ABSTRACT
The integrity of the bacterial cell envelope is essential to sustain life by countering the high turgor pressure of the cell and providing a barrier against chemical insults. In Bacillus subtilis, synthesis of both peptidoglycan and wall teichoic acids requires a common C55 lipid carrier, undecaprenyl-pyrophosphate (UPP), to ferry precursors across the cytoplasmic membrane. The synthesis and recycling of UPP requires a phosphatase to generate the monophosphate form Und-P, which is the substrate for peptidoglycan and wall teichoic acid synthases. Using an optimized clustered regularly interspaced short palindromic repeat (CRISPR) system with catalytically inactive (“dead”) CRISPR-associated protein 9 (dCas9)-based transcriptional repression system (CRISPR interference [CRISPRi]), we demonstrate that B. subtilis requires either of two UPP phosphatases, UppP or BcrC, for viability. We show that a third predicted lipid phosphatase (YodM), with homology to diacylglycerol pyrophosphatases, can also support growth when overexpressed. Depletion of UPP phosphatase activity leads to morphological defects consistent with a failure of cell envelope synthesis and strongly activates the σM-dependent cell envelope stress response, including bcrC, which encodes one of the two UPP phosphatases. These results highlight the utility of an optimized CRISPRi system for the investigation of synthetic lethal gene pairs, clarify the nature of the B. subtilis UPP-Pase enzymes, and provide further evidence linking the σM regulon to cell envelope homeostasis pathways.
IMPORTANCE The emergence of antibiotic resistance among bacterial pathogens is of critical concern and motivates efforts to develop new therapeutics and increase the utility of those already in use. The lipid II cycle is one of the most frequently targeted processes for antibiotics and has been intensively studied. Despite these efforts, some steps have remained poorly defined, partly due to genetic redundancy. CRISPRi provides a powerful tool to investigate the functions of essential genes and sets of genes. Here, we used an optimized CRISPRi system to demonstrate functional redundancy of two UPP phosphatases that are required for the conversion of the initially synthesized UPP lipid carrier to Und-P, the substrate for the synthesis of the initial lipid-linked precursors in peptidoglycan and wall teichoic acid synthesis.
INTRODUCTION
In bacterial peptidoglycan synthesis, a 55-carbon polyisoprenoid lipid carrier called undecaprenyl-pyrophosphate (UPP) is required to transport peptidoglycan precursor across the cell membrane (1). UPP is synthesized by UppS and then dephosphorylated by a UPP phosphatase (UPP-Pase) to Und-P (2). The MraY enzyme uses Und-P as a substrate, together with UDP–N-acetylmuramic acid (MurNAc)–pentapeptide, to synthesize lipid I, the first membrane-bound precursor of peptidoglycan synthesis (3). The addition of N-acetylglucosamine (GlcNAc) by MurG completes the synthesis of the critical intermediate, lipid II. Lipid II is a single GlcNAc-MurNAc-pentapeptide unit linked to UPP as the lipid carrier and serves as a substrate for penicillin-binding proteins (PBPs), which incorporate the disaccharide unit into the growing glycan strands.
Our understanding of this critical lipid II cycle is compromised by a lack of mechanistic understanding of key steps. The identity of the lipid II flippase required for the export of this essential precursor from the cytoplasmic to the external face of the membrane has been controversial. Recent results provide strong support for a role of MurJ as the lipid II flippase (4) and have further shown that this enzyme is redundant in function with a stress-regulated protein named alternate to MurJ (Amj) (5). The recycling of the UPP carrier to Und-P also involves redundant enzymes, perhaps to allow the conversion to occur on either the inner or outer face of the membrane. UppS synthesizes UPP on the cytosolic face of the membrane. Subsequent dephosphorylation of UPP, also presumed to occur on the cytosolic face of the inner membrane, generates Und-P as the substrate for MraY and TagO (6). After transporting cell wall precursors outside the membrane, the lipid carrier is released as UPP, which must again be recycled back to Und-P by the action of a UPP-Pase (7). This may occur by dephosphorylation on the outer face of the membrane or by flipping of the UPP across the membrane to serve as a substrate for a UPP-Pase active on the inner face of the membrane. It is assumed that the flipping of UPP (or Und-P) from the outer to the inner face of the membrane is facilitated by proteins, but the flippase has not been identified (6). It may be advantageous, in general, for cells to have UPP-Pase activity localized to both faces of the membrane to facilitate de novo Und-P synthesis (on the inner face) and UPP recycling (on the outer face), and this may account in part for the redundancy commonly observed in UPP-Pases (6–8).
In Gram-positive bacteria, the same UPP carrier is shared between the peptidoglycan and the wall teichoic acid (WTA) biosynthesis pathways. For WTA synthesis, Und-P serves as a substrate for TagO (9). As a result, mutations in later steps in WTA synthesis are lethal due to the sequestration of the limiting UPP carrier in dead-end products (10), and this observation has motivated the search for antibiotics active in late stages of WTA synthesis (11). A similar sequestration effect has been reported in Escherichia coli, when synthesis of the enterobacterial common antigen was impaired (1), and in Streptococcus pneumoniae mutants defective in synthesis of serotype 2 capsule (12).
As expected for a critical lipid carrier, the synthesis and recycling of UPP are essential, and therefore, steps in these processes are excellent targets for antibacterials. Recent in silico, in vitro, and in vivo approaches have identified inhibitors of UppS (13–15), including a method that used clustered regularly interspersed short palindromic repeat (CRISPR) interference (CRISPRi) to identify drug targets (16). We demonstrated previously that a ribosome-binding-site (RBS) mutation that decreased the expression of UppS led to vancomycin resistance and activation of the σM-dependent cell envelope stress response (17). Compounds that inhibit the recycling of UPP may also serve as effective antibiotics. The most widely used antibiotic of this class is bacitracin, which binds tightly to the pyrophosphate group on surface-exposed UPP to inhibit its dephosphorylation (18). Bacitracin also activates the σM stress response, which contributes to bacitracin resistance by increasing the synthesis of BcrC (19–21), a predicted UPP-Pase presumed to act on the outer face of the membrane to convert UPP (the target of bacitracin) into Und-P (22). Finally, a variety of structurally diverse antibiotics, including glycopeptides and lantibiotics, bind to lipid II, which serves to both inhibit cell wall synthesis and sequester the UPP carrier lipid (23).
The identity of the UPP-Pases has been clearly established in E. coli, where there are three UPP-pase enzymes (BacA, YbjG, and PgpB) and a fourth enzyme (YeiU, later renamed LpxT) that exhibits UPP-Pase activity in vitro (7). The BacA family includes the eponymous BacA protein, while YbjG, PgpB, and LpxT all belong to the type 2 phosphatidic acid phosphatase (PAP2) superfamily. BacA provides 75% of the cell's UPP-Pase activity, and overexpression of BacA makes cells bacitracin resistant (7). PgpB was originally identified in mutant cells lacking phosphatidylglycerol phosphate phosphatase activity (24) and has been shown to have broad substrate specificity (25, 26). The BacA, YbjG, and PgpB enzymes are functionally redundant; single mutants lacking any one of the three genes do not show significant growth defects. However, a triple mutant missing all three genes is not viable. Although LpxT displayed in vitro UPP-Pase activity, it could not support growth in the absence of at least one of the other three UPP-Pases (7). It was later found that LpxT transfers phosphate from UPP to lipid A to produce lipid A 1-diphosphate and in the process generates Und-P (27). In contrast, the number and identity of the UPP-Pases in Bacillus subtilis has not been resolved.
Genetic approaches to analyze the function of essential genes often rely on conditional mutants in which either gene expression or protein activity can be regulated. The optimization of conditional gene expression systems can be challenging (as reported here also), since leaky expression may suffice to support viability even in the absence of induction or the induced level may be insufficient to support viability. Recently, the CRISPR system, used by many bacterial and archaeal species to defend against foreign DNA, has been adapted as a powerful tool for conditionally regulating bacterial gene expression (16, 28–30).
Here, we report the use of an optimized CRISPRi system to investigate the essentiality of candidate UPP-Pases in B. subtilis. Using CRISPRi, we have identified UppP and BcrC as two functionally redundant UPP-Pases. We have also identified a third lipid phosphatase (YodM) that is capable of supporting growth in the absence of these two enzymes, but only when artificially overexpressed. Depletion of essential UPP-Pases, predicted to interrupt the synthesis of both peptidoglycan and WTA, results in cells that cannot maintain their rod shape and leads to the induction of the σM-dependent cell envelope stress response.
MATERIALS AND METHODS
Strains, plasmids, and growth condition.
