Abstract
Background and Aims Progress has been made in understanding the physiological and molecular basis of root response to mechanical stress, especially in the model plant Arabidopsis thaliana, in which bending causes the initiation of lateral root primordia toward the convex side of the bent root. In the case of woody roots, it has been reported that mechanical stress induces an asymmetric distribution of lateral roots and reaction wood formation, but the mechanisms underlying these responses are largely unknown. In the present work, the hypothesis was tested that bending could determine an asymmetric response in the two sides of the main root axis as cells are stretched on the convex side and compressed on the concave side.
Methods Woody taproots of 20 seedlings were bent to an angle of 90° using a steel net. Changes in the anatomy, lignin and phytohormone content and proteome expression in the two sides of the bent root were analysed; anatomical changes, including dissimilarities and similarities to those found in poplar bent woody stem, were also considered.
Key Results Compression forces at the concave side of poplar root induced the formation of reaction wood which presented a high lignin content and was associated with the induction of cambium cell activity. Auxin seemed to be the main hormone triggering lignin deposition and cell wall strengthening in the concave sides. Abscisic acid appeared to function in the water stress response induced by xylem structures and/or osmotic alterations in the compression sides, whereas gibberellins may control cell elongation and gravitropisms.
Conclusions Poplar root reaction wood showed characteristics different from those produced in bent stem. Besides providing biomechanical functions, a bent root ensures water uptake and transport in the deforming condition induced by tension and compression forces by two different strategies: an increase in xylem thickness in the compressed side, and lateral root formation in the tension side.
Keywords: Mechanical stress, phytohormones, proteomics, reaction wood, root anatomy
INTRODUCTION
Roots are important to plants as they perform fundamental functions, such as nutrient and water uptake, anchorage and mechanical support. Plants exhibit a remarkable root plasticity (reviewed in Hodge et al., 2009), whereby root undergo morphogenetic changes to react and/or to adapt to environmental stress conditions (Di Iorio et al., 2008; Montagnoli et al., 2012a, b, 2014; Foti et al., 2014).
Several factors, including alteration of gravity direction, touch, wind and bending, occurring in the environment may induce a mechanical stress condition and strongly affect plant stability.
Progress has been made in the understanding of the physiological, molecular and biochemical basis of root response to mechanical stress, especially in the herbaceous model plant Arabidopsis thaliana (Ditegou et al., 2008; Monshausen et al., 2009; Richter et al., 2009). Ditengou et al. (2008) and Richter et al. (2009) demonstrated that bending of arabidobsis roots causes the initiation of lateral root primordia towards the convex side of the bent rozot. It has been proposed that curve-related lateral root formation reflects differential dynamics of auxin transport/uptake in stretched and compressed cells (Laskowski et al., 2008), and that tension forces in the cell wall and/or the plasma membrane on the convex side of the bent root may trigger Ca2+ changes in the pericycle (Monshausen et al., 2009). However, despite this recent progress, the precise signalling/response pathways and molecular factors directing lateral root production to the convex side of curving roots remain to be determined in annual plants, whereas in the case of woody roots the regulatory mechanism underlying mechanical stress response is almost totally unknown.
In the root of a woody plant species such as Fraxinus ornus, Chiatante et al. (2007) observed that bending induces the emission of new lateral roots and the formation of a small amount of reaction wood. Reaction wood is formed as part of a developmental process, important to re-orient plant growth (Timell, 1986; Zobel and van Buijtenen, 1989; Plomion et al., 2001). In the stem of softwood species, reaction wood is generally called compression wood (CW) as it often appears in localized zones of the tree held in compression (the underside of a leaning stem). CW is highly lignified and contains less cellulose than normal wood; furthermore, tracheid length is reduced, the cell cross-sectional profile is rounder and the intercellular spaces are larger than in normal wood. In hardwood species, reaction wood is called tension wood (TW) as it tends to form in zones of the tree held in tension (the upper side of a leaning stem). The morphology, anatomy and ultrastructure of TW have been extensively studied (reviewed by Ruelle et al., 2014), and it is well known that TW is less lignified, has more longitudinally oriented cellulose microfibrils, and higher cellulose crystallinity and content than normal wood. Furthermore, the cell wall structure of TW exhibits the formation of a specialized gelatinous wall layer which has been proposed to be a low-cost, efficient strategy for the fast generation of tensile stress in broadleaved trees (Abedini et al., 2015). The wood produced on the side of a stem or branch opposite to the reaction wood is named opposite wood (OW) and is characterized by properties intermediate between normal and reaction wood (Timell, 1986). A large number of investigations have been carried out on TW formation in response to natural or artificial bending, providing important insights into the alteration of cell wall- and hormone-related genes (Déjardin et al., 2004; Lafarguette et al., 2004; Andersson-Gunneras et al., 2006). Data are also reported in the literature on signalling pathways involving post-translation protein regulation (Mauriat et al., 2015) and hormones (Andersson-Gunnerås et al., 2003; Israelsson et al., 2005; Gerttula et al., 2015). Despite this abundance of information regarding the stem, the exact mechanisms involved in the response of woody root to mechanical stress still remain to be investigated.
