ABSTRACT
Carotenoids constitute an important component of the defense system against photooxidative stress in bacteria. In Azospirillum brasilense Sp7, a nonphotosynthetic rhizobacterium, carotenoid synthesis is controlled by a pair of extracytoplasmic function sigma factors (RpoEs) and their cognate zinc-binding anti-sigma factors (ChrRs). Its genome harbors two copies of the gene encoding geranylgeranyl pyrophosphate synthase (CrtE), the first critical step in the carotenoid biosynthetic pathway in bacteria. Inactivation of each of two crtE paralogs found in A. brasilense caused reduction in carotenoid content, suggesting their involvement in carotenoid synthesis. However, the effect of crtE1 deletion was more pronounced than that of crtE2 deletion. Out of the five paralogs of rpoH in A. brasilense, overexpression of rpoH1 and rpoH2 enhanced carotenoid synthesis. Promoters of crtE2 and rpoH2 were found to be dependent on RpoH2 and RpoE1, respectively. Using a two-plasmid system in Escherichia coli, we have shown that the crtE2 gene of A. brasilense Sp7 is regulated by two cascades of sigma factors: one consisting of RpoE1and RpoH2 and the other consisting of RpoE2 and RpoH1. In addition, expression of crtE1 was upregulated indirectly by RpoE1 and RpoE2. This study shows, for the first time in any carotenoid-producing bacterium, that the regulation of carotenoid biosynthetic pathway involves a network of multiple cascades of alternative sigma factors.
IMPORTANCE Carotenoids play a very important role in coping with photooxidative stress in prokaryotes and eukaryotes. Although extracytoplasmic function (ECF) sigma factors are known to directly regulate the expression of carotenoid biosynthetic genes in bacteria, regulation of carotenoid biosynthesis by one or multiple cascades of sigma factors had not been reported. This study provides the first evidence of the involvement of multiple cascades of sigma factors in the regulation of carotenoid synthesis in any bacterium by showing the regulation of a gene encoding geranylgeranyl pyrophosphate synthase (crtE2) by RpoE1→RpoH2→CrtE2 and RpoE2→RpoH1→CrtE2 cascades in A. brasilense. It also provides an insight into existence of an additional cascade or cascades regulating expression of another paralog of crtE.
INTRODUCTION
Photosynthetic as well as nonphotosynthetic organisms encounter photooxidative stress caused by the generation of highly reactive singlet oxygen in the presence of light and oxygen. Singlet oxygen reacts with a wide range of cellular macromolecules, including proteins, lipids, DNA, and RNA, leading to the formation of reactive substances such as organic peroxides and sulfoxides (1). Mechanisms to cope with the photooxidative stress have been investigated in a range of photosynthetic and nonphotosynthetic microorganisms. These mechanisms include the use of quenchers, such as carotenoids, which interact either with excited photosensitizer molecules or singlet oxygen itself to prevent damage of cellular molecules (2). Carotenoids are a widely distributed class of structurally diverse yellow-, orange-, or red-pigmented compounds (tetraterpenoids) consisting of a polyene hydrocarbon chain derived from eight isoprene units. Modifications like cyclization and desaturations of C40 backbone result in a variety of divergent chemical structures produced in eukaryotes as well as prokaryotes (3).
Conversion of farnesyl pyrophosphate to geranylgeranyl pyrophosphate (GGPP) by GGPP synthase (CrtE) is a critical step before carotenoid biosynthesis begins in bacteria. The first two committed steps in the biosynthesis of carotenoids include conversion of GGPP into phytoene by a phytoene synthase (CrtB) followed by phytoene desaturation via phytoene dehydrogenase (CrtI). These three steps in the carotenoid biosynthetic pathway and associated enzymes are common in the carotenoid-producing organisms (4), suggesting evolutionary conservation of the early steps of this pathway (5). The regulation of their expression, however, may differ in different bacteria (6). While Rhodobacter capsulatus and Erwinia herbicola (7) produce carotenoids constitutively, Myxococcus xanthus (8, 9), Flavobacterium dehydrogenans (10), Sulfolobus spp. (11), and Streptomyces coelicolor (12) produce carotenoids in a photoinducible manner. In nonphotosynthetic bacteria, such as M. xanthus (a Gram-negative bacterium) (13) and S. coelicolor (a Gram-positive bacterium) (12), carotenoid synthesis is regulated by light via an extracytoplasmic function (ECF) sigma factor and its cognate anti-sigma factor. In Streptomyces griseus, carotenoid synthesis is also regulated by an ECF sigma factor, but the stimulus responsible for its induction is not known yet (14).
Azospirillum brasilense is a nonphotosynthetic, plant-growth-promoting rhizobacterium that belongs to the family Rhodospirillaceae of Alphaproteobacteria. Since it inhabits the rhizosphere as well as soil, it has to cope with fluctuations in its environment. It synthesizes bacterioruberin-type carotenoids (15). Recently, we reported that carotenoid synthesis is induced by light and regulated by a pair of ECF sigma factors (RpoE1 and RpoE2) in this bacterium (16–18). In addition, we have also shown that a heat shock sigma factor (RpoH2, or σH) is involved in the photooxidative stress response in this bacterium (19). These observations suggested that, besides RpoE1-ChrR1, RpoH2 might also be involved in the regulation of carotenoid biosynthesis in A. brasilense Sp7. Whether RpoE1 and RpoE2 regulate the carotenoid biosynthetic genes directly and whether RpoH2 is also involved in this regulation is not known yet. Using crtE, the first gene of the carotenoid biosynthetic pathway, we show here that the carotenoid biosynthetic pathway in A. brasilense is regulated by a network of multiple cascades of RpoE and RpoH sigma factors.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, and growth conditions.
