ABSTRACT
Neisseria gonorrhoeae (gonococci) and Neisseria meningitidis (meningococci) are human pathogens that cause gonorrhea and meningococcal meningitis, respectively. Both N. gonorrhoeae and N. meningitidis release a number of small peptidoglycan (PG) fragments, including proinflammatory PG monomers, although N. meningitidis releases fewer PG monomers. The PG fragments released by N. gonorrhoeae and N. meningitidis are generated in the periplasm during cell wall remodeling, and a majority of these fragments are transported into the cytoplasm by an inner membrane permease, AmpG; however, a portion of the PG fragments are released into the extracellular environment through unknown mechanisms. We previously reported that the expression of meningococcal ampG in N. gonorrhoeae reduced PG monomer release by gonococci. This finding suggested that the efficiency of AmpG-mediated PG fragment recycling regulates the amount of PG fragments released into the extracellular milieu. We determined that three AmpG residues near the C-terminal end of the protein modulate AmpG's efficiency. We also investigated the association between PG fragment recycling and release in two species of human-associated nonpathogenic Neisseria: N. sicca and N. mucosa. Both N. sicca and N. mucosa release lower levels of PG fragments and are more efficient at recycling PG fragments than N. gonorrhoeae. Our results suggest that N. gonorrhoeae has evolved to increase the amounts of toxic PG fragments released by reducing its PG recycling efficiency.
IMPORTANCE Neisseria gonorrhoeae and Neisseria meningitidis are human pathogens that cause highly inflammatory diseases, although N. meningitidis is also frequently found as a normal member of the nasopharyngeal microbiota. Nonpathogenic Neisseria, such as N. sicca and N. mucosa, also colonize the nasopharynx without causing disease. Although all four species release peptidoglycan fragments, N. gonorrhoeae is the least efficient at recycling and releases the largest amount of proinflammatory peptidoglycan monomers, partly due to differences in the recycling permease AmpG. Studying the interplay between bacterial physiology (peptidoglycan metabolism) and pathogenesis (release of toxic monomers) leads to an increased understanding of how different bacterial species maintain asymptomatic colonization or cause disease and may contribute to efforts to mitigate disease.
INTRODUCTION
Ten species in the genus Neisseria are associated with humans. Neisseria gonorrhoeae (gonococci [GC]) and Neisseria meningitidis (meningococci [MC]) are considered human-restricted pathogens, whereas N. cinerea, N. elongata, N. flavescens, N. lactamica, N. mucosa, N. polysaccharea, N. sicca, and N. subflava are considered nonpathogenic. The nonpathogenic species colonize the nasopharynges and oral cavities of healthy people (1–3). In rare cases, these species disseminate to cause endocarditis or septic infection in immunocompromised individuals or trauma patients (4). N. gonorrhoeae and N. meningitidis share many infection-related factors with the nonpathogenic species, including type IV pili, adhesins, and certain iron transport proteins (5). Unlike N. gonorrhoeae and N. meningitidis, nonpathogenic Neisseria spp. are considered to be noninflammatory, and they very rarely elicit a symptomatic inflammatory response (6).
N. gonorrhoeae commonly infects the genitourinary tract, causing urethritis in men and cervicitis in women. In women, the bacteria can spread to the uterus and Fallopian tubes, leading to highly inflammatory conditions, endometritis, pelvic inflammatory disease, and ectopic pregnancy. Gonococci can also disseminate to cause sepsis, tenosynovitis, and meningitis (7). Disease manifestations are due to the host inflammatory response. In pelvic inflammatory disease, the release of peptidoglycan (PG) fragments and endotoxin by gonococci in the Fallopian tubes induces an inflammatory response that kills the ciliated cells, and the cells come out of the epithelium and are sloughed off (8, 9). The loss of ciliated cells and the tissue damage results in tubal factor infertility or predisposes the woman to ectopic pregnancy.
N. gonorrhoeae is unusual among Gram-negative bacteria in that it releases significant amounts of PG fragments during growth (10). The most abundant fragments released are the PG monomers. These are disaccharide-tripeptide and disaccharide-tetrapeptide fragments carrying a 1,6-anhydro bond on the N-acetylmuramic acid residue (11). The disaccharide-tetrapeptide is identical to tracheal cytotoxin (TCT), the PG fragment released by Bordetella pertussis that induces death and sloughing of ciliated cells in the trachea (12–14). The disaccharide-tripeptide stimulates activation of the human pattern-recognition receptor NOD1 (15). When added to Fallopian tube tissue in organ culture, a mixture of the two monomers caused death and sloughing of ciliated cells, mimicking the tissue damage of pelvic inflammatory disease (8).
Although commonly considered a pathogen, N. meningitidis is a normal colonizer of the human nasopharynx and is carried asymptomatically by 10 to 40% of the population (16). The bacteria can spread to cause sepsis or meningitis, and approximately 550 cases of invasive meningococcal disease occur in the United States every year (17). In these invasive infections, meningococci elicit a large inflammatory response that frequently results in septic shock and the death of the patient within a few days of the onset of symptoms. However, N. meningitidis may not be inflammatory during the carriage state, only upregulating the expression of virulence factors required for invasion and immune evasion under certain conditions (18).
We have investigated the mechanisms involved in the generation and release of proinflammatory PG fragments by N. gonorrhoeae and N. meningitidis. The PG monomers are generated by lytic transglycosylases, which in Neisseria species, are predicted outer membrane lipoproteins (19, 20). As the bacteria grow and divide, they must degrade PG strands to make space for the incorporation of additional PG strands and remodel the cell wall to build and then split the septum for cell division and separation. Most of the PG fragments generated by these processes are taken up from the periplasm and transported to the cytoplasm by the inner membrane permease AmpG (21–25). However, in N. gonorrhoeae, 15% of the PG monomers escape from the cell and are released into the milieu (22). In comparison, only 4% of the PG monomers generated by N. meningitidis are released from the bacteria (23). We previously demonstrated that replacement of gonococcal ampG with meningococcal ampG led to reduced PG fragment release, suggesting that meningococcal AmpG is more efficient at PG fragment import (23).
In the present study, we examine the differences between gonococcal AmpG and meningococcal AmpG and characterize PG fragment release in N. sicca and N. mucosa. Replacement of meningococcal ampG with gonococcal ampG resulted in increased PG fragment release. Also, the nonpathogenic species exhibited highly efficient PG recycling and failed to release certain PG fragments that the pathogens do release, which may indicate additional differences in PG fragment degradation, recycling, and release in nonpathogenic neisseriae. Overall, these data show that Neisseria species that are usually asymptomatic colonizers, i.e., N. meningitidis, N. sicca, and N. mucosa, are more efficient at PG recycling than N. gonorrhoeae. Thus, N. gonorrhoeae has evolved an inefficient PG recycling system as it has moved to a proinflammatory infection lifestyle.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains used in this study are listed in Table 1. Neisseria strains (N. gonorrhoeae, N. meningitidis, N. sicca, and N. mucosa) were grown on either gonococcal base medium (GCB) agar plates (Difco) at 37°C with 5% CO2 or in gonococcal base liquid medium (GCBL) containing Kellogg's supplements (26) and 0.042% NaHCO3 (complete GCBL [cGCBL]) at 37°C with aeration. Escherichia coli cells were grown either on Luria-Bertani (LB) agar plates (Difco) at 37°C or in LB broth at 37°C with aeration. When necessary, media were supplemented with antibiotics for selection. Chloramphenicol was used at concentrations of 10 μg/ml (Neisseria spp.) or 25 μg/ml (E. coli), while erythromycin was used at concentrations of 10 μg/ml (Neisseria spp.) or 500 μg/ml (E. coli). Kanamycin was used at concentrations of 80 μg/ml (Neisseria spp.) or 40 μg/ml (E. coli).
TABLE 1.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| N. gonorrhoeae | ||
| MS11 | WT | 62 |
| EC505 | ΔampGGC ampGMC WT+ | 23 |
| EC508 | MS11 transformed with pEC016; ΔampGGC ampGGC Chimera 1+ | This study |
| EC509 | MS11 transformed with pEC017; ΔampGGC ampGGC Chimera 2+ | This study |
| EC510 | MS11 transformed with pEC018; ΔampGGC ampGGC Chimera 3+ | This study |
| EC511 | MS11 transformed with pEC019; ΔampGGC ampGGC Chimera 4+ | This study |
| EC512 | MS11 transformed with pEC028; ΔampGGC WT-FLAG3 | This study |
| EC515 | MS11 transformed with pEC037; ampGGCM391L | This study |
| EC516 | MS11 transformed with pEC038; ampGGCR398Q | This study |
| EC517 | MS11 transformed with pEC039; ampGGCI402A | This study |
| EC518 | MS11 transformed with pEC042; ampGGCM391L R398Q | This study |
| EC519 | MS11 transformed with pEC043; ampGGCM391L I402A | This study |
| EC521 | MS11 transformed with pEC054; ampGGCR398Q I402A | This study |
| EC523 | MS11 transformed with pEC058; ampGGCM391L R398Q I402A | This study |
| EC546 | MS11 transformed with pEC100; ampGGCM391L-FLAG3 | This study |
| EC548 | MS11 transformed with pEC102; ampGGCI402 M-FLAG3 | This study |
| EC549 | MS11 transformed with pEC103; ampGGCM391L R398Q I402 M-FLAG3 | This study |
| EC550 | MS11 transformed with pEC098; ΔampGGC ampGMC WT-FLAG+ | This study |
| N. meningitidis | ||
| ATCC 13102 Δcap | ATCC 13102 rpsL (K43R) siaD::cat (WT) | 23 |
| NM00268 | Serogroup B clinical isolate | 63 |
| EC1001 | ATCC 13102 Δcap transformed with pEC008; ΔampGMC ampGGC WT+ | This study |
| EC1008 | ATCC 13102 Δcap transformed with pEC029; ampGMC WT-FLAG3 | This study |
| N. sicca | ||
| ATCC 29256 | Pharyngeal mucosa isolate (WT) | N. Weyand |
| EC2004 | ATCC 29256 transformed with pEC081; ampGN. sicca::kan | This study |
| EC2004BC | ATCC 29256 transformed with EC2004 chromosomal DNA; ampGN. sicca::kan backcross | This study |
| N. mucosa | ||
| ATCC 25996 | Pharyngeal mucosa isolate (WT) | N. Weyand |
| EC2003 | ATCC 25996 transformed with pEC070; ampGN. mucosa::kan | This study |
| EC2003BC | ATCC 25996 transformed with EC2003 chromosomal DNA; ampGN. mucosa::kan | This study |
Strain construction.