In our previous studies, we have established that bending stress in poplar woody root induces specific responses in three different regions, namely the above bending sector (ABS), the bending sector (BS) and the below bending sector (BBS), which are subjected to different intensities of tension and compression forces and different directions of gravity. Indeed, we observed that in these three bending sectors lateral root emission and reaction wood formation are temporally and spatially modulated by a complex interplay among different signal transduction pathways involving reactive oxygen species (ROS), hormones and specific molecular factors regulating lignin deposition, cell wall integrity and lateral root formation (Trupiano et al., 2012a, b, 2013b, 2014; Rossi et al., 2015). However, according to the mechanical force distribution model proposed by Trupiano et al. (2012b), the convex and concave sides of each bent root sector undergo a different distribution of mechanical forces, whereby the convex side is subjected to tension forces, whereas the concave side is subjected to compression forces. As reported in the case of A. thaliana (Ditegou et al., 2008; Monshausen et al., 2009; Richter et al., 2009) and on the basis of our previous work, we hypothesize that in poplar woody taproot, bending may also determine an asymmetric response in convex and concave sides. Indeed, specific signal transduction pathways involving different molecular factors and phytohormone cross-talk could control the asymmetric distribution of lateral roots and reaction wood formation in stretched cells on the convex side and in compressed cells on the concave side. To test this hypothesis and to widen our knowledge on woody root biology, in the present work the response of the convex and concave sides of poplar bent woody taproot were separately analysed in terms of changes in anatomy, lignin and phytohormone content, and protein expression.
MATERIALS AND METHODS
Plant material and simulation of mechanical stress
Taproots of 22-year-old Populus nigra seedlings were subjected to bending stress as described in Scippa et al. (2008) and Trupiano et al. (2012a). Briefly, poplar taproots were tied around steel nets curved at a right angle (bent), while an equal number of taproots were linked to vertical steel nets (C, control). Afterwards, seedlings were grown in a greenhouse for 6 months under a controlled water regime, temperature and natural photoperiod. Taproots of control plants were randomly sampled at 12–27 cm from the base of the root collar zone (equivalent of the stem base), where secondary structure was well developed; each control sample was 5 cm long. In the case of bent roots, three different sectors were sampled, each 5 cm long: (1) the sector just above the bending zone, named the above bending sector (ABS) (12–17 cm distant from the root collar); (2) the sector representing the point of maximum radical bending, named the bending sector (BS) (17–22 cm distant from the root collar); and (3) the sector just below the bending zone, the below bending sector (BBS) (22–27 cm distant from the root collar). For lignin, hormone and protein content analyses, each region (ABS, BS and BBS) was further divided into two parts: the convex and concave sides (Supplementary Data Fig. S1). Sampled taproots from identical sides and sectors (C, ABS convex/concave, BS convex/concave and BBS convex/concave), freed of all laterals, were frozen in liquid N2 and stored at −20 °C until successive analysis.