The plasmids and strains used in this study are described in Table 1. Escherichia coli DH5α and E. coli S17-1 were grown in Luria Bertani (LB) medium at 37°C. A. brasilense was grown in minimal malate medium (20) as well as in LB medium at 30°C. Media used for bacterial growth were from Hi-Media (Mumbai, India), and enzymes used for DNA manipulation and cloning were from New England BioLabs. Primer sequences are given in Table S1 in the supplemental material.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | ΔlacU169 hsdR17 recA1 endA1 gyrA96 thiL relA1 | Gibco-BRL |
| S17-1 | Smr recA thi pro hsdR RP4-2(Tc::Mu Km::Tn7) | 24 |
| A. brasilense | ||
| Sp7 | Wild-type strain | 15 |
| Cd | Wild-type carotenoid-producing strain | 15 |
| Car-1 | Carotenoid-producing chrR1::Tn5 mutant of A. brasilense Sp7 | 16 |
| rpoE1::Km mutant | A. brasilense Sp7 rpoE1 gene disrupted by insertion of Kmr gene cassette | 25 |
| rpoH2::Km mutant | A. brasilense Sp7 rpoH2 gene disrupted by insertion of Kmr gene cassette | 19 |
| crtE1::Km mutant | A. brasilense Cd crtE1 gene disrupted by insertion of Kmr gene cassette | This work |
| crtE2::Km mutant | A. brasilense Cd crtE2 gene disrupted by insertion of Kmr gene cassette | This work |
| Plasmids | ||
| pSUP202 | ColE1 replicon, mobilizable, suicide vector for A. brasilense; Apr Cmr Tcr | 24 |
| pUC4K | Vector containing kanamycin resistance gene cassette | GE Healthcare |
| pAPD1 | crtE1::Km in pSUP202 | This work |
| pAPD2 | crtE2::Km in pSUP202 | This work |
| pCZ750 | Tetr Ampr; lacZ fusion reporter vector | 27 |
| pAPD3 | crtE1 promoter sequence of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAPD4 | crtE2 promoter sequence of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR1 | crtE2 promoter (deleted −35) region of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR2 | rpoH1 promoter sequence of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR3 | rpoH2 promoter sequence of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR4 | rpoH2 promoter −35 region (TGA to ACT) of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR5 | rpoH2 promoter −10 region (CG to GC) of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR6 | rpoH2 promoter (deleted −35) region of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR7 | rpoH3 promoter sequence of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR8 | rpoH4 promoter sequence of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pAKR9 | rpoH5 promoter sequence of A. brasilense Sp7 fused with lacZ in pCZ750 | This work |
| pMMB206 | Cmr; broad-host-range, low-copy-number expression vector | 26 |
| pAT5 | rpoE1 gene of A. brasilense Sp7 cloned in pMMB206 vector | 16 |
| pNG6 | rpoE2 gene of A. brasilense Sp7 cloned in pMMB206 vector | 17 |
| pAKR9 | rpoH1 gene of A. brasilense Sp7 cloned at EcoRI and PstI in pMMB206 vector | This work |
| pSNK10 | rpoH2 gene of A. brasilense Sp7 cloned in pMMB206 vector | 19 |
| pAKR11 | rpoH3 gene of A. brasilense Sp7 cloned in pMMB206 vector | This work |
| pAKR12 | rpoH4 gene of A. brasilense Sp7 cloned in pMMB206 vector | This work |
| pAKR13 | rpoH5 gene of A. brasilense Sp7 cloned in pMMB206 vector | This work |
Prediction of promoter regions of rpoH2 and crtE in A. brasilense.
Upstream nucleotide sequences of rpoH2 and crtE paralogs were analyzed by the online promoter prediction tool BPROM (21) to identify putative promoters. Upstream nucleotide sequences were also searched manually to find RpoE and RpoH2 promoter elements using the consensus sequences reported earlier in A. brasilense and related bacteria (22, 23).
Construction of mutants and RpoH-expressing plasmids.
The rpoH2, crtE1, and crtE2 genes of A. brasilense Sp7 were insertionally inactivated via allele replacement by using a suicide plasmid vector (pSUP202) as described earlier (24, 25). Entire coding regions of the rpoH paralogs were cloned in a broad-host-range expression vector (pMMB206) as described earlier (16, 26).
Determination of the TSS.
To determine the transcription start site (TSS), the 5′ ends of the mRNAs of crtE1, crtE2, and rpoH2 were identified by 5′ rapid amplification of cDNA ends (RACE) as described earlier (25). Briefly, 2 μg RNA was reverse transcribed to cDNA using gene-specific reverse primer 1 (GSP1), followed by cDNA purification and poly(dA) tailing. Poly(dA)-tailed cDNA was then PCR amplified using GSP2 and oligo(dT) anchor primers. The amplicon so obtained was submitted to the next round of nested PCR using anchor and GSP3 primers. The final PCR product was then cloned in the pGEM-T Easy vector (Promega), and nucleotide sequences were determined by the chain termination method.
Construction of promoter-lacZ fusions.
Upstream regions (approximately 500 bp) of the start codon of the selected genes were PCR amplified, and the amplicons were inserted into the pCZ750 vector (27) using XbaI and HindIII to construct promoter-lacZ transcriptional fusions. Constructs were confirmed by sequencing, and mobilized in A. brasilense by the biparental conjugation method (16). Nucleotide sequences of the −10 and −35 elements of the rpoH2 promoter (CG to GC and TGA to ACT, respectively) were mutated by the protocol for one-step site-directed mutagenesis (28). The −35 region of the crtE2 promoter was deleted by the overlap extension PCR technique (29).
Estimation of promoter activity.
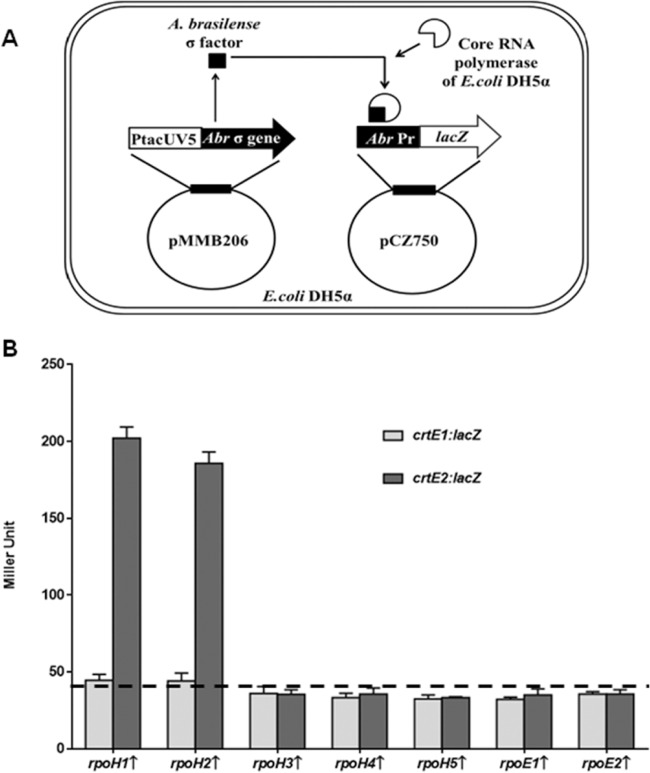
Promoter activity was monitored by measuring the β-galactosidase activity (30) of A. brasilense Sp7 and its mutants harboring different promoter-lacZ fusions. An E. coli DH5α-based two-plasmid system (31) was used to identify A. brasilense promoters that were directly activated by specific alternative sigma factors. We used this method by transforming E. coli DH5α with two recombinant plasmids—a pMMB206 derivative with a gene encoding an alternative sigma factor fused to the PtacUV5 promoter and a pCZ750 derivative harboring the promoter region of a target gene transcriptionally fused to the lacZ reporter—and measuring the β-galactosidase activity of the transformed cells.
