Abstract
Background Morphogenesis depends on the concerted modulation of cell proliferation and differentiation. Such modulation is dynamically adjusted in response to various external and internal signals via complex transcriptional regulatory networks that mediate between such signals and regulation of cell-cycle and cellular responses (proliferation, growth, differentiation). In plants, which are sessile, the proliferation/differentiation balance is plastically adjusted during their life cycle and transcriptional networks are important in this process. MADS-box genes are key developmental regulators in eukaryotes, but their role in cell proliferation and differentiation modulation in plants remains poorly studied.
Methods We characterize the XAL1 loss-of-function xal1-2 allele and overexpression lines using quantitative cellular and cytometry analyses to explore its role in cell cycle, proliferation, stem-cell patterning and transition to differentiation. We used quantitative PCR and cellular markers to explore if XAL1 regulates cell-cycle components and PLETHORA1 (PLT1) gene expression, as well as confocal microscopy to analyse stem-cell niche organization.
Key Results We previously showed that XAANTAL1 (XAL1/AGL12) is necessary for Arabidopsis root development as a promoter of cell proliferation in the root apical meristem. Here, we demonstrate that XAL1 positively regulates the expression of PLT1 and important components of the cell cycle: CYCD3;1, CYCA2;3, CYCB1;1, CDKB1;1 and CDT1a. In addition, we show that xal1-2 mutant plants have a premature transition to differentiation with root hairs appearing closer to the root tip, while endoreplication in these plants is partially compromised. Coincidently, the final size of cortex cells in the mutant is shorter than wild-type cells. Finally, XAL1 overexpression-lines corroborate that this transcription factor is able to promote cell proliferation at the stem-cell niche.
Conclusion XAL1 seems to be an important component of the networks that modulate cell proliferation/differentiation transition and stem-cell proliferation during Arabidopsis root development; it also regulates several cell-cycle components.
Keywords: MADS-box, XAANTAL1 (XAL1), cell-cycle, cyclins, CDKs, endoreplication, PLETHORA, proliferation/differentiation, root development, Arabidopsis thaliana
INTRODUCTION
Development depends on the dynamic spatio-temporal modulation of cell proliferation and differentiation during morphogenesis. When such modulation is perturbed, aberrant morphologies, such as tumours, may emerge (Dick and Rubin, 2013). In plants, tumours are rare, in comparison with in animals, probably due to the existence of plant cell walls (Sablowski and Carnier Dornelas, 2014), and also because plant morphogenesis has evolved to plastically adjust to environmental conditions, while maintaining patterns within functional limits (Lempe et al., 2013). In contrast to animals, which largely terminate development during embryogenesis, plants produce new organs during their whole life cycle. Plant growth and morphogenesis rely on two main meristems that maintain a pool of undifferentiated cells at the shoot (shoot apical meristem, SAM) and the root (root apical meristem, RAM) tips (Sarkar et al., 2007; Sablowski, 2011).
The root of Arabidopsis thaliana (Arabidopsis) has become a useful system to address the molecular genetic components of the networks underlying the modulation of cell proliferation and differentiation during development (Moubayidin et al., 2010). Within the root stem-cell niche (SCN), the stem or initial cells surrounding the organizer or quiescent centre (QC) eventually give rise to the cells of the differentiated tissues of the root, which from its outermost to inner layers are: epidermis, cortex, endodermis, pericycle and vascular cylinder (van den Berg et al., 1997). In addition, columella cells formed in the root apex are differentiated from the initials beneath the QC (Dolan et al., 1993). As the initial cells divide, daughter cells are displaced outside the SCN where they form the proliferation domain (PD) of the RAM, in which cells attain maximum division rates (Ivanov and Dubrovsky, 2013). After several division cycles, cells start dividing at lower rates at the transition domain (TD) within the meristem, and then stop dividing and start to enlarge at the elongation zone (EZ). Progressively, at the differentiation zone (DZ), cells acquire their final size and differentiated cellular features that are characteristic of each root tissue layer (Dolan et al., 1993; Dello Ioio et al., 2007; Ivanov and Dubrovsky, 2013).
Cell proliferation and differentiation are modulated by transcriptional regulatory networks that integrate external and internal signals (Boye and Nordstrom, 2003; Farkas et al., 2006; Slavov and Botstein, 2011). MADS-domain transcription factors have been shown to be key regulators of plant development (Alvarez-Buylla et al., 2000a; Messenguy and Dubois, 2003; Smaczniak et al., 2012), but their role in modulating cell proliferation and differentiation has not been fully addressed. In previous studies we showed that XAANTAL1 (XAL1/AGL12) and XAANTAL2 (XAL2/AGL14) two MADS-box factors, are necessary for normal root development and cell proliferation control (Tapia-Lopez et al., 2008; Garay-Arroyo et al., 2013). xal1 mutants have shorter roots than wild-type plants with fewer meristematic cells and longer cell-cycle duration, resulting in a diminished cell production rate. Moreover xal1 differentiated cells are smaller than in wild-type roots (Tapia-Lopez et al., 2008).
Understanding the specific role of MADS-domain transcription factors and the networks in which they participate in the modulation of proliferation/differentiation requires exploring if they regulate cell-cycle progression. The network underlying the cell-cycle is complex and seems also to be involved in regulating the transition of cells to endoreplication cycles during cell differentiation (Vanstraelen et al., 2009; Fox and Duronio, 2013; Edgar et al., 2014). Cell-cycle progression is regulated by cyclin-dependent kinases (CDKs), which associate with cyclins (CYCs) that confer substrate specificity (Lim and Kaldis, 2013). Different CDK/CYC complexes act throughout the cell cycle. The CDKA/CYCD complexes trigger the G1–S phase transition. After DNA replication during G2, CDKA and CDKB associated with A- and B-type CYCs induce G2/M transition and then, at later stages of the M phase, CYCA and CYCB must be degraded by APC/C complex to exit mitosis (Menges et al., 2005; Eloy et al., 2011).
