | 0.77 | 0.03 | −0.04 | 0.52 | 0.20 | 0.19 | 0.19 | −0.02 | ||||||
| G3 | DA | PD | T5 | T7 | T9 | T16 | T17 | T20 | T21 | T23 | T25 | |||
| b1 | 0.64 | 0.68 | 0.20 | 1.23 | 0.27 | 1.16 | 0.13 | 1.22 | 0.13 | 0.60 | 0.54 | |||
| bL | 0.23 | 0.40 | 0.04 | 2.13 | 0.10 | 0.69 | −0.43 | 1.05 | −0.43 | 0.33 | 0.11 | |||
| Ethnicity | AA | G1 | T2, T7, T8, T12, T13, T14, T16, T18, T19, T22, T24, T26, T27, T28 | |||||||||||
| PD | T1 | T4 | T5 | T6 | T9 | T15 | T20 | T23 | T25 | |||||
| G2 | b1 | 0.70 | 0.87 | 0.06 | 0.12 | 0.02 | 0.18 | 0.42 | 1.19 | 0.53 | 0.26 | |||
| bL | 0.60 | 0.94 | 0.04 | 0.06 | −0.03 | 0.12 | 0.53 | 1.10 | 0.42 | 0.18 | ||||
| DA | T3 | T10 | T11 | T17 | T21 | |||||||||
| G3 | b1 | 0.50 | 2.69 | 0.45 | 0.64 | 0.03 | 0.03 | |||||||
| bL | 0.33 | 2.23 | 0.71 | 0.96 | −0.22 | −0.22 | ||||||||
| Other | G1 | DA, PD, T1, T2, T4, T5, T7, T8, T9, T10, T11, T12, T13, T14, T15, T16, T17, T18, T19, T20, T21, T22, T24, T25, T26, T27, T28 | ||||||||||||
| T6 | T23 | |||||||||||||
| G2 | b1 | 0.02 | 0.53 | |||||||||||
| bL | −0.02 | 0.44 | ||||||||||||
| T3 | ||||||||||||||
| G3 | b1 | 2.69 | ||||||||||||
| bL | 2.14 | |||||||||||||
| kVp | G1 | T1, T6, T9, T10, T11, T12, T13, T14, T15, T16, T18, T20, T22, T24 | ||||||||||||
| DA | PD | T2 | T4 | T5 | T23 | T25 | T26 | T27 | T28 | |||||
| G2 | b1 | 0.47 | 0.67 | 0.71 | 0.05 | 0.13 | 0.52 | 0.27 | 0.27 | 0.27 | 0.02 | |||
| bL | 0.35 | 0.57 | 0.77 | 0.05 | 0.08 | 0.41 | 0.22 | 0.23 | 0.24 | −0.01 | ||||
| T3 | T7 | T8 | T17 | T19 | T21 | |||||||||
| G3 | b1 | 2.61 | 1.43 | 1.21 | 0.08 | 1.43 | 0.08 | |||||||
| bL | 2.06 | 2.34 | 1.45 | −0.26 | 1.78 | −0.26 | ||||||||
| mAs | G1 | T10, T11, T15, T17, T21, T24, T25, T26, T27, T28 | ||||||||||||
| T1 | T2 | T4 | T5 | T6 | T9 | T14 | T20 | T22 | ||||||
| G2 | b1 | 0.96 | 0.76 | 0.05 | 0.29 | 0.00 | 0.15 | 0.15 | 1.08 | 0.25 | ||||
| bL | 0.83 | 0.71 | 0.03 | 0.28 | −0.07 | 0.09 | 0.21 | 1.21 | 0.31 | |||||
| DA | PD | T3 | T7 | T8 | T12 | T13 | T16 | T18 | T19 | T23 | ||||
| G3 | b1 | 0.24 | 0.55 | 2.23 | 1.48 | 1.16 | 1.03 | 1.07 | 1.07 | 0.82 | 1.66 | 0.38 | ||
| bL | 0.56 | 0.78 | 2.73 | 2.02 | 1.38 | 0.49 | 0.40 | 0.05 | 0.51 | 1.19 | 0.65 | |||
For age, BMI, kVp, and mAs, b1 and bL correspond to the lowest and highest level, respectively. For ethnicity, b1 represents the White subgroup and bL corresponds to the African-American (AA) or to the Other or missing (Other) subgroups.