Real-time PCR.
Total RNA from A. brasilense strains was isolated, quantified and quality tested, as reported earlier (25). The cDNA was synthesized from 2 μg RNA using a cDNA synthesis kit (Fermentas, Germany). Real-time PCR was performed using SYBR green I (Roche) in a Light Cycler 480 II (Roche) according to the manufacturer's instructions, using rpoD as an endogenous control. The relative expression level was compared by the threshold cycle (2−ΔΔCT) method (32).
Extraction and estimation of carotenoids.
A. brasilense strains were grown in the LB medium up to the stationary phase. Pellets from equal volumes of cultures with equal optical densities at 600 nm (OD600) were washed twice with saline and resuspended in 5 ml 100% methanol, and carotenoids were extracted by shaking the suspension overnight at 180 rpm at 25°C in Oakridge tubes covered with aluminum foil (16). Concentrated methanolic extracts of carotenoids were spotted onto a thin-layer chromatography (TLC) plate, resolved, and visualized (15). Absorption spectra of the carotenoids extracted in methanol were recorded using a 300- to 800-nm range in a UV-visible (UV-Vis) spectrophotometer (V630, Jasco, Japan). Carotenoids were extracted and estimated three times. Student's t tests were performed for statistical comparisons, and P values of <0.05 were considered to show statistically significant differences in carotenoid contents.
RESULTS
Two CrtE paralogs are involved in carotenoid synthesis in A. brasilense.
In carotenoid-producing bacteria, the first step of the carotenoid biosynthetic pathway is catalyzed by GGPS (encoded by the crtE gene). Since the A. brasilense genome harbors two paralogs of this gene, crtE1 (locus tag AzoBR_180071) and crtE2 (locus tag AzoBR_p1140021), we examined their involvement in carotenoid synthesis by mutating each of them separately in a naturally carotenoid-overproducing strain, A. brasilense Cd. Qualitative analysis of carotenoids by TLC revealed 4 spots that were common in the extracts of A. brasilense Cd and its two crtE1::Km and Cd crtE2::Km mutants (data not shown). The intensity of the spots in case of the crtE1::Km mutant, however, was very weak in comparison to that detected in the crtE2::Km mutant or A. brasilense Cd. Absorption spectra of the methanolic extracts of the two mutants revealed that crtE1 inactivation led to an approximately 75% reduction in the carotenoid content, whereas crtE2 inactivation reduced carotenoids only by <25% (P < 0.03) (Fig. 1A; see Fig. S1 in the supplemental material), indicating involvement of both of the CrtE proteins in carotenoid biosynthesis.
FIG 1.
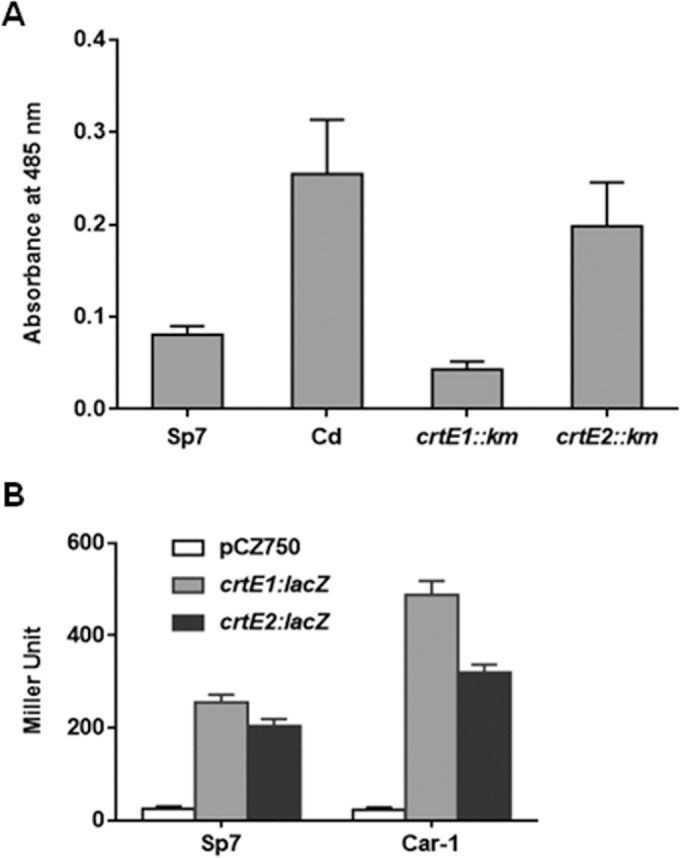
(A) Effect of crtE1 and crtE2 gene inactivation on carotenoid content in a carotenoid-overproducing strain, A. brasilense Cd. The carotenoid content was compared by measuring absorption maxima at 485 nm of the methanolic extracts of the wild-type or mutant strains. The carotenoid content of Cd is significantly different from those of the crtE1::Km and crtE2::Km mutants (P < 0.03). (B) β-Galactosidase activity due to lacZ fusions with the promoters of crtE1(pAPD3) and crtE2(pAPD4) in A. brasilense Sp7 and Car-1. Error bars show standard deviations from three replicates.
crtE promoters are regulated indirectly by RpoEs.
We have shown earlier that overexpression of rpoE1 or rpoE2 leads to enhancement in the carotenoid level in A. brasilense Sp7 (16, 17). To examine if crtE genes are regulated directly by RpoE1, we analyzed the effect of RpoE1 expression on the activation of crtE promoters by mobilizing crtE1::lacZ(pAPD3) and crtE2::lacZ(pAPD4) fusions in A. brasilense Sp7 and its anti-sigma mutant, Car-1 (a chrR1::Tn5 mutant overexpressing RpoE1 due to the inactivation of anti-sigma factor ChrR1), followed by estimating the β-galactosidase activity. Promoter assays revealed that the activities from both promoters in A. brasilense Sp7 were almost equal. Both promoters showed considerably higher activity in the Car-1 mutant than that observed in A. brasilense Sp7. Furthermore, the activity of the crtE1 promoter in the Car-1 mutant was relatively high (Fig. 1B). Although these results indicate an RpoE1-dependent regulation of crtE genes, they do not confirm if RpoE1 regulates the expression of crtE1 directly.