When RAM cells exit from the proliferative mitotic cycle to the EZ, plant cells may enter into endoreplicative cycles during which their DNA content and cell size increase as a result of DNA synthesis without mitotic cell division (Kondorosi et al., 2000; Inze and De Veylder, 2006; De Veylder et al., 2011; Edgar et al., 2014). Endoreplication is in part induced by inhibition of the activity of mitotic CDK–CYC activity by Kip-related proteins (KRPs) or SIAMESE (SIM) proteins (Morgan, 1997; Walker et al., 2000; Schnittger et al., 2002; Verkest et al., 2005; Boudolf et al., 2009).
Here we show that XAL1 is necessary to promote the transition to differentiation during root development as meristematic cells in the xal1-2 mutant prematurely transit to the EZ and next to the DZ but are unable to reach the final size of wild-type cells, probably because they attain a limited number of endoreplication cycles at the TD–EZ. We also found, in accordance with our previous data in which the mutant xal1 showed a longer cell-cycle (Tapia-Lopez et al., 2008), that XAL1 positively regulates CYCD3;1, CYCA2;3, CYCB1;1, CDKB1;1 and CHROMATIN LICENSING AND DNA REPLICATION FACTOR 1 (CDT1a; Castellano et al., 2004) expression, as well as PLETHORA1 (PLT1; Aida et al., 2004). XAL1 overexpression lines show a higher number of meristematic cells. Interestingly, some plants of these lines also showed altered SCN with abnormal cell divisions under and at the QC. Hence, XAL1 seems to be an important component of a network that underlies the modulation of cell proliferation/differentiation in root development, and is responsible for regulating some components of the mitotic and endoreplicative cycles (Ishida et al., 2009). Furthermore, our results suggest that such a network is involved in both SCN maintenance and apical–basal root development zonation (proliferation, elongation and differentiation) in Arabidopsis. This network also involves PLT1 (Aida et al., 2004; Galinha et al., 2007; Mahonen et al., 2014). Alternatively, XAL1 could be an important link between the two networks.
MATERIALS AND METHODS
Plant materials and growth conditions
The Arabidopsis plants used are Columbia-0 ecotype, except for the PLT1::GUS (Aida et al., 2004) reporter line which is in WS background. Seedlings were grown on vertical plates with 0·2× Murashige Skoog (Murashige and Skoog, 1962) salts (MP Biomedicals) and 1 % sucrose, as described by Tapia-Lopez et al. (2008).
XAL1 overexpression lines
XAL1 cDNA was amplified from the pSR102 clone (Rounsley et al., 1995) with the primers CB5F (5′-CGGATCCTCTATGGCTCGTGGAAAGATTCA-3′) and CB6R (5′-CCGGATCCGCTAGAACTGAAATATTTCAC-3′) that include BamH1 restriction sites. This DNA fragment was transferred to pGEM-T Easy (Promega, Madison, WI, USA) to generate the RT150 plasmid. After sequence confirmation the BamH1 cut fragment was cloned into the pBIN-JIT plasmid. Plants were transformed via Agrobacterium tumefaciens by floral deeping and plants resistant to kanamycin were selected.
RNA extraction and RT-qPCR
Plants were grown for 5 d post-germination (dpg) and roots from three independent biological replicates (25 plants each) were collected. Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and it was reverse-transcribed using Superscript II (Invitrogen). We amplified PDF2 (AT1G13320) and UPL7 (AT1G13320) as housekeeping control genes (Czechowski et al., 2005), and their stability across the compared samples was confirmed using NormFinder (http://moma.dk/normfinder-software; Vandesompele et al., 2002). Amplification efficiencies were analysed using real-time PCR Miner (Zhao and Fernald, 2005), and relative expression was calculated as in Perez-Ruiz et al. (2015). Primer sequences are included in Supplementary Data Table S1.
Microscopy
GUS-stained roots were cleared with Herr solution (Herr, 1971) and visualized by Nomarsky microscopy in an Olympus BX60. For quantification of CYCB1;1::GUS-stained cells, wild-type, xal1-2 and XAL1-OE 5.2.5 plants were grown for 1, 3, 5 and 7 dpg and 15 roots for each genotype were mounted on slides and all the spots visualized along the cortex tissue file were counted and used for further statistical analysis (Hacham et al., 2011). Root meristem organization was visualized using an Olympus FV1000 confocal microscope after roots were fixed and stained using the pseudo-Schiff protocol (Napsucialy-Mendivil et al., 2014).
Quantitative analysis of cellular parameters
For quantitative cellular analysis, roots were mounted in 30 % chloral hydrate and examined with Nomarsky optics. Measurements were performed according to Ivanov and Dubrovsky (1997) and Tapia-Lopez et al. (2008). Cell size profiles along the apical–basal axis of the root were obtained by cell size measurements along the cortex file of the root from QC to the adjacent cell of the first primordial hair cell.
Flow cytometry analysis
Arabidopsis complete roots of 1 and 3 dpg and root tips 1 cm long of 6 and 9 dpg were chopped and then incubated in Galbraith’s buffer (Galbraith et al., 1983). The extracts were filtered with nylon filters of 30 μm (Millipore, Billerica, MA, USA), and nuclei were stained with propidium iodide for 10 min and treated with RNAase (10 μg mL–1). Finally 10^000 events were measured, and DNA histograms were generated with the cytometer FACS Calibur package (Becton Dickinson, Franklin Lakes, NJ, USA).
