ABSTRACT
Quorum sensing (QS) is an important regulatory system in virulence expression and environmental adaptation in bacteria. The master QS regulators (MQSR) LuxR and AphA reciprocally control QS gene expression in vibrios. However, the molecular basis for the regulatory functions of AphA remains undefined. In this study, we characterized its regulatory roles in Vibrio alginolyticus, an important zoonotic pathogen causing diseases in marine animals as well as in humans. AphA is involved in the motility ability, biofilm formation, and in vivo survival of V. alginolyticus. Specifically, AphA is expressed at low-cell-density growth phases. In addition, AphA negatively regulates the expression of the main virulence factor, alkaline serine protease (Asp), through LuxR. Chromatin immunoprecipitation (ChIP) followed by high-throughput DNA sequencing (ChIP-seq) detected 49 enriched loci harboring AphA-binding peaks across the V. alginolyticus genome. An AphA-specific binding motif was identified and further confirmed by electrophoretic mobility shift assay (EMSA) and mutagenesis analysis. A quantitative real-time PCR (qRT-PCR) assay further validated the regulation of AphA on these genes. AphA binds directly to the aphA promoter and negatively regulates its own expression. Moreover, AphA directly regulates genes encoding adenylate cyclase, anti-σD, FabR, and the small RNA CsrB, revealing versatile regulatory roles of AphA in its physiology and virulence. Furthermore, our data indicated that AphA modulates motility through the coordinated function of LuxR and CsrB. Collectively, the findings of this work contribute to better understanding of the regulatory roles of AphA in QS and non-QS genes.
IMPORTANCE In this work, we determined that AphA, the master regulator of QS at low cell density, plays essential roles in expression of genes associated with physiology and virulence in V. alginolyticus, a Gram-negative pathogen for humans and marine animals. We further uncovered that 49 genes could be directly regulated by AphA and a 19-bp consensus binding sequence was identified. Among the 49 genes, the QS and other non-QS-associated genes were identified to be regulated by AphA. Besides, the small RNA CsrB was negatively regulated by AphA, and AphA regulate motility abilities through both CsrB and LuxR. Taken together, the findings of this study improve our understanding of the complex regulation network of AphA and QS.
INTRODUCTION
Bacteria are highly social organisms capable of sophisticated cooperative behaviors mediated by the quorum-sensing (QS) system, a cell density-dependent signaling circuit (1). In Vibrio harveyi, there are three well-defined QS signaling molecules, known as autoinduers (AIs), including acylated homoserine lactones (HAI-1), furanosyl borate diester (AI-2), and CAI-1 (2). The membrane sensors LuxN, LuxQ, and CqsS can detect their cognate AIs and transduce the signals required to trigger gene expression through phospho-relay cascades (2). At low cell densities (LCDs; low AI concentrations), phosphates are directed by kinases in upstream QS cascades to the pivotal regulator LuxO. This leads to repression of the master QS regulator (MQSR) LuxR by multiple small regulatory RNAs (Qrr1 to Qrr5) (2). In contrast, at high cell densities (HCDs; high AI concentrations), the phosphate circuit runs in the opposite direction, resulting in the dephosphorylation and inactivation of LuxO, which derepresses the expression of LuxR (2). The MQSR then binds to and regulates the expression of many (∼150) target genes, leading to diverse phenotypes, including bioluminescence, biofilm formation, and motility (3). V. harveyi and Vibrio cholerae also deploy other regulators, such as AphA (4), and various Qrr-mediated regulatory mechanisms (5) to optimize the output of QS regulation, thus demonstrating exquisite and sophisticated regulatory architectures in vibrio QS systems.
AphA was first identified as an activator of tcpP and tcpH expression and of virulence gene expression in V. cholerae (6). The crystal structure of AphA reveals that it belongs to the winged-helix DNA-binding transcriptional factor superfamily (7). AphA cooperates with AphB, a LysR family transcriptional factor, as well as with CRP (cyclic AMP [cAMP] receptor protein) on the promoter region of tcpPH to regulate virulence expression in V. cholerae (8–10). Interestingly, the aphA gene is situated between ppc (phosphoenolpyruvate carboxylase) and cysE (serine acetyltransferase) in the vibrio chromosome, a region containing genes involved in normal metabolic functions, suggesting that this gene is carried on the ancestral vibrio chromosome and not associated with a pathogenicity island (6). In V. cholerae, AphA is demonstrated to be involved in acetoin biosynthesis and motility/biofilm formation, which are not associated with the vibrio pathogenicity island (VPI) (11). The gene cluster cpsQ-mfpABC, which contributes to biofilm development in Vibrio parahaemolyticus, is also regulated by AphA (12).
One interesting aspect of the AphA regulatory network is that its expression is regulated by QS and it responds to cell density (13). At LCDs, AphA levels are relatively high and control the expression of ∼167 to 296 genes, whereas at HCDs, LuxR binds to the promoter of AphA and represses its expression (14). This results in the regulation of ∼625 genes in V. harveyi, rendering an overlap of 77 AphA- and LuxR-coregulated genes (4, 14). The reciprocal control of QS by the MQSR AphA and LuxR at LCDs and HCDs, respectively, optimizes the QS output in terms of temporal and strength and contributes to an exquisite and fine-tuned QS regulatory architecture. It will be intriguing to decipher the regulon and the molecular basis of the regulation of AphA across chromosomes. This will contribute to the understanding of how conserved regulatory systems coordinate the expression of QS and non-QS genes, as well as how the core-genome cross talks with the pan-genome via AphA.
Vibrio alginolyticus is a zoonotic pathogen that infects a wide range of sea animals and causes intra- and extraintestinal diseases in humans (15). The major virulence factors of V. alginolyticus are extracellular proteases, motility, siderophore-dependent iron uptake systems, biofilm, type III secretion systems (T3SS), and type VI secretion systems (T6SS) (16–22). These factors and some other virulence-related genes are closely regulated by the QS system in V. alginolyticus. In our recent investigation, AphA was identified and demonstrated to be involved in the regulation of LuxR expression (23). In this study, we further explored other genes regulated by AphA via chromatin immunoprecipitation (ChIP) followed by high-throughput DNA sequencing (ChIP-seq), and electrophoretic mobility shift assays (EMSA) and quantitative real-time PCR (qRT-PCR) experiments were used to verify the results. The AphA consensus sequence was identified by using Multiple EM for Motif Elicitation (MEME) (24). Overall, this work not only pinpoints the binding motif and the direct targets of AphA in vivo but also reveals that AphA plays important roles in the regulation of QS and other non-QS genes at LCDs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. V. alginolyticus strains and derivatives were normally grown at 30°C in Luria-Bertani (LB) medium supplemented with 3% NaCl (LBS). Escherichia coli strains were grown at 37°C in LB medium. When appropriate, the medium was supplemented with ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), kanamycin (100 μg/ml), isopropyl β-d-1-thiogalactopyranoside (1 mmol/ml), or l-arabinose (0.04%, wt/vol).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α λpir | Host for π-requiring plasmids | Laboratory collection |
| SM10 λpir | Host for π-requiring plasmids, conjugal donor | 22 |
| BL21(DE3) | Host strain for protein expression | Novagen |
| V. alginolyticus | ||
| EPGS | Wild type, isolated from the aquaculture farm of the South China Sea with CCTCC no. AB 209306; Ampr | Laboratory collection |
| WT lacZ+ | EPGS with lacZ inserted behind glmS; Ampr | 23 |
| EPGS aphA-Flag | EPGS with insertion of Flag tag fusion to aphA in genome; Ampr | This study |
| asp mutant | EPGS disrupted in asp; Ampr Cmr | 18 |
| ΔluxR mutant | EPGS with in-frame deletion in luxR; Ampr | 17 |
| ΔaphA mutant | EPGS with in-frame deletion in aphA; Ampr | This study |
| aphA+ strain | ΔaphA complemented strain harboring pBAD33::aphA-flag-1; Ampr Cmr | This study |
| ΔaphA ΔluxR mutant | EPGS with in-frame deletion in aphA and luxR; Ampr | This study |
| ΔcsrB mutant | EPGS with deletion in csrB; Ampr | This study |
| csrB+ strain | ΔcsrB complemented strain harboring pBAD33::csrB; Ampr Cmr | This study |
| ΔcsrB ΔluxR mutant | EPGS with deletion in csrB and luxR; Ampr | This study |
| ΔaphA csrB+ strain | EPGS with overexpression of csrB in ΔaphA mutant; Ampr | This study |
| ΔaphA/pBAD33 strain | ΔaphA; contains pBAD33 plasmid; Ampr Cmr | This study |
| ΔaphA/pBAD33::aphA strain | ΔaphA; pBAD33 derivative pBAD promoter-driven expression of AphA protein; Ampr Cmr | This study |
| ΔaphA/pBAD33::aphA-flag-2 strain | ΔaphA; pBAD33 derivative pBAD promoter-driven expression of AphA-Flag fusion protein; Ampr Cmr | This study |
| ΔaphA/pDM8::PaphA-luxAB strain | ΔaphA; pDM8 carrying the promoter region of aphA and luxAB; Ampr Cmr | This study |
| EPGS/pDM8::PaphA- luxAB | EPGS with pDM8 carrying the promoter region of aphA and luxAB; Ampr Cmr | This study |
| BL21/pET28a::aphA | BL21 with pET28a carrying the aphA ORFa; Kmr | This study |
| Plasmids | ||
| pDM4 | Suicide vector, pir dependent; R6K sacBR Cmr | 48 |
| pBAD33 | Ara-induced expression vector; Cmr | 49 |
| pBAD33-mob | pBAD33 with insertion of a mob gene; Cmr | This study |
| pET28a | Expression vector; Kmr | Novagen |
| pDM4::aphA | pDM4 with aphA fragment deletion of 4 to 573 nt; Cmr | This study |
| pDM4::csrB | pDM4 with csrB fragment deleted; Cmr | This study |
| pBAD33::csrB | pBAD33 derivative with csrB; Cmr | This study |
| pBAD33::aphA-flag-1 | pBAD33 derivative with PaphA-driven expression of AphA-Flag fusion protein; Cmr | This study |
| pBAD33::aphA | pBAD33 derivative pBAD promoter-driven expression of AphA protein; Cmr | This study |
| pBAD33::aphA-flag-2 | pBAD33 derivative pBAD promoter-driven expression of AphA-Flag fusion protein; Cmr | This study |
| pET28a::aphA | pET28a carrying the aphA ORF; Kmr | This study |
| pDM8::aphA-luxAB | pDM8 carrying the promoter region of aphA and luxAB; Cmr | This study |
ORF, open reading frame.
Construction of V. alginolyticus aphA-Flag fusion and csrB deletion mutant in the genome.