Root anatomy
Samples of controls and of each sector (ABS, BS and BBS) of the bent root were fixed in formalin–acetic acid–alcohol (FAA, 5:5:90), dehydrated through an ethanol series and embedded using the Technovit 7100 resin system (Heraeus Kulzer, Wehrheim, Germany) based on 2-hydroxyethyl-methacrylate. Root samples were sectioned into cross-sections (12 μm thick) using a sliding microtome. Finally, sections were stained in Toluidine Blue O (Parker et al., 1982) for 1 min. Sections were photographed using a Olympus BX63 light microscope equipped with a Olympus DP72 camera. Images were analysed by ImageJ 1.41o software (Wayne Rasbanb, National Institute of Health, USA). In order to define the convex and the concave sides precisely, a 45° rotated graphic crosswise object was applied, having the centre of the primary xylem stele as the anchor point (Supplementary Data Fig. S2) Measurements were carried out in the areas formed after the application of bending, i.e. the dark and light grey areas containing xylem and phloem, respectively, present in Fig. S2. For each root section (convex and concave sides), the following parameters were measured: (1) cambial cell number, which was calculated considering all cells as having a thin cell wall and a small radial diameter (Morel et al., 2015); (2) vessel wall thickness (μm); (3) fibre wall thickness (μm); (4) relative xylem thickness (%); and (5) relative phloem thickness (%). The latter two parameters represent the thickness of xylem and phloem, respectively, expressed as a percentage of the total root diameter. We also evaluated: (1) relative vessel area (%), corresponding to the vessel area measured in the xylem area analysed divided by the vessel area in the total xylem section; (2) specific vessel area (mm2 mm−2), which is the vessel area measured in the xylem area analysed divided by the xylem area analysed; (3) relative vessel number (number mm−2), corresponding to the number of vessels counted in the xylem area analysed divided by the number of vessels counted in the total xylem section; (4) specific vessel number (number mm−2), which is the number of vessels counted in the xylem area analysed divided by the xylem area analysed; and (5) mean vessel area (mm2), which represents the sum of the vessel areas divided by the number of vessels inside the xylem area analysed. As anatomical data did not follow a normal distribution, non-parametric statistics were applied. The Kruskal–Wallis multiple-comparison test was used to compare anatomical measurements among sectors for each side. The Mann–Whitney U-test was used for pairwise comparison of anatomical measurements between each bent root region and control roots. The latter test was also applied to compare convex and concave sides of each sector in bent roots and control roots. A 95 % significance level was applied to analysis with non-parametric methods. Statistical analysis was carried out using the statistical software package SPSS 17·0 (SPSS Inc., Chicago IL, USA).
Lignin content measurement
Lignin content in control and stressed root sections was measured using the thioglycolic acid method (Doster and Bostock, 1988) with some modifications, as already reported in Trupiano et al. (2012b). Lignin concentration was calculated by measuring the absorbance at 280 nm, using a specific absorbance coefficient of 6·0 L g−1 cm−1; the sample with the highest lignin content was used as a standard to normalize lignin content in other samples. Three biological replicates were used for statistical analysis (P < 0·01).
Hormone extraction and analysis
Indole-3-acetic acid (IAA), abscisic acid (ABA), gibberellins (GAs) and kinetin (Kin) in control and bent stressed root sections were extracted as reported in Trupiano et al. (2012b). Compounds were measured by reversed-phase high-pressure liquid chromatography (HPLC) performed on a Gemini-NX C18 (250 × 4·5 mm, 5 μm particle size) column (Phenomenex, Torrance, CA, USA) bearing a Security Guard® pre-column (Phenomenex), which was eluted with a gradient of acetonitrile containing 0·1 % (v/v) trifluoroacetic acid (solvent B) in aqueous 0·1 % (v/v) trifluoroacetic acid (solvent A), at 45 °C. Solvent B was ramped up from 15 to 30 % over 5 min, from 30 to 50 % over 5 min, from 50 to 80 % over 2 min, and then restored to starting conditions, at a flow rate of 1·5 mL min−1. Compounds were identified based on retention times, UV spectra and literature data using IAA (12886, Sigma Aldrich, Milan, Italy), ABA (A1049, Sigma), GA3 (G7645, Sigma), GA4 (G7276, Sigma) and Kin (K0753, Sigma) standards. Standard compounds were also used to build calibration curves (in the range 5–200 μg mL−1) at specific wavelengths (λIAA = 254 nm; λABA = 254 nm; λGAs = 205 nm; λKin = 269 nm). For quantitative analysis, two different extract amounts from unknown samples were injected in triplicate. Hormone concentration in plant tissues was expressed as μg of hormone per g of fresh tissue. The concentration of GAs was reported as the sum of the GA3 and GA4 content. Four independent extractions were used for statistical analysis (P < 0·05).