RESULTS
XAL1 is a positive transcriptional regulator of cell-cycle components
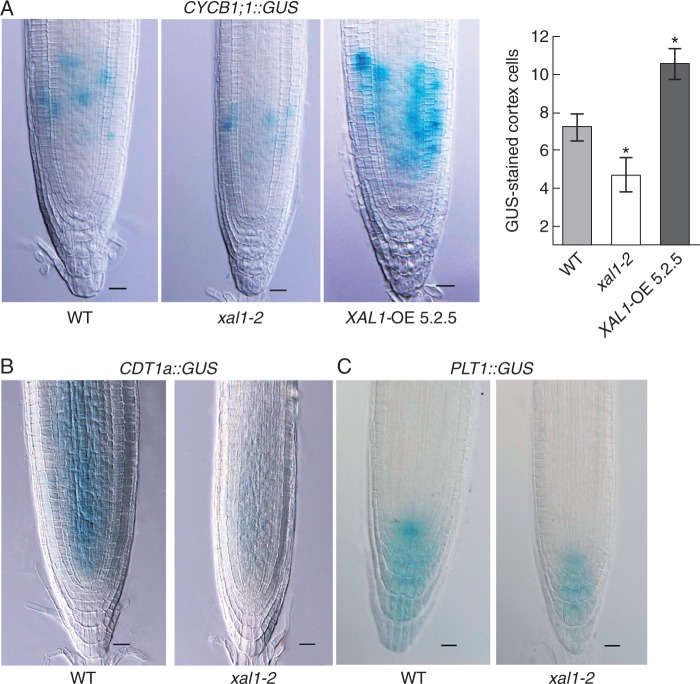
The fact that xal1-2 mutants have fewer root meristem proliferating cells and a longer cell-cycle duration in comparison with wild-type roots (Tapia-Lopez et al., 2008) suggest that XAL1 could be a regulator of cell-cycle components. Hence we assayed mRNA levels of several cell-cycle components in xal1-2 roots in comparison with wild-type (Fig. 1A). We found that cyclin CYCD3;1, which participates in the G1–S transition (Menges et al., 2006), as well as CYCA2;3 and CDKB1;1, which participate in the G2–M transition (Doerner et al., 1996; Menges et al., 2003; Li et al., 2005; Inze and De Veylder, 2006; Boudolf et al., 2009), are significantly down-regulated in xal1-2 in comparison with wild-type roots (Fig. 1B). However, other cell-cycle components as HISH4, CYCA2;1, CDKA and KRP2 are not significantly affected at their mRNA accumulation levels in the mutant background, indicating that XAL1 regulation is specific for some of the cell-cycle components (Fig. 1B). CDKB1;1 interacts with CYCB1;1 to perform its activity (Weingartner et al., 2004). Hence, it is not surprising that CYCB1;1::GUS (Colon-Carmona et al., 1999) was also diminished in the xal1-2 background and up-regulated in the overexpression line (XAL1-OE 5.2.5), in comparison with wild-type roots (Fig. 2A; Supplementary Data Fig. S1). We also found that CDT1a, an essential component for the pre-replication complex during the S phase (Castellano et al., 2004), was down-regulated in xal1-2 mutant compared with wild-type roots (Fig. 2B).
Fig. 1.
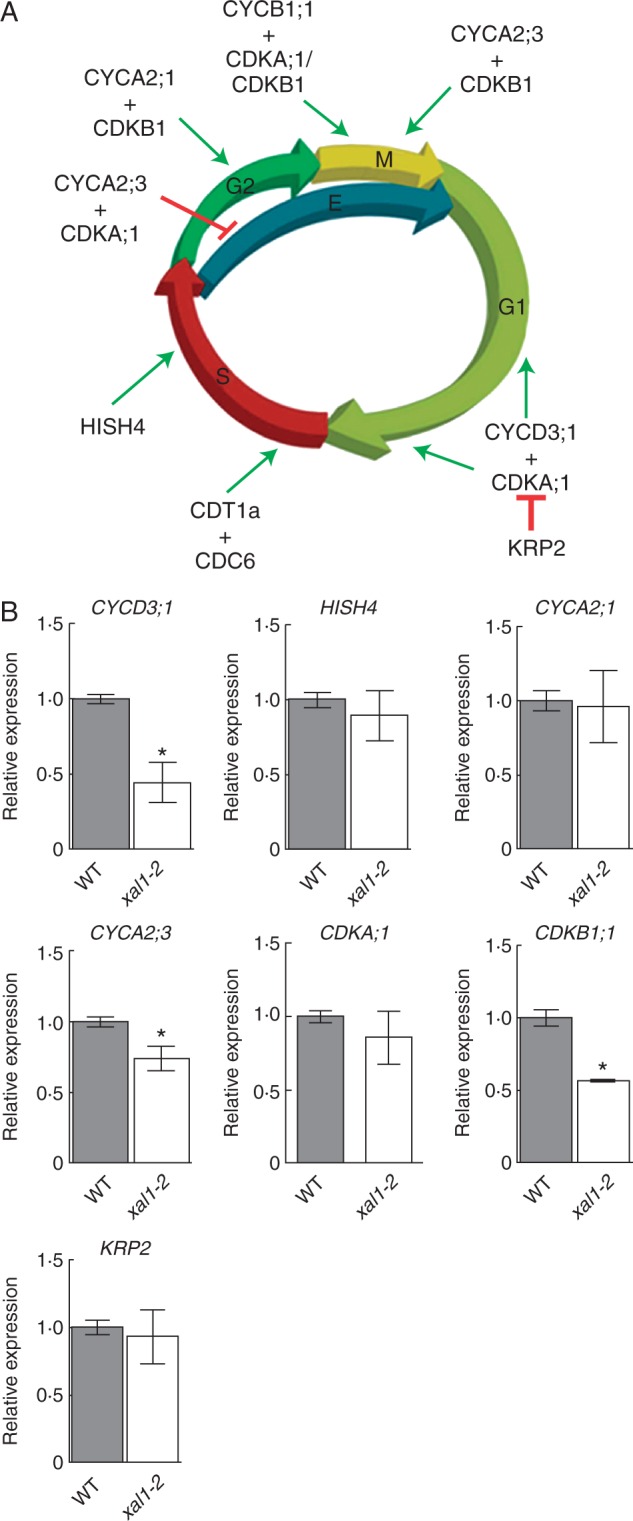
XAL1 is necessary for CYCD3;1, CYCA2;3 and CDKB1;1 transcriptional regulation. (A) Schematic representation of the participation of some genes in Arabidopsis cell-cycle transitions. The CYCD3;1/CDKA1 complex triggers G1–S phase by phosphorylation of RB-E2F pathway (not shown) and is essential for cell proliferation. This complex could be inhibited by KRP2 depending on hormonal conditions. CDT1a and CDC6 form the pre-replication complex performing in S phase. During G2 phase, cyclins CYCA2;3, CYCB1;1 and CYCB1;4 are associated with other CDK types (A or B), promoting transition to G2–M and their regulation is important for repressing the endoreplicative cycle. (B) Relative expression levels of some cell-cycle components in xal1-2 compared with wild-type (WT) roots at 5 dpg, showing that CYCD3;1, CYCA2;3 and CDKB1;1 are down-regulated in the mutant. Relative expression data are expressed as the mean ± s.d. and statistically significant differences from WT (*P < 0·05) were obtained by the Kruskal–Wallis test.