All strains and plasmids used in this work are listed in Table S4 in the supplemental material. Bacteria were grown in liquid lysogeny broth (LB) medium with shaking or on LB plates (1.5% agar; Difco) at 37°C unless otherwise stated. Plasmids were constructed using standard methods (31), and amplified in E. coli DH5α before being transformed into B. subtilis. For selection of transformants, 100 μg/ml ampicillin was used for E. coli. The antibiotics used for selection of B. subtilis transformants included kanamycin (15 μg/ml), spectinomycin (100 μg/ml), macrolide-lincosamide-streptogramin B (MLS) (includes 1 μg/ml erythromycin and 25 μg/ml lincomycin), neomycin (10 μg/ml), and chloramphenicol (10 μg/ml).
Genetic techniques.
Chromosomal and plasmid DNA transformation was performed as described previously (32). Markerless in-frame deletion mutants were constructed from BKE strains as described previously (5). Briefly, BKE strains were acquired from the Bacillus Genetics Stock Center, chromosomal DNA was extracted, and the mutation, containing an erm cassette, was transformed into our wild-type (WT) strain 168. The erm cassette was subsequently removed by the introduction of plasmid pDR244, which was later cured by growing at the nonpermissive temperature of 42°C. Gene deletions were confirmed by PCR screening using flanking primers. Transduction was performed to move bacteriophage SPβ derivatives into the 168 strain as previously described (33). Unless otherwise described, all PCR products were generated using B. subtilis strain 168 chromosomal DNA as the template. The DNA fragments used for gene overexpression were sequence verified. The null mutant construct was verified by PCR screen to have the correct size band.
Construction of the CRISPRi-based transcriptional repression system.
The CRISPRi system was based on plasmid vectors that replicate in E. coli and integrate into the B. subtilis chromosome. Briefly, the gene encoding catalytically inactive (“dead”) CRISPR-associated protein 9 (dCas9) is inserted into plasmid pAX01 to form pJPM1. pJMP1 is then integrated into the B. subtilis ganA gene by double-crossover recombination. single guide RNAs (sgRNAs) targeting bcrC or uppP were incorporated into integrative vectors by inverse PCR followed by cyclization by self-ligation. The expression of sgRNAs is constitutive, and the expression of dCas9 is induced by xylose. The sgRNAs were selected based on their specificity for and location within target genes. We identified all protospacer-adjacent motif (PAM) sites (NGG for Streptococcus pyogenes dCas9) within target genes and designed all possible sgRNAs for those targets. We then assigned each sgRNA a specificity score by aligning progressive truncations of the sgRNA sequences to the B. subtilis 168 genome using Bowtie (34); sgRNAs that retained specificity after several truncations received better scores. We then selected the most specific sgRNAs that targeted the nontemplate strand and were near the 5′ end of the gene, where CRISPR interference is thought to be most effective in bacteria (28, 30). After choosing the sgRNA sequences, the constructs were made by inverse PCR, using forward primers containing sequences specific to the genes of interest (base-pairing region) and a universal reverse primer which binds to the complementary strand of DNA adjacent to the base-pairing region. For example, bcrC1 (one of the two sgRNAs designed to bind close to the transcription start site), was made using pJMP3 as the template and primers 6411 CRISPRi bcrC-1 F and 6415 CRISPRi universal R. The inverse PCR product was self-ligated and transformed into E. coli DH5α. Plasmids extracted from successful transformations were confirmed by sequencing before being used to transform B. subtilis.
Construction of pSPβ-dCas9 was done by ligating a fragment containing the Pxyl-dCas9 construct from pJMP1 into the backbone of pJPM122, a plasmid that can integrate into the SPβ prophage in strain ZB307A, as described previously (35). To get the fragment containing Pxyl-dCas9, pJMP1 was first digested by SacII, treated with T4 DNA polymerase (NEB) to make a blunt end, and then digested by SalI. The DNA fragment containing Pxyl-dCas9 was gel purified. pJPM122 was digested with SalI and EcoRV, and the backbone was recovered from the agarose gel. The fragment containing Pxyl-dCas9 from pJMP1 was then ligated with pJPM122, forming plasmid pSPβ-dCas9. pSPβ-dCas9 was then used to transform B. subtilis ZB307A. The transformants were PCR screened to confirm that a double-crossover recombination event into the chromosome of ZB307A had occurred, and the verified transformants were named ZB307A/pSPβ-dCas9. Phage lysate from ZB307A/pSPβ-dCas9 was prepared using heat shock and was used to transduce B. subtilis 168 and its derivatives.
Disk diffusion assays.
Disk diffusion assays were performed as previously described (21). Overnight cultures of test strains were reinoculated into fresh LB liquid medium and grown to early log phase (optical density at 600 nm [OD600] of ∼0.4), and a 100-μl aliquot of the culture was mixed briefly with 4 ml LB soft agar (containing 0.75% agar, kept at 50°C) before being poured onto LB plates (containing 15 ml LB with 1.5% agar). The plates were then cooled and dried in a laminar airflow hood for 5 min. Filter paper disks were placed on the surface of the plates, and chemicals were added to the paper disks. The plates were then incubated at 37°C for 16 h before the diameter of the zone of inhibition was measured. The numbers reported are the zone diameter minus the 6.5-mm diameter of the filter paper disk. The chemicals added to the disks included the following: xylose (20 μl of a 50% solution), bacitracin (400 μg), ampicillin (1 mg), penicillin (1 mg), cefuroxime (10 mg), vancomycin (100 μg), fosfomycin (1 mg), d-cycloserine (100 μg), lysozyme (100 μg), daptomycin (100 μg), nisin (1 mg), SDS (10 μl of a 10% solution), Triton X-100 (10 μl of a 25% solution), EDTA (10 μl of a 0.5 M [pH 8.0] solution), novobiocin (100 μg).
Measurement of the fraction of suppressor mutants.
Strains were grown in liquid LB medium supplemented with 1% glucose overnight, and then the overnight culture was reinoculated into fresh LB liquid medium supplemented with 1% glucose and grown to early log phase (OD600 of ∼0.4). Serial dilutions were performed up to 10−6. Cultures with 10−4, 10−5, and 10−6 dilutions were plated on LB plates supplemented with 1% glucose, and cultures with no dilution and 10−1 and 10−2 dilutions were plated on LB plates supplemented with 2% xylose. Colonies were counted from plates having between 20 and 200 colonies, and CFU counts were calculated. The fraction of suppressor mutants is calculated by dividing the CFU count of a strain growing on xylose LB by the CFU count of the same strain growing on glucose LB. This experiment was done at least three times using biological replicates.
Phase-contrast time-lapse and fluorescence microscopy.
Strains were grown in CH medium (36) in the presence of 1% (wt/vol) glucose at 37°C. The cells were harvested when the OD of the culture reached ∼0.4. Then, cells were grown in CH medium in the presence of 2% (wt/vol) xylose in a microfluidic chamber. The phase-contrast time-lapse images were acquired on a Nikon N-STORM microscope equipped with a Plan Apo lambda 100× objective and a Hamamatsu ORCA complementary metal oxide semiconductor (CMOS) camera. Images were taken every 30 s. Images of FM5-95 (ThermoFisher) membrane staining were acquired on a GE DeltaVision Elite with DeltaVision connected to a PCO Edge scientific CMOS (sCMOS) camera. A 561-nm laser and a 60× 1.45 numeric aperture (NA) Plan Apo objective were used. The excitation filter was yellow fluorescent protein (YFP; with center wavelengths at 513-nm and 17-nm bandwidths), and the emission filter was mCherry (with center wavelengths at 632-nm and 60-nm bandwidths). The cells were stained with 1× FM5-69 (2 μM) mounted on 1% agarose in CH medium pads and immediately imaged with an exposure time of 2 s. All images were analyzed using Morphometrics (37) and a home-built MATLAB program.
Whole-genome sequencing and sequence analysis.
Chromosomal DNA of suppressor strains was extracted using the Qiagen DNeasy blood and tissue kit. DNA was then sent for sequencing using an Illumina HiSeq2500 instrument in high-output mode with single-end 100-bp reads. The sequencing results were analyzed using CLC workbench version 8.5.1. Single nucleotide variants (SNVs) were detected using default settings.
Microarray analysis.