To examine if the crtE genes are regulated directly by RpoE, we used a two-plasmid system (31). The principle of the two-plasmid system is based on the ability of the A. brasilense alternative sigma factors, expressed via an expression vector, to bind and recruit E. coli DH5α RNA polymerase to initiate transcription from the A. brasilense promoters fused to a lacZ reporter gene in a compatible vector (Fig. 2A). For this, we transformed crtE1::lacZ and crtE2::lacZ fusion plasmids into E. coli DH5α individually, and these transformants were then further transformed with rpoE1- and rpoE2-expressing plasmids pAT5 and pNG6, respectively. β-Galactosidase assays of these combinations showed that neither RpoE1 nor RpoE2 activated crtE1 or crtE2 promoters in E. coli DH5α (Fig. 2B), suggesting an indirect mode of RpoE-dependent regulation of crtE genes in A. brasilense.
FIG 2.
(A) Schematic presentation of the principle of the two-plasmid system. A gene encoding A. brasilense sigma factor (Abr σ) is cloned downstream of the inducible PtacUV5 promoter in the pMMB206 vector. Upon induction by IPTG, the A. brasilense sigma factor is expressed from the PtacUV5 promoter. The A. brasilense sigma factor expressed in E. coli then brings E. coli core RNA polymerase binding to the A. brasilense promoter (Abr Pr) cloned upstream of the lacZ reporter in pCZ750 vector. If the A. brasilense target promoter is recognized by the A. brasilense sigma factor expressed in E. coli DH5α, β-galactosidase will be expressed, which can be assayed. (B) Effect of the expression of rpoH and rpoE paralogs of A. brasilense on the β-galactosidase activity of E. coli DH5α harboring crtE1::lacZ(pAPD3) or crtE2::lacZ(pAPD4) fusions. Vertical arrows indicate expression of the genes. The dashed baseline shows the β-galactosidase activity of E. coli DH5α harboring empty vector. Error bars show standard deviations from three replicates.
crtE2 has an RpoH-dependent promoter.
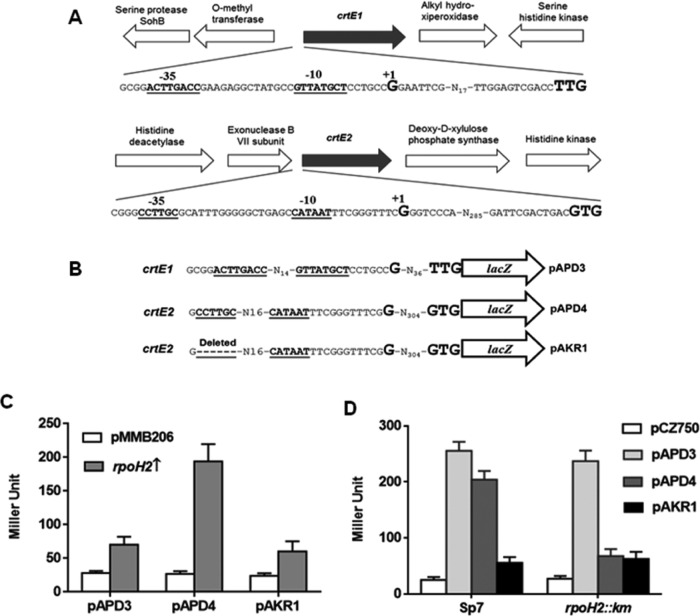
The absence of any direct regulation of crtE genes by RpoE prompted us to look for an RpoE-dependent intermediate regulator or regulators that might regulate crtE genes directly. In order to identify alternative sigma factors directly regulating crtE genes, we examined the upstream DNA regions of both crtE genes for recognizable promoter elements. This analysis failed to find any RpoE1-dependent promoter (25) in the upstream region of both crtE genes, which supports the lack of direct regulation of both of these genes by RpoE1. The upstream regions of crtE1 and crtE2, however, revealed the presence of CCTTGC-N17-CTATGC and CCTTGC-N17-CATAAT sequences, respectively, which showed strong resemblance to the RpoH-dependent promoter consensus. (Conserved residues are in boldface.) To validate this prediction, we determined the TSS to identify promoter elements of both crtE genes, which revealed that −35 (CCTTGC) and −10 (CTATGC) hexamers of only crtE2 match the RpoH-dependent consensus, while possible −35 (TTGACC) and −10 (TTATGC) hexamers of crtE1 did not match with RpoH-dependent promoter consensus (Fig. 3A and B). This indicated that crtE2 might be regulated in an RpoH-dependent manner.
FIG 3.
(A) Organization of A. brasilense crtE1 and crtE2 loci and possible −35 and −10 elements (boldface and underlined) of the crtE1 and crtE2 genes predicted on the basis of the identified TSS (shown as “+1” and in a larger and boldface font); start codons are shown in a larger and boldface font. (B) Representation of the promoter-lacZ fusions showing promoter elements of crtE1(pAPD3) and crtE2(pAPD4) as well as a derivative of the crtE2 promoter with a deletion of the −35 element (pAKR1). (C) Effect of rpoH2 overexpression (indicated by an upward arrow) on the β-galactosidase activity of A. brasilense crtE promoter-lacZ fusions using the two-plasmid system in E. coli DH5α. Error bars show standard deviations from three replicates. (D) β-Galactosidase activity of the lacZ fusions fused with the promoters of crtE1(pAPD3), crtE2(pAPD4), and the −35 element deletion derivative of the crtE2 promoter (pAKR1) in A. brasilense Sp7 and rpoH2::Km mutants.
RpoH1 and RpoH2 directly activate the crtE2 promoter.
Identification of an RpoH-dependent promoter in the crtE2 upstream region prompted us to use the two-plasmid system to examine which of the five RpoH paralogs, encoded in the genome of A. brasilense (19), directly regulate crtE genes. Since the rpoH gene of E. coli is expressed under a heat shock condition, expression of β-galactosidase in E. coli DH5α can be observed only if an A. brasilense RpoH protein is able to drive the expression of lacZ from the crtE1 or crtE2 promoters. Each RpoH paralog was expressed in E. coli DH5α harboring crtE1::lacZ or crtE2::lacZ to examine their involvement in the transcription of crtE genes. Promoter activity showed that only the crtE2::lacZ fusion was activated by both RpoH1 and RpoH2 (Fig. 2B); the crtE1 promoter does not seem to be activated by any of the RpoH paralogs. When we examined the ability of A. brasilense RpoH2 to activate a derivative of the promoter of the crtE2::lacZ fusion in which the −35 hexamer was deleted (Fig. 3B), only negligible β-galactosidase activity was seen (Fig. 3C). Since we have an rpoH2::Km mutant of A. brasilense we compared the activity of crtE2 promoter in the rpoH2::Km mutant and its parent. The promoter activity of crtE2 was very low in the rpoH2::Km mutant compared to that observed in the parent, showing an RpoH-dependent regulation of crtE2 in vivo (Fig. 3D). These observations confirmed that crtE2 is regulated directly by RpoH1 and RpoH2 and that the promoter elements identified for crtE2 are actually used for this regulation.
Expression of RpoH1 and RpoH2 enhances the carotenoid synthesis.