Fig. 2.
XAL1 positively regulates CYCB1, CDT1 and PLT1 in the RAM. (A) Lower and higher levels of pCYCB1;1::GUS expression in xal1-2 roots and XAL1-OE 5.2.5 respectively, compared with WT. The number of GUS-stained cortex cells is shown on the right. Data correspond to mean ± s.e. and statistically significant (*) differences from WT (P < 0·05) were determined with the Kruskal–Wallis test. (B, C) Expression of pCDT1a::GUS (B) and PLT1::GUS (C) is diminished in xal1-2 roots compared with WT roots. All plants are 5 dpg, n = 10 per line. Scale bars = 20 µm.
The transcription factors PLT1 and PLT2 are important for meristem function in a dose–response fashion in response to auxins regulating proliferation and endocycle onset (Aida et al., 2004; Galinha et al., 2007; Ishida et al., 2009; Zhou et al., 2011). XAL1 is also induced by auxin (Tapia-Lopez et al., 2008). Therefore, we tested if PLT1 is altered in xal1-2 roots. We found that PLT1::GUS (Aida et al., 2004) is down-regulated in xal1-2 roots in comparison with the wild-type (Fig. 2C), particularly at the QC and initial cells, indicating that XAL1 is a positive regulator of PLT1 as well.
XAL1 overexpression lines have increased cell proliferation in the root SCN
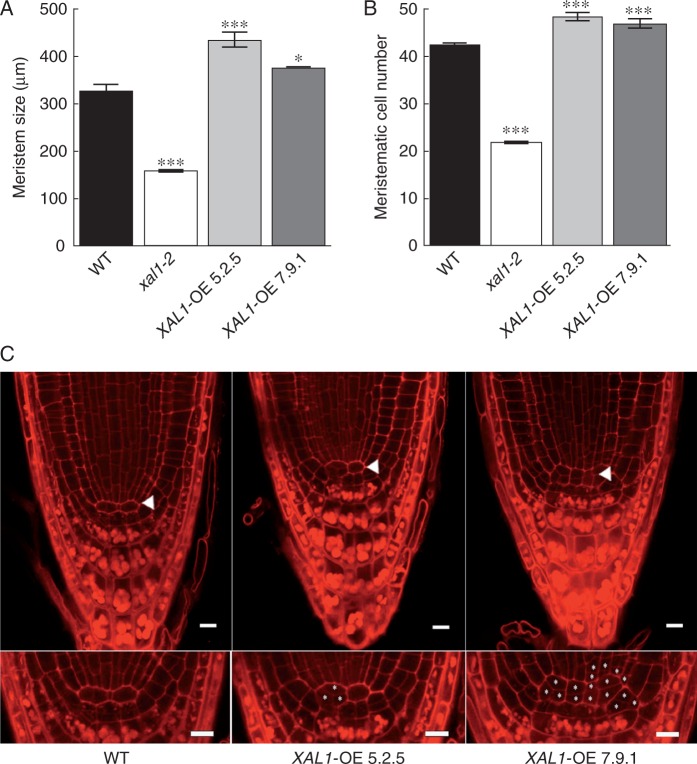
To corroborate the role of XAL1 in the establishment of SCN (Tapia-Lopez et al., 2008), we generated several XAL1 overexpression (OE) lines under the 35S promoter. We analysed seven 100 % kanamycin-resistant OE lines and focused on two of them, XAL1-OE 5.2.5 and XAL1-OE 7.9.1, that showed high XAL1 mRNA expression levels (Supplementary Data Fig. S2). Quantitative cellular analyses of these lines showed that XAL1 overexpression increases the meristem size and the number of meristematic cells at 5 dpg (Fig. 3A, B). In contrast, cell production rate and the final length of cortex cells in these overexpressing lines were similar to wild-type roots (Supplementary Data Fig. S3). Interestingly, over 50 % of the plants of both OE lines showed roots with cellular pattern alterations in the SCN with increased cell divisions (Fig. 3C).
Fig. 3.
Overexpression of XAL1 is sufficient to promote root cell proliferation. (A, B) Meristem size (A) and meristematic cell number (B) are higher in the overexpression lines (XAL1-OE 5.2.5 and XAL1-OE 7.9.1) while xal1-2 has both parameters diminished compared with WT roots at 5 dpg. Data correspond to mean ± s.e. Statistical significance (***P < 0·001, *P < 0·05) was obtained using the Kruskal–Wallis test (n = 30). (C) XAL1-overexpression increases cell divisions at the SCN. Confocal microscopy showing atypical divisions of the QC (arrowheads) and initial cells (labelled cells in the insets) in 50 % of the XAL1-OE 5.2.5 and 7.9.1 plants compared with WT roots. Seedlings of 3 dpg were stained with the pseudo-Schiff technique. n = 35 (WT), 20 (XAL1-OE 5.2.5) and 32 (XAL1-OE 7.9.1) plants. Scale bars = 10 μm.
XAL1 participates in modulating the transition to cell differentiation in roots
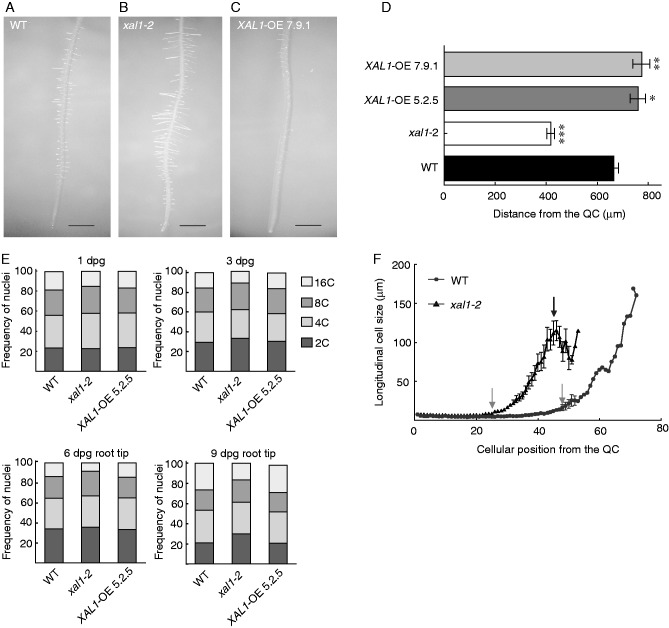
To test if XAL1 is involved in regulation of the cell transition from the RAM to the EZ and DZ, we analysed the distance from the QC at which root hairs, which are clear markers of differentiation (Foreman and Dolan, 2001), first appear in XAL1 loss- and gain-of-function lines. We found that root hairs appeared at shorter distances from the QC in xal1-2 (Fig. 4A, B, D), while these appeared farther away from the QC in the XAL1-OE 7.9.1 line in comparison with wild-type plants (Fig. 4A, C, D). Interestingly, root hairs in the mutant are longer and in the OE line are shorter than in wild-type roots (Fig. 4A–C).