The wild-type and depletion strains were grown separately in liquid LB medium to an OD600 of ∼0.4 and then each diluted 100-fold into two 250-ml flasks each containing 50 ml of fresh LB medium. After 1.5 h of initial growth, one of the two flasks of each strain was supplemented with 50% xylose to a final concentration of 2%, while the other flask was left untreated as a control. After another 2.5 h of growth, the cells were collected and total RNA was extracted using a phenol-chloroform-based method. The total RNA was quantified, and the quality of RNA was tested using a Bioanalyzer instrument. The RNA quality numbers for all four RNA samples were 10 out of 10. The total RNA was then treated with Turbo-DNase (Thermo Fisher Scientific) to remove any possible DNA contamination and reverse transcribed into cDNA, using the SuperScript indirect cDNA labeling system (Thermo Fisher Scientific). The cDNA was aliquoted and labeled with either fluorescent dye 555 or dye 647 and hybridized to microarray slides containing 60-nucleotide (nt) oligonucleotides for each coding region according to laboratory protocol (31). The dye swap was performed to reduce systematic errors caused by differences in labeling and hybridization efficiency for different dyes. For instance, in one hybridization, the cDNA of the WT strain is labeled with dye 555 and the cDNA of the depletion strain is labeled with dye 647. In the dye swap experiment, the cDNA from the WT will then be labeled with dye 647 and that of the depletion strain with dye 555.
Comparison of transcriptomes among different strains and conditions was performed by comparing the fluorescent signals from different pairs. Among the treated group, the transcriptomes of the WT and depletion strains were compared to reveal changes in gene transcription, and the transcriptomes of untreated WT and depletion strains were used as controls. The fold change for each gene is the average of the results of two experiments containing a dye swap. Up- and downregulated genes after UPP-Pase depletion were defined as those with an average fold change of at least threefold and at least twofold in each hybridization. It is also crucial that only signals well above the background level are considered trustworthy. Gene function and regulon information was acquired from SubtiWiki (http://subtiwiki.uni-goettingen.de/ [38]).
Cluster 3.0 (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm) and Java Treeview (https://sourceforge.net/projects/jtreeview/) were used to generate heat maps. Briefly, we chose genes regulated by extracytoplasmic function (ECF) sigma factors and two-component systems involved in cell envelope homeostasis (e.g., WalR, BceR, LiaR, PsdR, and YvrHb). The bcrC gene was not included in the list of genes (even though it is controlled by multiple ECF sigma factors), as it was artificially repressed using CRISPRi. Microarray fold change data were log10 transformed and used for hierarchical clustering (using uncentered correlation and complete linkage functions), showing genes that were upregulated as red and genes that were downregulated as green.
Accession number(s).
The transcriptome results have been deposited in the NCBI GEO database under accession number GSE85492.
RESULTS
UppP and BcrC are functionally redundant UPP-Pases.
We identified B. subtilis UppP, BcrC, and YodM as homologs of known UPP-Pases in E. coli using BLASTp with default parameters. UppP is a homolog of BacA, whereas BcrC and YodM are homologs of YbjG, PgpB, and LpxT, members of the PAP2 superfamily (see Table S1 in the supplemental material). BcrC was previously shown to have UPP-Pase activity and to be regulated by σM (19, 22). UppP (YubB) was also speculated to function as a UPP-Pase based on homology but played a less important role than BcrC in bacitracin resistance (22). Mutations that affect UPP levels were later found to affect the sensitivity of B. subtilis to rare earth metals and, based on this phenotype, it was also suggested that BcrC was the major enzyme, with UppP having a minor role (39). However, this assay may report specifically on the level of extracellular UPP that is accessible for binding by rare earth metals.
To investigate functional redundancy among these candidate UPP-Pases, we constructed single mutants with each UPP-Pase gene replaced by an antibiotic cassette, followed by transformation to make mutants missing two or all three UPP-Pases. While we could construct yodM bcrC and yodM uppP double mutants, we were unable to obtain a bcrC uppP double mutant in multiple attempts, despite previous suggestions that a double mutant had been obtained (22, 39). For example, when chromosomal DNA from a bcrC::mls (macrolide-lincosamide-streptogramin B) strain was used to transform a uppP::spc (spectinomycin) strain, we recovered MLSr transformants, but only if the cells simultaneously acquired a functional copy of uppP (and therefore lost Spcr). This phenomenon, in which a cell acquires two unlinked markers by transformation, is referred to as congression. Overall, our findings indicate that bcrC and uppP are a synthetic lethal gene pair and likely encode the only functional UPP-Pases in B. subtilis, at least under our laboratory conditions.
The YodM protein can provide UPP-Pase activity when overexpressed.
To characterize the physiological consequences of limiting UPP-Pase activity, we sought to generate conditional depletion strains. We introduced ectopic copies of bcrC, uppP, or yodM under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter Pspac(hy). Next, we attempted to sequentially delete bcrC and uppP under conditions where the ectopic gene was expressed. However, due to either leakiness or insufficient expression, we were unable to construct a depletion strain conditionally expressing bcrC or uppP, even after attempts to modify the ribosome-binding site to change the translational efficiency (see Table S2 in the supplemental material) (40).
One interesting result that emerged from this exercise is that YodM, if overexpressed, can support growth even in the absence of BcrC and UppP. In this strain, the native yodM RBS was replaced with a strong RBS (40) and the start codon was changed from TTG to ATG. The resulting Pspac(hy)-yodM construct allowed the generation of a bcrC uppP double mutant strain that could grow in the presence but not in the absence of IPTG (see Fig. S1 in the supplemental material). Note that this strain has two copies of yodM, a native copy and an ectopic copy controlled by Pspac(hy) and optimized for expression. Because of the homology between YodM and E. coli UPP-Pases and the fact that overexpression of YodM can rescue the growth defect of the bcrC uppP double mutant, we infer that YodM has some UPP-Pase activity and that this activity is sufficient to support growth in the absence of the other two UPP-Pases when it is expressed at an artificially high level. However, YodM is not able to support the growth of a bcrC uppP double mutant at its normal expression levels. Recently, we have learned that similar results were independently obtained by the group of Thorsten Mascher (personal communication). Overall, we conclude that BcrC and UppP are the two primary UPP-Pases, at least under our laboratory conditions.
Demonstration of essentiality of UPP-Pases using CRISPRi.
We next explored the use of CRISPRi as a tool to monitor the effects of UPP-Pase depletion on cell physiology. The CRISPRi system uses a catalytically inactive form of the Cas9 endonuclease (dCas9) and a single guide RNA (sgRNA) (28, 41). The sgRNA contains a customizable 20-nt base-pairing region to direct dCas9 to specific DNA sequences, and the dCas9-sgRNA complex binds DNA and serves as a road block to transcription. In the system used in this work, the dCas9 gene is expressed from a xylose-inducible promoter and the sgRNAs are constitutively expressed (16). Based on previous studies in E. coli (28) and mycobacteria (42), we anticipated that the strength of transcriptional repression would be increased by promoter-proximal targeted complexes and by the use of two sgRNAs. We therefore designed two sgRNAs to bind close to the transcription start site (bcrC1 and uppP1) (predicted to have a strong repressive effect) and two sgRNAs to bind further downstream (bcrC2 and uppP2) (predicted to have weaker repressive effects).
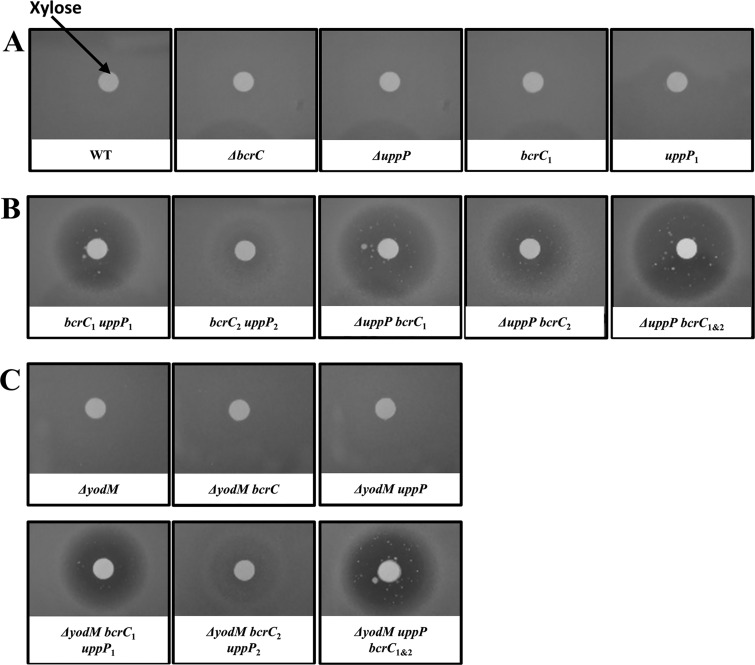
The conditional depletion strains did not show any growth defect in the absence of the dCas9 inducer xylose but were growth impaired in medium with xylose. In order to illustrate the functional redundancy of BcrC and UppP, we used disk diffusion assays as a measure of xylose sensitivity. B. subtilis is not sensitive to xylose per se and can use it as a carbon source (43). As expected based on the phenotype of the null mutant strains, depletion of either uppP or bcrC using CRISPRi did not lead to xylose sensitivity (Fig. 1A). However, strains in which both genes were targeted for transcriptional repression showed a notable sensitivity to xylose, as shown by a clear zone of inhibition around the paper disk containing xylose (Fig. 1B). A similar effect was noted in strains where uppP was deleted and bcrC was targeted for transcriptional repression. Strains containing the weak sgRNAs bcrC2 and uppP2 showed a small zone of inhibition, demonstrating that these cells are less xylose sensitive and suggesting that bcrC2 and uppP2 provide less efficient transcriptional repression (Fig. 1B). This is consistent with the previous observation that sgRNAs farther from the 5′ end of the transcription start site caused weaker repression (42). In the uppP null mutant, depleting bcrC led to xylose sensitivity. The use of the promoter-proximal sgRNA bcrC1 led to a high level of xylose sensitivity, and this was modestly increased with the inclusion of the second sgRNA, bcrC2. We were not able to construct a strain with bcrC deleted and containing uppP1, possibly due to leaky expression of dCas9, which has been shown to have moderate repressive activity even in the absence of xylose induction (16).