The construction of mutant strains followed a previous strategy developed with the suicide plasmid pDM4 (22). To construct the aphA-Flag fusion, PCR was performed to amplify sequences upstream and downstream of the termination codon of aphA (TAA), and the Flag DNA fragment was inserted in the front of TAA. The primers used are listed in Table 2. The PCR products were cloned into the XbaI site of pDM4 by isothermal assembly, as previously described (25). The recombinant plasmid was conjugated into wild-type (WT) strain EPGS with selection for ampicillin and chloramphenicol resistance. Then the correct colony was screened for sucrose (12%) sensitivity, which typically indicates a double-crossover event and, thus, the occurrence of gene replacement. Additionally, the aphA-Flag fusion strain was confirmed by PCR and sequencing. The construction of a csrB deletion mutant followed the same strategy.
TABLE 2.
Primers used in this study
| Primer name | Primer sequencea (5′–3′) | Target or purpose |
|---|---|---|
| AphA-FlagP1 | GAGCTCAGGTTACCCGCATGCAAGATCTATTTACCACACGTAATCTTAACTGT | aph-Flag |
| AphA-FlagP2 | GTCGTCATCCTTGGGTGGCGGTGGCTCAGATTAACCAATCACTTCAAGTTCAGTTA | aph-Flag |
| AphA-FlagP3 | AGCCACCGCCACCCAAGGATGACGACGATAAGTAAACTCCTTTTGCTTTGATA | aph-Flag |
| AphA-FlagP4 | CCCTCGAGTACGCGTCACTAGTGGGGCCCTCACAGAACAATGTGAAAGTAC | aph-Flag |
| AphA-FlagOut-1F | ACAAGTTTATTGACCATTTGGA | aph-Flag |
| AphA-FlagOut-2R | GTCATCCTTGTAATCTGAGCCAC | aph-Flag |
| aphA-Flag-F | CCATACCCGTTTTTTTGGGCTAGCGAATTCTCGAACTCGGCAAGTTGCCACTTGG | aphA complement |
| aphA-Flag-R1 | GGTCAGCATGGGTACCTTTCTCCTCTTTAATTACTTGTCGTCGTCGTCCTTGTAGTC | aphA complement |
| aphA-Flag-R2 | CTTGTCGTCGTCGTCCTTGTAGTCTGAGCCACCGCCACCACCAATCACTTCAAGTTCAGTTAG | aphA complement |
| aphA-Flag-F1 | CCATACCCGTTTTTTTGGGCTAGCGAATTCTTGTTTACAAGTTTATTGACCATT | aphA-Flag fusion |
| aphA-R | GGTCAGCATGGGTACCTTTCTCCTCTTTAATTAACCAATCACTTCAAGTTCAGT | aphA protein |
| aphApET28a-F | ATCGGATCCATGTTTACAAGTTTATTGACCATTT | Protein cloning |
| aphApET28a-R | ATATGTCGACACCAATCACTTCAAGTTCAGTT | Protein cloning |
| aphA-luxAB-1 | ATTGCTGCAGGTCGACGGATCCGGGGAATTCGCTTACGCAATATCTACGTTGAGC | Pasp-luxAB |
| aphA-luxAB-2 | ATTTGTTGGTAGCTACTTATACTTTCAGCTGCGCTC | Pasp-luxAB |
| aphA-luxAB-3 | GTATAAGTAGCTACCAACAAATAAGGAAATGTTATG | Pasp-luxAB |
| aphA-luxAB-4 | GTCGACCTGCAGCCCAAGCTTATCGATTCGTTACGAGTGGTATTTGACGATGTTG | Pasp-luxAB |
| csrB1 | GAGCTCAGGTTACCCGCATGCAAGATCTATGGATGACAAGCCTTACATGGCA | csrB mutant |
| csrB2 | AACTTTAGGCACTCTCTTAGCGTTCAAAAAAG | csrB mutant |
| csrB3 | GCTAAGAGAGTGCCTAAAGTTTTTCTCAGTAGATC | csrB mutant |
| csrB4 | CCCTCGAGTACGCGTCACTAGTGGGGCCCTTTCTCGTGTGATGCATCCAAAGTT | csrB mutant |
| csrBout-F | ACCGATTTCTCTTTCCGCACG | csrB mutant |
| csrBout-R | AGTGGTCGATTTAGCCAAACTG | csrB mutant |
| csrBcom-F | CCATACCCGTTTTTTTGGGCTAGCGAATTCTCTTTTTTGAACGCTAAGAGAG | csrB complement |
| csrBcom-R | GGTCAGCATGGGTACCTTTCTCCTCTTTAAACAGATTGCCTTTAACGGTGAAGG | csrB complement |
| aphA-F | TGCCTGCAGGTCGACGATGATAATCGCACCAAAACAAAGG | EMSA |
| aphA-R | CTTATACTTTCAGCTGCGCTCT | EMSA |
| ΔaphA1 | TAATATTGATATCTCTCGCTACTAC | EMSA |
| ΔaphA2 | GTATATAAATAGTAGGACAAAATTACCCTAAAA | EMSA |
| ΔaphA3 | TTTGTCCTACTATTTATATACTACTTGCCTGCTG | EMSA |
| ΔaphA4 | GCTGCGACCAGCGTCTGTAATGG | EMSA |
| 09797-F | TGCCTGCAGGTCGACGATGGTTTATCCTACCATTCTCAT | EMSA |
| 09797-R | GGGGAATTCCTTGGTTGAACA | EMSA |
| 00030-F | TGCCTGCAGGTCGACGATTGCTCTGCAGACCCGAAGGGC | EMSA |
| 00030-R | GCCCTCATGATCTGATCGCCC | EMSA |
| 09557-F | TGCCTGCAGGTCGACGATTACCGATTACGATCACGTCAT | EMSA |
| 09557-R | TTCTGTAGGGTACAAGAGCCTC | EMSA |
| sRNA-EMSAF | TGCCTGCAGGTCGACGATGAGTCTTTCGCTCGACCCCTTGTGC | EMSA |
| sRNA-EMSAR | CGTGTCAGCATCCTTCCGACAGGTT | EMSA |
| Control-F | TGCCTGCAGGTCGACGATGACACAGTGCTGGCGATATAGCA | EMSA |
| Control-R | TAAGTGATTTTGTGCGCATTGGT | EMSA |
| 04316-F | CCCATCTCCTTCCGAACTGC | qRT-PCR |
| 04316-R | TTAGTCACGACAAAGCCGGT | qRT-PCR |
| 04332-F | TTTCTCGAGCAACAGCGGTA | qRT-PCR |
| 04332-R | TTCGTACCTCGGTCGAAACC | qRT-PCR |
| 04385-F | CGTTACGACGCGCAAATTGA | qRT-PCR |
| 04385-R | ATTGGGATGCCAGTGTCGTT | qRT-PCR |
| sRNA-F | GGAACAGGAAACACCACGGA | qRT-PCR |
| sRNA-R | GGTGTCCATTTCCCGTCCTT | qRT-PCR |
| 16S RNA-F | AAAGCACTTTCAGTCGTGAGGAA | qRT-PCR |
| 16S RNA-R | TGCGCTTTACGCCCAGTAAT | qRT-PCR |
Underlining indicates the overhang sequence for isothermal assembly or the restriction enzyme cut site.
Construction of the promoter-reporter plasmids.
Plasmid pDM8 was used to construct the promoter-luxAB fusion of aphA. The aphA promoter regions were amplified by PCR using the primers shown in Table 2 and then cloned into the SmaI site in pDM8 by isothermal assembly (25). The recombinant plasmid was introduced into the wild type and the ΔaphA strain by conjugation. The recombinant plasmids were confirmed by DNA sequencing.
qRT-PCR.
Equal amounts of RNA (1 μg) were used to generate cDNA (Toyobo, Tsuruga, Japan) using random primers. Three independent experiments were performed, each in triplicate, with specific primer pairs (Table 2) on an Applied Biosystems 7500 real-time system (Applied Biosystems, Foster City, CA), and transcript levels were normalized to 16S RNA in each sample by the threshold cycle (ΔΔCT) method.
Western blot assay.
For the immunoblotting assay, supernatants and bacterial cell pellets were harvested at the same optical density at 600 nm (OD600). Then, 15 μl of each sample was loaded onto a 12% denaturing polyacrylamide gel; proteins were resolved by electrophoresis and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). The membranes were blocked with a 10% skim milk powder solution, incubated with a 1:2,000 dilution of Asp-specific, LuxR-specific (GL Peptide Ltd., Shanghai, China), or Flag-specific (Sigma-Aldrich, St. Louis, MO) antiserum, and then incubated with a 1:2,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA). Finally, the blots were visualized with an ECL reagent (Thermo Fisher Scientific Inc., Waltham, MA).
Motility and biofilm assay.
The motility and biofilm assay was performed as previous described (22). The overnight-cultured WT, ΔaphA, and aphA+ strains were diluted to an OD600 of 1.0 and then spotted to the LBS containing 0.3% (swimming) and 1.5% (swarming) agar. The cultures (50 μl) were diluted to 5 ml of LBS medium in glass tubes and incubated at 30°C without shaking for 48 h. Total biofilm was measured by 2% crystal violet staining. The experiments were performed at least three times, and one representative result is shown.
ECP activity assay (HPA).
Extracellular protease (ECP) activity was determined using hide powder azure (HPA) digestion as previously described (22). Briefly, strains were grown in LBS medium at 30°C for 9 h. The cell density was measured at 600 nm. The bacterial cultures were centrifuged, and then the supernatants were filtered through 0.22-μm-pore-size filters (Millipore). After that, 1-ml quantities of filtered supernatants were mixed with 1 ml of phosphate-buffered saline (PBS; pH 7.2) and 10 mg of HPA (Sigma-Aldrich). The mixture was incubated with shaking at 37°C for 2 h. After stopping the reaction by adding 10% trichloroacetic acid, total protease activity was measured at 600 nm. ECP activity was normalized by dividing total activity by the OD600 for each strain.
EMSA.
Purified 6×His-tagged AphA was incubated with different Cy5-labeled DNA probes (Table 2) in 20 μl of loading buffer (10 mM NaCl, 0.1 mM dithiothreitol [DTT], 0.1 mM EDTA, 10 mM Tris [pH 7.4]). After incubation at 25°C for 30 min, the samples were resolved by 6% polyacrylamide gel electrophoresis in 0.5× Tris-boric acid-EDTA (TBE) buffer on ice at 100 V for 120 min. The gels were then scanned with an FLA 9500 apparatus (GE Healthcare, Uppsala, Sweden).
ChIP-seq analysis.