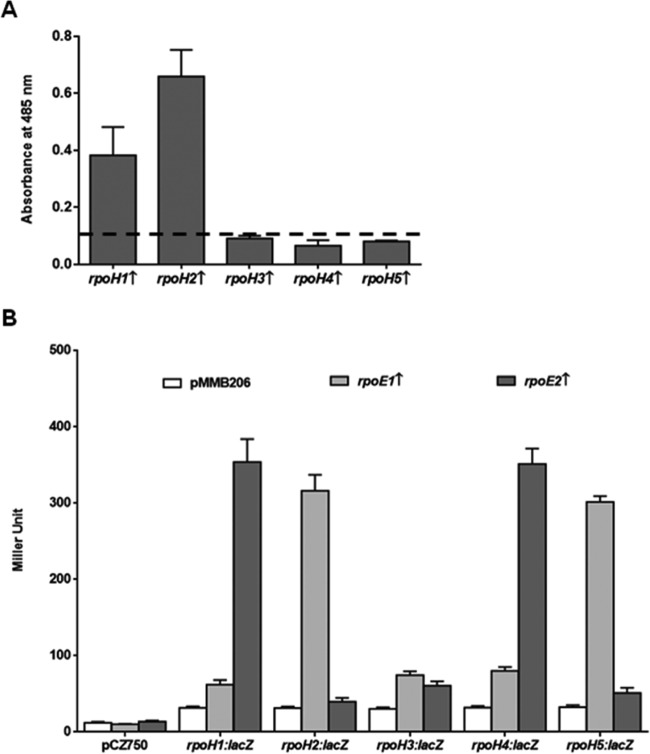
RpoH1- and RpoH2-dependent regulation of crtE2 suggested a positive role of these RpoHs in the regulation of carotenoid synthesis of A. brasilense. To confirm this, we expressed each of the five RpoH sigma factors in A. brasilense Sp7 and estimated their effects on the carotenoid content. Figure 4A shows that the overexpression of rpoH1 and rpoH2 caused very significant increases in carotenoid content in A. brasilense Sp7; RpoH2 caused maximum synthesis of carotenoids. The carotenoid contents in the strain derivatives of A. brasilense Sp7 overexpressing rpoH3, rpoH4, and rpoH5 were not significantly different from that in A. brasilense Sp7. These results confirm the role of RpoH1 and RpoH2 in the regulation of carotenoid synthesis in A. brasilense.
FIG 4.
(A) Effect of overexpression of the rpoH paralogs (indicated by upward arrows) on the carotenoid content of A. brasilense Sp7. Carotenoid content was compared by measuring absorption maxima at 485 nm of the methanolic extracts of the strains. The dashed baseline shows the carotenoid content in A. brasilense Sp7 harboring the empty vector. (B) Effect of overexpression of rpoE1(pAT5) or rpoE2(pNG6) on the β-galactosidase activity from lacZ fusions with the promoters of five rpoH paralogs of A. brasilense using the two-plasmid system in E. coli DH5α. pCZ750 and pMMB206 are the vectors used to construct lacZ fusions and express sigma factor genes, respectively. Error bars show standard deviations from three replicates.
RpoE1 and RpoE2 directly regulate rpoH promoters.
After establishing a direct regulation of crtE2 by RpoH1 as well as RpoH2, we hypothesized that carotenoid synthesis in A. brasilense might be indirectly regulated by RpoEs, using RpoH1 or RpoH2 as an intermediate regulator. To confirm this hypothesis, we used the two-plasmid system once again to examine whether RpoE1 and RpoE2 directly regulate the rpoH paralogs. Figure 4B shows that the activities of the rpoH1::lacZ and the rpoH4::lacZ fusions, respectively, were upregulated about 3-fold by RpoE2. Similarly, the activities of rpoH2::lacZ and rpoH5::lacZ fusions were upregulated by more than 3-fold by RpoE1. These observations indicate that rpoH1 and rpoH4 promoters are directly regulated by RpoE2, whereas rpoH2 and rpoH5 promoters are directly regulated by RpoE1. The activity of the rpoH3::lacZ fusion, however, does not seem to be affected by the expression of RpoE1 or RpoE2.
rpoH2 has an RpoE1-dependent promoter.
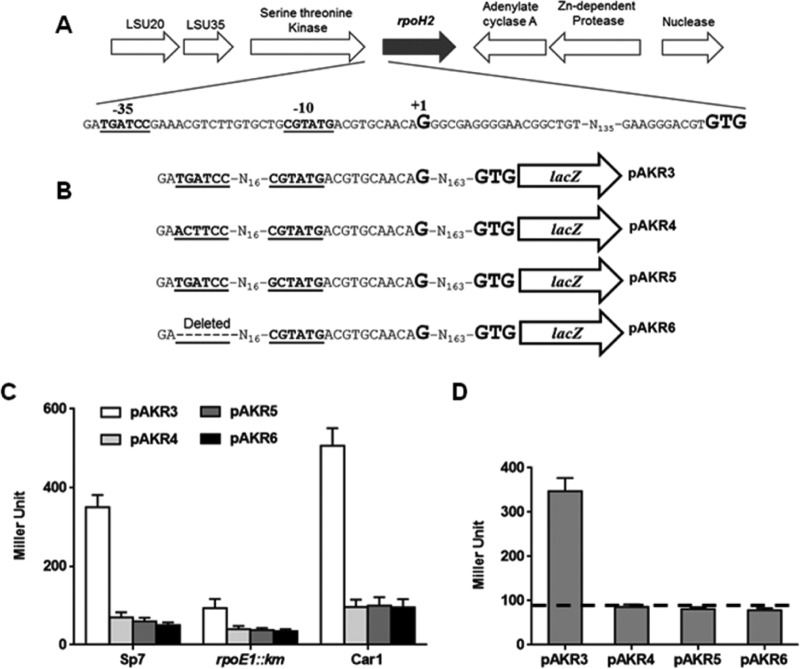
Since RpoH2 directly regulates crtE2, and the expression of rpoH2 seems to be directly regulated by RpoE1, we attempted to examine whether rpoH2 harbors an RpoE1-dependent promoter. Determination of the rpoH2 TSS revealed a “G” located 163 nucleotides upstream of the start codon and TGATCC and CGTATG as possible −35 and −10 elements, respectively, with a space of 16 nucleotides (Fig. 5A). The promoter elements of rpoH2 predicted on the basis of TSS matched well with the earlier reported promoter sequence recognized by the RpoE1 sigma factor (25). In order to prove further that the predicted −35 and −10 elements are indeed utilized by the RpoE1 sigma factor to drive the transcription of rpoH2, we generated three site-directed mutants of the rpoH2 promoter. In one, TGA of the −35 element was replaced with ACT. In the second, CG of the −10 element was replaced by GC. Finally, in the third, the −35 element was deleted completely (Fig. 5B). The lacZ fusion with the rpoH2 native promoter gave the maximum and minimum activities in the Car-1 and rpoE1::Km mutants, respectively, indicating dependence of rpoH2 promoter on RpoE1. The derivatives of the rpoH2::lacZ fusion carrying three different mutant versions of the promoters showed very low activity even in the Car-1 mutant, clearly indicating that the identified −35 and −10 elements were the essential promoter elements of the RpoE1-dependent regulation of rpoH2.