Mutant or complemented strains of N. gonorrhoeae, N. meningitidis, N. sicca, and N. mucosa were generated using spot transformation (27). Briefly, 1 to 20 μg of linearized plasmid DNA or chromosomal DNA were spotted onto GCB plates. Then, three to ten piliated colonies were streaked over the spots, followed by incubation overnight at 37°C with 5% CO2. Transformants were screened by colony PCR and restriction enzyme digestion where applicable and then confirmed by sequencing (28).
Plasmid construction.
All plasmids used in this study are listed in Table 2, and all primers used to generate the constructs are listed in Table 3. Specific details of plasmid construction are described in the supplemental material. pIDN3 is a cloning plasmid that contains the gonococcal and meningococcal DNA uptake sequence (GCCGTCTGAA) and was used as a vector backbone to generate most of the plasmids used in this study (29, 30). However, transformation into N. sicca and N. mucosa may have greater efficiency with an alternate DNA uptake sequence (GTCGTCTGAA), which is more commonly found in N. sicca ATCC 29256 and N. mucosa ATCC 25996 (5, 31). Thus, we constructed pEC026, a derivative of pIDN3 that contains the alternate DNA uptake sequence to be used as a vector backbone for transformations into N. sicca and N. mucosa. To facilitate screening of transformants, we introduced a silent mutation at base 993 (L331, CTG→CTA) of gonococcal and meningococcal ampG to generate an NheI site. For clarity and simplicity, constructs that have the WT ampG coding sequence are referred to as ampGGC WT or ampGMC WT, while constructs with the screening site are referred to as ampGGC or ampGMC.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pIDN3 | Cloning plasmid containing GC/MC DNA uptake sequences | 30 |
| pHSS6 | Cloning plasmid, source of kanR | 64 |
| pEC026 | pIDN3 containing N. sicca/N. mucosa DNA uptake sequences | This study |
| pEC005 | ampGMCp-ampGGC WT cloned into pIDN3 | This study |
| pEC006 | ampGGCp-ampGMC WT cloned into pIDN3 | 23 |
| pEC007 | ampGGCp-ampGGC cloned into pIDN3 | This study |
| pEC008 | ampGMCp-ampGGC cloned into pIDN3 | This study |
| pEC013 | ampGGCp ampGMC cloned into pIDN3 | This study |
| pEC016 | ampGGCp ampGChimera 1 cloned into pIDN3 | This study |
| pEC017 | ampGGCp ampGChimera 2 cloned into pIDN3 | This study |
| pEC018 | mpGGCp ampGChimera 3 cloned into pIDN3 | This study |
| pEC019 | ampGGCp ampGChimera 4 cloned into pIDN3 | This study |
| pEC028 | ampGGC-FLAG3 cloned into pIDN3 | This study |
| pEC029 | ampGMC-FLAG3 cloned into pIDN3 | This study |
| pEC037 | ampGGCp-ampGGCM391L cloned into pIDN3 | This study |
| pEC038 | ampGGCp-ampGGCR398Q cloned into pIDN3 | This study |
| pEC039 | ampGGCp-ampGGCI402A cloned into pIDN3 | This study |
| pEC042 | ampGGCp-ampGGCM391L R398Q cloned into pIDN3 | This study |
| pEC043 | ampGGCp-ampGGCM391L I402A cloned into pIDN3 | This study |
| pEC054 | ampGGCp-ampGGCR398Q I402A cloned into pIDN3 | This study |
| pEC058 | ampGGCp-ampGGCM391L cloned into pIDN3 | This study |
| pEC063 | ampGN. sicca cloned into pEC026 | This study |
| pEC064 | ampGN. mucosa cloned into pIDN3 | This study |
| pEC067 | ampGN. mucosa::kan cloned into pIDN3 | This study |
| pEC070 | ampGN. mucosa::kan cloned into pEC026 | This study |
| pEC081 | ampGN. sicca::kan cloned into pEC026 | This study |
| pEC098 | ampGMC WT-FLAG3 cloned into pIDN3 | This study |
| pEC100 | ampGGCM391L-FLAG3 cloned into pIDN3 | This study |
| pEC102 | ampGGCI402A-FLAG3 cloned into pIDN3 | This study |
| pEC103 | ampGGCM391L R398Q I402A-FLAG3 cloned into pIDN3 | This study |
TABLE 3.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| MC ampG sacI F3 | ATTCAGAGCTCCATCGGCGGCATCATCAAAC |
| SacI ampG F | CGGGAGCTCGCGATATTTGCTACAATAGGC |
| XbaI ampG R | GCCCTCTAGACACAATATCAGGTAAACGCTCC |
| ampG 5′ flank R | AGCCTATTGTAGCAAATATCGCC |
| ampG F2 | GGCGATATTTGCTACAATAGGCT |
| ampG 3′ flank F | TCAAACTGGAGCGTTTACCTGATATTG |
| ampG1325R | CAATATCAGGTAAACGCTCCAGTTTGA |
| ampG 3′ flank R BamHI | CTCAGGATCCGTTCTTTATATGAGCGGCAGG |
| ampG 990-bp NheI F | GTAAGCTAGCGGCAGTTATCGGCGCGGAAG |
| ampG 990-bp NheI R | CTAAGCTAGCATCAGCCTCTCGCCTGTG |
| ampG internal 1 F | CAGCGAGCAGGTGGATTTGAAG |
| ampG internal 1 R | CTTCAAATCCACCTGCTCGCTG |
| ampG internal 2 F | GGATATGGGTTTCAGCAAGAC |
| ampG internal 2 R | GTCTTGCTGAAACCCATATCC |
| Alt-DUS AvrII F | GGCTGCCTAGGTTCAGACGACAAGCTAATT |
| Alt-DUS AvrII R | GGTTGCCTAGGTTCAGACGACATGCAGC |
| ampG-FLAG3 F | CAGGCAGAGGTTCCGCTGGCTCCGCT |
| ampGend-FLAG3 R | CCTGCATCCTTATGAGAAAGTAAGTTC |
| (GC) FLAG3-ampGend F | TTCTCATAAGAGAAAACCCAGGATGCAGG |
| (GC) FLAG3-ampGend R | AGCGGAACCTCTGCCTGCATCCTGGG |
| (MC) FLAG3-ampGend F | TTCTCATAAGAGAAAACTCAGGATGCAGG |
| (MC) FLAG3-ampGend R | AGCGGAACCTCTGCCTGCATCCTGAG |
| MS11 AmpG R398Q F | GTACCGTTTTTCCAGTTGTGTTTCATAC |
| MS11 AmpG R398Q R | GTATGAAACACAACTGGAAAAACGGTAC |
| MS11 AmpG M391L F | CTGATCGAATGGCTGGGTTATGTACCG |
| MS11 AmpG M391L R | CGGTACATAACCCAGCCATTCGATCAG |
| MS11 AmpG I402A F | GGCTGTGTTTCGCACTTGCCCTG |
| MS11 AmpG I402A R | CAGGGCAAGTGCGAAACACAGCC |
| MS11 AmpG R398Q I402A F | CAGTTGTGTTTCGCACTTGCCCTG |
| MS11 AmpG R398Q I402A R | CAGGGCAAGTGCGAAACACAACTG |
| NSi ampG SacIF | CAGGAGAGCTCGTACTGCTCATCCATTATGAC |
| NSi ampG down BamHI R2 | CATTAGGATCCCAATCGGCGTGTCTGCGATG |
| NMu ampG SacIF | CGCCGAGAGCTCGATGTTGTTCTCCCATTATGAC |
| NMu ampG BamHI R | GACGAGGATCCCTACCGATACATTCAAACG |
| rmp-RT-F | CGAAGGCCATACCGACTTTATGG |
| rmp-RT-R | GTTGCTGACCAGGTTGTTTGC |
| ampG-RT-F | GTGCGTGCTGCTGTTTATC |
| ampG-RT-R | GTCTTGCTGAAACCCATATCC |
| gdh-F2 | GTAGCGATGAGTAGTATTAC |
| gdh-R1 | GCCGTACTATTTGTACTGTC |
| gdh-R2 | GTGATTTCAGACGGCATATC |
| gdh-internal-F | GGCAAAGAAAGCCTGC |
The chimeric ampG constructs (pEC016 to pEC019) were generated with pEC013 as a base. The ampG coding sequence was divided into four unequal quarters (also called ampG regions 1 to 4), in which each region contains at least one nonsynonymous nucleotide polymorphism in GC and MC. The ampG coding region is 1,284 bp long. Region 1 encompassed bp 1 to 150, while region 2 contained bp 151 to 788. Region 3 is comprised of bp 789 to 992, while region 4 included bp 993 to 1284. The chimeric ampG constructs also contained an ∼600-bp ampGGC 5′ and 3′ flanking region to facilitate double-crossover homologous recombination when transformed into Neisseria.