RNA extraction and expression analysis of ACO
To measure the 1-aminocyclopropane-1-carboxylate oxidase (ACO) gene expression level in control and stressed root sections, total RNA was extracted from root tissue (0·07 g) using the mirPremier® microRNA Isolation Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Extracted total RNA samples were retro-transcribed using the ImProm-II™ Reverse Transcription System kit (Promega, Madison, WI, USA) and oligo(dT)15 primers. Gene-specific primers were used for amplification of the ACO gene (F5′-TTCAGGTTGAGAACCATGGAC-3′; R5′-GGGATCTTTATCCATCCTCCA-3′). Conditions for reverse transcription–PCR (RT–PCR) analysis were as follows: 95 °C for 4 min; 38 cycles of 45 s at 95 °C, 45 s at 50 °C and 50 s at 72 °C; followed by a final extension of 7 min at 72 °C. PCRs were performed with GoTaq® G2 Flexi DNA Polymerase (Promega) in a 25 μL final volume. Three independent biological replicates were used for each sample. PCR products were separated on a 1·5 % (w/v) agarose gel in 1× TBE (Tris-borate/EDTA) buffer, with three technical replications. Cyclophilin gene expression levels (F5'-GGCTAATTTTGCCGATGAGA-3′; R5′-ACGTCCATCCCTTCAACAAC-3′) were used to normalize data. Images of gels were acquired by ChemiDoc (Bio-Rad Hercules, CA, USA) using Quantity One software (Bio-Rad) and analysed using ImageJ 1.41o software.
Cloning and sequencing
To determine the sequence of the ACO gene evaluated by RT–PCR analysis, a corresponding gel slice was purified using the Wizard® SV Gel and PCR Clean-Up System (Promega) according to the manufacturer’s instructions. The purified product was then analysed and quantified on a 1·5 % agarose gel and subsequently cloned using pGEM®-T Easy Vector System I (Promega) according to the manufacturer’s instructions. The cloning reaction was incubated overnight at 4 °C and then used to transform Escherichia coli chemocompetent cells. Plasmids were isolated from individual colonies and sequenced by BMR genomics (http://www.bmr-genomics.it/). The obtained sequence was matched and identified in the Phytozome V.9 database (http://www.phytozome.org).
Protein extraction and separation
Total proteins of control and stressed root samples were extracted from 2 g of root tissue following a phenol-based protocol (Mihr and Braun, 2003), as previously described by Scippa et al. (2008). The Bradford assay (Bradford, 1976) was used to quantify protein concentration, using bovine serum albumin (BSA) as standard.
For isoelectric focusing (IEF) analysis, immobilized pH gradient (IPG) strips (17 cm; pH 3–10 non-linear; Bio-Rad) were rehydrated overnight with 300 mL of rehydration buffer [6 m urea, 2 % (w/v) CHAPS, 0·5 % (v/v) Triton X-100, 20 mm dithiothreitol (DTT) and 1 % (w/v) carrier ampholytes pH 3–10] and 700 μg of total proteins. IEF was performed in a PROTEAN IEF Cell (Bio-Rad) set up with the following program: (1) 250 V for 90 min in linear mode; (2) 500 V for 90 min in linear mode; (3) 1000 V for 180 min in linear mode; and (4) 8000 V in rapid mode until 56 kVh is reached. After IEF, the IPG strips were equilibrated in 10 mL of equilibration buffer [50 mm Tris–HCl, pH 8·8, 6 m urea, 30 % (w/v) glycerol, 2 % (w/v) SDS] supplemented with 1 % (w/v) DTT for 20 min; then, they were treated with 10 mL of equilibration buffer containing 2·5 % (w/v) iodoacetamide, for 20 min. The latter two treatments allowed the reduction and alkylation of proteins, respectively.
Proteins were separated in the second dimension by 12 % polyacrylamide gel (17 cm × 24 cm × 1 mm) electrophoresis (SDS–PAGE); in detail, analysis was performed in a PROTEAN (Bio-Rad) vertical apparatus containing 25 mm Tris–HCl, pH 8·3, 1·92 m glycine, 1 % (w/v) SDS as running buffer. A constant voltage of 70 V was applied for 16 h, until the dye front reached the bottom of the gel. For each sample, three replicates were run. Finally, separated proteins were fixed by treating gels with 40 % (v/v) methanol, 7 % (v/v) acetic acid, for 30 min, and then visualized by staining with Coomassie Brilliant Blue G-250 (Bio-Rad). Gels were scanned using a GS-800 calibrated densitometer (Bio-Rad); corresponding digital images were recorded and analysed using PDQuest software (Bio-Rad). Finally, statistical analysis was conducted applying a Student’s t-test (P < 0·01). A 2-fold change (<0·5 and >2) of normalized spot densities was considered indicative of differential expression between samples.