FIG 1.
Growth inhibition by depletion of BcrC and UppP. Photos of disk diffusion assay of WT and mutant strains. Cells were grown on LB plates with xylose on the filter paper disk to induce dCas9 and inhibit transcription of bcrC and/or uppP. (A) WT and mutants with either bcrC or uppP null mutations or containing sgRNA targeting only one UPP-Pase are not sensitive to the induction of dCas9 by xylose. (B) Mutants containing sgRNAs for both bcrC and uppP (bcrC1 uppP1 or bcrC2 uppP2) and mutants with a uppP null mutation and sgRNA(s) for bcrC (ΔuppP bcrC1, ΔuppP bcrC2, and ΔuppP bcrC1 bcrC2 [ΔuppP bcrC1&2]) are sensitive to xylose. (C) Mutant strains show similar phenotypes in a ΔyodM background.
Optimization of CRISPRi to reduce suppressor formation in conditional depletion strains.
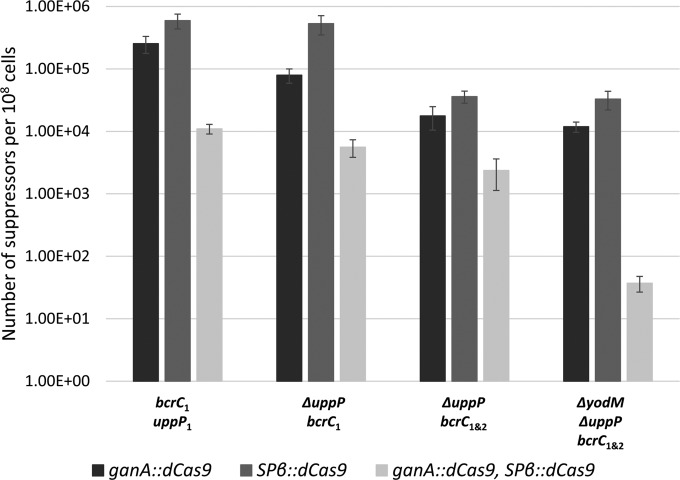
It is apparent that suppressor mutations arise frequently in the zone of xylose-dependent growth inhibition (Fig. 1B). Whole-genome sequencing of 10 independent suppressor strains revealed frameshift mutations in the dCas9 gene (nine strains) and a single base pair mutation in the bcrC gene which altered the protospacer-adjacent motif (PAM) sequence for sgRNA bcrC1 and thus prevented binding of the dCas9-sgRNA complex (one strain). To facilitate further physiological studies, we sought to first reduce the frequency of suppressor mutants. Since our sequencing results indicate that inactivation of the gene encoding dCas9 is the single most frequent class of suppressor mutation, we constructed a merodiploid strain containing a second copy of dCas9 integrated into the SPβ prophage. We then monitored the frequency with which suppressor mutations arise by plating the depletion strain on LB medium supplemented with 1% glucose or 2% xylose and comparing the CFU counts. As expected, strains with two copies of the gene encoding dCas9 displayed a reduction (approximately 10-fold) in the frequency of suppressor mutations (Fig. 2). We also note that suppressor mutation formation was somewhat higher in strains with dCas9 expressed only from the SPβ prophage locus rather than the ganA locus (Fig. 2; see also Fig. S2A in the supplemental material). Since it is also possible to isolate suppressor mutations that mutate either the sgRNA or its target, we reasoned that having two sgRNAs targeted to the same gene would further decrease the frequency of suppressor mutants. In general, the frequency of suppressor mutations was reduced in the uppP null strain carrying both bcrC1 and bcrC2 sgRNAs relative to their frequency in the strain with only bcrC1; however, these effects were rather modest. We hypothesized that another possible class of suppressor mutations might result from upregulation of yodM. Indeed, the introduction of a yodM null mutation into the uppP bcrC1 bcrC2 strain greatly reduced the frequency of suppressor mutants, but only if dCas9 was present in two copies (Fig. 2; see also Fig. S2B and C). These results indicate that inactivation of dCas9 is the most frequent cause of suppression (as inferred from the whole-genome sequencing studies) and that the use of two sgRNAs and elimination of known or predicted pathways of suppression (such as upregulation of yodM) may be required for a significant further reduction in suppressor mutation occurrence.
FIG 2.
Reduction of suppressor mutations in UPP-Pase depletion strains. Fractions of suppressor mutants in populations of various depletion strains. UPP-Pase depletion strains cannot grow on LB plates containing 2% xylose unless they have a suppressor mutation. Cells were plated on LB plates with or without 2% xylose, and the number of cells on each type of plate was counted and back calculated to CFU/ml. The fraction of suppressor mutants in each population was calculated by dividing the CFU count of suppressors by the CFU count of total cells. Error bars show standard deviations.
We then isolated two independent suppressor mutants from the optimized strain (HB17235) and performed whole-genome sequencing. While we might have expected to discover mutations revealing a novel UPP-Pase(s), each suppressor mutant contained a frameshift mutation (base deletion) in a homopolymeric stretch within both dCas9 genes (in one suppressor mutant, the sequence was TTCCTTTGAAAAAAA, and in the other, it was CAGTATCAAAAAAAA). The fact that both dCas9 genes in each suppressor mutant have the same point mutation in each copy of the gene leads us to speculate that the first mutation may have resulted from a relatively frequent strand slippage event in the homopolymeric sequence, followed by a gene conversion event to generate the double mutant.
One of our goals in generating depletion strains for UPP-Pases was to monitor the terminal phenotype, and this is complicated if suppressor mutants arise at a high frequency. To determine whether our optimized CRISPRi system had sufficiently reduced the appearance of suppressor mutants to allow growth studies, we inoculated ∼4 × 104 cells into 200 μl of xylose-containing medium using an automated BioScreen growth analyzer. For each strain, we monitored 30 replicate cultures (3 biological replicates containing 10 technical replicates each). In this assay, the majority of the cultures for each strain grew after a long lag phase (see Table S3 in the supplemental material), suggesting that suppressor mutations were arising and might therefore complicate the analysis of the terminal phenotype. The single exception was our optimized strain (HB17235), which is a dCas9 merodiploid lacking both UppP and YodM and with two sgRNAs targeting bcrC. With this strain, there was outgrowth in less than 10% of the wells.
Dynamics of UPP-Pase depletion.
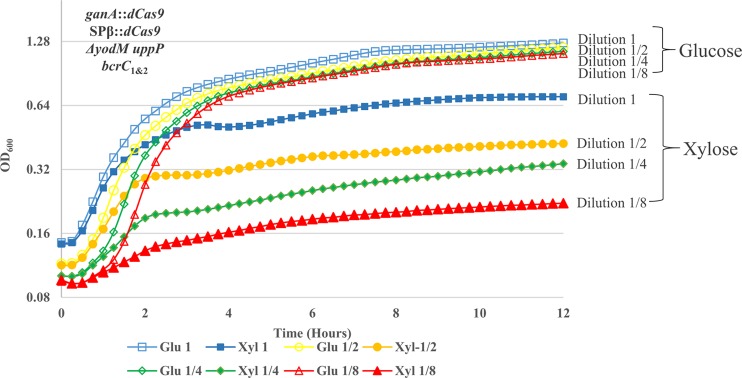
To determine how long it takes for the existing BcrC protein to be depleted in the optimized (HB17235) strain and thereby limit growth, we monitored growth curves as a function of the initial inoculum in LB with either 1% glucose or 2% xylose. With a starting culture at an initial OD600 of 0.14, the culture reached a final OD600 of 0.7 in the presence of 2% xylose, while it could reach an OD600 of ∼1.2 with glucose (Fig. 3). With each twofold dilution of the initial inoculum, the final OD600 reached under depletion conditions was also decreased by nearly twofold, consistent with a growth-limiting depletion of preexisting BcrC after 3 to 4 doublings.