Characterization of released PG fragments.
Metabolic labeling of PG using [6-3H]glucosamine was performed as described by Rosenthal and Dziarski (32) with modifications from Cloud and Dillard (33). Quantitative PG fragment release analysis was performed as described by Garcia and Dillard (22). Briefly, Neisseria strains were pulse-labeled using 10 μCi/ml [6-3H]glucosamine in GCBL lacking glucose and supplemented with 0.042% NaHCO3 and pyruvate as a carbon source to label the sugar backbone or using 25 μCi/ml [2,6-3H]diaminopimelic acid in Dulbecco modified Eagle medium lacking cysteine supplemented with 100 μg/ml methionine and 100 μg/ml threonine to label the peptide stems. For quantitative PG fragment release, an aliquot of the culture was removed after labeling for determination of the number of radioactive counts per minute (cpm) by liquid scintillation counting. The number of cpm was then normalized to obtain equal numbers of cpm in the bacteria in each culture. Pulse-labeling was then followed by a 2-h (N. meningitidis) or a 2.5-h (N. gonorrhoeae, N. sicca, and N. mucosa) chase period in cGCBL to achieve an equal number of generations. At the end of the chase period, culture supernatant was obtained by centrifugation at 3,000 × g for 10 min and filter sterilization of the supernatant using a 0.22-μm-pore size filter. Radiolabeled PG fragments in the supernatant were separated by size using tandem size-exclusion chromatography and detected by liquid scintillation counting. The relative amounts of PG fragments released were determined by calculating the area under the curve.
Immunoblotting and detection of AmpG-FLAG3.
Portions (10 μg) of whole-cell lysates were electrophoresed on 12% SDS-PAGE gels. The proteins were then transferred onto polyvinylidene fluoride membrane (Bio-Rad) either at 100 V for 1 h or at 20 V overnight. The membranes were blocked with 5% milk in Tris-buffered saline (TBS) for 1 h at room temperature and then incubated with anti-FLAG M2 primary antibody (Sigma-Aldrich) in TBS with 0.05% Tween 20 (TTBS) and 5% milk either for 1 h at room temperature or overnight at 4°C. Membranes were washed four times with TTBS for 5 min each at room temperature, incubated with goat anti-mouse IgG–horseradish peroxidase secondary antibody (Santa Cruz) in TTBS for 1 h, and then washed five times with TTBS for 5 min each. Blots were developed using an Immun-Star horseradish peroxidase substrate kit (Bio-Rad), and imaged using the Odyssey Fc Imagining System (LI-COR). Band intensities and protein concentrations were determined using Odyssey Fc.
Quantitative RT-PCR.
Quantitative reverse transcription-PCR (RT-PCR) was performed as described by Salgado-Pabón et al. (34). Briefly, gonococcal strains were grown in cGCBL until mid-log phase. RNA from 2 ml of culture was isolated using TRIzol reagent and treated with Turbo DNase to remove DNA contaminants (Life Technologies). Reverse transcription was then performed using an iScript cDNA synthesis kit (Bio-Rad). The resulting cDNA samples were used for quantitative real-time PCR using iQ SYBR green supermix (Bio-Rad) with the primers ampG-RT-F (GTGCGTGCTGCTGTTTATC) and ampG-RT-R (GTCTTGCTGAAACCCATATCC) to measure ampG transcript levels and the primers rmp-RT-F (CGAAGGCCATACCGACTTTATGG) and rmp-RT-R (GTTGCTGACCAGGTTGTTTGC) to measure rmp transcript levels as a control. Rmp was chosen as a control because it is a constitutively expressed protein that is not regulated by iron levels, and rmp levels have been used to normalize RT-PCR data (35–37). Quantitative RT-PCR results were analyzed using the StepOnePlus System (Applied Biosciences). Statistical analyses were performed using a Student two-tailed t test.
Model of gonococcal AmpG structure.
The predicted structure of gonococcal AmpG was modeled using the I-TASSER server (38–41) with multiple threading templates and using Phyre2 with a multitemplate/ab initio template (42). The structures of the following proteins were used as the templates for I-TASSER: E. coli glycerol-3-transporter GlpT (PDB ID 1PW4), MdfA multidrug transporter (PDB ID 4ZOW), E. coli YajR transporter (PDB ID 3WDO), and E. coli lactose permease LacY (PDB ID 1PV6). The structures of the following proteins were used as the templates for Phyre2: human glucose transporter GLUT3/SLC2A3 (PDB ID 5C6C), E. coli glycerol-3-phosphate transporter GlpT (PDB ID 1PW4), E. coli YajR transporter (PDB ID 3WDO), E. coli lactose permease LacY (PDB ID 1PV7), a eukaryotic phosphate transporter (PDB ID 4J05), and a Staphylococcus epidermidis glucose transporter (PDB ID 4LDS).
RESULTS
Meningococcal AmpG is more efficient at PG fragment recycling compared to gonococcal AmpG.
We previously generated a gonococcal strain that expresses meningococcal ampG (EC505) and characterized the PG fragment profile of this gene replacement mutant (23). The native gonococcal ampG (ampGGC WT) was replaced with meningococcal ampG (ampGMC WT) coding region through double-crossover homologous recombination to generate EC505. Using metabolic labeling of PG with [6-3H]glucosamine and quantitative fragment release in three independent experiments, we determined that EC505 released 52% PG monomers and 33% disaccharide compared to wild-type (WT) N. gonorrhoeae (MS11) (Fig. 1A and C) in agreement with previous observations (23). We used a similar strategy to generate a meningococcal strain that expresses gonococcal ampG (EC1001) and determined that EC1001 released ∼39% more PG monomers than WT MC (ATCC 13102) (Fig. 1B and C). The differences in the amounts of PG monomers released in the gene replacement mutants compared to WT GC and WT MC are not identical to each other or to the differences seen between WT GC and WT MC (2.8-fold less in WT MC). This discrepancy is likely due to the increased degradation of PG fragments in MC compared to GC, as previously described (23). Our results suggest that meningococcal AmpG is more efficient at PG fragment recycling than is gonococcal AmpG. Thus, the expression of meningococcal AmpG by N. gonorrhoeae reduced the amount of proinflammatory PG monomers released into the extracellular milieu and vice versa.
FIG 1.

Expression of non-native Neisseria ampG in N. gonorrhoeae and N. meningitidis altered peptidoglycan fragment release. Released [3H]glucosamine-labeled PG fragments were separated by size-exclusion chromatography and detected by liquid scintillation counting to generate a PG fragment release profile. The symbols for PG sugars and amino acids are based on those used by Jacobs et al. (24). (A) Comparison of WT GC (MS11) to a gonococcal ampG replacement mutant expressing ampGMC WT (ampGGC WT– ampGMC WT+; EC505). (B) Comparison of WT MC (ATCC 13102) to a meningococcal ampG replacement mutant expressing ampGGC WT (EC1001). (C) Quantification of the amount of PG fragments released by the ampG replacement mutants compared to WT in three independent experiments. An asterisk indicates the amount of PG fragments released by the gene replacement mutant was significantly different compared to the WT as determined by a Student two-tailed t test (P < 0.05).