In-gel protein digestion, mass spectrometry analysis and identification
Protein spots of interest were excised from gels and triturated. After a washing step with water, proteins were reduced, S-alkylated and digested with trypsin as previously reported (Vascotto et al., 2006). Digest aliquots were removed and subjected to a desalting/concentration step on μZipTipC18 (Millipore Corp., Bedford, MA, USA) using 5 % formic acid/50 % acetonitrile as eluent before matrix-assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF-MS) or nano-liquid chromatography-electrospray ionization-linear ion trap-tandem mass spectrometry (nanoLC-ESI-LIT-MS/MS) analysis. The Mascot software package (Matrix Science, UK) was used to identify spots unambiguously as described in detail in Trupiano et al. (2012a).
RESULTS
Root anatomical analysis
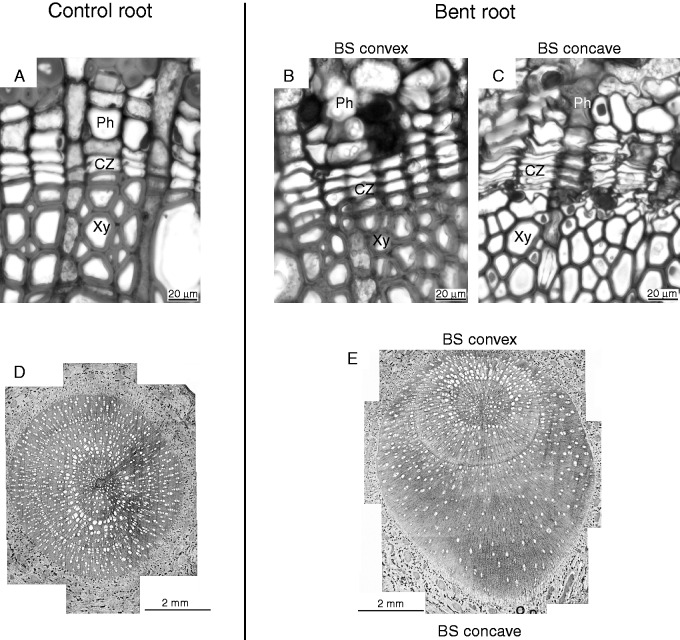
Compared with the control, different intensities of mechanical forces asymmetrically affected wood formation and cambium cell activity in the convex and concave sides of the three bent poplar woody root sectors (Fig. 1A). Indeed, the highest number of cambial cells was determined in BS concave, whereas in BS convex and in both sides of ABS and BBS it was similar to that of control roots (P < 0·05, Table 1). Moreover, quantitative anatomical analysis revealed differences (P < 0·05) in almost all measured traits between convex and concave sides of each bent sector (ABS, BS and BBS). In BS concave, two types of secondary xylem were measured, which were characterized by differences in both fibre and vessel wall thickness (μm). In particular, fibre wall thickness measured in cells in contact with the cambial zone was found to be significantly lower (P < 0·05) than that determined in fibre cells at a distal position (Fig. 1; Table 1). The fibre wall thickness value in BS convex was comparable with that of convex and concave sides of the other two sectors, and with that of control roots, where both vessels and fibres in contact with the cambial zone were fully differentiated (Table 1). Although not significant, vessel wall thickness was smaller in cells in contact with the cambial zone.
Fig. 1.
Photographs of the control and of the convex and concave sides of a bent taproot sector. Root sections were stained in Toluidine Blue O. Cross-section of the cambial zone in control (A), bending sector (BS) convex (B) and BS concave (C) roots. Scale bar = 20 μm. Entire cross-section of control (D) and a bent taproot sector (E); in the case of the bent taproot, the BS was analysed. Scale bar = 2 mm.
Table 1.