APPENDIX C: CORRELATIONS WITH GAIL RISK, BREAST DENSITY, AND BILATERAL SYMMETRY
TABLE VI.
Parenchymal texture feature correlations with established risk factors and assessment of bilateral breast symmetry. Correlation coefficients with the Gail lifetime risk score (rG), breast percent density PD (rPD), and (c) per-woman (i.e., intraclass) correlation (ICC), for raw and processed digital mammograms.
| Vendor 1 | Vendor 2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw images | Processed images | Raw images | Processed images | |||||||||
| Feature | rG | rPD | ICC | rG | rPD | ICC | rG | rPD | ICC | rG | rPD | ICC |
| T1 | −0.08 | −0.05 | 0.69 | a | −0.07 | 0.69 | −0.16 | −0.47 | 0.82 | −0.09 | −0.57 | 0.86 |
| T2 | a | a | 0.12 | a | a | 0.12 | a | −0.17 | 0.11 | a | −0.27 | 0.24 |
| T3 | a | −0.04 | 0.28 | −0.05 | a | 0.38 | 0.03 | −0.09 | 0.36 | 0.04 | 0.04 | 0.74 |
| T4 | 0.04 | a | 0.85 | 0.05 | −0.07 | 0.82 | 0.17 | −0.08 | 0.83 | 0.29 | 0.56 | 0.84 |
| T5 | −0.05 | a | 0.89 | a | −0.21 | 0.77 | 0.07 | −0.33 | 0.85 | 0.19 | 0.33 | 0.55 |
| T6 | 0.08 | 0.08 | 0.92 | 0.08 | 0.30 | 0.88 | −0.03 | 0.41 | 0.91 | −0.19 | −0.49 | 0.81 |
| T7 | 0.12 | 0.13 | 0.93 | 0.14 | 0.32 | 0.88 | 0.05 | 0.54 | 0.94 | 0.07 | 0.49 | 0.84 |
| T8 | a | 0.10 | 0.93 | a | 0.15 | 0.83 | −0.08 | 0.29 | 0.92 | −0.17 | −0.13 | 0.84 |
| T9 | a | a | 0.90 | a | −0.18 | 0.85 | 0.10 | −0.25 | 0.88 | 0.24 | 0.40 | 0.85 |
| T10 | 0.04 | 0.15 | 0.53 | 0.04 | a | 0.31 | 0.04 | −0.11 | 0.58 | 0.11 | 0.03 | 0.82 |
| T11 | a | 0.11 | 0.60 | a | −0.04 | 0.31 | 0.05 | −0.19 | 0.66 | 0.12 | 0.06 | 0.82 |
| T12 | 0.13 | 0.17 | 0.87 | 0.18 | 0.30 | 0.91 | 0.03 | 0.51 | 0.91 | a | 0.09 | 0.77 |
| T13 | 0.12 | 0.16 | 0.88 | 0.16 | 0.28 | 0.92 | a | 0.50 | 0.92 | −0.04 | a | 0.77 |
| T14 | 0.22 | 0.11 | 0.77 | 0.15 | 0.34 | 0.73 | 0.22 | 0.40 | 0.84 | 0.13 | 0.58 | 0.88 |
| T15 | −0.24 | −0.07 | 0.57 | a | −0.06 | 0.47 | −0.21 | −0.30 | 0.72 | a | −0.14 | 0.70 |
| T16 | 0.07 | 0.12 | 0.90 | 0.10 | 0.22 | 0.92 | a | 0.43 | 0.92 | −0.12 | −0.26 | 0.78 |
| T17 | 0.15 | 0.23 | 0.71 | 0.19 | 0.22 | 0.79 | 0.04 | 0.49 | 0.82 | 0.19 | 0.36 | 0.79 |
| T18 | a | 0.06 | 0.76 | a | −0.09 | 0.43 | 0.08 | −0.23 | 0.80 | 0.17 | 0.21 | 0.80 |
| T19 | 0.10 | 0.09 | 0.93 | 0.12 | 0.23 | 0.74 | a | 0.50 | 0.93 | −0.05 | 0.06 | 0.81 |
| T20 | −0.22 | −0.22 | 0.80 | −0.21 | −0.33 | 0.75 | −0.16 | −0.45 | 0.83 | −0.04 | −0.22 | 0.77 |
| T21 | 0.15 | 0.23 | 0.71 | 0.19 | 0.22 | 0.79 | 0.04 | 0.49 | 0.82 | 0.19 | 0.36 | 0.79 |
| T22 | −0.21 | −0.07 | 0.77 | −0.14 | −0.22 | 0.73 | −0.19 | −0.49 | 0.84 | −0.07 | −0.44 | 0.85 |
| T23 | 0.19 | 0.21 | 0.88 | 0.23 | 0.27 | 0.85 | 0.04 | 0.46 | 0.90 | 0.11 | 0.47 | 0.78 |
| T24 | 0.07 | 0.13 | 0.80 | 0.09 | −0.05 | 0.66 | 0.15 | −0.22 | 0.71 | 0.06 | 0.17 | 0.11 |
| T25 | 0.12 | 0.04 | 0.73 | a | a | 0.58 | 0.18 | −0.04 | 0.80 | 0.25 | 0.72 | 0.90 |
| T26 | −0.14 | −0.09 | 0.85 | −0.15 | −0.17 | 0.76 | −0.23 | −0.15 | 0.86 | −0.23 | −0.70 | 0.92 |
| T27 | −0.15 | −0.09 | 0.85 | −0.15 | −0.17 | 0.76 | −0.23 | −0.15 | 0.86 | −0.23 | −0.70 | 0.92 |
| T28 | −0.07 | −0.13 | 0.81 | −0.10 | 0.05 | 0.65 | −0.15 | 0.22 | 0.73 | −0.07 | −0.21 | 0.19 |