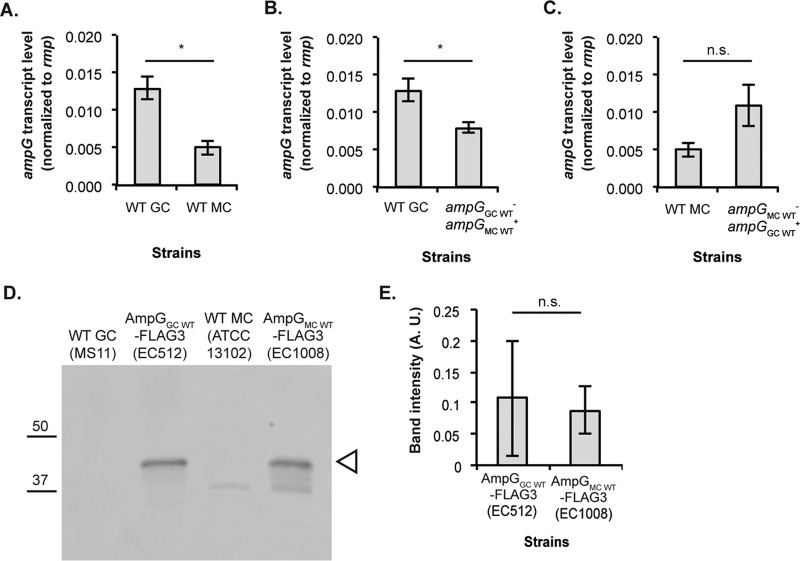
AmpG from gonococcal strain MS11 and meningococcal strain ATCC 13102 have 97% identity and differ only by nine amino acid residues (see Fig. 3A; see also Fig. S1 in the supplemental material). We sought to determine whether the difference in PG recycling efficiency is caused by differences in ampG expression levels or whether small differences in protein sequence impact AmpG function. We performed quantitative RT-PCR on RNA samples isolated from WT GC (MS11), WT MC (ATCC 13102), GC expressing meningococcal ampG (EC505), and MC expressing gonococcal ampG (EC1001). If the difference in recycling efficiency is a direct consequence of differences in ampG expression, we would expect to see higher levels of ampG transcript expressed by strains that release lower levels of PG fragments, such as ATCC 13102 and EC505, compared to strains that release higher levels of PG fragments, such as MS11 and EC1001. Interestingly and in contrast to this hypothesis, bacterial strains that are more efficient at recycling produced lower levels of ampG transcript than strains that are less efficient at recycling. Gonococcal strain MS11 produced higher levels of ampG transcript than meningococcal strain ATCC 13102 (Fig. 2A). EC505, which is more efficient at recycling than MS11 produced lower levels of ampG transcript than MS11 (Fig. 2B). ATCC 13102, which is more efficient at recycling than EC1001, did not show increased ampG transcript compared to the latter strain (Fig. 2C).
FIG 3.
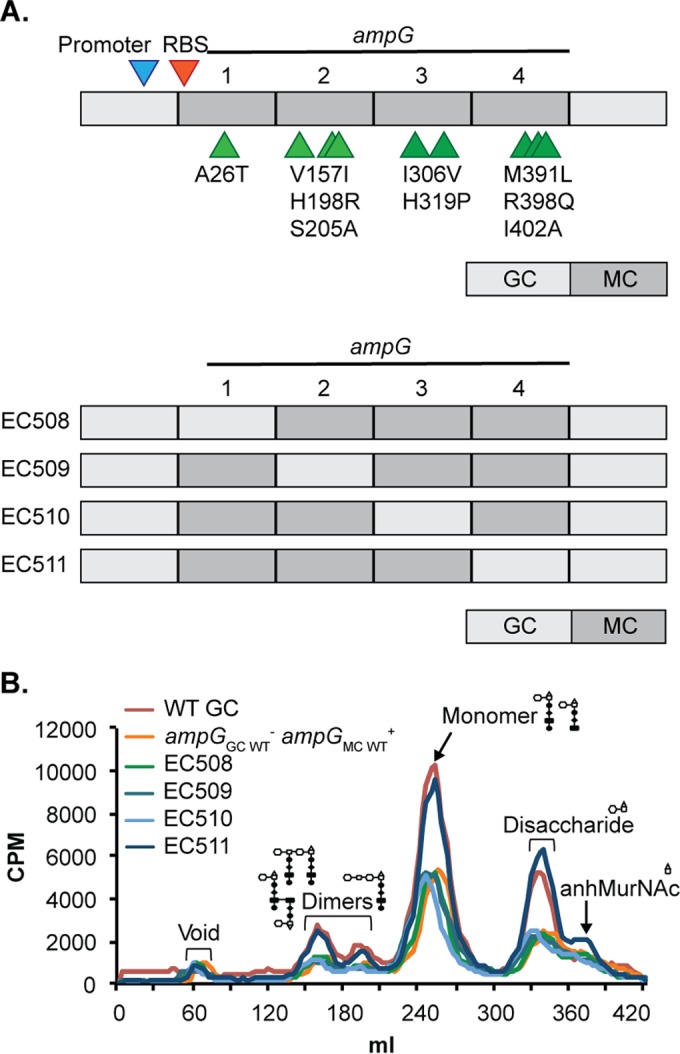
Residues near the C-terminal end of AmpG (AmpG region 4) modulated AmpG recycling efficiency. (A) Cartoon depiction of the AmpG replacement and AmpG chimera constructs expressed in N. gonorrhoeae (not drawn to scale). The AmpG replacement construct (top) was used as a base to generate the chimera constructs. Residues that differ between GC AmpG and MC AmpG are indicated in the order gonococcal residue, residue number, and meningococcal residue. Each chimera construct contained approximately one-quarter gonococcal ampG coding region and three-quarters meningococcal ampG coding region and contained a mixture of gonococcal and meningococcal residues. (B) PG fragment release profiles for N. gonorrhoeae strains expressing different versions of ampG.
FIG 2.
Neisseria strains that were more efficient at recycling PG fragments did not express higher levels of ampG. Transcript levels for ampG were determined comparing WT MC (ATCC 13102) to WT GC (MS11) (A), ΔampGGC WT ampGMC WT+ (EC505) to WT GC (B), and WT MC to ΔampGMC WT ampGGC WT+ (EC1001) (C). RT-PCR results are from three biological replicates with technical triplicates. (D) Protein levels of AmpG-FLAG3 were determined for GC and MC strains by Western blotting. WT gonococci and meningococci that did not express FLAG3 tagged AmpG protein were included as negative controls. (E) Quantification of the AmpG-FLAG3 bands from three independent experiments was performed using LiCor Odyssey Fc. Statistical significance was determined by using a Student two-tailed t test. An asterisk indicates statistical significance, with P < 0.05, whereas n.s. indicates not significant.
To determine levels of AmpG protein in WT gonococci and in WT meningococci, we raised polyclonal antibodies against a short AmpG epitope (FRREILSDEELGLG) (GenScript). Unfortunately, this antibody was not specific enough to detect AmpG levels in an immunoblot (data not shown). As an alternative, we generated strains expressing AmpG fused to a C-terminal triple FLAG tag [(DYKDDDDK)3] and performed immunoblotting with anti-FLAG M2 primary antibody. There was no significant difference in the amount of AmpG-FLAG3 expressed by WT gonococci and WT meningococci (Fig. 2D and E). Taken together, these results suggested that the difference in PG fragment release between N. gonorrhoeae and N. meningitidis was not due to higher ampG expression levels or AmpG protein levels in strains that are more efficient at recycling.
Three residues near the C-terminal end of AmpG modulate AmpG recycling efficiency.
Although AmpG sequences from N. gonorrhoeae strain MS11 and N. meningitidis ATCC 13102 are 97% identical, the nine amino acid residues that differ may impact protein function. To determine which residues affect AmpG efficiency, we designed four chimeric ampG constructs to be expressed in N. gonorrhoeae (Fig. 3A). We divided AmpG into four unequal regions—region 1 (N-terminal end, bp 1 to 150), region 2 (mid gene, closer to the N-terminal end, bp 151 to 788), region 3 (mid gene, closer to the C-terminal end, bp 789 to 992), and region 4 (C-terminal end, bp 993 to 1284)—in which each region contained at least one residue that differs between MS11 and ATCC 13102. Each chimeric gene construct is comprised of approximately one-quarter gonococcal ampG coding region and approximately three-quarters meningococcal ampG coding region, so that each chimeric protein expressed would contain a mixture of gonococcal and meningococcal AmpG residues. We would expect to see a WT GC-like phenotype for PG fragment release in strains that express the gonococcal region(s) that codes for AmpG residues important for function, while the other strains would phenocopy a strain that expresses ampGMC WT (EC505). Only expression of a chimeric AmpG protein with meningococcal regions 1 to 3 and gonococcal region 4 (EC511) resulted in a WT GC-like phenotype (Fig. 3B). This strain showed a large increase in release of PG monomers, as well as increased release of the other small PG fragments, compared to strains that express the other chimeric AmpG proteins with gonococcal ampG region 1, 2, or 3. We also produced a GC strain expressing ampG carrying gonococcal regions 1 to 3 and meningococcal region 4. This strain phenocopied EC505, indicating that the six changes in these three regions do not decrease AmpG function (data not shown). Our results suggest that residues in AmpG region 4 modulate AmpG efficiency.
Three residues in AmpG region 4 that differ between gonococcal and meningococcal AmpG are residues 391 (methionine in GC and leucine in MC), 398 (arginine in GC and glutamine in MC), and 402 (isoleucine in GC and alanine in MC). To determine which residues are most important for modulating AmpG function, we utilized site-directed mutagenesis to perform single, double, and triple substitutions of gonococcal AmpG residues 391, 398, and 402 with the corresponding meningococcal residues. The expression of ampGGCM391L and ampGGCI402A reduced PG monomer release in N. gonorrhoeae, although not to the levels seen in the gene replacement mutant, EC505 (Fig. 4A). Expression of ampGGCR398Q resulted in a WT GC-like phenotype (Fig. 4A).
FIG 4.