Root anatomical analysis
| ABS |
BS |
BBS |
Control |
|||||
|---|---|---|---|---|---|---|---|---|
| Convex side | Concave side | Convex side | Concave side | Convex side | Concave side | Left side | Right side | |
| CCN | 3·8 (0·7)A | 4·9 (1·0)a | 4·5 (0·9)A | 8·1 (1·1)b | 4·0 (0·5)A | 4·8 (0·8)a | 3·4 (0·5)A | 3·5 (0·5)a |
| VWT | 2·7 (0·7)A | 3·0 (0·9)a | 3·2 (0·4)A n.p. | 3·0 (0·3)*a | 2·9 (0·8)A | 3·2 (0·6)a | 3·3 (0·4)A | 3·5 (0·3)a |
| 3·8 (0·8)†a | ||||||||
| FWT | 2·6 (0·8)A | 2·8 (0·8)a | 2·4 (0·5)A n.p. | 0·85 (0·3)b | 2·7 (0·4)A | 2·6 (0·3)a | 3·0 (0·1)A | 2·9 (0·1)a |
| 2·4 (0·5)†a | ||||||||
| RXT | 23·7 (5·8)AB | 36·4 (4·0)b | 16·6 (2·1)B | 55·3 (6·2)c | 24·6 (1·6)A | 33·3 (4·7)ab | 26·1 (1·8)A | 28·8 (1·8)a |
| RPT | 18·2 (4·2)AB | 21·5 (2·8)a | 10·3 (0·7)C | 17·5 (0·3)b | 17·1 (1·6)B | 22·3 (4·7)ab | 22·6 (1·5)A | 22·5 (1·8)a |
| RVA | 1·84 (0·37)AB | 4·48 (0·74)a | 0·62 (0·16)B | 2·65 (1·09)a | 3·32 (0·66)A | 3·49 (0·37)a | 2·94 (0·27)A | 3·49 (0·71)a |
| SVA | 0·30 (0·03)AB | 0·28 (0·02)a | 0·23 (0·03)B | 0·17 (0·03)b | 0·35 (0·01)A | 0·30 (0·03)a | 0·29 (0·03)A | 0·27 (0·01)a |
| RVN | 7·3 (0·8)AB | 12·1 (3·1)a | 5·0 (1·0)B | 7·5 (2·3)a | 10·4 (1·8)A | 11·0 (1·6)a | 10·7 (1·3)A | 12·5 (2·7)a |
| SVN | 126 (23)AB | 82 (4)a | 207 (63)B | 51 (2)b | 111 (7)A | 95 (8)a | 106 (13)A | 99 (6)a |
| MVA | 0·0025 (0·0004)A | 0·0034 (0·0003)a | 0·0016 (0·0008)B | 0·0034 (0·0005)a | 0·0032 (0·0001)A | 0·0032 (0·0002)a | 0·0028 (0·0001)A | 0·0028 (0·0000)a |
Cross-sections of control root and of each sector (ABS, BS and BBS) of the bent root. The following parameters were analysed: CCN, cambial cell number; VWT, vessel wall thickness (μm); FWT, fibre wall thickness (μm); RXT, relative xylem thickness (%); RPT, relative phloem thickness (%); RVA, relative vessel area (%); SVA, specific vessel area (%); RVN, relative vessel number (number of cells mm−2); SVN, specific vessel number (number of cells mm−2); MVA, mean vessel area (mm2). ABS, above bending sector; BS, bending sector; BBS, below bending sector.
*Partially differentiated tissue adjacent to the cambial zone.
†Fully differentiated tissues.
n.p., absence of partially differentiated tissue.
The side of unstressed root corresponding to the concave side of bent taproot was named ‘right’, while that corresponding to the convex side was named ‘left’.
Lower case letters indicate significant differences (Mann–Whitney U-test, P < 0·05) between the right side of control and the concave side of the three bent sectors. Upper case letters indicate significant differences (Mann–Whitney U-test, P < 0·05) between the left side of the control and the convex side of the three bent sectors. Bold values indicate significant differences (Mann–Whitney U-test, P < 0·05) between the convex and concave sides of the same sector. Values marked with the same letter are not statistically significant (Mann–Whitney U-test, P < 0·05).
Significant differences were measured in the relative secondary xylem thickness. In particular, its value was significantly higher in BS concave compared with all other sectors, i.e. almost 2-fold higher than that of the control and >3-fold higher than that of BS convex (Table 1). Compared with the control, relative xylem thickness values were higher in ABS and BBS concave, and similar in ABS and BBS convex. The lowest value compared with all other sectors and with the control root was found in BS convex.
Relative phloem thickness showed the lowest values in both sides of BS, compared with all other sectors and the control root. Moreover, its value was significantly lower in BS convex than in BS concave. In the case of ABS and BBS, there were no differences in relative phloem thickness values between the convex and concave sides (Table 1).
Relative vessel area showed the lowest value in BS convex, whereas specific vessel area presented similar values in both sides for all the three bent sectors (Table 1). Furthermore, the specific vessel area value measured in BS concave was significantly lower than those measured in both ABS and BBS concave and in control roots; it was significantly lower in BS convex compared with BBS convex, but similar in ABS convex and control roots (Table 1).
The relative vessel number in ABS was significantly higher in the concave compared with the convex side; it was significantly lower in BS convex than in BBS convex and control roots, whereas it showed similar values in the concave sides of all bent sectors and control root (Table 1). Only in the case of BS was the specific vessel number significantly higher in the convex than in the concave side. Furthermore, the specific vessel number measured in BS concave was significantly lower than its counterparts measured in the concave side of all bent sectors and control roots. The specific vessel number measured in BS convex was significantly higher than in BBS convex and control roots (Table 1).