The pBAD33Flag::AphA and pBAD33Flag plasmids, encoding AphA-Flag and the Flag tag alone, respectively, were transferred to the ΔaphA strain for ChIP assays as previously described (23). Overnight cultures of each strain in LBS medium were diluted (1:100) in 50 ml of fresh LBS medium with 0.04% l-arabinose. After 2 h of growth with shaking, the protein-DNA complexes in the bacterial cells were cross-linked in vivo with 1% formaldehyde at room temperature for 10 min. Cross-linking was stopped by addition of 125 mM glycine. Bacteria were then washed twice with cold PBS and resuspended in 5 ml of SDS lysis buffer (50 mM HEPES-KOH [pH 7.5], 0.1% sodium deoxycholate, 150 mM NaCl, 0.1% SDS, 1 mM EDTA [pH 8], 1% Triton X-100, protease inhibitors). Then the bacteria were sonicated, and the DNA was fragmented to 100 to 500 bp at 200 W. Insoluble cellular debris was removed by centrifugation, and the supernatant was used as the input sample in IP experiments as follows. Both input and IP samples were washed with 50 μl of protein G beads for 1 h. Then IP samples were incubated overnight with 50 μl of Flag-labeled beads (Sigma-Aldrich). After incubation, the beads were washed twice with a low-salt wash buffer (10 mM Tris-HCl [pH 8], 150 mM NaCl, 0.1% SDS, 1 mM EDTA [pH 8], 1% Triton X-100), twice with 1 ml of a high-salt wash buffer (10 mM Tris-HCl [pH 8], 500 mM NaCl, 0.1% SDS, 1 mM EDTA [pH 8], 1% Triton X-100), twice with 1 ml of LiCl wash buffer (10 mM Tris-HCl [pH 8], 250 mM LiCl, 1 mM EDTA [pH 8], 0.5% Triton X-100, 0.5% sodium deoxycholate), and twice with 1 ml of TE buffer. The beads were resuspended in 200 μl of elution buffer (50 mM Tris HCl [pH 8], 10 mM EDTA, 1% SDS), incubated for 2 h at 65°C, and centrifuged at 5,000 × g for 1 min. The supernatants containing the immunoprecipitated DNA were collected, and 8 μl of 5 M NaCl was added to all of the tubes (IPs and inputs). The tubes were incubated at 65°C overnight to reverse the DNA-protein cross-links. Then, 1 μl of RNase A was added, and the solutions were incubated for 30 min at 37°C. Four microliters of 0.5 M EDTA, 8 μl of 1 M Tris-HCl, and 1 μl of proteinase K were added to each tube and incubated at 45°C for 2 h. The DNA was purified using phenol-chloroform.
DNA fragments were used for library construction with the VAHTS Turbo DNA library prep kit (Vazyme, Nanjing, China). The number of reads per microliter of each library was determined by quantitative real-time-PCR against a standard curve and sequenced with a MiSeq sequencer (Illumina, San Diego, CA). ChIP-seq reads were mapped to the V. alginolyticus EPGS genome using Bowtie 2 (26). The enriched peaks were identified using MACS software (26), followed by MEME analysis to generate the AphA-binding motif (27). The KEGG pathway analysis was performed with the Kobas 2.0 to illustrate the enriched gene function (28).
CI assay.
The competitive index (CI) of the WT, ΔaphA, and aphA+ strains was determined using the WT strain harboring a lacZ gene behind the glmS locus (Table 1). The infection dose was 105 CFU/fish; seven zebra fish were sacrificed at 24 h postinfection and then plated to the LBS containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and Amp. The plates were incubated overnight at 30°C. The WT strain was differentiated from the mutant strain based on the production of blue and white colonies (blue for WT strains). The ratios of the WT counts to the ΔaphA and aphA+ strain counts were used to determine the CI.
All animal experiments presented here were approved by the Animal Care Committee of the East China University of Science and Technology (2006272). The experimental animal care and use guidelines from the Ministry of Science and Technology of China (MOST-2011-02) were strictly adhered to.
RESULTS
Cell density-dependent expression of AphA in V. alginolyticus.
To investigate the function of AphA and its relationship to the QS system and virulence in V. alginolyticus, we cloned and identified aphA from the genome of V. alginolyticus EPGS. In our recent investigation, AphA was identified and determined to bind directly to the luxR promoter and repress the expression of LuxR in V. alginolyticus (23). In this study, we set out to investigate AphA production throughout growth phases by assaying the relative transcript and protein production profiles of AphA at different stages. Transcriptional levels of aphA were decreased as the cells grew and the densities increased, as determined with a transcriptional fusion construct of PaphA-luxAB (Fig. 1A). The transcriptional activity of PaphA was highest at the early exponential growth stage; however, its transcriptional activity was reduced dramatically when the cells were approaching late exponential or stationary growth phases. Western blot analysis with a strain carrying a translational fusion of aphA-Flag in the aphA locus indicated that AphA was expressed at LCDs (Fig. 1B). Taken together, these data indicate that the expression of AphA is cell density dependent, and the production of AphA is highest at LCDs.
FIG 1.
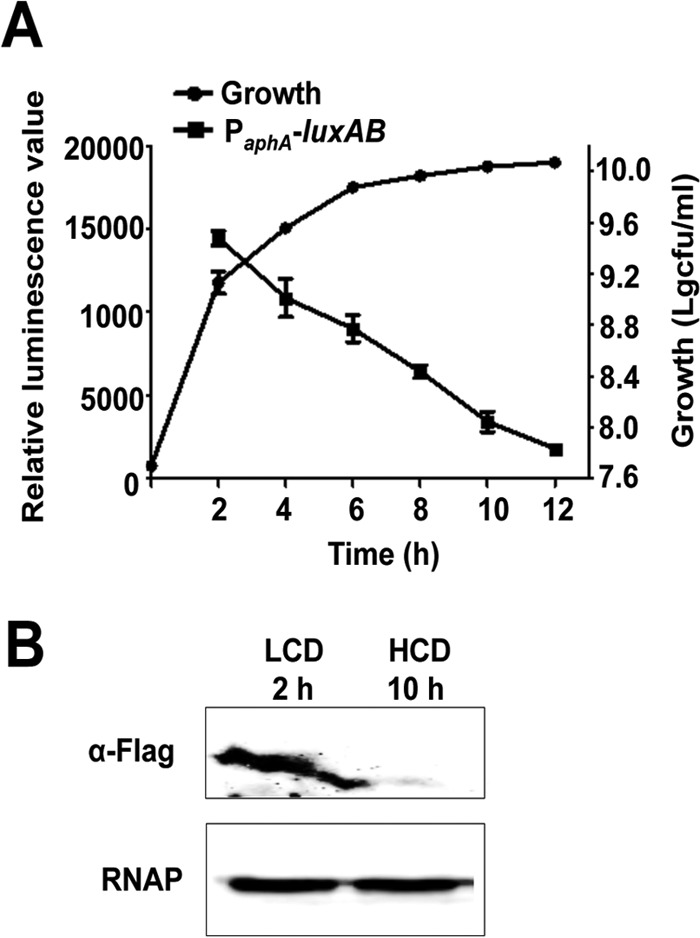
Low-cell-density-dependent expression of aphA in V. alginolyticus. (A) Growth of V. alginolyticus and transcriptional levels of aphA at different growth phases were determined. The cultures were sampled at various time points and plate counted after serial dilutions with fresh LBS medium. To assay the transcriptional activity of aphA, a pDM8 derivative plasmid carrying the aphA promoter region (PaphA) fused to promoterless luxAB (pDM8-PaphA-luxAB) was conjugated into the WT strain and the fluorescence was assayed. (B) Western blot analysis of AphA expression. Flag-tagged AphA was expressed from the WT strain carrying the aphA-Flag translational fusion at the aphA locus (AphA-Flag/EPGS) (Table 1) and blotted against a Flag-specific antibody to determine the expression levels of AphA in WT cells. RNAP was used as a loading control for the cell lysates.
Essential roles of AphA in motility, biofilm production, and in vivo survival of zebra fish.
Two flagellar systems have been identified in V. alginolyticus. A single polar flagellum serves to swim, and numerous lateral flagella serve to swarm (29). Both the swarming and swimming abilities were significantly reduced in the ΔaphA strain; these abilities were restored to that of the WT in the complemented strain (Fig. 2A). Furthermore, biofilm formation was significantly decreased in the ΔaphA strain compared to that in the WT strain, and the complemented strain restored biofilm formation (Fig. 2B). To further test how AphA affects in vivo growth of the bacterium, the WT and ΔaphA strains were mixed in a 1:1 ratio and inoculated into zebra fish. A profound competitive defect was observed in in vivo growth (∼50-fold) of the ΔaphA mutant strain in comparison to that of the WT strain (Fig. 2C); no competitive defect was observed for either the WT or ΔaphA strain in LBS cultures. These results suggest that AphA plays important roles in the regulation of motility, biofilm formation, and in vivo survival in zebra fish. The roles in controlling motility and biofilm by AphA have also been observed in V. parahaemolyticus and V. cholerae (30–32), suggesting an AphA-dependent common regulatory network in these aspects in vibrios.
FIG 2.
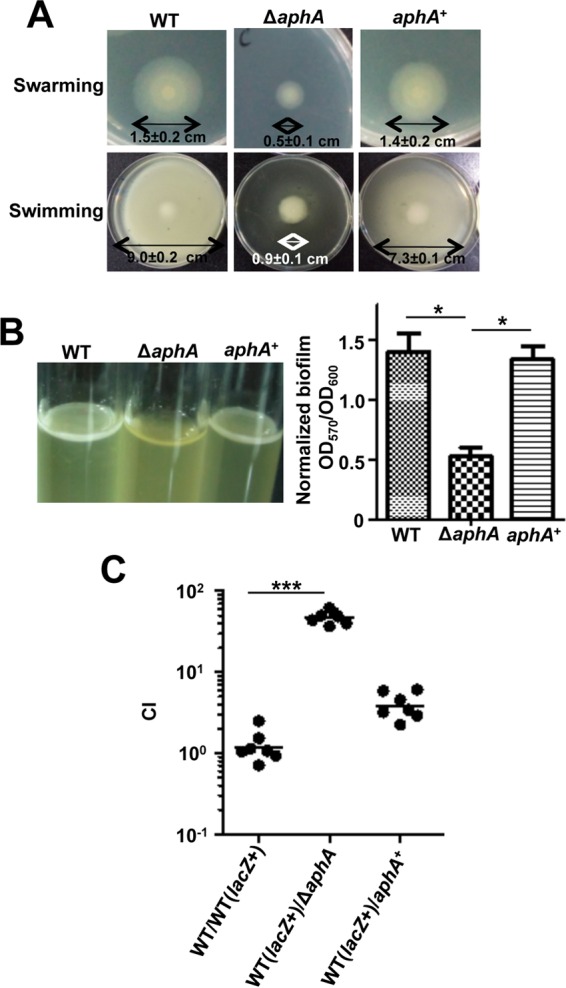
Essential roles of AphA in motility, biofilm formation, and in vivo survival of V. alginolyticus. (A) Motility assays of WT, ΔaphA, and aphA+ complemented strains. Swarming motility assays were performed on LBS plates with 1.5% agar, whereas swimming motility assays were performed with 0.3% agar. The diameters of the colonies are shown for the motility assays. The data are presented as means ± SD (n = 3). (B) Biofilm formation in glass tubes after 48 h was quantitatively assayed with crystal violet staining of the biofilm cells. The colorimetric assay of the released crystal violet from the biofilm cells was measured at OD570 and normalized with the OD600 of the cultures. (C) CI assays for the ΔaphA strain against WT (lacZ+), the WT strain carrying lacZ following the glmS locus. *, P < 0.05; ***, P < 0.001 (Student's t test).