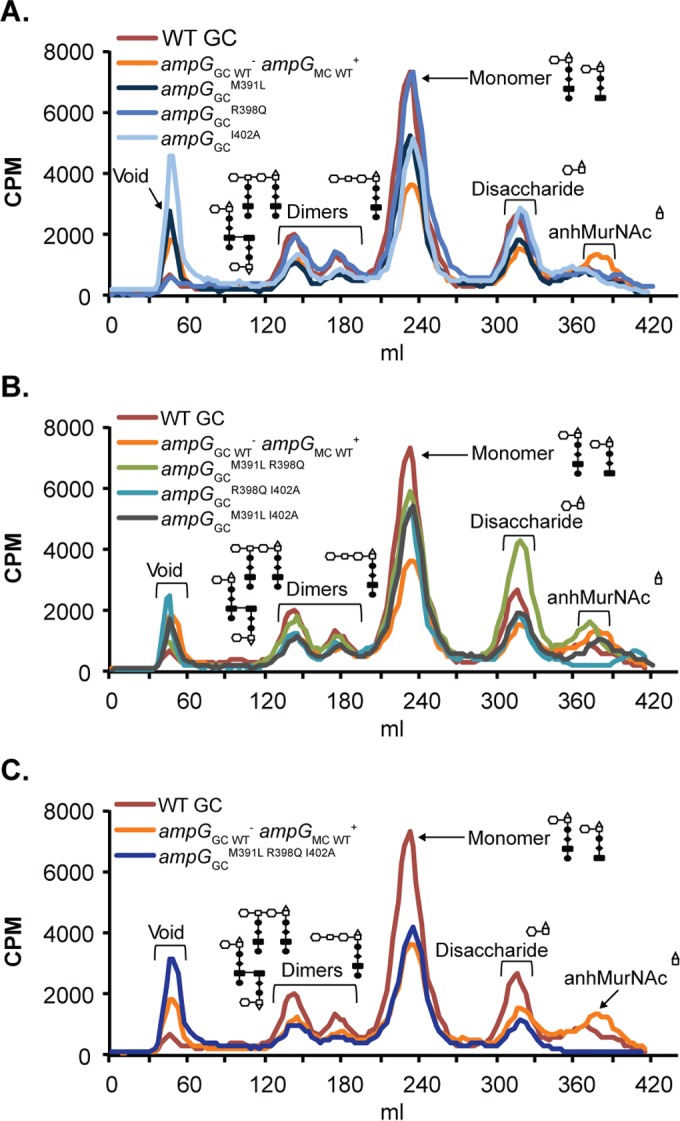
AmpG residues 391, 398, and 402 worked cooperatively to modulate AmpG recycling efficiency. PG fragment release profiles are shown for single substitutions of AmpG residues 391, 398, and 402 (EC515, EC516, and EC517) compared to the whole gene replacement mutant (EC505) and the wild type (MS11) (A), double substitutions of AmpG residues 391, 398, and 402 (EC518, EC519, and EC521) compared to the WT and EC505 (B), and triple substitutions of AmpG residues 391, 398, and 402 (EC523) compared to the WT and EC505 (C).
We next sought to determine whether double substitutions of residues 391 and 402 from the gonococcal to the meningococcal residues would result in an additive effect, leading to PG monomer release levels similar to that of gonococci expressing meningococcal ampG. Gonococcal strains that expressed ampGGCM391L I402A phenocopied strains that expressed the ampGGCM391L and ampGGCI402A single substitution mutants, releasing an intermediate level of PG monomers (Fig. 4B). Double substitutions of any of the three residues resulted in PG monomer release levels similar to that of gonococcal strains expressing ampGGCM391L or ampGGCI402A, suggesting that double mutations did not have an additive effect on PG recycling efficiency (Fig. 4B). Substitutions of all three residues 391, 398, and 402 from the gonococcal to the meningococcal residues resulted in PG monomer release levels similar to that of gonococci expressing meningococcal ampG (Fig. 4C). Our results suggested that residues 391, 398, and 402 work cooperatively to modulate AmpG function.
AmpG residues 391, 398, and 402 do not regulate levels of AmpG protein.
We hypothesized that substitutions of residues 391, 398, and 402 from the gonococcal to the meningococcal variants might stabilize the protein. Thus, increased recycling efficiency in the gonococcal strain that expressed ampGGCM391L R398Q I402A could be a result of increased AmpG protein levels. To test this idea, we tagged various gonococcal ampG substitution mutants that were more efficient at recycling compared to WT GC with the C-terminal triple FLAG epitope and measured AmpG protein levels by immunoblotting. There was no significant difference in the amounts of AmpG-FLAG3 protein in any of the mutant strains tested (Fig. 5). Thus, strains that expressed ampG variants that are more efficient at recycling PG fragments did not produce more AmpG-FLAG3 protein compared to WT gonococci. The immunoblot results suggested that substitutions of residues 391, 398, and 402 from the gonococcal to the meningococcal variants do not increase AmpG stability and levels.
FIG 5.
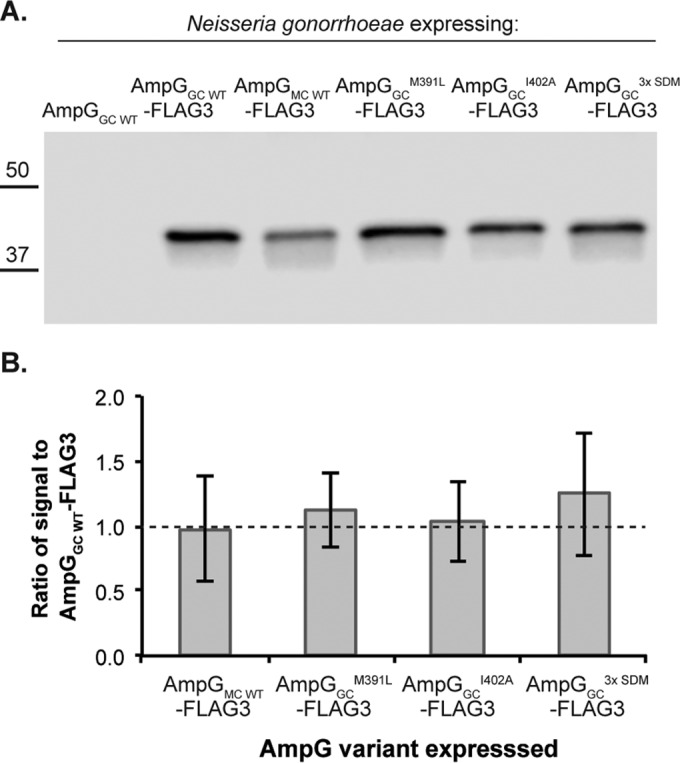
Increased recycling efficiency of gonococcal ampG mutants was not a consequence of increased AmpG protein levels. (A) AmpG was tagged with a C-terminal triple FLAG (FLAG3) epitope to determine AmpG levels made by N. gonorrhoeae via immunoblotting, comparing the levels of AmpG-FLAG3 expressed by different ampG mutant strains that have GC WT-like (EC512) or more efficient PG recycling (EC546, EC548, and EC550). (B) Quantification of band intensities from three independent experiments using LiCor Odyssey Fc.
N. sicca and N. mucosa are more efficient at PG recycling and release lower levels of PG fragments than N. gonorrhoeae.
There are eight species of human-associated, nonpathogenic Neisseria that asymptomatically colonize the human nasopharyngeal and oropharyngeal spaces. These strains include N. sicca, N. mucosa, N. lactamica, N. polysaccharea, N. subflava, N. flavescens, N. cinerea, and N. elongata (43). We hypothesized that nonpathogenic neisseriae would release lower levels of PG fragments to evade immune clearance and maintain asymptomatic carriage in human hosts. We found that both N. sicca and N. mucosa released lower levels of PG monomers than did N. gonorrhoeae (Fig. 6). Intriguingly, both N. sicca and N. mucosa also released very small amounts or possibly no PG dimers (Fig. 6).
FIG 6.
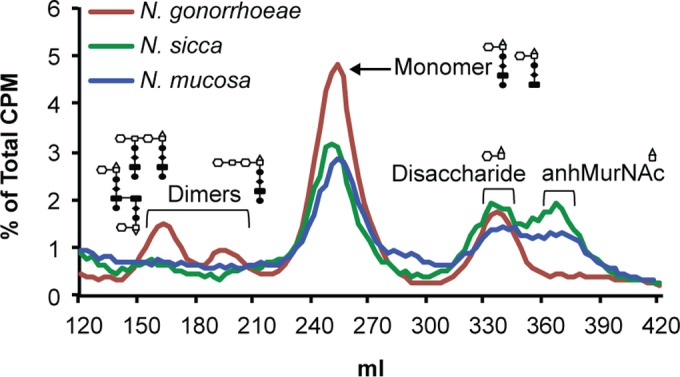
PG fragment release from nonpathogenic N. sicca (ATCC 29256) and N. mucosa (ATCC 25996) compared to N. gonorrhoeae (MS11).
To determine AmpG recycling efficiency in N. sicca and in N. mucosa, we compared the amounts of PG fragments released by WT and an ampG mutant that is unable to recycle PG fragments. We mutated N. sicca and N. mucosa ampG by interrupting the ampG coding sequence with a kanamycin resistance cassette. Since there are currently no complementation constructs available for N. sicca and N. mucosa, we generated backcrossed strains by transforming WT N. sicca and N. mucosa with chromosomal DNA isolated from the ampG deletion mutants. We calculated the recycling efficiency in N. sicca and N. mucosa by determining the area under the monomer curve for WT and ampG mutants. Both N. sicca and N. mucosa released 5% and recycled 95% of PG monomers liberated during PG turnover (Fig. 7). This level of PG monomer release is very similar to that of N. meningitidis, which releases 4% of PG monomers (23). Free disaccharide release was also increased in the N. sicca and N. mucosa ampG mutants, suggesting that the permease also transports these PG molecules, a finding in agreement with previous reports (23, 44).