Nonstatistically significant correlations at the α = 0.05 level.
CONFLICT OF INTEREST DISCLOSURE
Dr. Conant reports membership on the Hologic, Inc., Scientific Advisory Board. Since the original work was conducted, Dr. Keller has accepted a new position with Hologic, Inc. The other authors have no relevant conflicts of interest to disclose.
REFERENCES
- 1.Ferlay J., Soerjomataram I., Ervik M., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D. M., Forman D., and Bray F., GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 (International Agency for Research on Cancer, Lyon, France, 2013), http://globocan.iarc.fr, accessed 28 January 2016. [Google Scholar]
- 2.Rahib L., Smith B. D., Aizenberg R., Rosenzweig A. B., Fleshman J. M., and Matrisian L. M., “Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States,” Cancer Res. , 2913–2921 (2014). 10.1158/0008-5472.CAN-14-0155 [DOI] [PubMed] [Google Scholar]
- 3.Løberg M., Lousdal M. L., Bretthauer M., and Kalager M., “Benefits and harms of mammography screening,” Breast Cancer Res. (1), 63 (2015). 10.1186/s13058-015-0525-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tice J. A. and Kerlikowske K., “Screening and prevention of breast cancer in primary care,” Primary Care: Clinics Off. Pract. (3), 533–558 (2009). 10.1016/j.pop.2009.04.003 [DOI] [PubMed] [Google Scholar]
- 5.Wang A. T., Vachon C. M., Brandt K. R., and Ghosh K., “Breast density and breast cancer risk: A practical review,” Mayo Clin. Proc. (4), 548–557 (2014). 10.1016/j.mayocp.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 6.Sartor H., Zackrisson S., Elebro K., Hartman L., and Borgquist S., “Mammographic density in relation to tumor biomarkers, molecular subtypes, and mode of detection in breast cancer,” Cancer Causes Control (6), 931–939 (2015). 10.1007/s10552-015-0576-6 [DOI] [PubMed] [Google Scholar]
- 7.Manduca A., Carston M. J., Heine J. J., Scott C. G., Pankratz V. S., Brandt K. R., Sellers T. A., Vachon C. M., and Cerhan J. R., “Texture features from mammographic images and risk of breast cancer,” Cancer Epidemiol., Biomarkers Prev. , 837–845 (2009). 10.1158/1055-9965.EPI-08-0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onega T. et al. , “Breast cancer screening in an era of personalized regimens: A conceptual model and National Cancer Institute initiative for risk-based and preference-based approaches at a population level,” Cancer (19), 2955–2964 (2014). 10.1002/cncr.28771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roubidoux M. A., Bailey J. E., Wray L. A., and Helvie M. A., “Invasive cancers detected after breast cancer screening yielded a negative result: Relationship of mammographic density to tumor prognostic factors,” Radiology (1), 42–48 (2004). 10.1148/radiol.2301020589 [DOI] [PubMed] [Google Scholar]
- 10.Boyd N. F., Martin L. J., Bronskill M., Yaffe M. J., Duric N., and Minkin S., “Breast tissue composition and susceptibility to breast cancer,” J. Natl. Cancer Inst. (16), 1224–1237 (2010). 10.1093/jnci/djq239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byng J. W., Boyd N. F., Fishell E., Jong R. A., and Yaffe M. J., “The quantitative analysis of mammographic densities,” Phys. Med. Biol. (10), 1629–1638 (1994). 10.1088/0031-9155/39/10/008 [DOI] [PubMed] [Google Scholar]