AphA represses exotoxin asp expression.
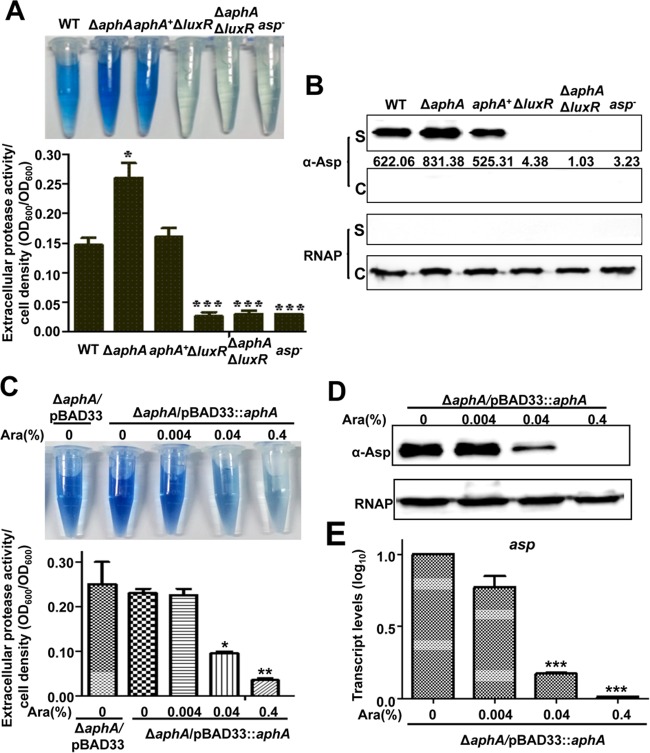
Alkaline serine protease (Asp) is the main extracellular toxin (exotoxin) of V. alginolyticus, and it is closely regulated by QS. In this study, we examined whether AphA could regulate the expression of Asp. A notable increase in Asp activity was displayed in the ΔaphA strain, whereas that of the aphA complemented strain decreased to WT levels (Fig. 3A). Western blot analysis using an Asp-specific antibody also demonstrated an increased level of Asp production in the supernatant pellets of this mutant strain. The complemented strain restored extracellular Asp production to WT levels (Fig. 3B).
FIG 3.
AphA negatively regulates Asp expression. (A) HPA digestion assays for extracellular Asp activity in different strains. After 9 h, culture supernatants were used to measure the protease activities of each strain, which were normalized by cell density. (B) Western blotting with concentrated supernatants (S) and cellular pellets (C) was performed using an Asp-specific antiserum. RNAP was used as a loading control for the blots. (C to E) HPA digestion assays (C) and Western blotting (D) for the extracellular Asp activity and qRT-PCR analysis of asp expression (E) in the ΔaphA/pBAD33::aphA strain with different concentrations of l-arabinose (Ara). The results are shown as means ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test) compared to values for corresponding samples from the WT or the inducible strain without addition of arabinose.
We further tested the AphA regulation of exotoxin Asp with an l-arabinose-inducible AphA strain. The results showed that low concentrations of l-arabinose, which generated low levels of AphA, led to Asp activities that were similar to those in the samples without l-arabinose or with the strain harboring pBAD33-empty plasmids (Fig. 3C). In contrast, high AphA levels, induced by high l-arabinose concentrations, strongly repressed Asp production (Fig. 3C and D). Finally, qRT-PCR analysis also confirmed a significant repression of asp transcription in the AphA-overexpressing strain (Fig. 3E). Taken together, our data suggest that AphA represses the expression of Asp.
AphA regulates Asp expression via LuxR.
We had previously reported that the MQSR LuxR positively regulates the expression of Asp (17, 18) and that AphA represses LuxR expression by binding directly to the luxR promoter region (23). In this study, we tested whether AphA regulates the expression of Asp in a LuxR-dependent manner. For this reason, we constructed a ΔaphA ΔluxR double-mutant strain. The HPA digestion assay and Western blot analysis showed that the ΔaphA ΔluxR strain exhibited no expression of Asp, like the ΔluxR strain (Fig. 3A), suggesting that AphA regulates Asp expression via LuxR. Then we detected the expression levels of LuxR induced by different concentrations of l-arabinose in the l-arabinose-driven AphA overexpression strain. The results showed that a low concentration of l-arabinose produced no or low levels of AphA, whereas it increased the expression of LuxR (Fig. 4A). Increased levels of AphA led to a significant reduction in LuxR expression (Fig. 4A). Additionally, qRT-PCR analysis also confirmed a significant repression of luxR transcription in the AphA overexpression strain (Fig. 4B). Taken together, our results demonstrated that AphA regulates the expression of Asp via LuxR in V. alginolyticus.
FIG 4.
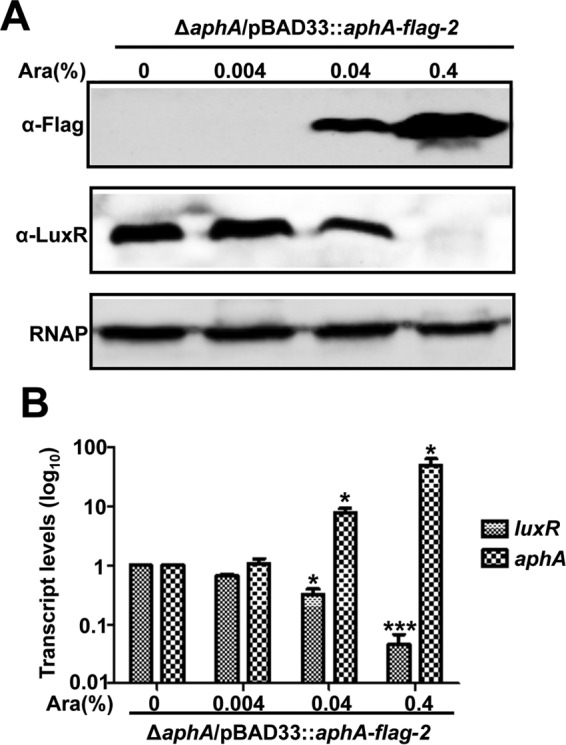
AphA regulates Asp expression via LuxR. (A) Western blot assay of LuxR expression with various concentrations of AphA (induced with l-arabinose). Bacterial cells of the ΔaphA strain harboring plasmid pBAD33::aphA-flag-2 were cultured in LBS medium for 9 h, harvested, and blotted with specific anti-Flag and anti-LuxR antibodies. (B) qRT-PCR analysis of the transcriptional level of aphA and luxR in the ΔaphA strain harboring pBAD33::aphA-flag-2 with different concentrations of l-arabinose. The bacterium was cultured in LBS for 9 h, and mRNA transcripts of aphA and luxR were detected by qRT-PCR. The 16S rRNA gene was selected as a control. The results are shown as means ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test) compared to the value without addition of l-arabinose.
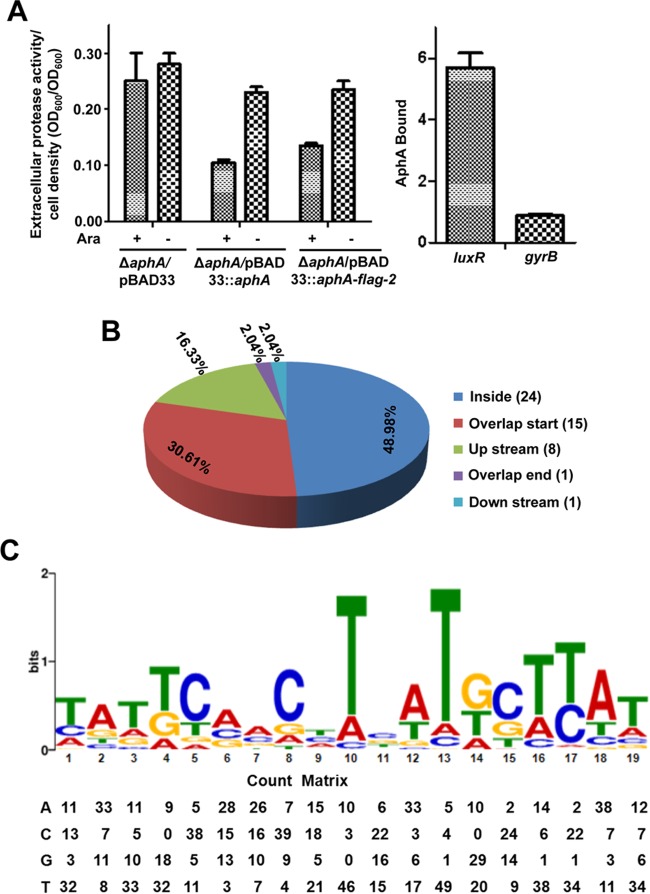
Genome-wide screen of AphA-binding regions by ChIP-seq.
As shown above, AphA is the master regulator of QS and virulence. We further investigated all the possible AphA-binding loci on the chromosomes of V. alginolyticus using ChIP-seq experiments with a Flag-tagged AphA, which was overexpressed from plasmid pBAD33 in the ΔaphA strain. As a control, first we verified that Flag-AphA behaves similarly to WT AphA in vivo (Fig. 5A) by assaying their activities in the repression of Asp production and by measuring Flag-AphA occupancy at the luxR promoter region that contains a known AphA binding site in V. alginolyticus (23). Our ChIP-qPCR analysis revealed an ∼5.5-fold enrichment of AphA binding at the promoters of luxR but not at gyrB, a promoter that is not bound by AphA (Fig. 5A).
FIG 5.
ChIP-seq reveals in vivo binding sites of AphA in the V. alginolyticus chromosomes. (A) Behaviors of AphA and Flag-tagged AphA (AphA-Flag) expressed with a pBAD33 plasmid with (+) or without (−) addition of l-arabinose on the production of Asp activities (left) or on the binding of the luxR promoter region in V. alginolyticus using ChIP-qPCR analysis (right). The DNA region in gyrB, which does not bind to AphA, was used as a negative control. (B) Pie chart of 49 enriched AphA-binding peaks indicating the locations of binding sites in various genes. (C) The most significant motif derived from a ChIP-seq binding sequence generated by the MEME tool (24, 27). The height of each letter represents the relative frequency of each base at a different position in the consensus sequence. A position frequency matrix describes the alignment of AphA sites and indicates the frequency of each nucleotide at each position.