The mean vessel area was significantly larger in the concave side compared with the convex side, only in the case of BS. It was significantly smaller in BS convex compared with the convex side of all other bent sectors and control roots. The mean vessel area was similar in the concave side of all bent sectors and control roots (Table 1).
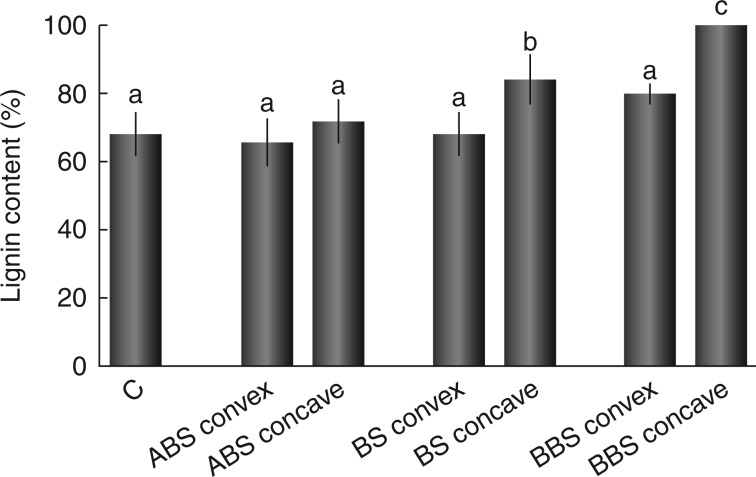
Lignin content
Lignin quantification showed comparable values ( not statistically significant) among control, the concave side of ABS and the convex side of all bent root regions (ABS, BS and BBS) (Fig. 2). In BS and BBS, lignin content was significantly higher (P < 0·01) in the concave sides compared with the corresponding regions of the convex side, as shown in Fig. 2.
Fig. 2.
Lignin content in control and convex and concave sides of three bent sectors. Lignin content is expressed as a percentage of the value measured in BBS concave, considered as 100 %. Data represent the mean of three independent extractions ± s.d. Values marked with the same letter are not statistically significant (t-test, P < 0·01). ABS, above bending sector; BS, bending sector; BBS, below bending sector.
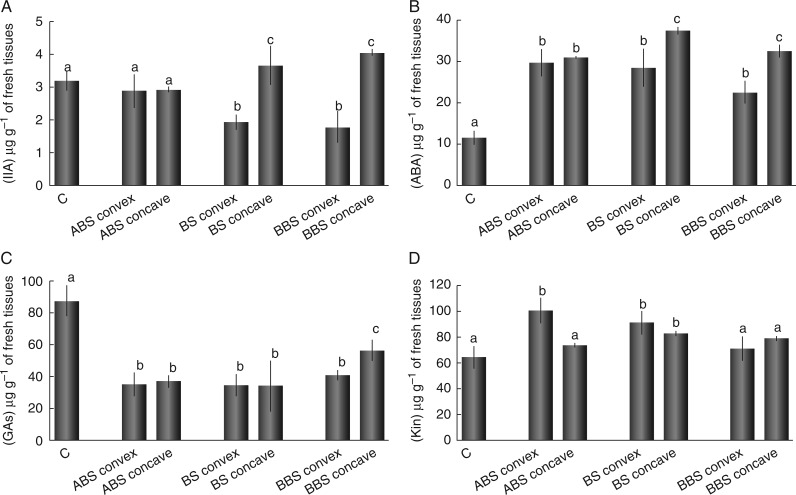
Distribution of hormones along the bent taproot
The content of the main endogenous plant hormones (IAA, ABA, GAs and Kin) measured in the control and in the different portions of bent roots (ABS convex/concave, BS convex/concave and BBS convex/concave) is reported in Fig. 3. IAA showed an asymmetric distribution between the convex and concave sides of BS and BBS; indeed, compared with the control, IAA levels were significantly (P < 0·05) lower in the convex and higher in the concave side, whereas they remained unchanged in both sides of ABS (Fig. 3A). ABA levels were always higher (P < 0·05) in both sides of all the three bent sectors, compared with the control; in BS and BBS they were higher in the concave side than in the corresponding convex side (Fig. 3B). The GA concentrations were significantly lower in both sides of all three bent sectors compared with control roots (Fig. 3C). Differences between convex and concave sides were significant (P < 0·05) only in BBS, with a higher content of these hormones in the concave than in the convex side (Fig. 3C). Kin levels were found to be unchanged in ABS concave and in both sides of BBS, compared with control root, while they were higher (P < 0·05) than the control in ABS convex and in both sides of BS (Fig. 3D). Differences between convex and concave sides were significant (P < 0·05) only in ABS, with a higher content in the convex side.