FIG 7.
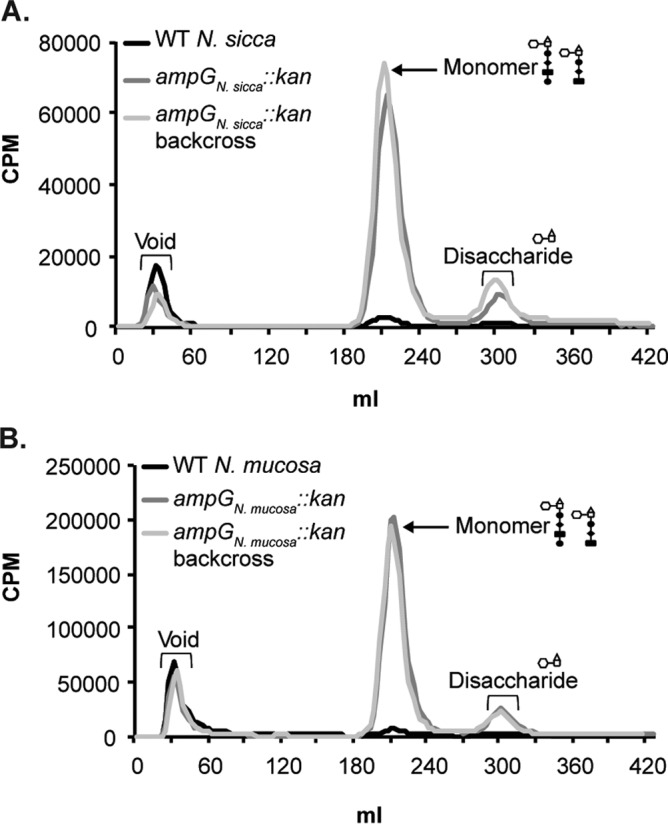
N. sicca and N. mucosa possess functional AmpG proteins. PG fragment release was examined for mutants carrying a kanamycin resistance cassette interrupting ampG in N. sicca (EC2004) (A) and N. mucosa (EC2003) (B) compared to WT N. sicca and N. mucosa. PG fragment release profiles for backcrossed mutants (EC2004BC and EC2003BC) are also shown. Quantification of the peaks was performed with data obtained from three independent experiments.
Bioinformatic analyses demonstrate that all gonococci encode M391, R398, and I402 in ampG.
We compiled and aligned ampG alleles expressed by 31 strains from nine species of Neisseria and found that all gonococcal strains surveyed have methionine, arginine, and isoleucine at AmpG positions 391, 398, and 402 (see Fig. S1 in the supplemental material). A query of the sequences at the Neisseria multilocus sequence typing website (http://pubmlst.org/neisseria) and the Meningitis Research Foundation meningococcus genome library database (http://meningitis.org/research/genome) revealed that although no gonococcal strains (out of 1,847 sequences) had leucine, glutamine, and alanine at the three positions, there were two strains of N. polysaccharea (out of 19 sequences) (45), eight strains of N. lactamica (out of 130 sequences), and around 420 meningococcal strains (out of 7,141 sequences), predominantly of the ST-269 subtype and, to a lesser extent, the ST41/44 subtypes, that had methionine, arginine, and isoleucine at AmpG residues 391, 398, and 402 (data not shown). These three amino acid changes were found in 5.88% of meningococcal strains. One example each of N. polysaccharea (strain 12030-2014), N. lactamica (strain 049-12), and N. meningitidis (strain M10-240473) are shown in Fig. S1 in the supplemental material.
We also sequenced ampG from several meningococcal clinical isolates and found an isolate, N. meningitidis strain NM00268, that codes for the gonococcus-like residues arginine and isoleucine at AmpG positions 398 and 402 (see Fig. S1 in the supplemental material). NM00268 labels poorly with [3H]glucosamine and thus was labeled with [3H]diaminopimelic acid instead. In accordance with our model, NM00268 released ∼1.7 times more PG monomers than did ATCC 13102 (Fig. 8), providing support to our hypothesis that having gonococcus-like residues at AmpG positions 391, 398, and/or 402 contributes to increased PG monomer release. There were no significant differences in the amounts of peptides released by NM00268 and ATCC 13102.
FIG 8.
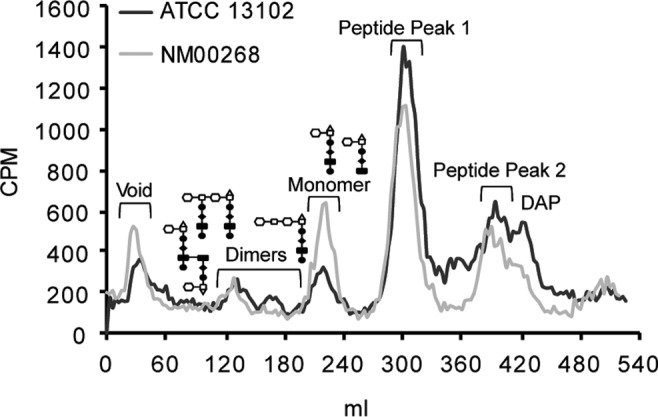
PG fragment release from a meningococcal strain with naturally occurring GC-like AmpG residues 398 and 402 (NM00268). ATCC 13102 and NM00268 were labeled with [3H]diaminopimelic acid, which labels the peptide stem of PG fragments. Quantification of the area under the curve was performed with data from three independent experiments.
We constructed a neighbor-joining tree based on AmpG sequences and found that although gonococcal strains tend to cluster together, strains of N. lactamica and N. polysaccharea that expressed GC-like AmpG residues 391, 398, and 402 did not cluster with N. gonorrhoeae or with each other. Although N. meningitidis strains NM00268 and M10-240473 cluster close to each other, they did not cluster with N. gonorrhoeae or with N. lactamica strain 049-12 or N. polysaccharea strain 12030-2014. In addition, N. polysaccharea strain 12030-2014 did not cluster well with other strains from the same species, suggesting that the AmpG sequences in these nongonococcal strains evolved independently or resulted from horizontal gene transfer events, creating mosaic AmpG sequences, as is seen for N. meningitidis PBP2 (46). As a control, we also constructed a neighbor-joining tree based on Gdh sequences (see Fig. S2 in the supplemental material). With the exception of N. meningitidis strain 8013, all other strains clustered with members of the same species. Overall, these results demonstrated that while M391, R398, and I402 are present in a small fraction of meningococcal or nonpathogenic Neisseria strains, these AmpG-crippling mutations are universally present in N. gonorrhoeae, making it likely that all N. gonorrhoeae isolates release high levels of PG fragments.
AmpG residues 391, 398, and 402 are predicted to be located on a transmembrane helix near the periplasmic face of the protein.
We used I-TASSER and Phyre2 servers to predict the structure of AmpG and obtained two different putative AmpG structures (38–42). The model of AmpG structure obtained by I-TASSER showed an inward-facing conformation, in which irregularly arranged helices surround a substrate binding cavity that opens toward the cytoplasm (Fig. 9A). On the other hand, the predicted structure of AmpG using Phyre2 showed an occluded conformation that may be a transitional state between the inward-facing and the outward (periplasmic)-facing conformations during transport (Fig. 9B). In both models, AmpG residues 391, 398, and 402 are located near the periplasmic face of the protein at the start of the last transmembrane helix.
FIG 9.
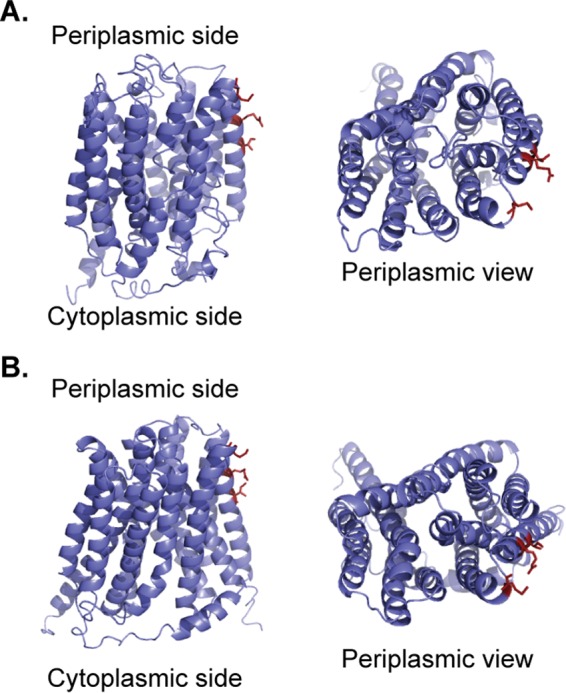
Prediction of gonococcal AmpG structure. The predicted structure of gonococcal AmpG was determined using I-TASSER server with multiple threading templates (A) and using Phyre2 with a multitemplate/ab initio template (B). Side view (left) and the view from the periplasmic face (right) of the AmpG structure, with residues 391, 398, and 402 displayed as dark red sticks. Residues 391, 398, and 402 are located close to the periplasmic face of the protein.