FIG 3.
Growth of UPP-Pase depletion strain under depletion/nondepletion conditions with different initial culture population size. Depletion strain (ganA::dCas9 SPβ::dCas9 ΔyodM uppP bcrC1 bcrC2) growing at logarithmic phase was diluted and reinoculated into liquid LB medium supplemented with either 1% (wt/vol) glucose or 2% (wt/vol) xylose. Growth was carried at 37°C with shaking. OD600 was measured every 15 min. The starting culture with an OD600 of ∼0.1 was designated dilution 1, and further twofold dilutions were performed until the smallest inoculum contained 1/8 the amount of the cells in dilution 1. Different initial inoculums affected the final OD of cells growing in LB xylose medium, while they had no significant effect on cells growing in LB glucose medium.
Changes in cell morphology upon depletion of UPP-Pases.
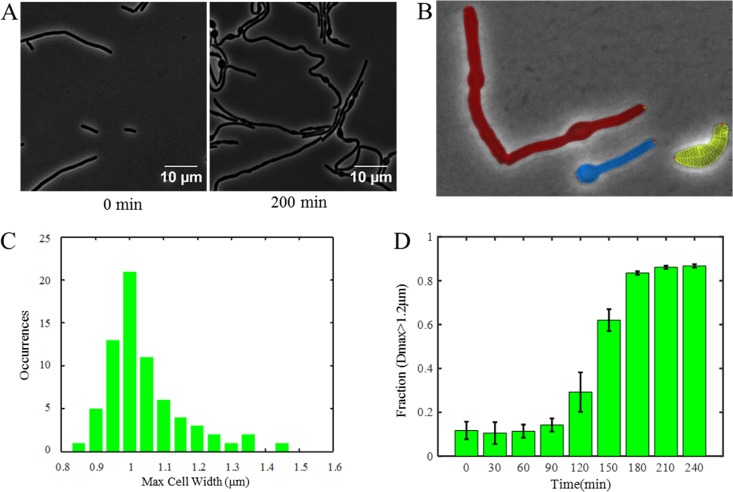
Defects in cell wall synthesis often cause morphological changes as the integrity of the cell wall is compromised. For example, depletion of MreB and its homologs leads to a loss of the rod shape and the formation of bulged cells (44), and cells missing all class A PBPs form longer and bent cells with abnormal protrusions into the cell plasma (45). Cells with UppS expression repressed by CRISPRi also show bulged cells without significant lysis (16). We therefore anticipated a similar phenotype from the UPP-Pase depletion strain. Cells were grown in a microfluidic chamber in CH medium supplemented with 2% xylose, and images were taken every 30 s. At the beginning of the experiment, cells did not show any obvious morphological defect relative to the WT. After about 2 h, cells grown with xylose began to form bulges (Fig. 4; see also Movie S1 in the supplemental material). In comparison, WT cells did not exhibit any growth defect in the presence of 2% xylose (see Movie S2). Surprisingly, few cell lysis events were observed during the time lapse experiment. This is similar to what was observed in the UppS depletion strain (16, 46) and a strain depleted of lipid II flippases (5). The lantibiotic NAI-107 (which targets UPP-linked cell wall precursors like lipids I and II and precursors of WTA) leads to a similar terminal phenotype (47).
FIG 4.
UPP-Pase depletion leads to curved and bulged cells. Microscopy images of depletion strains under depletion conditions. Cells were observed by phase-contrast microscopy under ×100 magnification (each pixel size is 65 nm). (A) Depletion strain (HB17235) growing in CH medium supplemented with 2% xylose at 37°C. Cells showed normal morphology at the beginning of the experiment (0 min) and showed curved and bulged morphology as UPP-Pases were depleted (200 min). (B) Example showing how cell width was measured. Lines were drawn across the cell width, and the cell width at each line was measured. The maximum width of each cell was used for quantification. (C) Distribution of maximum cell widths of depletion strain at the beginning of treatment (0 min). The majority of cells had a maximum width of 0.95 to 1.05 μm, while approximately 10% of cells had a maximum width greater than 1.2 μm. (D) Changes in fraction of cells having maximum cell width (Dmax; maximum diameter of cells) greater than 1.2 μm through time. The fraction increased significantly after 120 min and reached its peak after 180 min, indicating that more and more cells began to form bulges as UPP-Pases were depleted. Error bars show standard deviations.
To quantify the fraction of cells forming bulges when UPP-Pases were depleted, we observed more than 200 cells at each time point and used Morphometrics and an in-house MATLAB script to measure the cell width along the whole length of the cell and determine the maximum cell width for each cell (Fig. 4B). Before the depletion of UPP-Pases, the majority of cells exhibited a maximum width of between 0.95 and 1.05 μm (Fig. 4C) and only ∼10% of cells had a width of >1.2 μm (Fig. 4C and D). As the cells grew under depletion conditions, the percentage of cells with a maximal width above 1.2 μm increased significantly (Fig. 4D). Septum formation was still present in the depletion strain, as visualized with the membrane stain FM5-95, even when cells exhibited bulging (see Fig. S3 in the supplemental material).
The UPP-Pase depletion strain is sensitive to cell envelope stress.
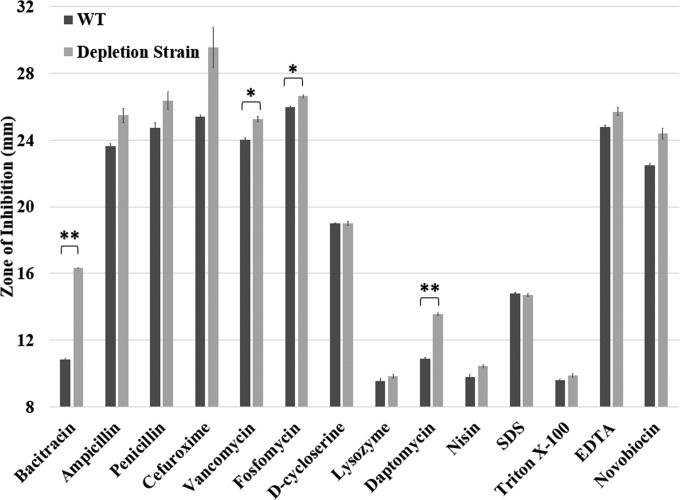
It has been reported that leaky expression of dCas9 in the absence of xylose may lead to ∼3-fold repression of target genes (16). We therefore hypothesized that our depletion strain may have reduced expression of UPP-Pases even in the absence of inducer. Indeed, the depletion strain exhibited more lysis than the WT when stored on plates. To test this hypothesis, we used disk diffusion assays under noninducing conditions to monitor sensitivity to cell envelope stress conditions (Fig. 5). In fact, the depletion strain was significantly (P < 0.01) more sensitive to both bacitracin, which binds UPP and thereby inhibits UPP-Pase activity, and daptomycin. A more modest level of sensitivity (P < 0.05) was noted for the peptidoglycan synthesis inhibitors vancomycin and fosfomycin. The depletion strain was not significantly more sensitive to several other tested stress conditions, including compounds that elicit membrane stress (nisin, SDS, and Triton X-100). The reason for the increased sensitivity to some but not other peptidoglycan synthesis inhibitors is presently unclear, but interpreting such effects can be complex, as discussed previously (17).
FIG 5.
Sensitivity of depletion strain against stresses under partial depletion conditions. Cells were grown on LB plates with chemicals added onto the filter paper disk. The sensitivity of the depletion strain was measured as the clear zone of inhibition around the paper disk. Chemicals used in this assay include bacitracin at 400 μg, ampicillin at 1 mg, penicillin at 1 mg, cefuroxime at 10 mg, vancomycin at 100 μg, fosfomycin at 1 mg, d-cycloserine at 100 μg, lysozyme at 100 μg, daptomycin at 100 μg, nisin at 1 mg, SDS at 10 μl of 10% solution, Triton X-100 at 10 μl of 25% solution, EDTA at 10 μl of 0.5 M (pH 8.0) solution, and novobiocin at 100 μg. The data are expressed as mean results ± standard errors (n = 3). Statistical analysis was performed using the t test with two samples assuming unequal variances. *, two-tailed P value of <0.05; **, two-tailed P value of <0.01.