Fig. 4.
XAL1 regulates cell transitions to differentiation in Arabidopsis root. (A–C) xal1-2 (B) has larger root hairs and these appear at shorter distances from the root-tip, while XAL1-OE 7.9.1 (C) has shorter root hairs that first appear at more distant positions with respect to the QC in comparison to WT (A). Roots of 5 dpg plants. Scale bars = 1 cm. (D) Distance to the first root hair from the QC in WT, xal1-2 and both overexpression lines, 5.2.5 and 7.9.1, in 5 dpg seedlings. Data correspond to mean ± s.e. and statistically significant differences (*P < 0·05, **0·01, ***0·001) were obtained with the Kruskal–Wallis test (n = 15). (E) Ploidy distribution of nuclei DNA content of WT, xal1-2 and XAL1-OE 5.2.5 from 1 and 3 dpg whole roots and 6 and 9 dpg root-tips (1 cm long). (F) Cell size profiles of cortex root cells of xal1-2 and WT roots from the QC to the first root hair in 5 dpg roots. Grey arrows show the point at which cells start to elongate, and the black arrow indicates the point at which cortex cells stop growing. The last point in the curve corresponds to the first cell that presents a root hair. Data correspond to mean ± s.e. (n = 20).
In the transition between the RAM and the EZ, cells exit from the mitotic cycle and go into endoreplicative cycles (Vanstraelen et al., 2009). In fact, differentiated cells, such as those with already formed root hairs, show the highest levels of endoreplication in Arabidopsis roots (Abel et al., 1995; Guimil and Dunand, 2007; Takahashi et al., 2013). Hence, we hypothesized that xal1-2 roots, in which cells prematurely transit into the EZ and DZ, should also prematurely enter the endoreplication cycle. We used flow cytometry analysis to determine the ploidy levels found in xal1-2 and the XAL1-OE 5.2.5 line compared with wild-type complete roots at 1 and 3 dpg, and root tips (1 cm long) at 6 and 9 dpg to avoid interference caused by lateral roots at these ages (Fig. 4E). Our data showed that 16C ploidy was reduced in xal1-2 compared with wild-type roots, at 3, 6 and 9 dpg (Fig. 4E). These results suggest that the lack of XAL1 causes diminished proportions of cells with higher ploidy levels, at the same time that it causes a premature differentiation of cells, with root hairs formed at shorter distances to the QC than in wild-type roots (Fig. 4D). Ploidy levels in the XAL1-OE 5.2.5 line are similar to those of wild-type roots (Fig. 4E), although in the former, root hairs appeared farther away from the QC than in wild-type roots (Fig. 4D).
We have previously reported (Tapia-Lopez et al., 2008) and corroborated here (Supplementary Data Fig. S2B) that totally differentiated cells in xal1-2 roots are shorter than wild-type cells, so it is possible that in this mutant, during transition to differentiation, cells are unable to attain normal sizes due to relatively lower ploidy levels (16C) compared with wild-type roots. We measured all the cortex lineage cells from the initial to the first differentiated cell that presented a root hair in xal1-2 and wild-type roots (Fig. 4F). We found that xal1-2 cells started to elongate sooner than wild-type cells, but at the equivalent wild-type TD distance from the QC they stopped growing. These data further suggest that xal1-2 cells transit faster to the elongation and differentiation zones, in comparison with wild-type root cells, but they are unable to endoreplicate at the same rate as in wild-type roots, in correlation with the smaller sizes that they attain in comparison with cells in wild-type roots.
DISCUSSION
We had previously shown that XAL1 is necessary for normal root growth and development by positively regulating cell proliferation rates and cell elongation, and modulating cell-cycle duration in Arabidopsis roots (Tapia-Lopez et al., 2008). Hence, we tested here if XAL1 regulates cell-cycle regulators.
XAL1 regulates some cell-cycle components
We found that expression levels of G1–S factors CYCD3;1 and CDT1a (Menges et al., 2006) and G2–M check-point factors CYCA2;3, CYCB1;1 and CDKB1;1 (Doerner et al., 1996; Menges et al., 2003; Li et al., 2005; Inze and De Veylder, 2006; Boudolf et al., 2009) are significantly diminished in xal1-2 mutant roots in comparison with wild-type (Figs 1 and 2). Also, we found that overexpression of XAL1 results in up-regulation of CYCB1;1::GUS expression (Fig. 2A; Fig. S1). In agreement with these results, xal1 mutant alleles have shorter meristems with fewer meristematic cells and the XAL1-OE lines have a higher meristematic cell number than wild-type plants (Fig. 3A, B; Tapia-López et al., 2008).
CYCD3;1 forms a complex with CDKA;1 to phosphorylate RBR1, which in turn releases it from E2F transcription factors. This process regulates entry to the cell-cycle and it is required for cell-cycle transitions (Nakagami et al., 1999, 2002; Uemukai et al., 2005; Magyar et al., 2012). The triple mutant cycd3;1 cycd3;2 cycd3;3 has smaller organs with fewer cells (Dewitte et al., 2007), thus confirming the role of CYCD3 proteins in cell proliferation. Therefore, it is possible that XAL1, a MADS-box transcription factor, promotes cell proliferation by up-regulating CDKB1;1, CYCB1;1, CYCA2;3 and CYCD3;1 at least. Also, lower levels of CDT1a in xal1-2, a component of the pre-replication complex (Castellano et al., 2004; Sanchez et al., 2012), could explain the longer cell-cycle observed in xal1 root meristem (Tapia-Lopez et al., 2008).