- 12.Keller B. M., Nathan D. L., Wang Y., Zheng Y., Gee J. C., Conant E. F., and Kontos D., “Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c-means clustering and support vector machine segmentation,” Med. Phys. (8), 4903–4917 (2012). 10.1118/1.4736530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Highnam R., Brady J. M., Yaffe M., Karssemeijer N., and Harvey J., Digital Mammography, Robust Breast Composition Measurement—Volpara™ (Springer LNCS, Berlin Heidelberg, 2010), Vol. 6136, pp. 342–349. [Google Scholar]
- 14.Zheng Y., Wang Y., Keller B. M., Conant E., Gee J. C., and Kontos D., “A fully-automated software pipeline for integrating breast density and parenchymal texture analysis for digital mammograms: Parameter optimization in a case-control breast cancer risk assessment study,” Proc. SPIE , 86701B (2013). 10.1117/12.2008155 [DOI] [Google Scholar]
- 15.Zheng Y., Wang Y., Keller B. M., Ray S., Wang Y., Conant E., Gee J. C., and Kontos D., “Parenchymal texture analysis in digital mammography: Optimization of a fully-automated pipeline for breast cancer risk assessment,” Med. Phys. , 4149–4160 (2015). 10.1118/1.4921996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heine J. J. and Velthuizen R. P., “Spectral analysis of full field digital mammography data,” Med. Phys. (5), 647–661 (2002). 10.1118/1.1445410 [DOI] [PubMed] [Google Scholar]
- 17.Lu L. J., Nishino T. K., Khamapirad T., Grady J. J., M. H. Leonard, Jr., and Brunder D. G., “Computing mammographic density from a multiple regression model constructed with image-acquisition parameters from a full-field digital mammographic unit,” Phys. Med. Biol. (16), 4905–4921 (2007). 10.1088/0031-9155/52/16/013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan Y., Giger M. L., Li H., and Sennett C., “Correlative feature analysis on FFDM,” Med. Phys. (12), 5490–5500 (2008). 10.1118/1.3005641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heine J. J., Cao K., Rollison D. E., Tiffenberg G., and Thomas J. A., “A quantitative description of the percentage of breast density measurement using full-field digital mammography,” Acad. Radiol. (5), 556–564 (2011). 10.1016/j.acra.2010.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gierach G. L., Li H., Loud J. T., Greene M. H., Chow C. K., Lan L., Prindiville S. A., Eng-Wong J., Soballe P. W., Giambartolomei C., Mai P. L., Galbo C. E., Nichols K., Calzone K. A., Olopade O. I., Gail M. H., and Giger M. L., “Relationships between computer-extracted mammographic texture pattern features and BRCA1/2 mutation status: A cross-sectional study,” Breast Cancer Res. (4), 424 (2014). 10.1186/s13058-014-0424-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrish O. W., Tucker L., Black R., Willsher P., Duffy S. W., and Gilbert F. J., “Mammographic breast density: Comparison of methods for quantitative evaluation,” Radiology (2), 356–365 (2015). 10.1148/radiol.14141508 [DOI] [PubMed] [Google Scholar]