FIG 5.
(A) Organization of the A. brasilense rpoH2 locus. Shown are the −35 and −10 elements (in boldface and underlined) of the rpoH2 gene predicted after determination of the transcription start site (shown as “+1” in boldface and larger font). The rpoH2 start codon (GTG) is shown in boldface and larger font. (B) Schematic diagram showing the rpoH2::lacZ fusion (pAKR3) and its derivatives with the mutated −35 element (pAKR4) and mutated −10 element (pAKR5), as well as with deletion of the −35 element (pAKR5). (C) β-Galactosidase activity due to the rpoH2::lacZ fusion and its derivatives in A. brasilense Sp7 and the rpoE1::Km and Car-1 (chrR1::Tn5) mutants. (D) Effect of overexpression of A. brasilense rpoE1 on the β-galactosidase activity from the A. brasilense rpoH2 promoter-lacZ fusion and its mutant derivatives using the two-plasmid system in E. coli DH5α. The dashed baseline shows the β-galactosidase activity in E. coli DH5α expressing A. brasilense rpoE1 and the promoterless lacZ gene on pCZ750.
To prove the RpoE1-dependent direct regulation of rpoH2 promoter and that the −35 and −10 elements are real promoter elements, we used the two-plasmid system to examine the ability of RpoE1 to directly activate the lacZ-fused native rpoH2 promoter and any of the rpoH2 promoter mutant versions. Figure 5D shows that RpoE1 was able to directly activate lacZ-fused rpoH2 native promoter, whereas it failed to activate any of the mutant versions of the rpoH2 promoter. These observations further established that TGATCC and CGTATG are the −35 and −10 elements of the rpoH2 promoter, respectively, and these elements are utilized as promoters by RpoE1 for the transcription of rpoH2. We also examined transcript levels of rpoH2 in A. brasilense Sp7 and the rpoE1::Km and Car-1 mutants by real-time PCR and reproducibly observed that rpoH2 expression was upregulated >5-fold in Car-1, but downregulated by about 0.5-fold in the rpoE1::Km mutant, in comparison to that in A. brasilense Sp7 (see Fig. S2 in the supplemental material). This result further corroborated our inference about an RpoE1-dependent regulation of rpoH2, as the rpoH2 transcript level was maximum in Car-1 as it overexpresses RpoE1 due to autoregulation in the absence of ChrR.
DISCUSSION
In our studies on the regulation of carotenoid biosynthesis in A. brasilense, we have shown earlier that two ECF sigma factors, RpoE1 and RpoE2, positively regulate carotenoid synthesis in A. brasilense Sp7 (16–18, 25). Whether carotenoid biosynthetic genes are controlled directly or indirectly by RpoEs was not known. In this study, we have shown that carotenoid synthesis in A. brasilense is controlled by RpoE indirectly via RpoH. Although RpoH2 acts as a component of the RpoE→RpoH2 cascade regulating photooxidative stress response in Rhodobacter sphaeroides, carotenoid biosynthetic genes were not regulated by this cascade. Carotenoid biosynthetic genes were shown to be directly regulated by ECF sigma factors in S. coelicolor and M. xanthus. While ECF sigma factor σLitS was shown to regulate the expression of carotenoid biosynthetic genes directly in S. coelicolor (12), the ECF sigma factor CarQ, essential for light-induced carotenogenesis in M. xanthus, was shown to directly activate the promoters of two carotenoid biosynthetic genes (33). This study, however, provides the first evidence of the regulation of carotenoid biosynthesis by a cascade of RpoE-RpoH sigma factors in any bacterium.
We used a two-plasmid system to identify promoters of A. brasilense genes that are recognized by the alternative sigma factors of A. brasilense in E. coli. Such a system was used earlier to identify Mycobacterium tuberculosis genes directly regulated by σF and Staphylococcus aureus genes activated by σB (34, 35). The advantage of using a two-plasmid system lies in E. coli providing a noise-free genetic background to analyze the expression of genes, as native RpoH and RpoE sigma factors of E. coli are expressed only under heat shock conditions.
Observations that both of the crtE genes are involved in carotenoid biosynthesis and that RpoE1 and RpoE2 sigma factors activate the promoters of the two crtE genes in A. brasilense suggest that RpoEs may control carotenoid biosynthesis in A. brasilense, most likely by the regulation of crtE genes. The inability of RpoE1/RpoE2, however, to activate either of the crtE promoters in the E. coli background suggests that crtE genes are indirectly regulated by these sigma factors. A clue about the regulation of crtE genes was provided by the in silico analysis of their upstream regions, which predicted RpoH-dependent promoters and hence indicated RpoH-mediated regulation of these genes. Confirmation of the promoters of crtE on the basis of TSS revealed that the prediction was, at least, valid for crtE2. By showing that crtE2 expression was directly regulated by RpoH1 and RpoH2, our data from two-plasmid system-based promoter activation indicated involvement of RpoH1 and RpoH2 in the regulation of carotenoid biosynthesis in A. brasilense. This was further corroborated by an increase in the carotenoid content due to the overexpression of RpoH in A. brasilense.
On the basis of these as well as our previous observations (16, 18), it can be hypothesized that the RpoEs may indirectly regulate crtE2 via RpoH1/RpoH2 to control carotenoid synthesis in A. brasilense, and if so, these RpoHs should be regulated directly or indirectly by RpoEs. Our data from the two-plasmid system reveal direct regulation of rpoH2 and rpoH5 by RpoE1 and of rpoH1 and rpoH4 by RpoE2 and confirm the occurrence of at least two cascades regulating crtE2, which include RpoE1→RpoH2→CrtE2 and RpoE2→RpoH1→CrtE2. Furthermore, mutagenesis of crtE2 and rpoH2 promoters followed by their promoter activity assays provided another level of evidence for RpoE1→RpoH2→CrtE2 cascade by revealing that (i) RpoE1 activates the native promoter of rpoH2 but not those having mutations in −10 and −35 elements of rpoH2 promoter and, similarly, (ii) deletion of the −35 element of the crtE2 promoter abolishes regulation by RpoH2.