Depletion of UPP-Pases triggers a cell envelope stress response.
To further characterize the effects of UPP-Pase depletion, we monitored changes in the transcriptome 2.5 h after the addition of xylose to deplete BcrC, a time known to coincide with the onset of visible morphological changes and decreased growth (Fig. 3 and 4). In the depletion strain, >150 genes were upregulated >3-fold and nearly 300 genes were downregulated >3-fold (see Data Set S1 in the supplemental material). The set of upregulated genes includes known cell envelope stress response genes, including, most notably, the σM regulon. In contrast, downregulated genes include many genes and regulons consistent with catabolite repression in the xylose-treated cultures and with entry of the untreated cells into transition phase during the 2.5 h of incubation. The upregulation of the flagellar and motility systems controlled by σD in the control cells as they enter into transition phase likely explains the apparent downregulation of these genes in the depletion strain.
To gain further insights into the effects of UPP-Pase depletion on cell envelope stress responses, we used hierarchical clustering to compare the expression of genes regulated by ECF sigma factors and the WalR, BceR, PsdR, LiaR, and YvrHb two-component system regulons (Fig. 6). The microarray data generated in this study (UPP-Pase depletion), as well as data for cells treated with targocil (46), bacitracin (48), d-cycloserine (49), Triton X-100 (49), and vancomycin (50) (see Data Set S1 in the supplemental material) were used for clustering and heat map generation as previously described (50). Based on the observations described above, we anticipated that the effects of UPP-Pase depletion (occurring over 2.5 h) might not be directly comparable to those elicited by relatively short treatments with cell envelope antibiotics. Nevertheless, we noted several striking similarities. Specifically, the σM regulon is selectively induced by UPP-Pase depletion, with comparatively weak effects noted for other ECF σ factor stress responses. Induction of the σM regulon is consistent with the inclusion of the BcrC UPP-Pase as part of this regulon (19), as well as the previously noted induction in cells treated with bacitracin (48) or containing mutations that decrease UPP synthesis (17). In contrast with σM, the σW regulon, most strongly activated by membrane stress conditions (50), was downregulated in the depleted cells compared to its expression in the control.
FIG 6.
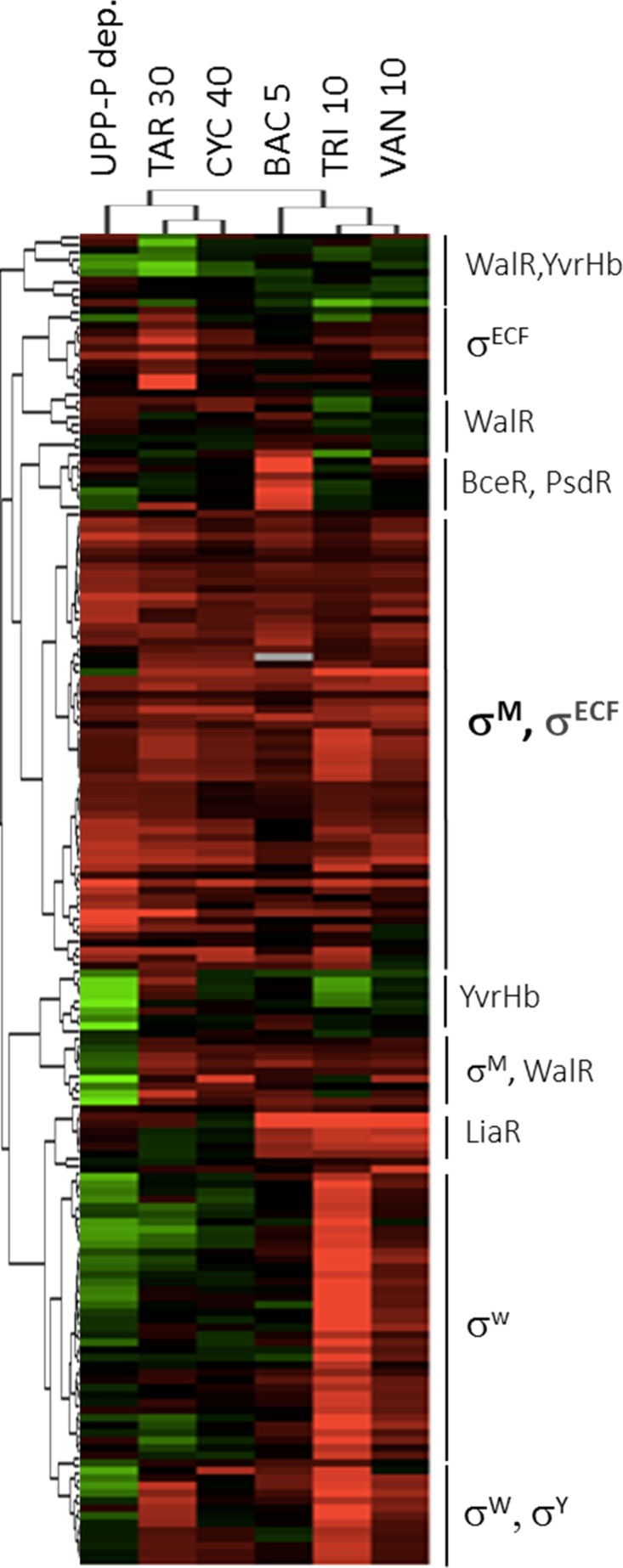
UPP-Pase depletion induces the σM cell envelope stress response. Hierarchical clustering was used to generate a heat map of upregulation (red) or downregulation (green) of known cell envelope stress-responsive genes (genes and regulons are indicated to the right). Cells were either depleted of UPP-Pase (compared to non-depletion strain control cells) or treated with targocil (TAR), d-cycloserine (CYC), bacitracin (BAC), Triton X-100 (TRI), or vancomycin (VAN). Numbers indicate the minutes of treatment with each antibiotic.
In addition to the clustering based on genes, we also determined the clustering based on treatments. Among the antibiotics used for comparison, the treatment condition most similar to UPP-Pase depletion is targocil, which inhibits WTA export and therefore also causes a gradual depletion of the Und-P pool. d-cycloserine, an inhibitor of early steps in peptidoglycan synthesis that converge with Und-P to allow lipid II synthesis, also leads to a similar set of stress responses. We might have expected bacitracin (which binds directly to UPP) to trigger a response similar to the response to UPP-Pase depletion. However, despite several similarities, bacitracin additionally elicits strong (albeit transient) induction of the BceR and LiaR regulons after 5 min of treatment (51). This activation can happen at bacitracin concentrations well below the MIC and does not depend on cell wall damage (52). The responses to stress conditions that were least similar to the response to UPP-Pase depletion were those elicited by Triton X-100 and vancomycin, which both led to the induction of the large σW regulon under the conditions tested.
DISCUSSION
UPP is a crucial lipid carrier in the synthesis of components of the cell envelope, including both peptidoglycan and WTA. In peptidoglycan synthesis, UPP serves as a lipid carrier for the disaccharide-pentapeptide precursor in the central intermediate lipid II. Although initially synthesized as the pyrophosphate form (UPP), dephosphorylation is required to generate the substrate (Und-P) for MraY. Similarly, in B. subtilis strain 168, Und-P is required for WTA synthesis, which is initiated by the transfer of GlcNac-1-phosphate from UDP-GlcNac to Und-P by TagO to generate the initial membrane-bound precursor. The peptidoglycan precursor, lipid II, is flipped across the membrane by either of two flippases, MurJ or Amj, where it serves as the substrate for synthesis of the glycan polymer by PBPs. In contrast, synthesis of the WTA polymer occurs in the cytosol and the completed polymer (a glycerol-phosphate alternating copolymer in B. subtilis) is exported and then attached to peptidoglycan. In both peptidoglycan and WTA synthesis, the UPP carrier is released on the extracytoplasmic face of the membrane (by PBPs or the TagTUV family of WTA attachases) and recycled (9).
Disruption of cell envelope synthesis is one of the most effective strategies for inhibiting bacterial growth, and numerous antibiotics target these processes. The lipid II cycle can be disrupted by compounds that inhibit UPP synthesis or that bind directly to lipid II (nisin, vancomycin, and ramoplanin) and to UPP itself (bacitracin). Other peptidoglycan synthesis inhibitors target the PBPs and thereby inhibit the transpeptidation (β-lactams) or transglycosylation (moenomycin) reactions. It is also possible to disrupt the lipid II cycle by blocking the synthesis of WTA at late steps, thereby leading to sequestration of this limiting lipid carrier (11).