- 22.Keller B. M., McCarthy A. M., Chen J., Armstrong K., Conant E. F., Domchek S. M., and Kontos D., “Associations between breast density and a panel of single nucleotide polymorphisms linked to breast cancer risk: A cohort study with digital mammography,” BMC Cancer , 143 (2015). 10.1186/s12885-015-1159-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kontos D., Ikejimba L. C., Bakic P. R., Troxel A. B., Conant E. F., and Maidment A. D., “Analysis of parenchymal texture with digital breast tomosynthesis: Comparison with digital mammography and implications for cancer risk assessment,” Radiology (1), 80–91 (2011). 10.1148/radiol.11100966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daye D., Keller B., Conant E. F., Chen J., Schnall M. D., Maidment A. D., and Kontos D., “Mammographic parenchymal patterns as an imaging marker of endogenous hormonal exposure: A preliminary study in a high-risk population,” Acad. Radiol. (5), 635–646 (2013). 10.1016/j.acra.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eng A., Gallant Z., Shepherd J., McCormack V., Li J., Dowsett M., Vinnicombe S., Allen S., and dos-Santos-Silva I., “Digital mammographic density and breast cancer risk: A case-control study of six alternative density assessment methods,” Breast Cancer Res. (5), 439 (2014). 10.1186/s13058-014-0439-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bick U. and Diekmann F., Digital Mammography (Springer, Berlin Heidelberg, 2010), pp. 69–83. [Google Scholar]
- 27.Li Q. and Nishikawa R. M., Computer-Aided Detection and Diagnosis in Medical Imaging (CRC, UK, 2015), pp. 21–56. [Google Scholar]
- 28.Vachon C. M., Fowler E. E., Tiffenberg G., Scott C. G., Pankratz V. S., Sellers T. A., and Heine J. J., “Comparison of percent density from raw and processed full-field digital mammography data,” Breast Cancer Res. (1), R1 (2013). 10.1186/bcr3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moschidis E., Chen X., Taylor C., and Astley S. M., “Texture-based breast cancer prediction in full-field digital mammograms using the dual-tree complex wavelet transform and random forest classification,” in Breast Imaging (Springer International Publishing, Switzerland, 2014), pp. 209–216. [Google Scholar]
- 30.Fowler E. E., Vachon C. M., Scott C. G., Sellers T. A., and Heine J. J., “Automated percentage of breast density measurements for full-field digital mammography applications,” Acad. Radiol. (8), 958–970 (2014). 10.1016/j.acra.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller B. M., Nathan D. L., Gavenonis S. C., Chen J., Conant E. F., and Kontos D., “Reader variability in breast density estimation from full-field digital mammograms: The effect of image post processing on relative and absolute measures,” Acad. Radiol. (5), 560–568 (2013). 10.1016/j.acra.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller B. M., Oustimov A., Wang Y., Chen J., Acciavatti R. J., Zheng Y., Ray S., Gee J. C., Maidment A. D. A., and Kontos D., “Parenchymal texture analysis in digital mammography: Robust texture feature identification and equivalence across devices,” J. Med. Imaging (2), 024501 (2015). 10.1117/1.jmi.2.2.024501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller B. M., Chen J., Daye D., Conant E. F., and Kontos D., “Preliminary evaluation of the publicly available laboratory for breast radiodensity assessment (libra) software tool: Comparison of fully-automated area and volumetric density measures in a case-control study with digital mammography,” Breast Cancer Res. (1), 117 (2015). 