By demonstrating a major role of crtE1 in carotenoid biosynthesis in A. brasilense and strong upregulation of the crtE1 promoter in the Car-1 mutant, in which RpoE1 is constitutively overexpressed, our observations clearly indicate the existence of an even more important RpoE-dependent carotenoid regulatory cascade in A. brasilense. Although crtE1 is also regulated indirectly by RpoE1, the regulatory cascade involved in this case seems to be RpoH independent, as none of the RpoH paralogs could directly activate the crtE1 promoter in the E. coli background. These observations suggest RpoE-mediated regulation of the two crtE paralogs through different regulatory cascades involving different types of intermediate alternative sigma factors to control carotenoid biosynthesis in A. brasilense. This provides an insight into the flexibility of regulation of carotenoid biosynthesis in A. brasilense to integrate and respond to different kinds of stimuli which may require carotenoids to cope with the fluctuations in its environment. For example, in Salmonella enterica serovar Typhimurium, a cascade of sigma factors linking σE, σH, and σS allows the integration of diverse environmental signals to result in the expression of a common stress response (36).
Although alternative sigma factors can act independently, a considerable level of overlap between the genes controlled by alternative sigma factors was demonstrated in Borrelia burgdorferi (37, 38). Our previous study showed that A. brasilense RpoE1 and RpoE2 are highly homologous, and hence, they not only regulate carotenoid synthesis but also regulate the expression of several common genes in A. brasilense (18). Data from this work show that although RpoE1 and RpoE2 both regulate carotenoid biosynthesis in A. brasilense, they bring about this regulation through different RpoH sigma factors (RpoE1 regulating RpoH2/5 and RpoE2 regulating RpoH1/4), indicating recognition of different promoters by the two RpoEs. An overlapping function of these RpoEs is manifested by RpoH1- and RpoH2-mediated regulation of a common gene (crtE2). A similar overlap of functions between RpoH1 and RpoH2 has also been demonstrated in R. sphaeroides (39) and Sinorhizobium meliloti (40). Furthermore, RpoE1- and -2-mediated regulation of RpoH4 and -5, which are not involved in carotenoid regulation, provides an insight into the utilization of RpoE1→RpoH4 and RpoE2→RpoH5 regulatory cascades for responding to other stresses, such as salinity and organic stresses, in which involvement of RpoE has been shown (25).
Since A. brasilense is a nonphotosynthetic bacterium, it may not be as vulnerable to photooxidative stress/damage as photosynthetic bacteria like R. sphaeroides. However, being a soil bacterium, it may be exposed to the damaging effects of light while swimming under submerged soil conditions. Inducibility of carotenoid synthesis in response to light might thus be a desirable and beneficial trait in A. brasilense. On the basis of this study and our earlier observations, we propose a model of regulation of carotenoid biosynthesis in A. brasilense (Fig. 6). In the absence of light, RpoE sigma factor remains bound to its cognate anti-sigma factor (ChrR). The singlet oxygen produced in the presence of light may react with the two paralogs of ChrR and alter their conformation to release their cognate RpoE sigma factors, which can express their target genes, including rpoH2 and rpoH1. RpoH2 and RpoH1, in turn, express their own target genes, including crtE2, which encodes geranylgeranyl pyrophosphate synthetase to catalyze the first step of the carotenoid biosynthesis. However, identification of the RpoE-regulated alternative sigma factor that directly regulates the expression of crtE1 is needed to develop a better understanding of carotenoid biosynthesis in A. brasilense.
FIG 6.
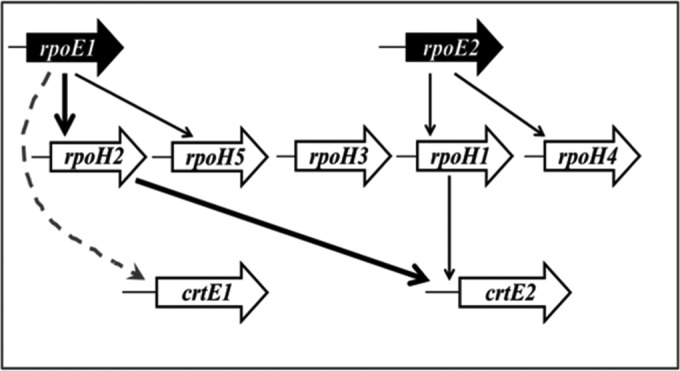
Proposed network of alternative sigma factors regulating the expression of the crtE2 gene of the carotenoid biosynthetic pathway in A. brasilense Sp7. In response to appropriate stimuli, RpoE1 regulates RpoH2 and RpoH5, while RpoE2 regulates RpoH1 and RpoH4. RpoH1 and RpoH2 initiate the transcription of a common gene, crtE2, to regulate carotenoid biosynthesis in A. brasilense. The crtE1 promoter is also activated in an RpoE1-dependent manner but via an unknown intermediate regulator or regulators. Thick arrows indicate the cascade established in this study by mutation studies as well as by using a two-plasmid system. Thin arrows show the cascades inferred from promoter activity assays using the two-plasmid system. The dotted arrow indicates an indirect regulation of crtE1 by RpoE1 via an unknown intermediate sigma factor or factors.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Department of Biotechnology, New Delhi, to A.K.T. A.K.R., A.P.D., S.K., and M.N.M. were supported by fellowships from ICMR and CSIR.
We thank Lacy Daniels (Texas) for reading the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00460-16.