DISCUSSION
The release of PG fragments is not unique to Neisseria, although few genera other than Neisseria release mainly toxic anhydro-PG monomers. PG moieties released by bacteria have been implicated in the resuscitation of dormant mycobacteria, the development of Myxococcus fruiting bodies, the germination of Bacillus subtilis spores, and the establishment of mutualism between Bacillus cereus and Flavobacterium johnsoniae (reviewed in references 47, 48 and 49). Nonetheless, the release of PG monomers by bacteria tends to lead to inflammation and the death of animal host cells, whether or not this interaction leads to beneficial or detrimental effects at the organismal level. Tetrapeptide monomer (also known as TCT) and lipopolysaccharide released by Vibrio fischeri work synergistically to induce the regression of ciliated epithelial cells near the light organ of the Hawaiian bobtail squid to allow the establishment of squid-Vibrio symbiosis (21, 50, 51). The production of PG fragments is also thought to be important for the pathogenesis of multiple bacterial species, including but not limited to human pathogens such as Helicobacter pylori and Shigella flexneri, as well as plant pathogens such as Pseudomonas syringae and Erwinia amylovora (reviewed in reference 48).
In addition to N. gonorrhoeae, the effects of released PG fragments on host fitness is most well studied with respect to the human pathogen Bordetella pertussis, which causes whooping cough. Unlike Neisseria, which releases a mixture of tripeptide monomer and TCT, B. pertussis releases exclusively TCT. TCT causes the sloughing and death of ciliated tracheal cells in ex vivo hamster tracheal tissue studies (14, 52, 53). An insertion element (IS491) located ∼90 bp upstream of B. pertussis ampG reduced ampG expression in B. pertussis and results in high levels of TCT released (54). When IS491 is deleted, or when E. coli ampG is expressed in B. pertussis instead, the amount of TCT released is significantly lowered (54). Collectively, these findings suggest that both human pathogens B. pertussis and N. gonorrhoeae evolved different strategies to reduce PG fragment recycling efficiency to release more PG monomers. This process generates an inflammatory environment that may be favorable for bacterial growth and invasion.
In this work, we showed that N. gonorrhoeae releases more PG monomer and is less efficient at recycling PG monomers than N. meningitidis, N. sicca, and N. mucosa, three species of Neisseria that can asymptomatically colonize the human nasopharyngeal space. With N. gonorrhoeae and N. meningitidis, the difference in the recycling efficiency is not due to higher expression of ampG in N. meningitidis compared to N. gonorrhoeae. In fact, gonococcal and meningococcal strains that are more efficient at recycling consistently produced lower levels of ampG transcript than strains that are less efficient at recycling. Furthermore, we did not see significant differences in the amounts of various AmpG-FLAG3 proteins expressed by N. gonorrhoeae, and amino acid substitutions to make gonococcal AmpG more like meningococcal AmpG did not increase AmpG protein levels. These data indicated that it is not reduced amounts of ampG transcript or AmpG protein that makes N. gonorrhoeae deficient at recycling but rather the reduced function of gonococcal AmpG in facilitating PG fragment recycling.
We also showed that reduced recycling efficiency in N. gonorrhoeae can be accounted for by the amino acid identities of residues 391, 398, and 402, which are close to the C-terminal end of AmpG (Fig. 9). Although we do not yet understand how these three residues modulate AmpG function (other than that the three residues do not change AmpG protein levels), we do have several hypotheses. One hypothesis is that residues 391, 398, and 402 may directly bind to PG and that the gonococcal residues are either less able to bind PG fragments or bind PG fragments too tightly, making the transport of PG fragments less efficient compared to the meningococcal, N. sicca, or N. mucosa AmpG counterparts.
The E. coli AmpG homolog is powered by proton motive force (44), although it is unknown whether AmpG functions as a H+/PG fragment symporter or whether AmpG interacts with a proton-transducing protein that powers the permease. It is also unknown whether PG-degrading enzymes work together in a complex to remodel the PG layer and whether such complexes colocalize or interact with AmpG to ensure efficient recycling. Lytic transglycosylases are PG-degrading enzymes that cleave the glycan backbone to generate PG monomers (20). N. gonorrhoeae has two lytic transglycosylases, LtgA and LtgD, that generate all or nearly all the PG monomers released by the bacterium. Deletion of ltgD leads to a larger reduction in the amount of PG monomers released compared to the deletion of ltgA (62% reduction versus 38% reduction) (19). However, LtgA generates more PG monomers than LtgD, and LtgA-generated monomers are preferentially taken up into the cytoplasm for recycling (R. E. Schaub et al., unpublished data). Thus, it is also possible that residues 391, 398, and 402 facilitate protein-protein interaction with a hypothetical accessory protein(s) or with PG-degrading enzymes such as LtgA in the periplasm to power AmpG function or ensure efficient PG recycling.
Another hypothesis is that the residues at these positions are important for facilitating conformational changes required for the import of PG fragments into the cytoplasm. AmpG belongs to major facilitator superfamily (MFS). MFS proteins are typically membrane transport proteins with 12 or 14 transmembrane α-helices that can function as uniporters, symporters, and antiporters and can be found in bacteria, eukaryotes, and archaea (55, 56). The most well-studied MFS protein, LacY, is a lactose/H+ symporter that can assume one of at least two conformations, as determined by X-ray crystallography studies. LacY can assume a conformation with 2-fold pseudosymmetry with a large aqueous, substrate-binding cavity that opens toward the cytoplasm (PDB IDs 1PV7 and 2V8N) (57, 58). It has been proposed that LacY can assume a similar conformation in which the aqueous cavity opens toward the periplasm for substrate binding (59). LacY can also form an occluded conformation with a narrow cavity that opens slightly toward the periplasm that is thought to be an intermediate conformation during substrate transport (PDB IDs 4OAA and 4ZYR) (60, 61). Given that the two predicted AmpG structures resembled the two structurally determined conformations of LacY, AmpG may function similarly to LacY. As such, residues at positions 391, 398, and 402 might impact the rate of conformational changes required for transport. The crystal structure of AmpG and the exact mechanism of action that AmpG uses to transport PG fragments are currently unknown. A crystal structure of AmpG would help inform studies of AmpG's mechanism of action and provide insight into how residues 391, 398, and 402 impact AmpG efficiency.
We hypothesize that the differences in AmpG function and PG fragment release between the asymptomatic colonizers and N. gonorrhoeae contribute to the differences in the inflammatory responses to these species at their different infection sites. It should be noted that ampG is not the only factor affecting PG fragment release. Comparing N. meningitidis to N. gonorrhoeae, expression of meningococcal ampG in gonococci results in a nearly 2-fold decrease in PG release, but the expression of gonococcal ampG in meningococci only resulted in a 39% increase in PG monomer release (Fig. 1). These results suggest that additional features of PG fragment metabolism in N. gonorrhoeae may favor PG fragment release and that N. meningitidis and the nonpathogenic Neisseria species may have additional mechanisms for increasing PG fragment recycling and diminishing PG fragment release. Increased PG fragment breakdown by N. meningitidis, as well as N. mucosa and N. sicca, compared to N. gonorrhoeae can be seen in the PG fragment release profiles (Fig. 1 and 6) (23). Fewer PG dimers and monomers are released, but more anhMurNAc is released compared to N. gonorrhoeae. In addition to the reduced PG fragment release we have shown here, nonpathogenic Neisseria spp. are also known to produce a lipid A structure that is less inflammatory (6). Together with differences in the responsiveness of the different tissues infected by these species, the differences in lipid A and PG fragment release may explain how nonpathogenic Neisseria spp. are able to maintain asymptomatic colonization, whereas N. gonorrhoeae usually induces a strong inflammatory response and disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nate Weyand for the gift of N. sicca ATCC 29256 and N. mucosa ATCC 25996. We are grateful to Katie Hackett, Jon Lenz, and Ryan Schaub for experimental support and discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00437-16.