Sequence reads were obtained from three independent ChIP-seq assays and mapped to the V. alginolyticus genome. We identified 49 enriched loci harboring AphA-binding peaks (Table 3), which were enriched >4-fold (P < 0.001) over those in control samples from the WT/pBAD33-FLAG strain (WT strain expressing only the Flag tag). These 49 loci are located across the genome, specifically in intergenic regions (51.02%) and within coding regions (48.98%), suggesting that AphA is a global transcriptional regulator in V. alginolyticus (Fig. 5B). Using the MEME suite, a 19-bp AphA consensus sequence (TATTCGN7GCTTAT) was identified (Fig. 5C). This sequence matches the motif in the luxR promoter region, which was revealed by the DNase I footprinting assay (23), and shows high similarity to the binding sites for AphA previously identified in other vibrios (32). The count matrix was provided to describe the alignment of AphA binding sites and represents the relative frequency of each base at a different position in the consensus sequence (Fig. 5C).
TABLE 3.
AphA ChIP-seq data
| Gene IDa | Annotation | AphA box-like sequence | Avg fold enrichment | Microarray (V. harveyi)b | P value |
|---|---|---|---|---|---|
| VEPGS-03659 | Excinuclease ATPase subunit | TCATTCAAGATGATTCATT | 8.8 | N | <0.001 |
| VEPGS-03814 | NADPH:quinone reductase and related Zn-dependent oxidoreductases | TCGGAGAACCTTATGTTTA | 8.45 | N | <0.001 |
| VEPGS-03822 | Alanine-alpha-ketoisovalerate (or valine-pyruvate) aminotransferase | AAAGGGAAGCTTATGCTTC | 8 | N | <0.001 |
| VEPGS-03826 | Type IIA topoisomerase (DNA gyrase/topoisomerase II, topoisomerase IV), B subunit | TCTTCACGCTCAGAGTTAA | 8.1 | N | <0.001 |
| VEPGS-03844 | FoF1-type ATP synthase, subunit b | TGATTAACCTAGTTGACCA | 7.75 | N | <0.001 |
| VEPGS-03847 | FoF1-type ATP synthase, gamma subunit | TATTCAACAAGTTTGTAAA | 8.3 | N | <0.001 |
| VEPGS-03856 | Dihydroxyacid dehydratase/phosphogluconate dehydratase | TGATTCGTCATCTTGCTCG | 8.15 | N | <0.001 |
| VEPGS-03880 | Zn finger domain associated with topoisomerase type I | ATGTGACCATCATTGCTGT | 8.1 | N | <0.001 |
| VEPGS-03964 | Aspartate ammonia-lyase | TGGTCTCCTTCTTTTTTAG | 13.6 | N | <0.001 |
| VEPGS-04037 | Exoribonuclease R | TCGTTTGACGACATGGATT | 8.65 | Y | <0.001 |
| VEPGS-04051 | Iron-sulfur cluster-binding protein | CTTTCAATCTCTTGCATAT | 8.85 | N | <0.001 |
| VEPGS-04106 | Predicted transcriptional regulators (AphA) | TATTCCACTTCATGCTTAT | 14.62 | Y | <0.001 |
| VEPGS-04284 | Small RNA, CsrB family | CATTGAATTTAATACTCAT | 8.75 | N | <0.001 |
| VEPGS-04291 | Putative multidrug resistance protein | TCATTCCGGCTTTAGCTCC | 7.7 | N | <0.001 |
| VEPGS-04316 | Regulator of sigma D (anti-σD) | TTATCGTTAAGTAAATGTG | 7.9 | N | <0.001 |
| VEPGS-04317 | DNA-directed RNA polymerase beta′ subunit | TATAGATCGTTAAGATCAG | 7.55 | Y | <0.001 |
| VEPGS-04318 | DNA-directed RNA polymerase beta subunit | ACAGGCGAGATCATTGTTA | 7.55 | N | <0.001 |
| VEPGS-04329 | Vitamin B12 receptor | TATTCGCACTCTTGTCTTT | 7.95 | N | <0.001 |
| VEPGS-04332 | Transcriptional regulator (FabR) | TATGTCTATAAATTGCATC | 12.6 | N | <0.001 |
| VEPGS-04337 | Glycerol-3-phosphate acyltransferase | TAATTGCCCAAATGCTCAT | 8.1 | N | <0.001 |
| VEPGS-04385 | Adenylate cyclase | TATGGTTTTGTATCCAGAT | 7.7 | N | <0.001 |
| VEPGS-04400 | Predicted transcriptional regulators | TTTTACCCTCTAATCTTCT | 8.35 | N | <0.001 |
| VEPGS-04448 | d-Erythrose 4-phosphate dehydrogenase | TATGCAACTTAGTGTCCAC | 11.45 | N | <0.001 |
| VEPGS-04453 | UDP-N-acetylmuramate dehydrogenase | ACCGATCCCCTGATTCACA | 9.75 | N | <0.001 |
| VEPGS-04457 | Putative translation factor | AACGCGGCATCAAAGTTAT | 7.9 | N | <0.001 |
| VEPGS-04459 | Shikimate 5-dehydrogenase | TGCACCTGTTGATGGTTTT | 8.85 | N | <0.001 |
| VEPGS-01281 | Transcriptional regulators of sugar metabolism | TATAAAGCGTTTTGCCTTT | 8.8 | N | <0.001 |
| VEPGS-01343 | 5′-Nucleotidase/2′,3′-cyclic phosphodiesterase and related esterases | TCTGCGCCTTCTTTCTTCA | 9.65 | N | <0.001 |
| VEPGS-01349 | Hypothetical protein | AAAGTGGCCTGATGGACAT | 8.1 | N | <0.001 |
| VEPGS-01356 | 50S ribosomal protein L15p | TAGTTGCCATGGTGTTCTT | 7.6 | N | <0.001 |
| VEPGS-01395 | UDP-glucose 6-dehydrogenase | TGATAAGCTTTCTGCTTAT | 8.45 | N | <0.001 |
| VEPGS-01401 | Hypothetical protein | CGTATCACCTGTTACTGAT | 7.95 | N | <0.001 |
| VEPGS-01451 | ABC-type oligopeptide transport system, periplasmic component | AAAGCCAAATCGTGCTTTT | 8.4 | N | <0.001 |
| VEPGS-01456 | Biopolymer transport proteins | TAGTTCAGTCAGCTGCTTA | 7.85 | N | <0.001 |
| VEPGS-01492 | Disulfide bond chaperones of the HSP33family | TGCTTTGCCTTCATGCTCA | 7.75 | N | <0.001 |
| VEPGS-01503 | Signal transduction histidine kinase, nitrogen specific | TGAACAATCACTTTCTCAA | 8.75 | N | <0.001 |
| VEPGS-01511 | Cytochrome c553 | TATAAAGCGTTTTGCCTTT | 7.9 | N | <0.001 |
| VEPGS-01513 | 5′-3′ exonuclease (including N-terminal domain of polymerase I) | TAAGCTGATTTTTGCCTTT | 7.65 | Y | <0.001 |
| VEPGS-01530 | Hypothetical protein | ATATGCCACTCCTTATTTA | 7.65 | N | <0.001 |
| VEPGS-01564 | Hypothetical protein | ACTGGCAACCTGATTGTTC | 8.6 | N | <0.001 |
| VEPGS-01566 | Peptide ABC transporter, permease protein | TCTATCGACATCATGGATT | 8 | N | <0.001 |
| VEPGS-01569 | Peptide ABC transporter, ATP-binding protein | CATTAACCATATTATTCAT | 8.95 | N | <0.001 |
| VEPGS-01574 | Transcriptional regulator | TTGGTATCTTGATACTCGT | 8.7 | N | <0.001 |
| VEPGS-01579 | Transcriptional regulator | TCTTTGGCTTTATTTTCAT | 8.75 | N | <0.001 |
| VEPGS-01835 | OpaR (LuxR) | TAGTGTATCACCACGTTCA | 4.11 | N | <0.001 |
| VEPGS-02615 | RNA polymerase sigma factor | TATTCGCACTCTTGTCTTT | 7.75 | N | <0.001 |
| VEPGS-02635 | Glutamine synthetase adenylyltransferase | CCTTCACGATCTTTCACAT | 8.2 | N | <0.001 |
| VEPGS-02651 | Cytochrome b subunit of the bc complex | TACTCTACATCACGCATAA | 7.95 | N | <0.001 |
| VEPGS-02767 | Hypothetical protein | TTATTAAACTATATTCATA | 15.5 | N | <0.001 |
In the enriched list (Table 3), the luxR promoter region was identified to be a binding substrate of AphA enriched 4.1-fold (P < 0.001), validating the approach and as reported for other vibrios (14). Furthermore, the genes corresponding to 4 loci (aphA, exoribonuclease R, DNA-directed RNA polymerase beta′ subunit, and 5′-3′ exonuclease) in the enriched list have also been shown to be regulated by AphA in V. harveyi (4, 14), further validating our ChIP-seq methodology. The other 44 genes were, for the first time, identified to be directly regulated by AphA (Table 3). Several transcriptional regulator genes (n = 8), and genes including those for sigma D, anti-sigma D, type I/IIA topoisomerases, RNA polymerase (RNAP) beta subunit, a putative translation factor related to gene transcription/translation processes, and FabR (involved in the unsaturated fatty acids biosynthesis) were enriched in the ChIP-seq peaks (Table 3). In the list were also the genes related to adenylate cyclase (related to cAMP generation) and FoF1-type ATP synthase, which is involved in virulence regulation networks in vibrios (9, 33). In addition, AphA seemed to directly bind to a small RNA (sRNA) gene that belongs to the CsrB/RsmB family. Finally, many metabolism-associated genes seemed to also be regulated by AphA. Collectively, these newly identified target genes strongly suggest that AphA is an MQSR in multiple virulence and metabolic pathways at LCDs.
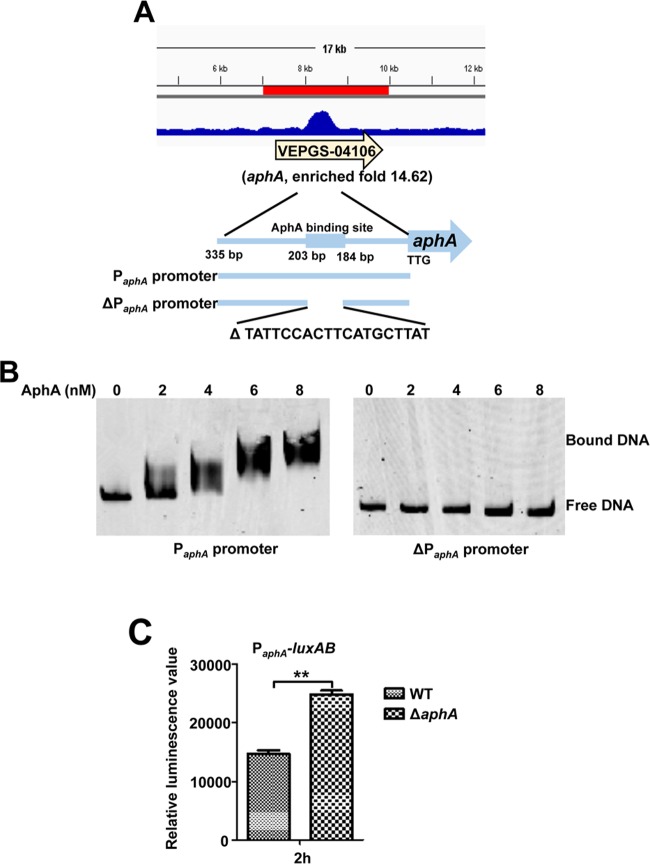
AphA represses its own expression.