10.1186/s13058-015-0626-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ojala T., Pietikainen M., and Maenpaa T., “Multiresolution gray-scale and rotation invariant texture classification with local binary patterns,” IEEE Trans. Pattern Anal. Mach. Intell. (7), 971–987 (2002). 10.1109/TPAMI.2002.1017623 [DOI] [Google Scholar]
- 35.Caldwell C. B., Stapleton S. J., Holdsworth D. W., Jong R. A., Weiser W. J., Cooke G., and Yaffe M. J., “Characterisation of mammographic parenchymal pattern by fractal dimension,” Phys. Med. Biol. (2), 235–247 (1990). 10.1088/0031-9155/35/2/004 [DOI] [PubMed] [Google Scholar]
- 36.Haralick R. M., Shanmugam K., and Dinstein I. H., “Textural features for image classification,” IEEE Trans. Syst. Man Cybern. (6), 610–621 (1973). 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- 37.Materka A. and Strzelecki M., “Texture analysis methods—A review,” COST B11 Report (Technical University of Lodz, Institute of Electronics, Brussels, 1998), pp. 9–11. [Google Scholar]
- 38.Galloway M. M., “Texture analysis using gray level run lengths,” Comput. Graphics Image Process. , 172–179 (1975). 10.1016/S0146-664X(75)80008-6 [DOI] [Google Scholar]
- 39.Chu A., Sehgal C., and Greenleaf J., “Use of gray value distribution of run lengths for texture analysis,” Pattern Recognit. Lett. (6), 415–419 (1990). 10.1016/0167-8655(90)90112-F [DOI] [Google Scholar]
- 40.Mohd Khuzi A., Besar R., Wan Zaki W., and Ahmad N., “Identification of masses in digital mammogram using gray level co-occurrence matrices,” Biomed. Imaging Intervention J. (3), e17 (2009). 10.2349/biij.5.3.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huo Z., Giger M. L., Olopade O. I., Wolverton D. E., Weber B. L., Metz C. E., Zhong W., and Cummings S. A., “Computerized analysis of digitized mammograms of BRCA1 and BRCA2 gene mutation carriers,” Radiology (2), 519–526 (2002). 10.1148/radiol.2252010845 [DOI] [PubMed] [Google Scholar]
- 42.Pinherio J. C. and Bates D. M., Mixed-Effects Models in S and S-PLUS, Statistics and Computing Series (Springer, New York, NY, 2004). [Google Scholar]
- 43.Aiken L. S. and West S. G., Multiple Regression: Testing and Interpreting Interactions (Sage, Thousand Oaks, 1991). [Google Scholar]
- 44.NCI Breast Cancer Risk Assessment Tool, http://www.cancer.gov/bcrisktool/, accessed 28 January 2016.
- 45.Damases C. N., Brennan P. C., and McEntee M. F., “Mammographic density measurements are not affected by mammography system,” J. Med. Imaging (1), 015501 (2015). 10.1117/1.JMI.2.1.015501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller B. M., Chen J., Conant E. F., and Kontos D., “Breast density and parenchymal texture measures as potential risk factors for estrogen-receptor positive breast cancer,” Proc. SPIE , 90351D (2014). 10.1117/12.2043710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Food U. S. and Drug Administration, “Mammography quality standards act and program,” http://www.fda.gov/Radiation-EmittingProducts/MammographyQualityStandardsActandProgram/, accessed 28 January 2016.