REFERENCES
- 1.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu G, Tang CM, Exley RM. 2015. Nonpathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology 161:1297–1312. doi: 10.1099/mic.0.000086. [DOI] [PubMed] [Google Scholar]
- 4.Johnson AP. 1983. The pathogenic potential of commensal species of Neisseria. J Clin Pathol 36:213–223. doi: 10.1136/jcp.36.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marri PR, Paniscus M, Weyand NJ, Rendon MA, Calton CM, Hernandez DR, Higashi DL, Sodergren E, Weinstock GM, Rounsley SD, So M. 2010. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One 5:e11835. doi: 10.1371/journal.pone.0011835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John CM, Liu M, Phillips NJ, Yang Z, Funk CR, Zimmerman LI, Griffiss M, Stein DC, Jarvis GA. 2012. Lack of lipid A pyrophosphorylation and functional lptA reduces inflammation by Neisseria commensals. Infect Immun 80:4014–4026. doi: 10.1128/IAI.00506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayor MT, Roett MA, Uduhiri KA. 2012. Diagnosis and management of gonococcal infections. Am Fam Physician 86:931–938. [PubMed] [Google Scholar]
- 8.Melly MA, McGee ZA, Rosenthal RS. 1984. Ability of monomeric peptidoglycan fragments from Neisseria gonorrhoeae to damage human Fallopian tube. J Infect Dis 149:378–386. doi: 10.1093/infdis/149.3.378. [DOI] [PubMed] [Google Scholar]
- 9.McGee ZA, Jensen RL, Clements CM, Taylor-Robinson D, Johnson AP, Gregg CR. 1999. Gonococcal infection of human Fallopian tube mucosa in organ culture: relationship of mucosal tissue TNF-α concentration to sloughing of ciliated cells. Sex Transm Dis 26:160–165. doi: 10.1097/00007435-199903000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Rosenthal RS. 1979. Release of soluble peptidoglycan from growing gonococci: hexaminidase and amidase activities. Infect Immun 24:869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha RK, Rosenthal RS. 1980. Release of soluble peptidoglycan from growing gonococci: demonstration of anhydro-muramyl-containing fragments. Infect Immun 29:914–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenthal RS, Nogami W, Cookson BT, Goldman WE, Folkening WJ. 1987. Major fragment of soluble peptidoglycan released from growing Bordetella pertussis is tracheal cytotoxin. Infect Immun 55:2117–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman WE, Klapper DG, Baseman JB. 1982. Detection, isolation, and analysis of a released Bordetella pertussis product toxic to cultured tracheal cells. Infect Immun 36:782–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cookson BT, Cho H, Herwaldt LA, Goldman WE. 1989. Biological activities and chemical composition of purified tracheal cytotoxin of Bordetella pertussis. Infect Immun 57:2223–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girardin SE, Boneca IG, Carneiro LAM, Antignac A, Jehanno M, Viala J, Tedin K, Taha M-K, Labigne A, Zahringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. 2003. Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 16.Stephens DS, Greenwood B, Brandtzaeg P. 2007. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369:2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention. 2015. Meningococcal disease: technical and clinical information. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/meningococcal/clinical-info.html. [Google Scholar]
- 18.Loh E, Kugelberg E, Tracy A, Zhang Q, Gollan B, Ewles H, Chalmers R, Pelicic V, Tang CM. 2013. Temperature triggers immune evasion by Neisseria meningitidis. Nature 502:237–240. doi: 10.1038/nature12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cloud-Hansen KA, Hackett KT, Garcia DL, Dillard JP. 2008. Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J Bacteriol 190:5989–5994. doi: 10.1128/JB.00506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan YA, Hackett KT, Dillard JP. 2012. The lytic transglycosylases of Neisseria gonorrhoeae. Microb Drug Resist 18:271–279. doi: 10.1089/mdr.2012.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. 2009. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol 191:2012–2022. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia DL, Dillard JP. 2008. Mutations in ampG or ampD affect peptidoglycan fragment release from Neisseria gonorrhoeae. J Bacteriol 190:3799–3807. doi: 10.1128/JB.01194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodhams KL, Chan JM, Lenz JD, Hackett KT, Dillard JP. 2013. Peptidoglycan fragment release from Neisseria meningitidis. Infect Immun 81:3490–3498. doi: 10.1128/IAI.00279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs C, Huang L, Bartowsky E, Normark S, Park JT. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J 13:4684–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korfmann G, Sanders CC. 1989. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother 33:1946–1951. doi: 10.1128/AAC.33.11.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellogg DS, Peacock WL, Deacon WE, Brown L, Pirkle CI. 1963. Neisseria gonorrhoeae: Virulence genetically linked to clonal variation. J Bacteriol 85:1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dillard JP. 2011. Genetic manipulation of Neisseria gonorrhoeae. Curr Protoc Microbiol Chapter 4:Unit 4A.2. doi: 10.1002/9780471729259.mc04a02s23. [DOI] [PubMed] [Google Scholar]
- 28.Wright CJ, Jerse AE, Cohen MS, Cannon JG, Seifert HS. 1994. Nonrepresentative PCR amplification of variable gene sequences in clinical specimens containing dilute, complex mixture of microorganisms. J Clin Microbiol 32:464–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman SD, Scocca JJ. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton HL, Schwartz KJ, Dillard JP. 2001. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J Bacteriol 183:4718–4726. doi: 10.1128/JB.183.16.4718-4726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frye SA, Nilsen M, Tonjum T, Ambur OH. 2013. Dialects of the DNA uptake sequence in Neisseriaceae. PLoS Genet 9:e1003458. doi: 10.1371/journal.pgen.1003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenthal RS, Dziarski R. 1994. Isolation of peptidoglycan and soluble peptidoglycan fragments. Methods Enzymol 235:253–285. doi: 10.1016/0076-6879(94)35146-5. [DOI] [PubMed] [Google Scholar]
- 33.Cloud KA, Dillard JP. 2002. A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production. Infect Immun 70:2752–2757. doi: 10.1128/IAI.70.6.2752-2757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salgado-Pabón W, Du Y, Hackett KT, Lyons KM, Arvidson CG, Dillard JP. 2010. Increased expression of the type IV secretion system in piliated Neisseria gonorrhoeae variants. J Bacteriol 192:1912–1920. doi: 10.1128/JB.01357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaik YB, Grogan S, Davey M, Sebastian S, Goswami S, Szmigielski B, Genco CA. 2007. Expression of the iron-activated nspA and secY genes in Neisseria meningitidis group B by Fur-dependent and -independent mechanisms. J Bacteriol 189:663–669. doi: 10.1128/JB.01638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agarwal S, King CA, Klein EK, Soper DE, Rice PA, Wetzler LM, Genco CA. 2005. The gonococcal Fur-regulated tbpA and tbpB genes are expressed during natural mucosal gonococcal infection. Infect Immun 73:4281–4287. doi: 10.1128/IAI.73.7.4281-4287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grifantini R, Sebastian S, Frigimelica E, Draghi M, Bartolini E, Muzzi A, Rappuoli R, Grandi G, Genco CA. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc Natl Acad Sci U S A 100:9542–9547. doi: 10.1073/pnas.1033001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction, and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tønjum T. 2015. Neisseria, p 1–48. In Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH (ed), Manual of clinical microbiology, 5th ed American Society for Microbiology, Washington, DC. [Google Scholar]
- 44.Cheng Q, Park JT. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J Bacteriol 184:6434–6436. doi: 10.1128/JB.184.23.6434-6436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jolley KA, Maiden MCJ. 2010. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowler LD, Zhang QY, Riou JY, Spratt BG. 1994. Interspecies recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitidis: natural events and laboratory simulation. J Bacteriol 176:333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertsche U, Mayer C, Götz F, Gust AA. 2015. Peptidoglycan perception: sensing bacteria by their common envelope structure. Int J Med Microbiol 305:217–223. doi: 10.1016/j.ijmm.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 48.Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. 2006. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol 4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 49.Dworkin J. 2014. The medium is the message: interspecies and interkingdom signaling by peptidoglycan and related bacterial glycans. Annu Rev Microbiol 68:137–154. doi: 10.1146/annurev-micro-091213-112844. [DOI] [PubMed] [Google Scholar]
- 50.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 51.Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, McFall-Ngai MJ. 2009. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol 11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole PJ, Wilson R. 1994. Effect of tracheal cytotoxin from Bordetella pertussis on human neutrophil function in vitro. Infect Immun 62:639–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luker KE, Tyler AN, Marshall GR, Goldman WE. 1995. Tracheal cytotoxin structural requirements for respiratory epithelial damage in pertussis. Mol Microbiol 16:733–743. doi: 10.1111/j.1365-2958.1995.tb02434.x. [DOI] [PubMed] [Google Scholar]
- 54.Lyon RS. 2001. Tracheal cytotoxin production by the Bordetellae. PhD thesis. Washington University, St. Louis, MO. [Google Scholar]
- 55.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH. 2012. The major facilitator superfamily (MFS) revisited. FEBS J 279:2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan N. 2015. Structural biology of the major facilitator superfamily transporters. Annu Rev Biophys 44:257–283. doi: 10.1146/annurev-biophys-060414-033901. [DOI] [PubMed] [Google Scholar]
- 57.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 58.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. 2007. Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A 104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smirnova I, Kasho V, Choe J-Y, Altenbach C, Hubbell WL, Kaback HR. 2007. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci U S A 104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar H, Kasho V, Smirnova I, Finer-Moore JS, Kaback HR, Stroud RM. 2014. Structure of sugar-bound LacY. Proc Natl Acad Sci U S A 111:1784–1788. doi: 10.1073/pnas.1324141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar H, Finer-Moore JS, Kaback HR, Stroud RM. 2015. Structure of LacY with an α-substituted galactoside: connecting the binding site to the protonation site. Proc Natl Acad Sci U S A 112:9004–9009. doi: 10.1073/pnas.1509854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swanson J. 1972. Studies on gonococcus infection. II. Freeze-fracture, freeze-etch studies on gonococci. J Exp Med 136:1258–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woodhams KL, Benet ZL, Blonsky SE, Hackett KT, Dillard JP. 2012. Prevalence and detailed mapping of the gonococcal genetic island in Neisseria meningitidis. J Bacteriol 194:2275–2285. doi: 10.1128/JB.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehr IJ, Long CD, Serkin CD, Seifert HS. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.