In V. cholerae, V. harveyi, and V. parahaemolyticus, AphA has been established to repress its own expression (4, 34, 35). Among the 49 enriched loci that were identified in the ChIP-seq experiment (Table 3), aphA (VEPGS-04106) represented one of the highest peaks (14.62-fold) in the V. alginolyticus genome bound by AphA (Fig. 6A). As expected, the AphA protein bound directly to the aphA promoter region in a concentration-dependent manner in the EMSA in the presence of high concentrations (10-fold) of nonspecific poly(dI·dC) competitor (Fig. 6B). BLAST analysis showed that the aphA promoter contains the specific binding site of the AphA protein (Table 3) and the absence of this binding site abolished the capacity of AphA to bind to its own promoter (Fig. 6A and B). Given this direct interaction, we next attempted to determine whether aphA expression is regulated by AphA in vivo. To this end, we compared the transcriptional levels of aphA in the WT and ΔaphA strains with the transcriptional fusion construct PaphA-luxAB (Table 1) at 2 h of culture. The luminescence in the ΔaphA mutant was significantly higher than that in the WT (Fig. 6C). These results showed that AphA binds directly to its own promoter to negatively regulate the expression of AphA at LCDs in V. alginolyticus, which represents further evidence that AphA regulates itself.
FIG 6.
AphA binds directly to its own promoter for autorepression. (A) AphA binds to its own promoter region as illustrated with the peak (enriched 14.6-fold) identified by the ChIP-seq experiment. The AphA binding site and the probes (335-bp PaphA promoter and 316-bp ΔPaphA promoter with deletion of AphA binding site ranging from bp 184 to 203 relative to start site TTG) used for EMSAs in panel B are illustrated. (B) EMSA of the aphA promoter region (left) or the aphA promoter region with deletion of the specific AphA-binding box (ΔPaphA) (right) with purified AphA. The amounts of AphA protein used are indicated, and 20 ng of each Cy5-labeled probe was added to the EMSA mixtures. The specificity of the shifts was verified by adding a 10-fold excess of nonspecific competitor DNA [poly(dI·dC)] to the EMSA mixtures. (C) PaphA-luxAB transcriptional analysis in WT and ΔaphA strains. The WT and ΔaphA strain carrying the PaphA-luxAB reporter plasmid were cultured in LBS for 2 h and assayed for luminescence. The results are shown as means ± SD (n = 3). **, P < 0.01 (Student's t test) compared to the value for the WT strain harboring PaphA-luxAB.
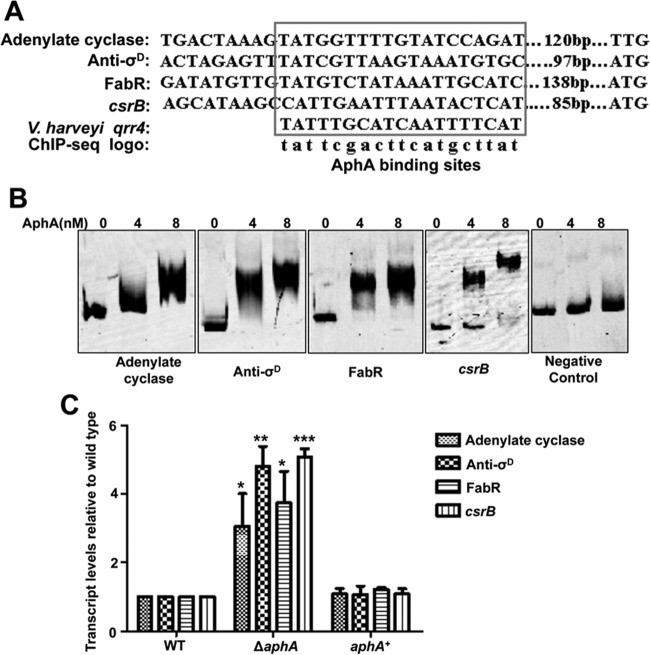
Multiple roles of AphA in adenylate cyclase, anti-σD, FabR, and CsrB expression.
We further analyzed the ChIP-seq data by choosing genes of interest in the list (VEPGS-04385 for adenylate cyclase, VEPGS-04316 for anti-σD, VEPGS-04332 for FabR, and VEPGS-04284 for small RNA CsrB) (Table 3). In V. harveyi, the AphA protein can bind directly to the promoter region of Qrr4 (4). An alignment of the AphA protein binding site in these four promoters and Qrr4 of V. harveyi is shown in Fig. 7A, and these binding sites are similar to the AphA logo identified by ChIP-seq. Next, we used EMSA to test the binding of AphA to these four promoters in vitro. AphA bound efficiently to all four probes in a concentration-dependent manner in the presence of high concentrations (10-fold excess of the probe DNA) of a nonspecific competitor, whereas the negative control (gyrB) remained unbound even at the highest protein concentrations (Fig. 7B). Then, qRT-PCR was performed in the WT, ΔaphA, and aphA+ complemented strains. The levels of expression of adenylate cyclase, anti-σD, FabR, and the putative small RNA CsrB (see below) were significantly increased, 3- to 5-fold, in the ΔaphA strain compared to the WT (Fig. 7C); furthermore, the complemented strain was restored to WT levels. Taken together, these results confirmed the ChIP-seq data and suggested multiple novel roles of AphA in various cellular processes of V. alginolyticus.
FIG 7.
Validation of ChIP-seq results in vitro and in vivo. (A) Alignment of the selected promoter region of genes containing an AphA-binding box (revealed by ChIP-seq analysis) and as established previously (4). The conserved consensus motif is boxed, and their positions relative to the start codon are shown. (B) AphA specifically binds to the selected target regions, as revealed by EMSA. The promoter regions of adenylate cyclase, anti-σD, FabR, and CsrB were chosen and the PCR products containing the indicated fragment were added to the reaction mixtures with different concentrations of AphA protein and a 10-fold excess of nonspecific competitor [poly(dI·dC)]. The negative control (gyrB gene fragment) showed no binding to AphA. (C) qRT-PCR analysis of the transcripts of selected genes. The WT, ΔaphA, and, aphA+ strains were cultured in LBS for 2 h, and mRNA transcripts were detected by qRT-PCR. The 16S rRNA gene was selected as a control. The results are shown as means ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test) compared to the corresponding WT results.
AphA regulates motility through CsrB and LuxR.
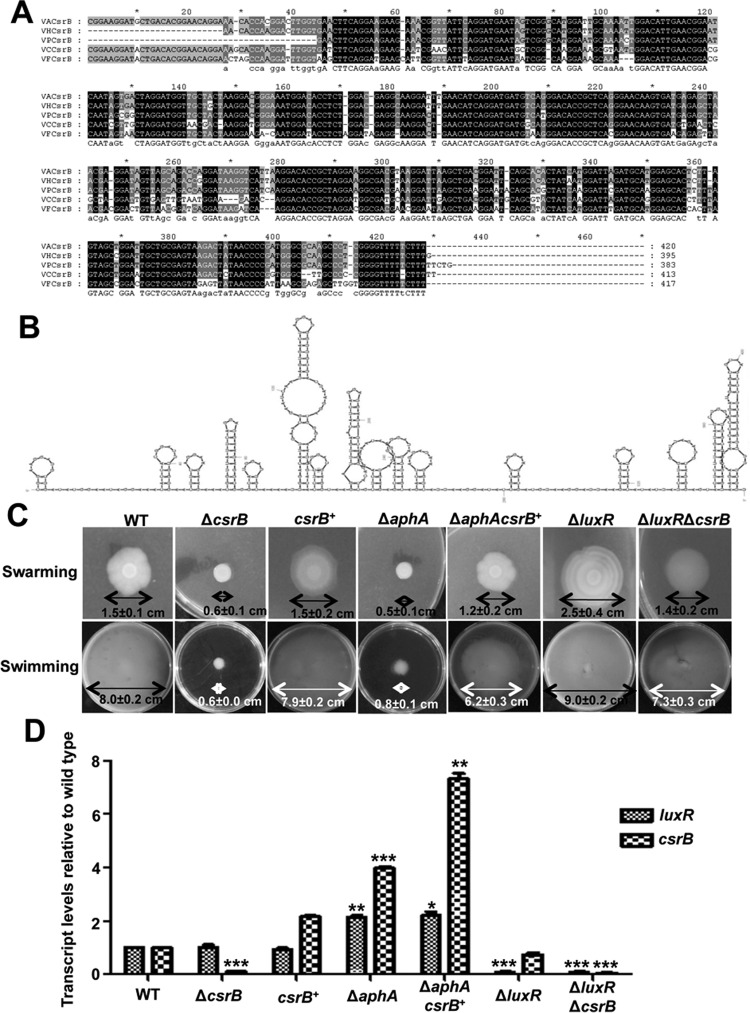
We were intrigued that the sRNA CsrB was bound directly and negatively regulated by AphA in the bacteria (Fig. 7). Bioinformatics analyses indicated that the CsrB gene of V. alginolyticus shared 98, 95, 87, and 84% identities to the homologous genes present in V. harveyi, V. parahaemolyticus, V. cholerae, and V. fischeri, respectively (Fig. 8A). CsrB had been identified by Northern blotting in V. cholerae and V. fischeri (36, 37) with functions similar to the CsrB families described for E. coli (acting redundantly to titrate the activity of the global regulator protein CsrA) (38, 39). The predicted secondary structure of CsrB (40) is shown in Fig. 8B; specifically, CsrB has 13 AGGA motifs and 10 ARGGA motifs (R stands for G, C, and T) in loop regions of stem-loops or in single-stranded regions, which are the hallmark sites of the interaction with CsrA in E. coli (41, 42). In V. cholerae and V. fischeri, CsrB has 18 and 21 AGGA/ARGGA motifs, respectively (36, 37).
FIG 8.