Down-regulation of CYCA2;3, CYCB1;1 and CDKB1;1 in xal1-2 should also favour endocycle entry (Schnittger et al., 2002; Boudolf et al., 2004; Sablowski and Carnier Dornelas, 2014), but as mentioned above, premature enlargement of the meristematic cells does not affect the first rounds of endoreplication. Also we did not find alterations in KRP2 or CDKA;1 mRNA expression levels (Fig. 1). These latter components participate in endocycle/mitosis decisions, indicating that at least their transcriptional regulation is independent of XAL1.
Our results suggest that XAL1 regulates both cell proliferation at the meristem and endocycle maintenance during differentiation and it seems that this transcription factor is one of the interconnecting players of the networks underlying these two processes, or they are both regulated by the same network in which XAL1 participates.
MADS-domain factors seem to share some functions among all eukaryotes as suggested by the high conservation of the DNA-binding domain sequence in all lineages of the MADS-domain protein family (Alvarez-Buylla et al., 2000b). In animals, MEF-related MADS-domain factors, as is XAL1, have been also identified as critical components of the mechanisms involved in cell proliferation/differentiation decisions in myoblasts by regulating E2F (Naya and Olson, 1999).
XAL1 mediates the transition to cell differentiation
We analysed cells transition from the RAM to EZ to the DZ in loss- and gain-of-function lines of XAL1. Cells in the xal1-2 mutant showed premature transition to the elongation and differentiation zones, while this transition was delayed in the XAL1-OE lines (Fig. 4), where we found smaller and larger meristems in the XAL1 loss- and gain-of-function lines, respectively (Fig. 3A, B). Concordantly, we found that in xal1-2, root cells start to elongate and differentiate at positions closer to the QC, in comparison with wild-type roots, and we observed the opposite in the XAL1-OE lines (Fig. 4). These results imply that XAL1 participates in the network that underlies the correct timing at which cells transit to a differentiation state.
Some mutants with short root phenotypes that are involved in DNA replication mechanisms show smaller meristems and differentiated cells similar to xal1, but with overall diminished ploidy levels and an accumulation of 2–4C cells in comparison with wild-type roots (Breuer et al., 2007; Dittmer et al., 2007; Kirik et al., 2007; Zhou et al., 2011). Here we demonstrated premature and delayed transitions to differentiation in the mutant and OE lines of XAL1, respectively. Therefore, we expected to find a correlation with larger proportions of cells with higher ploidy levels in the former and lower in the latter, in agreement with studies that have demonstrated that prematurely differentiated cells present higher ploidy levels in their nuclei (Perilli et al., 2012). Contrary to this expectation, we found lower proportions of 16C in the xal1 mutant and no change in ploidy levels in the OE line compared with wild-type roots (Fig. 4E), thus indicating that XAL1 is necessary for maintaining normal high ploidy levels in the root cells but is not sufficient to alter them. It has been established that there is a correlation between cell size and high ploidy levels (Kondorosi et al., 2000; Sugimoto-Shirasu and Roberts, 2003; Inze and De Veylder, 2006). Our results indicate that cells in xal1-2 start elongating earlier than wild-type cells and as a consequence they differentiate earlier as well, affecting their normal final size (Fig. 4F; Supplementary Data Fig. S3B). Mutants in components of the topoisomerase VI and II are unable to progress into endoreplicative cycles beyond 8C ploidy. Thus, mutants such as rhl1, rhl2, bin4 and mid showed very few root hairs and less developed trichomes than wild-type plants (Sugimoto-Shirasu et al., 2002; Kirik et al., 2007). As endoreplication and higher ploidy levels have been related to cell differentiation as in trichome cells (Walker et al., 2000; Pattanaik et al., 2014), it is surprising that the xal1 mutant with low ploidy levels is able to develop root hairs that are rather longer, and not shorter, than wild-type roots. Also, it was unexpected that in the OE lines ploidy levels were not significantly different from those observed in wild-type roots, although as XAL1 is a MADS-domain transcriptional factor it is possible that its function requires the participation of additional MADS-domain proteins. This is the case in floral organ development during which the overexpression of single MADS-domain proteins is not sufficient to cause the conversion of leaves into floral organs (Honma and Goto, 2001). Overexpression of XAL1 is probably not sufficient to alter cell growth and endoreplication.
In conclusion, the data presented in this study suggest that XAL1 participates in the temporal pattern of cell-fate decisions, particularly during the elongation/differentiation transition, probably by affecting endoreplication maintenance and compromising the final cell size of cortical cells (Tapia-Lopez et al., 2008).
Several signals including plant hormones alter root development, particularly proliferation profiles along the root, and the rate and distance from the QC at which cells transit from the proliferation to elongation/differentiation zones are affected by auxin and cytokinin concentrations, at least (Dello Ioio et al., 2007, 2008; Moubayidin et al., 2010; Takahashi et al., 2013). Indeed, auxins and cytokinins have been associated with all or most of these phenotypes in Arabidopsis root development (Dello Ioio et al., 2008; Moubayidin et al., 2010; Su et al., 2011). The xal1 mutant and XAL1-OE lines have similar phenotypes to those described above. We also know that XAL1 is positively regulated by auxins (Tapia-Lopez et al., 2008) and the PLT1 gene is induced by XAL1 (Fig. 2C). Hence, these hormone networks and the XAL1 regulatory module are probably interconnected and together underlie proper root development.