A common feature of many different compounds that inhibit the lipid II cycle is that they invoke cell envelope stress responses, including activation of the σM regulon (53). A major function of the σM regulon is to increase the expression of key enzymes involved in cell envelope synthesis and to upregulate alternative enzymes to replace those that might be inhibited. Notable examples of σM-dependent compensation reactions include upregulation of the lipid II flippase Amj (which substitutes for MurJ) (5), the lipoteichoic acid synthase LtaSa (YfnI, which substitutes for LtaS), and BcrC (which substitutes for UppP) (19, 50). Similarly, antibiotics that inhibit the lipid II cycle, including bacitracin (48), vancomycin (50), moenomycin (54), and the WTA-targeting targocil (46), all activate the σM regulon (53).
In the work presented here, we set out to explore the role of three genes encoding candidate UPP-Pases in the lipid II cycle in B. subtilis. The CRISPRi system has recently been adapted as a tool to repress bacterial gene expression and facilitate the analysis of essential gene functions (16). Our inability to construct a uppP bcrC double mutant suggested that these two genes might be a synthetic lethal pair (in contrast with a previous report [22, 39]), although they could both be deleted when a third candidate phosphatase, YodM, was overexpressed. The ability of YodM to rescue cell growth in a uppP bcrC double mutant and its homology to other UPP-Pases support a possible role for YodM as a UPP-Pase that is active under growth conditions not yet identified. Alternatively, this may be an adventitious reaction that only occurs when this enzyme is artificially overexpressed. This may be similar to the case in E. coli, where LpxT transfers phosphate from UPP to lipid A and, as a product, also generates Und-P (27). The efficiency of recycling UPP by this reaction is not enough to support cell growth, but this enzyme indeed has UPP-Pase activity. To test the hypothesis that uppP and bcrC are a synthetic lethal pair and to explore the physiological consequences of UPP-Pase depletion, we optimized a previously described CRISPRi repression system (16) to minimize the appearance of suppressor mutations. We used two copies of dCas9 to reduce the occurrence of suppressor mutations. The frequency of suppressor mutations varies significantly between loci (unpublished results) and may be affected by the length of time required to deplete the cell of essential proteins. Whereas one sgRNA generally suffices for gene knockdown (16), we included two sgRNAs targeting the bcrC gene. The presence of two sgRNAs led to a modest increase in repression efficiency, as judged by xylose sensitivity (Fig. 1B), and also slightly reduced the frequency of suppressor mutations (Fig. 2). In this system, the greatest reduction in suppressor mutation frequency required the deletion of yodM, which eliminates the only other known protein with UPP-Pase activity (Fig. 2). In general, optimization of CRISPRi for knockdown of essential genes (or sets of genes) may benefit from the use of a dCas9 merodiploid, since the inactivation of dCas9 is one frequent cause of suppression. Further modifications worth exploring include the use of two sgRNAs per gene (although this effect may be minor) and the elimination of known or predicted pathways of suppression.
The two independent suppressor mutants analyzed from our optimized strain each contained mutations in both copies of the dCas9 gene. The occurrence of frameshift mutations in runs of adenines suggests that these may be hot spots for mutagenesis, and the fact that each strain contains the same point mutation in both dCas9 genes suggests a gene conversion event after the first mutation. One way to further reduce the fraction of suppressor mutants might be to use a dCas9 gene with a different coding sequence as the second copy. For example, swapping alternate but still preferred codons for B. subtilis (55) could be used to reduce the number of homopolymeric sequences and also to reduce sequence-sequence similarity between two dCas9 genes. Nevertheless, the fraction of suppressor mutants in the optimized strain used in this study was sufficiently low to carry out the physiological assays performed here.
Depletion of UPP-Pases leads to a cessation of growth after ∼3 to 4 doublings. Even mild depletion (due to the leaky expression of dCas9 in the depletion strain) leads to an increase in sensitivity to bacitracin. As UPP is further depleted, the cells begin to display morphological abnormalities, including a prominent bulging of the cells. This is consistent with the morphological changes induced by other genetic and chemical perturbations known to affect peptidoglycan synthesis. Concomitant with the onset of morphological defects resulting from depletion of UPP, the cells strongly activate the σM cell envelope stress response, consistent with the effects of other antibiotics that target the lipid II cycle (53).
In conclusion, our results establish that B. subtilis requires either of two UPP-Pases for viability, UppP and BcrC. Because of the nature of Und-P synthesis and recycling, it is reasonable to speculate that one UPP-Pase has a cytosol-facing active site and functions to convert UPP produced by UppS into Und-P for use in cell wall synthesis, while the other one recycles UPP produced as a product of the transglycosylase reaction catalyzed by class A PBPs. We speculate that the former activity is UppP, which might account for the minor role of this enzyme with respect to sensitivity to rare earth metals (39). After each addition to the growing peptidoglycan (or WTA) polymer, UPP is released and must be dephosphorylated and flipped back to the cytoplasmic face of the membrane for reuse. This is the likely role for BcrC, which by reducing surface-exposed UPP can prevent the binding of bacitracin to its target. In contrast, all four of the E. coli UPP-Pases may have active sites on the outer face of the membrane. These include the PAP2 family UPP-Pases PgpB (26), YbjG and YeiU (56), and based on protein modeling, BacA (57). Manat and coworkers provided supporting evidence for this model using truncated BacA-β-lactamase reporters (58).
Regardless of whether the two UPP-Pases in B. subtilis have their active sites on the same or opposite faces of the membrane, the fact that B. subtilis can survive with only one UPP-Pase suggests that the UPP-Pase function only needs to be localized to one face to support viability. This suggests either that de novo-synthesized UPP is dephosphorylated inside the cell and the UPP product released by the transglycosylase reaction can be flipped from outside to inside or that UPP synthesized in the cytoplasm is first flipped to the outside to be dephosphorylated and then flipped back to the cytoplasmic face to support peptidoglycan (MraY) and WTA (TagO) synthesis. The flipping of Und-P or UPP is unlikely to be spontaneous, although the identity of any proteins that help catalyze this reaction is not yet clear. Ongoing efforts to define this and other poorly understood steps in the lipid II cycle will help to identify new candidates for the targeting of antibacterials and will refine our understanding of the complex mechanisms of acclimation to antibiotics in bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pete Chandrangsu and Ahmed Gaballa for help with the experimental design and interpretation.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00507-16.