An AphA-controlled sRNA csrB gene is involved in the mobility of V. alginolyticus. (A) Alignment of DNA sequences for the csrB sRNAs in V. alginolyticus (VA), V. cholerae (VC), V. fischeri (VF), V. parahaemolyticus (VP), and V. harveyi (VH). (B) Secondary structure of CsrB in V. alginolyticus. The predicted secondary structure for CsrB was obtained using MFOLD (40) in V. alginolyticus showing multiple AGGA and ARGGA motifs in the loop regions or in the single-stranded regions. (C) Motility assays of the WT, various mutant strains, and strains overexpressing csrB (csrB+). Swarming motility assays were performed on LBS plates with 1.5% agar, whereas swimming motility assays were performed with 0.3% agar. (D) qRT-PCR analysis of luxR and csrB transcripts. The strains were cultured in LBS for 10 h, and the mRNA transcripts were detected by qRT-PCR. The 16S rRNA gene was selected as a control. The results are shown as means ± SD (n = 3). *, P < 0.05; **, P < 0.0; ***, P < 0.001 (Student's t test) compared to the corresponding WT results.
Further analysis showed that CsrB positively regulates motility in V. alginolyticus, as deletion of csrB in the bacterium significantly downregulated both swarming and swimming abilities and complemented csrB restored motility in the ΔcsrB strain (Fig. 8C). Overexpression of CsrB in the ΔaphA mutant strain (ΔaphA csrB+) dramatically enhanced the motility of the ΔaphA strain (Fig. 8C). Because AphA controls motility capacities (Fig. 2A and 8C), and AphA can negatively regulate the expression of CsrB, we inferred that AphA might regulate motility through CsrB and another regulator(s). LuxR negatively regulates motility in V. alginolyticus (Fig. 8C) (17), and AphA directly represses the expression of LuxR (23). Therefore, we further explored how LuxR and CsrB coordinate the motility capacities with a ΔluxR ΔcrsB double-mutant strain. The ΔluxR ΔcrsB strain partially restored the motility capacities of the ΔcrsB strain to the WT level, but the activity was weaker than that of the ΔluxR strain. These data suggested that both LuxR and CsrB modulate motility, with stronger inhibition by LuxR and relatively weaker activation by CsrB. We further investigated the transcription of luxR and csrB in different strains at HCDs (10-h LBS culture) using qRT-PCR. The results indicated that AphA negatively regulated both LuxR and CsrB, but LuxR and CsrB appeared to not regulate each other (Fig. 8D). Taken together, these results demonstrated that AphA can regulate motility through CsrB and LuxR.
DISCUSSION
QS systems are intricately regulated as the global regulatory networks of gene expression in bacteria. In V. alginolyticus, a V. harveyi-like QS signaling system has been characterized, which involved pivotal LuxO and LuxR regulators (16–22, 29). AphA has been reported as another master regulator of QS in V. harveyi, V. cholerae, and V. parahaemolyticus (3, 4, 34). AphA is present at LCDs and regulates the expression of ∼167 genes in V. harveyi (14). The molecular mechanisms of QS systems on the virulence and their dependence on AphA remain undefined for V. alginolyticus. We have recently found that LuxR and AphA can bind directly to the luxR promoter and repressed the expression of LuxR in V. alginolyticus (23). In this study, we characterized the aphA gene of V. alginolyticus and identified the direct targets of AphA by ChIP-seq. An AphA-dependent binding motif was disclosed that revealed novel regulation targets of AphA in the bacterium.
In V. alginolyticus, motility and biofilm formation have been proposed as important virulence-associated factors that could be regulated by QS (22, 29, 43). Here we report that AphA, another master regulator of QS, could also modulate motility, biofilm formation, and in vivo survival of the bacterium in zebra fish (Fig. 2). In V. cholerae, AphA enhances biofilm formation by activating expression of biofilm regulator VpsT, and cyclic di-GMP (c-di-GMP) and QS are also involved in the regulation of biofilm by controlling the expression of VpsT and AphA (31, 32). AphA might be also involved in biofilm formation through the similar regulatory network in V. alginolyticus. Furthermore, AphA could regulate the extracellular protein Asp through LuxR (Fig. 3 and 4). AphA as a global regulator protein has been well studied for its role in controlling virulence gene expression in V. cholerae, V. harveyi, and V. parahaemolyticus (9, 14, 30). AphA binds directly to the Qrr4 promoter to repress its transcription; the same mechanism of repression is expected for Qrr2 and Qrr3 in V. harveyi (14). At LCDs, the Qrr sRNAs are constitutively produced and activate aphA expression. This activation maybe due to Qrr sRNAs bound to the target mRNA, which induces a conformation in the aphA 5′ untranslated region (UTR) conducive to ribosome binding in V. harveyi (4). Then AphA directly binds to its own promoter and autorepresses its transcription in both V. cholerae and V. harveyi (4). LuxR can also directly bind to the aphA promoter in V. cholerae, V. harveyi, and V. parahaemolyticus (4, 13, 44). AphA can cooperate with AphB to regulate the expression of V. cholerae virulence factors, toxin-coregulated pilus (TCP) and cholera toxin (CT) (10). The research on AphA-coordinated AphB binding to specific gene promoters in V. alginolyticus is under way, which will further illuminate the complex regulatory networks and roles of AphA in vibrios.
Given that AphA is a transcription factor, it will be intriguing to examine how it controls the expression of additional genes in V. alginolyticus. We performed the ChIP-seq analysis to identify all potential in vivo binding sites of AphA in V. alginolyticus. Our analysis demonstrated that AphA is produced at LCDs (Fig. 1B); therefore, ChIP-seq samples were collected after a 2-h incubation period, and 49 AphA-binding peaks were detected. Previous microarray analyses established that ∼300 genes are regulated by AphA at LCDs in V. harveyi, a subset of which was also controlled by LuxR (4). Other research also showed that AphA regulates 167 genes, LuxR regulates 625 genes, and they coregulate 77 genes in V. harveyi (14). BLAST indicated that among the 49 detected peaks, 4 loci overlapped in the ChIP-seq data of V. alginolyticus and microarray analyses of V. harveyi, including VEPGS-04037 (exoribonuclease R), VEPGS-04106 (AphA), VEPGS-04317 (DNA-directed RNA polymerase beta′ subunit), and VEPGS-01513 (exonuclease). Because AphA can directly bind to its own promoter, which has been demonstrated for other vibrios (4, 34, 35), the overlapped list of genes validated, to some extent, our ChIP-seq analysis pipeline. Additionally, it suggested the versatile roles of AphA in different bacterial contexts. Work with different bacteria will help elucidate its functions on QS and non-QS regulations.
In this study, we defined a consensus AphA-binding sequence (TATTCGN7GCTTAT), which is consistent with the motif identified by DNase I footprinting assays of the luxR promoter in V. parahaemolyticus, V. cholerae, and V. alginolyticus (4, 23, 34). In another study, the authors collected nine known or predicted direct targets of AphA from different vibrios and generated an AphA 20-bp consensus, ATATGCAN6TGCATAT, that contained imperfect inverted repeats of ATATGCA with a 6-nucleotide (nt) centered spacer (34). Our ChIP-seq-generated consensus sequence was similar but not completely identical to this consensus, which may also reflect that the binding sites of AphA vary in different bacteria or the preferential binding activities differ in the bacteria. The research with V. cholerae showed that the site of binding of AphA to the vpsT promoter was a 7-nt centered spacer, while the sites of binding of AphA to promoters of tcpPH and alsRD were 6-nt centered spacers (32). The above-mentioned discrepancies in the length of the consensus and spacer sequence may be due to the difference in modulators (e.g., AphB) in terms of modulator species and cooperative strength with AphA that facilitates AphA binding to specific promoter regions. In V. harveyi and V. parahaemolyticus, AphA can directly bind to the promoters for Qrr2 to -4 (4, 34), but these targets were missing in our ChIP-seq data. However, the biochemical and bioinformatic analyses also showed that AphA could directly regulate the expression of putative Qrr4 in V. alginolyticus. Generally, ChIP-seq analysis may lose some specific targets because of the detection limits of ChIP with specific antibodies.
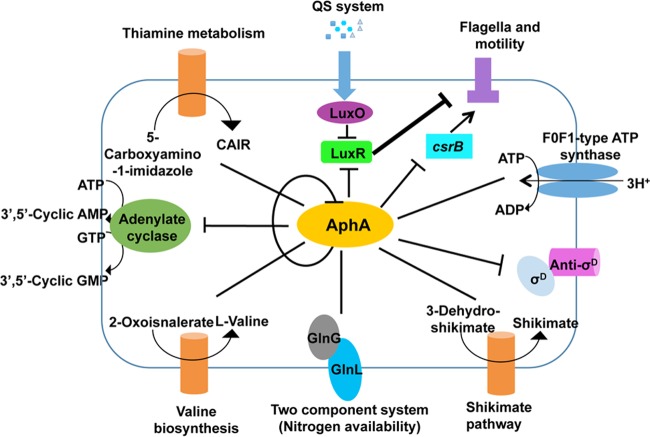
In accordance with the results of ChIP-seq, we used the Kobas 2.0 (28) to analyze the possible pathways of genes containing an AphA-binding motif (Fig. 9). Among the major AphA targets are several transcription factors, such as aphA, which inhibits its own expression (Fig. 9). VEPGS-04385 is predicted as an adenylate cyclase, a protein that transforms ATP to cAMP and GMP to cGMP (Fig. 9). In V. cholerae, the cAMP-CRP complex binds to sites in the tcpPH promoter that overlap AphA and AphB binding sites, thus competing with the capacity of these two activators to bind (10). In V. vulnificus, the cAMP-CRP complex could directly bind to the promoter regions of rpoS and result in the repression of rpoS, hence modulating numerous cellular processes (33). ChIP-seq data (Table 3) also showed that AphA could regulate thiamine metabolism, valine biosynthesis, and the shikimate pathway (Fig. 9), indicating the profound roles of AphA in a variety of metabolic pathways. The results also indicated that AphA could regulate the GlnL/GlnG two-complement system involved in the nitrogen metabolism regulation (Fig. 9). In addition, we also found that AphA could regulate anti-σD, taking part in the regulation of σD and FoF1-type ATP synthase expression (Table 3).
FIG 9.
Schematic summarization of the regulatory roles of AphA in V. alginolyticus. The KEGG pathway analysis was performed with the Kobas 2.0 (28). The various pathways and their respective cellular locations, as well as the regulatory roles of AphA, are illustrated with arrows (activation) or bar-ended lines (repression) and are discussed in the text. The inferred stronger inhibition of motilities by LuxR outcompetes CsrB-mediated activation of motilities and is depicted with a thicker bar-ended line. See the related text for detailed explanation.