XAL1 participates in cell division regulation at the SCN
The XAL1 loss-of-function mutant (Tapia-Lopez et al., 2008) and OE lines presented abnormal stem-cell proliferation (Fig. 3). Therefore, it is likely that XAL1 is a component of the complex regulatory network that underlies stem-cell divisions in the Arabidopsis roots. Among other transcription factors, the PLT genes are important components of such a network (Azpeitia et al., 2013; Davila-Velderrain et al., 2014a, b). PLTs are necessary for stem-cell maintenance and cell division in the root (Aida et al., 2004; Galinha et al., 2007) and also their proteins accumulation level define the location of developmental zones (proliferation, elongation and differentiation) (Mahonen et al., 2014). Interestingly, here we have shown that XAL1 is a positive regulator of PLT1 (Fig. 2C). PLT1 is an AP2 transcription factor family member that regulates CYCB1;1 transcription during root development (Aida et al., 2004). PLT1 presents nine CArG boxes in its regulatory region that suggest that XAL1 could directly bind the PLT1 promoter [plant cis-acting regulatory DNA elements (PLACE), http://www.dna.affrc.go.jp/PLACE/]. Interestingly, the plt1 plt2 double mutant has reduced cell division rate in the root meristem and also shows diminished levels of CYCB1;1::GUS expression (Galinha et al., 2007), as was found for xal1-2. Furthermore, in a recent genomic study that inferred regulatory networks for Arabidopsis root from available microarray data, we found interactions between XAL1 and PLT genes (Chavez Montes et al., 2014).
In conclusion, our study further documents that XAL1 is an important component of the network underlying cell proliferation and elongation/differentiation transitions and overall cell proliferation at the root stem-cell niche. Moreover, XAL1 participates in the maintenance of cell endoreplication. The results presented here for this MADS-box factor together with previous studies (Tapia-Lopez et al., 2008; Chavez Montes et al., 2014) suggest that the regulatory networks in which this, and probably other MADS-domain proteins participate, underlie all such dynamics. The role of XAL1 in cell proliferation, cell-cycle duration, cell transition to elongation/differentiation and endocycle performance may be explained, at least in part, by its regulatory role of some relevant cell-cycle components.
SUPPLEMENTARY DATA
Supplementary information is available online at www.aob.oxfordjournal.org and consist of the following. Fig. S1: pCYCB1;1::GUS mitosis marker in WT, xal1-2 and XAL1-OE 5.2.5 line at 1, 3 and 7 dpg. Fig. S2: XAL1 mRNA accumulation in XAL1-overexpression lines. Fig. S3: Cell production rate and mature cortical cell length in XAL1 mutant and overexpression lines. Table S1: Primer sequences used for quantitative and semi-quantitative RT-PCR.
ACKNOWLEDGEMENTS
This work constitutes a partial fulfilment of the Graduate Program ‘Doctorado en Ciencias Biomédicas’ of the Universidad Nacional Autónoma de México in which Karla V. García-Cruz developed this project. We acknowledge the Consejo Nacional de Ciencia y Tecnología (CONACYT, México) that provided her scholarship. This work was supported by CONACYT: 240180 and 180380; PAPIIT, UNAM: IN203214-3; IN203113-3; IN203814-3; IN211516 and BFU2012-34821 from MINECO (Spain) to C.G., and an institutional grant from Fundación Ramón Areces to Centro de Biología Molecular Severo Ochoa. We thank David Cruz Sánchez and Diana Romo for technical and logistical support, respectively.
LITERATURE CITED
- Abel S, Nguyen MD, Theologis A. 1995. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. Journal of Molecular Biology 251: 533–549. [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, et al. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, et al. 2000a. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. The Plant Journal 24: 457-66. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, et al. 2000b. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proceedings of the National Academy of Sciences USA 97: 5328-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpeitia E, Weinstein N, Benitez M, Mendoza L, Alvarez-Buylla ER. 2013. Finding missing interactions of the Arabidopsis thaliana root stem cell niche gene regulatory network. Frontiers in Plant Science 4: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, et al. 2004. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. The Plant Cell 16: 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, et al. 2009. CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiology 150: 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E, Nordstrom K. 2003. Coupling the cell cycle to cell growth. EMBO Reports 4: 757–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Stacey NJ, West CE, et al. 2007. BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell 19: 3655–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, Boniotti MB, Caro E, Schnittger A, Gutierrez C. 2004. DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. The Plant Cell 16: 2380–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez Montes RA, Coello G, Gonzalez-Aguilera KL, Marsch-Martinez N, de Folter S, Alvarez-Buylla ER. 2014. ARACNe-based inference, using curated microarray data, of Arabidopsis thaliana root transcriptional regulatory networks. BMC Plant Biology 14: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P. 1999. Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant Journal 20: 503–508. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila-Velderrain J, Martínez-García JC, Alvarez-Buylla ER. 2014a. Epigenetic landscape models: the post genomic era. bioRxiv 004192; doi: http://dx.doi.org/10.1101/004192. [Google Scholar]
- Davila-Velderrain J, Servin-Marquez A, Alvarez-Buylla ER. 2014b. Molecular evolution constraints in the floral organ specification gene regulatory network module across 18 angiosperm genomes. Molecular Biology and Evolution 31: 560–573. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A. 2011. Molecular control and function of endoreplication in development and physiology. Trends in Plant Science 16: 624–634. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, et al. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, et al. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, et al. 2007. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proceedings of the National Academy of Sciences USA 104: 14537-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Rubin SM. 2013. Molecular mechanisms underlying RB protein function. Nature Reviews Molecular Cell Biology 14: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. 2007. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. The Plant Cell 19: 2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner P, Jorgensen JE, You R, Steppuhn J, Lamb C. 1996. Control of root growth and development by cyclin expression. Nature 380: 520–523. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, et al. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C. 2014. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nature Reviews Molecular Cell Biology 15: 197–210. [DOI] [PubMed] [Google Scholar]
- Eloy NB, de Freitas Lima M, Van Damme D, et al. 2011. The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. The Plant Journal 68: 351–563. [DOI] [PubMed] [Google Scholar]
- Farkas IJ, Wu C, Chennubhotla C, Bahar I, Oltvai ZN. 2006. Topological basis of signal integration in the transcriptional-regulatory network of the yeast, Saccharomyces cerevisiae. BMC Bioinformatics 7: 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Dolan L. 2001. Root hairs as a model system for studying plant cell growth. Annals of Botany 88: 1–7. [Google Scholar]