REFERENCES
- 1.Anthony JR, Warczak KL, Donohue TJ. 2005. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci U S A 102:6502–6507. doi: 10.1073/pnas.0502225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuss AM, Glaeser J, Klug G. 2009. RpoH(II) activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J Bacteriol 191:220–230. doi: 10.1128/JB.00925-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong GA, Alberti M, Hearst JE. 1990. Conserved enzymes mediate the early reactions of carotenoid biosynthesis in nonphotosynthetic and photosynthetic prokaryotes. Proc Natl Acad Sci U S A 87:9975–9979. doi: 10.1073/pnas.87.24.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodwin TW. 1980. The biochemistry of the carotenoids, vol 1 Chapman and Hall, Ltd., London, United Kingdom. [Google Scholar]
- 5.Kim SW, Keasling JD. 2001. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng 72:408–415. doi:. [DOI] [PubMed] [Google Scholar]
- 6.Misawa N, Satomi Y, Kondo K, Yokoyama A, Kajiwara S, Saito T, Ohtani T, Miki W. 1995. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol 177:6575–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sieiro C, Poza M, de Miguel T, Villa TG. 2003. Genetic basis of microbial carotenogenesis. Int Microbiol 6:11–16. [DOI] [PubMed] [Google Scholar]
- 8.Burchard RP, Dworkin M. 1966. Light-induced lysis and carotenogenesis in Myxococcus xanthus. J Bacteriol 91:535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galbis-Martínez L, Galbis-Martínez M, Murillo FJ, Fontes M. 2008. An anti-antisigma factor in the response of the bacterium Myxococcus xanthus to blue light. Microbiology 154:895–904. doi: 10.1099/mic.0.2007/013359-0. [DOI] [PubMed] [Google Scholar]
- 10.Weeks OB, Garner RJ. 1967. Biosynthesis of carotenoids in Flavobacterium dehydrogenans Arnaudi. Arch Biochem Biophys 121:35–49. doi: 10.1016/0003-9861(67)90007-0. [DOI] [PubMed] [Google Scholar]
- 11.Grogan DW. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J Bacteriol 171:6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takano H, Obitsu S, Beppu T, Ueda K. 2005. Light-induced carotenogenesis in Streptomyces coelicolor A3(2): identification of an extracytoplasmic function sigma factor that directs photodependent transcription of the carotenoid biosynthesis gene cluster. J Bacteriol 187:1825–1832. doi: 10.1128/JB.187.5.1825-1832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitworth DE, Hodgson DA. 2001. Light-induced carotenogenesis in Myxococcus xanthus: evidence that CarS acts as an anti-repressor of CarA. Mol Microbiol 42:809–819. [DOI] [PubMed] [Google Scholar]
- 14.Lee HS, Ohnishi Y, Horinouchi S. 2001. A sigmaB-like factor responsible for carotenoid biosynthesis in Streptomyces griseus. J Mol Microbiol Biotechnol 3:95–101. [PubMed] [Google Scholar]
- 15.Nur I, Yuval LS, Okon Y, Henis Y. 1981. Carotenoid composition and function in nitrogen-fixing bacteria of the genus Azospirillum. J Gen Microbiol 122:27–32. [Google Scholar]
- 16.Thirunavukkarasu N, Mishra MN, Spaepen S, Vanderleyden J, Gross CA, Tripathi AK. 2008. An extra-cytoplasmic function sigma factor and anti-sigma factor control carotenoid biosynthesis in Azospirillum brasilense. Microbiology 154:2096–2105. doi: 10.1099/mic.0.2008/016428-0. [DOI] [PubMed] [Google Scholar]
- 17.Gupta N, Kumar S, Mishra MN, Tripathi AK. 2013. A constitutively expressed pair of rpoE2–chrR2 in Azospirillum brasilense Sp7 is required for survival under antibiotic and oxidative stress. Microbiology 159:205–218. doi: 10.1099/mic.0.061937-0. [DOI] [PubMed] [Google Scholar]
- 18.Gupta N, Gupta A, Kumar S, Mishra R, Singh C, Tripathi AK. 2014. Cross-talk between cognate and noncognate RpoE sigma factors and Zn(2+)-binding anti-sigma factors regulates photooxidative stress response in Azospirillum brasilense. Antioxid Redox Signal 20:42–59. doi: 10.1089/ars.2013.5314. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S, Rai AK, Mishra MN, Shukla M, Singh PK, Tripathi AK. 2012. RpoH2 sigma factor controls the photooxidative stress response in a non-photosynthetic rhizobacterium, Azospirillum brasilense Sp7. Microbiology 158:2891–2902. doi: 10.1099/mic.0.062380-0. [DOI] [PubMed] [Google Scholar]
- 20.Vanstockem M, Michiels K, Vanderleyden J, Van Gool AP. 1987. Transposon mutagenesis of Azospirillum brasilense and Azospirillum lipoferum: physical analysis of Tn5 and Tn5-mob insertion mutants. Appl Environ Microbiol 53:410–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW. (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 22.Nuss AM, Glaeser J, Berghoff BA, Klug G. 2010. Overlapping alternative sigma factor regulons in response to singlet oxygen in Rhodobacter sphaeroides. J Bacteriol 192:2613–2623. doi: 10.1128/JB.01605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLellan SR, MacLean AM, Finan TM. 2006. Promoter prediction in the rhizobia. Microbiology 152:1751–1763. doi: 10.1099/mic.0.28743-0. [DOI] [PubMed] [Google Scholar]
- 24.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 25.Mishra MN, Kumar S, Gupta N, Kaur S, Gupta A, Tripathi AK. 2011. An extracytoplasmic function sigma factor cotranscribed with its cognate anti-sigma factor confers tolerance to NaCl, ethanol and methylene blue in Azospirillum brasilense Sp7. Microbiology 157:988–999. doi: 10.1099/mic.0.046672-0. [DOI] [PubMed] [Google Scholar]
- 26.Morales VM, Bäckman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47. doi: 10.1016/0378-1119(91)90007-X. [DOI] [PubMed] [Google Scholar]
- 27.Blanvillain SD, Boulanger A, Lautier M, Guynet C, Denancé N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng L, Baumann U, Reymond JL. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryksin AV, Matsumura I. 2010. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 31.Homerova D, Halgasova L, Kormanec J. 2008. Cascade of extracytoplasmic function sigma factors in Mycobacterium tuberculosis: identification of a sigmaJ-dependent promoter upstream of sigI. FEMS Microbiol Lett 280:120–126. doi: 10.1111/j.1574-6968.2007.01054.x. [DOI] [PubMed] [Google Scholar]
- 32.Pfaffli MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browning DF, Whitworth DE, Hodgson DA. 2003. Light-induced carotenogenesis in Myxococcus xanthus: functional characterization of the ECF sigma factor CarQ and antisigma factor CarR. Mol Microbiol 48:237–251. doi: 10.1046/j.1365-2958.2003.03431.x. [DOI] [PubMed] [Google Scholar]
- 34.Homerova D, Surdova K, Mikusova K, Kormanec J. 2007. Identification of promoters recognized by RNA polymerase containing Mycobacterium tuberculosis stress-response sigma factor sigma(F). Arch Microbiol 187:185–197. doi: 10.1007/s00203-006-0185-6. [DOI] [PubMed] [Google Scholar]
- 35.Bischoff M, Entenza JM, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol 183:5171–5179. doi: 10.1128/JB.183.17.5171-5179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bang IS, Frye JG, McClelland M, Velayudhan J, Fang FC. 2005. Alternative sigma factor interactions in Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol Microbiol 56:811–823. doi: 10.1111/j.1365-2958.2005.04580.x. [DOI] [PubMed] [Google Scholar]
- 37.Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A 98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A 102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dufour YS, Imam S, Koo BM, Green HA, Donohue TJ. 2012. Convergence of the transcriptional responses to heat shock and singlet oxygen stresses. PLoS Genet 8:e1002929. doi: 10.1371/journal.pgen.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnett MJ, Bittner AN, Toman CJ, Oke V, Long SR. 2012. Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J Bacteriol 194:4983–4994. doi: 10.1128/JB.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.