A small RNA belonging to the CsrB/RsmB family in vibrios (36, 37) was also determined to be regulated by AphA (Fig. 9). AphA binding to the corresponding binding sites was confirmed in vitro and in vivo by EMSA and qRT-PCR (Fig. 7). Moreover, our data also showed that AphA could modulate motility via LuxR and CsrB, probably in a mutually independent manner (Fig. 8). In V. cholerae, the VarS/VarA system could regulate the expression of CsrB and then control the QS through CsrA, a global regulatory protein (36). The VarS/VarA-CsrA/B/C/D system is involved in the control of HapA expression and biofilm production through HapR in V. cholerae (45, 46). In this study, we found that CsrB could be involved in the regulation of the motility in V. alginolyticus, and it predicted the targets of CsrB by sRNA Target (47). A total of 23 flagellar genes were predicted as targets of CsrB, including the structural genes and regulatory genes of both lateral and polar flagella. CsrB could directly influence these genes' expression to modulate motility. We assumed that AphA controls motility through both LuxR- and CsrB-mediated independent pathways, with the following observations: (i) LuxR and CsrB appear to be mutually independent, as deletion in luxR did not affect csrB expression and vice versa (Fig. 8D); (ii) both LuxR and CsrB control motility through repression and activation on the motility-associated pathway, respectively (Fig. 8C) (17); (iii) AphA tightly regulates motility (Fig. 8C) and controls the expression of LuxR and CsrB at LCDs (Fig. 7B and C) (23); and (iv) the ΔluxR ΔcsrB double mutant shows faster motility than that of the ΔcsrB mutant, which suggests that the stronger derepression of motilities by LuxR deletion outcompetes the repression rendered by CsrB abrogation and thus promotes motility in the bacterium (Fig. 8C). Taken together, these data suggested that LuxR acts as a stronger regulator than CsrB does in terms of motility capacities, and both are controlled by AphA in V. alginolyticus (Fig. 9). The molecular mechanisms for how CsrB expression is controlled by AphA and mediates motility, possibly independent of LuxR, need to be further elucidated in light of spatial-temporal regulation as well as involvement of other factors in V. alginolyticus.
In summary, the results presented in this work provide an improved understanding of the AphA-LuxR-centered complex regulatory network of vibrios QS. AphA can regulate the extracellular protein Asp through LuxR. Additionally, AphA can bind directly to the luxR and aphA promoter regions to then repress the expression of LuxR and AphA. AphA also plays essential roles in biofilm formation, motility, and survival in zebra fish. AphA motility modulation might be dependent on the action of CsrB and LuxR. Our ChIP-seq results also provided useful information that would allow us to further characterize other non-QS targets and processes regulated by AphA in the future.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (no. 31372560 and 31301059) and the Ministry of Agriculture of China (no. CARS-50 and nyhyzx-201303047).
REFERENCES
- 1.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 2.Ng W-L, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL. 2013. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. mBio 4:e00378–13. doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. 2011. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev 25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng L, Rutherford ST, Papenfort K, Bagert JD, van Kessel JC, Tirrell DA, Wingreen NS, Bassler BL. 2015. A Qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell 160:228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skorupski K, Taylor RK. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol 31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 7.De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K. 2005. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J Biol Chem 280:13779–13783. doi: 10.1074/jbc.M413781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovacikova G, Skorupski K. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol 181:4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacikova G, Skorupski K. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol 41:393–407. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 10.Kovacikova G, Lin W, Skorupski K. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol Microbiol 53:129–142. doi: 10.1111/j.1365-2958.2004.04121.x. [DOI] [PubMed] [Google Scholar]
- 11.Kovacikova G, Lin W, Skorupski K. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol Microbiol 57:420–433. doi: 10.1111/j.1365-2958.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhou DS, Yan XJ, Qu F, Wang L, Zhang YQ, Hou J, Hu Y, Li J, Xin S, Qiu J, Yang R, Mao P. 2013. Quorum sensing modulates transcription of cpsQ-mfpABC and mfpABC in Vibrio parahaemolyticus. Int J Food Microbiol 166:458–463. doi: 10.1016/j.ijfoodmicro.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Kovacikova G, Skorupski K. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol 44:1135–1147. [DOI] [PubMed] [Google Scholar]
- 14.van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. 2013. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol 195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Austin B. 2010. Vibrios as causal agents of zoonoses. Vet Microbiol 140:310–317. doi: 10.1016/j.vetmic.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Wang QY, Liu Q, Ma Y, Rui HP, Zhang YX. 2007. LuxO controls extracellular protease, haemolytic activities and siderophore production in fish pathogen Vibrio alginolyticus. J Appl Microbiol 103:1525–1534. doi: 10.1111/j.1365-2672.2007.03380.x. [DOI] [PubMed] [Google Scholar]
- 17.Rui HP, Liu Q, Ma Y, Wang QY, Zhang YX. 2008. Roles of LuxR in regulating extracellular alkaline serine protease A, extracellular polysaccharide and mobility of Vibrio alginolyticus. FEMS Microbiol Lett 285:155–162. doi: 10.1111/j.1574-6968.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- 18.Rui HP, Liu Q, Wang QY, Ma Y, Liu H, Shi CB, Zhang YX. 2009. Role of alkaline serine protease, Asp, in Vibrio alginolyticus virulence and regulation of its expression by LuxO-LuxR regulatory system. J Microbiol Biotechnol 19:431–438. doi: 10.4014/jmb.0807.404. [DOI] [PubMed] [Google Scholar]
- 19.Cao XD, Wang QY, Liu Q, Liu H, He HH, Zhang YX. 2010. Vibrio alginolyticus MviN is a luxO-regulated protein and affects cytotoxicity towards epithelioma papulosum cyprini (EPC) cells. J Microbiol Biotechnol 20:271–280. [PubMed] [Google Scholar]
- 20.Cao XD, Wang QY, Liu Q, Rui HP, Liu H, Zhang YX. 2011. Identification of a luxO-regulated extracellular protein Pep and its roles in motility in Vibrio alginolyticus. Microb Pathog 50:123–131. doi: 10.1016/j.micpath.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, Zhang LP, Ren CH, Zhao JJ, Chen C, Jiang X, Luo P, Hu CQ. 2011. Autophagy is induced by the type III secretion system of Vibrio alginolyticus in several mammalian cell lines. Arch Microbiol 193:53–61. doi: 10.1007/s00203-010-0646-9. [DOI] [PubMed] [Google Scholar]
- 22.Sheng LL, Lv YZ, Liu Q, Wang QY, Zhang YX. 2013. Connecting type VI secretion, quorum sensing, and c-di-GMP production in fish pathogen Vibrio alginolyticus through phosphatase PppA. Vet Microbiol 162:652–662. doi: 10.1016/j.vetmic.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Gu D, Guo M, Yang MJ, Zhang YX, Zhou XH, Wang QY. 2016. A σE-mediated temperature gauge controls a switch from LuxR-mediated virulence gene expression to thermal stress adaptation in Vibrio alginolyticus. PLoS Pathog 12:e1005645. doi: 10.1371/journal.ppat.1005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 26.Feng JX, Liu T, Qin B, Zhang Y, Liu XL. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philip M, Timothy LB. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 2712:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie C, Mao XZ, Huang JJ, Ding Y, Wu JM, Dong S, Kong L, Gao G, Li CY, Wei LP. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Y, Wang QY, Liu Q, Ma Y, Cao XD, Guan LY, Zhang YX. 2008. Involvement of LuxS in the regulation of motility and flagella biogenesis in Vibrio alginolyticus. Biosci Biotechnol Biochem 72:1063–1071. doi: 10.1271/bbb.70812. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Ling Y, Jiang HJ, Qiu YF, Qiu JF, Chen HP, Yang RF, Zhou DS. 2013. AphA is required for biofilm formation, motility, and virulence in pandemic Vibrio parahaemolyticus. Int J Food Microbiol 160:245–251. doi: 10.1016/j.ijfoodmicro.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol 193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang MH, Frey EM, Liu Z, Bishar R, Zhu J. 2010. The virulence transcriptional activator AphA enhances biofilm formation by Vibrio cholerae by activating expression of the biofilm regulator VpsT. Infect Immun 78:697–703. doi: 10.1128/IAI.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HJ, Park SJ, Choi SH, Lee KH. 2008. Vibrio vulnificus rpoS expression is repressed by direct binding of cAMP-cAMP receptor protein complex to its two promoter regions. J Biol Chem 283:30438–30450. doi: 10.1074/jbc.M802219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun FJ, Zhang YQ, Wang L, Yan XJ, Tan YF, Guo ZB, Qiu JF, Yang RF, Xia PY, Zhou DS. 2012. Molecular characterization of direct target genes and cis-acting consensus recognized by quorum-sensing regulator AphA in Vibrio parahaemolyticus. PLoS One 7:e44210. doi: 10.1371/journal.pone.0044210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin W, Kovacikova G, Skorupski K. 2007. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol Microbiol 64:953–967. doi: 10.1111/j.1365-2958.2007.05693.x. [DOI] [PubMed] [Google Scholar]
- 36.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol 58:1186–1203. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 37.Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV. 2006. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res 34:3361–3369. doi: 10.1093/nar/gkl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pernestig AK, Melefors O, Georgellis D. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J Biol Chem 276:225–231. doi: 10.1074/jbc.M001550200. [DOI] [PubMed] [Google Scholar]
- 40.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu MY, Gui G, Wei B, Preston JF, Oakford L, Yuksel U. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem 272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 42.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol 48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 43.Liu H, Wang QY, Liu Q, Cao XD, Shi CB, Zhang YX. 2011. Roles of Hfq in the stress adaptation and virulence in fish pathogen Vibrio alginolyticus and its potential application as a target for live attenuated vaccine. Appl Microbiol Biotechnol 91:353–364. doi: 10.1007/s00253-011-3286-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang YQ, Qiu YF, Tan YF, Guo ZB, Yang RF, Zhou DS. 2012. Transcriptional regulation of opaR, qrr2-4 and aphA by the master quorum-sensing regulator OpaR in Vibrio parahaemolyticus. PLoS One 7:e34622. doi: 10.1371/journal.pone.0034622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang JY, Jung KT, Yoo CK, Rhie G. 2010. Regulation of hemagglutinin/protease expression by the VarS/VarA-CsrA/B/C/D system in Vibrio cholerae. Microb Pathog 48:245–250. doi: 10.1016/j.micpath.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Tsou AM, Liu Z, Cai T, Zhu J. 2011. The VarS/VarA two-component system modulates the activity of the Vibrio cholerae quorum-sensing transcriptional regulator HapR. Microbiology 157:1620–1628. doi: 10.1099/mic.0.046235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y, Zhao YL, Cha L, Ying XM, Wang LG, Shao NS, Li WJ. 2009. sRNATarget: a web server for prediction of bacterial sRNA targets. Bioinformation 3:364–366. doi: 10.6026/97320630003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang SY, Lauritz J, Jass J, Milton DL. 2002. A ToxR homolog from Vibrio anguillarum serotype O1 regulates its own production, bile resistance, and biofilm formation. J Bacteriol 184:1630–1639. doi: 10.1128/JB.184.6.1630-1639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]