- Fox DT, Duronio RJ. 2013. Endoreplication and polyploidy: insights into development and disease. Development 140: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 3: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, et al. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Ortiz-Moreno E, de la Paz Sanchez M, et al. 2013. The MADS transcription factor XAL2/AGL14 modulates auxin transport during Arabidopsis root development by regulating PIN expression. EMBO Journal 32: 2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimil S, Dunand C. 2007. Cell growth and differentiation in Arabidopsis epidermal cells. Journal of Experimental Botany 58: 3829–3840. [DOI] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, et al. 2011. Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr JM. 1971. A new clearing-squash technique for the study of ovule development in angiosperms. American Journal of Botany 58: 785–790. [Google Scholar]
- Honma T, Goto K. 2001. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529. [DOI] [PubMed] [Google Scholar]
- Inze D, De Veylder L. 2006. Cell cycle regulation in plant development. Annual Review of Genetics 40: 77–105. [DOI] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, et al. 2009. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov V, Dubrovsky J. 1997. Estimation of the cell cycle duration in the root apical meristem: model of linkage between cell-cycle, rate of cell production and rate of root growth. International Journal of Plant Sciences 158: 757–763. [Google Scholar]
- Ivanov VB, Dubrovsky JG. 2013. Longitudinal zonation pattern in plant roots: conflicts and solutions. Trends in Plant Science 18: 237–243. [DOI] [PubMed] [Google Scholar]
- Kirik V, Schrader A, Uhrig JF, Hulskamp M. 2007. MIDGET unravels functions of the Arabidopsis topoisomerase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. The Plant Cell 19: 3100–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E, Roudier F, Gendreau E. 2000. Plant cell-size control: growing by ploidy? Current Opinions in Plant Biology 3: 488–492. [DOI] [PubMed] [Google Scholar]
- Lempe J, Lachowiec J, Sullivan AM, Queitsch C. 2013. Molecular mechanisms of robustness in plants. Current Opinions in Plant Biology 16: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P. 2005. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proceedings of the National Academy of Sciences USA 102: 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Kaldis P. 2013. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 140: 3079–3093. [DOI] [PubMed] [Google Scholar]
- Magyar Z, Horvath B, Khan S, et al. 2012. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO Journal 31: 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen AP, ten Tusscher K, Siligato R, et al. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA. 2003. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Molecular Biology 53: 423–442. [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JA. 2005. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. The Plant Journal 41: 546–566. [DOI] [PubMed] [Google Scholar]
- Menges M, Samland AK, Planchais S, Murray JA. 2006. The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. The Plant Cell 18: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E. 2003. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316: 1–21. [DOI] [PubMed] [Google Scholar]
- Morgan DO. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annul Review of Cell Development Biology 13: 261–91. [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Current Biology 20: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15: 473–497. [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A. 1999. Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. The Plant Journal 18: 243–252. [DOI] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A. 2002. Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. The Plant Cell 14: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napsucialy-Mendivil S, Alvarez-Venegas R, Shishkova S, Dubrovsky JG. 2014. Arabidopsis homolog of trithorax1 (ATX1) is required for cell production, patterning, and morphogenesis in root development. Journal of Experimental Botany 65: 6373–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Olson E. 1999. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Current Opinion in Cell Biology 11: 683–688. [DOI] [PubMed] [Google Scholar]
- Pattanaik S, Patra B, Singh SK, Yuan L. 2014. An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Frontiers in Plant Science 5: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ruiz RV, Garcia-Ponce B, Marsch-Martinez N, et al. 2015. XAANTAL2 (AGL14) is an important component of the complex gene regulatory network that underlies Arabidopsis shoot apical meristem transitions. Molecular Plant 8: 796–813. [DOI] [PubMed] [Google Scholar]
- Perilli S, Di Mambro R, Sabatini S. 2012. Growth and development of the root apical meristem. Current Opinions in Plant Biology 15: 17–23. [DOI] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF. 1995. Diverse roles for MADS box genes in Arabidopsis development. The Plant Cell 7: 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. 2011. Plant stem cell niches: from signalling to execution. Current Opinions in Plant Biology 14: 4–9. [DOI] [PubMed] [Google Scholar]
- Sablowski R, Carnier Dornelas M. 2014. Interplay between cell growth and cell cycle in plants. Journal of Experimental Botany 65: 2703–2714. [DOI] [PubMed] [Google Scholar]
- Sanchez ML, Costas C, Sequeira-Mendes J, Gutierrez C. 2012. Regulating DNA replication in plants. Cold Spring Harbor Perspectives in Biology 4: a010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, et al. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. [DOI] [PubMed] [Google Scholar]
- Schnittger A, Schobinger U, Stierhof YD, Hulskamp M. 2002. Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Current Biology 12: 415–420. [DOI] [PubMed] [Google Scholar]
- Slavov N, Botstein D. 2011. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Molecular Biology of the Cell 22: 1997–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Angenent GC, Kaufmann K. 2012. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139: 3081–3098. [DOI] [PubMed] [Google Scholar]
- Su YH, Liu YB, Zhang XS. 2011. Auxin-cytokinin interaction regulates meristem development. Molecular Plant 4: 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. 2003. “Big it up”: endoreduplication and cell-size control in plants. Current Opinions in Plant Biology 6: 544–553. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC. 2002. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Current Biology 12: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kajihara T, Okamura C, et al. 2013. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Current Biology 23: 1812–1817. [DOI] [PubMed] [Google Scholar]
- Tapia-Lopez R, Garcia-Ponce B, Dubrovsky JG, et al. 2008. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiology 146: 1182–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemukai K, Iwakawa H, Kosugi S, et al. 2005. Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: cyclin D expression overcomes this repressor activity. Plant Molecular Biology 57: 83–100. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, et al. 2009. APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proceedings of the National Academy of Sciences USA 106: 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, et al. 2005. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. The Plant Cell 17: 1723–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC. 2000. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127: 3931–3940. [DOI] [PubMed] [Google Scholar]
- Weingartner M, Criqui MC, Meszaros T, et al. 2004. Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell 16: 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. 2005. Comprehensive algorithm for quantitative real-time polymerase chain reaction. Journal of Computational Biology 12: 1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li Q, Chen X, et al. 2011. The Arabidopsis RETARDED ROOT GROWTH gene encodes a mitochondria-localized protein that is required for cell division in the root meristem. Plant Physiology 157: 1793–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.