REFERENCES
- 1.Jorgenson MA, Kannan S, Laubacher ME, Young KD. 2016. Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol Microbiol 100:1–14. doi: 10.1111/mmi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 3.Lovering AL, Safadi SS, Strynadka NC. 2012. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem 81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 4.Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. 2014. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. 2015. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A 112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manat G, Roure S, Auger R, Bouhss A, Barreteau H, Mengin-Lecreulx D, Touze T. 2014. Deciphering the metabolism of undecaprenyl-phosphate: the bacterial cell-wall unit carrier at the membrane frontier. Microb Drug Resist 20:199–214. doi: 10.1089/mdr.2014.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Ghachi M, Derbise A, Bouhss A, Mengin-Lecreulx D. 2005. Identification of multiple genes encoding membrane proteins with undecaprenyl pyrophosphate phosphatase (UppP) activity in Escherichia coli. J Biol Chem 280:18689–18695. doi: 10.1074/jbc.M412277200. [DOI] [PubMed] [Google Scholar]
- 8.Bickford JS, Nick HS. 2013. Conservation of the PTEN catalytic motif in the bacterial undecaprenyl pyrophosphate phosphatase, BacA/UppP. Microbiology 159:2444–2455. doi: 10.1099/mic.0.070474-0. [DOI] [PubMed] [Google Scholar]
- 9.Brown S, Santa Maria JP Jr, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Elia MA, Millar KE, Beveridge TJ, Brown ED. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol 188:8313–8316. doi: 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasquina LW, Santa Maria JP, Walker S. 2013. Teichoic acid biosynthesis as an antibiotic target. Curr Opin Microbiol 16:531–537. doi: 10.1016/j.mib.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xayarath B, Yother J. 2007. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J Bacteriol 189:3369–3381. doi: 10.1128/JB.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durrant JD, Cao R, Gorfe AA, Zhu W, Li J, Sankovsky A, Oldfield E, McCammon JA. 2011. Non-bisphosphonate inhibitors of isoprenoid biosynthesis identified via computer-aided drug design. Chem Biol Drug Des 78:323–332. doi: 10.1111/j.1747-0285.2011.01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu W, Zhang Y, Sinko W, Hensler ME, Olson J, Molohon KJ, Lindert S, Cao R, Li K, Wang K, Wang Y, Liu YL, Sankovsky A, de Oliveira CA, Mitchell DA, Nizet V, McCammon JA, Oldfield E. 2013. Antibacterial drug leads targeting isoprenoid biosynthesis. Proc Natl Acad Sci U S A 110:123–128. doi: 10.1073/pnas.1219899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farha MA, Czarny TL, Myers CL, Worrall LJ, French S, Conrady DG, Wang Y, Oldfield E, Strynadka NC, Brown ED. 2015. Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase. Proc Natl Acad Sci U S A 112:11048–11053. doi: 10.1073/pnas.1511751112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA. 2016. A comprehensive, CRISPR-based approach to functional analysis of essential genes in bacteria. Cell 165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YH, Helmann JD. 2013. Reducing the level of undecaprenyl pyrophosphate synthase has complex effects on susceptibility to cell wall antibiotics. Antimicrob Agents Chemother 57:4267–4275. doi: 10.1128/AAC.00794-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Economou NJ, Cocklin S, Loll PJ. 2013. High-resolution crystal structure reveals molecular details of target recognition by bacitracin. Proc Natl Acad Sci U S A 110:14207–14212. doi: 10.1073/pnas.1308268110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao M, Helmann JD. 2002. Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J Bacteriol 184:6123–6129. doi: 10.1128/JB.184.22.6123-6129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohki R, Tateno K, Okada Y, Okajima H, Asai K, Sadaie Y, Murata M, Aiso T. 2003. A bacitracin-resistant Bacillus subtilis gene encodes a homologue of the membrane-spanning subunit of the Bacillus licheniformis ABC transporter. J Bacteriol 185:51–59. doi: 10.1128/JB.185.1.51-59.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kingston AW, Zhao H, Cook GM, Helmann JD. 2014. Accumulation of heptaprenyl diphosphate sensitizes Bacillus subtilis to bacitracin: implications for the mechanism of resistance mediated by the BceAB transporter. Mol Microbiol 93:37–49. doi: 10.1111/mmi.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernard R, El Ghachi M, Mengin-Lecreulx D, Chippaux M, Denizot F. 2005. BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance. J Biol Chem 280:28852–28857. doi: 10.1074/jbc.M413750200. [DOI] [PubMed] [Google Scholar]
- 23.Schneider T, Sahl HG. 2010. Lipid II and other bactoprenol-bound cell wall precursors as drug targets. Curr Opin Investig Drugs 11:157–164. [PubMed] [Google Scholar]
- 24.Icho T, Raetz CR. 1983. Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J Bacteriol 153:722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan J, Jiang D, Zhao Y, Liu J, Zhang XC. 2014. Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B. Proc Natl Acad Sci U S A 111:7636–7640. doi: 10.1073/pnas.1403097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Touzé T, Blanot D, Mengin-Lecreulx D. 2008. Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. J Biol Chem 283:16573–16583. doi: 10.1074/jbc.M800394200. [DOI] [PubMed] [Google Scholar]
- 27.Touzé T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. 2008. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol 67:264–277. doi: 10.1111/j.1365-2958.2007.06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters JM, Silvis MR, Zhao D, Hawkins JS, Gross CA, Qi LS. 2015. Bacterial CRISPR: accomplishments and prospects. Curr Opin Microbiol 27:121–126. doi: 10.1016/j.mib.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Y, Asai K, Sadaie Y, Helmann JD. 2010. Transcriptomic and phenotypic characterization of a Bacillus subtilis strain without extracytoplasmic function sigma factors. J Bacteriol 192:5736–5745. doi: 10.1128/JB.00826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, Helmann JD. 2012. Analysis of the role of Bacillus subtilis sigma(M) in beta-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol 83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. Wiley New York, NY. [Google Scholar]
- 34.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slack FJ, Mueller JP, Sonenshein AL. 1993. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J Bacteriol 175:4605–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sterlini JM, Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, Huang KC. 2014. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci U S A 111:E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michna RH, Commichau FM, Todter D, Zschiedrich CP, Stulke J. 2014. SubtiWiki—a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res 42:D692–D698. doi: 10.1093/nar/gkt1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inaoka T, Ochi K. 2012. Undecaprenyl pyrophosphate involvement in susceptibility of Bacillus subtilis to rare earth elements. J Bacteriol 194:5632–5637. doi: 10.1128/JB.01147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vellanoweth RL, Rabinowitz JC. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol 6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 41.Hawkins JS, Wong S, Peters JM, Almeida R, Qi LS. 2015. Targeted transcriptional repression in bacteria using CRISPR interference (CRISPRi). Methods Mol Biol 1311:349–362. doi: 10.1007/978-1-4939-2687-9_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choudhary E, Thakur P, Pareek M, Agarwal N. 2015. Gene silencing by CRISPR interference in mycobacteria. Nat Commun 6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 43.Schmiedel D, Hillen W. 1996. A Bacillus subtilis 168 mutant with increased xylose uptake can utilize xylose as sole carbon source. FEMS Microbiol Lett 135:175–178. doi: 10.1111/j.1574-6968.1996.tb07985.x. [DOI] [Google Scholar]
- 44.Formstone A, Errington J. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol 55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- 45.McPherson DC, Popham DL. 2003. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol 185:1423–1431. doi: 10.1128/JB.185.4.1423-1431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schirner K, Eun YJ, Dion M, Luo Y, Helmann JD, Garner EC, Walker S. 2015. Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat Chem Biol 11:38–45. doi: 10.1038/nchembio.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Münch D, Muller A, Schneider T, Kohl B, Wenzel M, Bandow JE, Maffioli S, Sosio M, Donadio S, Wimmer R, Sahl HG. 2014. The lantibiotic NAI-107 binds to bactoprenol-bound cell wall precursors and impairs membrane functions. J Biol Chem 289:12063–12076. doi: 10.1074/jbc.M113.537449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50:1591–1604. doi: 10.1046/j.1365-2958.2003.03786.x. [DOI] [PubMed] [Google Scholar]
- 49.Hutter B, Schaab C, Albrecht S, Borgmann M, Brunner NA, Freiberg C, Ziegelbauer K, Rock CO, Ivanov I, Loferer H. 2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob Agents Chemother 48:2838–2844. doi: 10.1128/AAC.48.8.2838-2844.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eiamphungporn W, Helmann JD. 2008. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol 67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Radeck J, Gebhard S, Orchard PS, Kirchner M, Bauer S, Mascher T, Fritz G. 2016. Anatomy of the bacitracin resistance network in Bacillus subtilis. Mol Microbiol 100:607–620. doi: 10.1111/mmi.13336. [DOI] [PubMed] [Google Scholar]
- 52.Fritz G, Dintner S, Treichel NS, Radeck J, Gerland U, Mascher T, Gebhard S. 2015. A new way of sensing: need-based activation of antibiotic resistance by a flux-sensing mechanism. mBio 6:e00975. doi: 10.1128/mBio.00975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helmann JD. 2016. Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr Opin Microbiol 30:122–132. doi: 10.1016/j.mib.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salzberg LI, Luo Y, Hachmann AB, Mascher T, Helmann JD. 2011. The Bacillus subtilis GntR family repressor YtrA responds to cell wall antibiotics. J Bacteriol 193:5793–5801. doi: 10.1128/JB.05862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moszer I, Rocha EP, Danchin A. 1999. Codon usage and lateral gene transfer in Bacillus subtilis. Curr Opin Microbiol 2:524–528. doi: 10.1016/S1369-5274(99)00011-9. [DOI] [PubMed] [Google Scholar]
- 56.Tatar LD, Marolda CL, Polischuk AN, van Leeuwen D, Valvano MA. 2007. An Escherichia coli undecaprenyl-pyrophosphate phosphatase implicated in undecaprenyl phosphate recycling. Microbiology 153:2518–2529. doi: 10.1099/mic.0.2007/006312-0. [DOI] [PubMed] [Google Scholar]
- 57.Chang HY, Chou CC, Hsu MF, Wang AH. 2014. Proposed carrier lipid-binding site of undecaprenyl pyrophosphate phosphatase from Escherichia coli. J Biol Chem 289:18719–18735. doi: 10.1074/jbc.M114.575076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manat G, El Ghachi M, Auger R, Baouche K, Olatunji S, Kerff F, Touze T, Mengin-Lecreulx D, Bouhss A. 2015. Membrane topology and biochemical characterization of the Escherichia coli BacA undecaprenyl-pyrophosphate phosphatase. PLoS One 10:e0142870. doi: 10.1371/journal.pone.0142870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.