Fig. 3.
Content of phytohormones in control and convex and concave sides of three bent sectors. Concentrations represent the amount of each phytohormone in root tissues (μg g−1 of fresh weight) as analysed by HPLC. Data represent the mean of four independent extractions ± s.d. Values marked with the same letter are not statistically significant (t-test, P < 0·05). IAA, indole-3-acetic acid; ABA, abscisic acid; GAs, gibberellins (GA3 + GA4); Kin, kinetin. ABS, above bending sector; BS, bending sector; BBS, below bending sector.
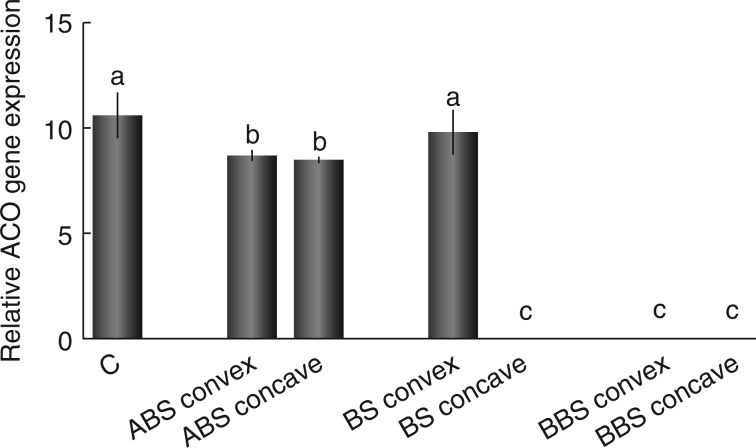
ACO gene expression along the bent taproot
The ACO gene expression level analysed by RT–PCR was used as an indirect indicator of ethylene content in the control and in the different sectors of bent root (ABS convex/concave, BS convex/concave and BBS convex/concave). Data showed a significant reduction of ACO mRNA level (Potri.006G151600; Supplementary Data Fig. S3) in the convex and the concave side of ABS, with respect to the control (P < 0·05); however, no change was observed between the two sides of this region (Fig. 4). Moreover, the ACO expression level was unchanged in BS convex, compared with the control, while it was found to be completely absent in BS concave and BBS convex and concave (Fig. 4).
Fig. 4.
Relative ACO gene expression levels measured in control and in convex and concave sides of three bent sectors. Data were normalized to cyclophilin gene expression values and represent the mean of three independent extractions ± s.d. Values marked with the same letter are not statistically significant (t-test, P < 0·05). ABS, above bending sector; BS, bending sector; BBS, below bending sector.
Proteomic analysis
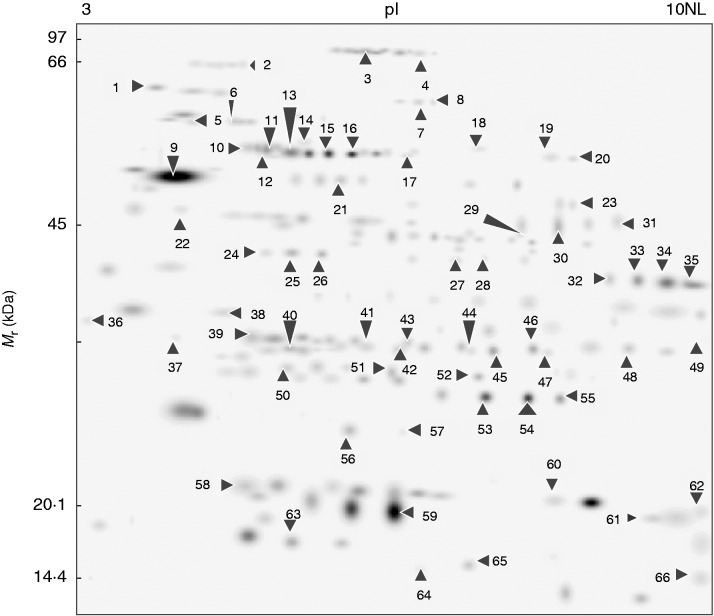
Highly reproducible proteomic 2-D electrophoresis maps were obtained for the control and the convex and concave sides of each bent-stressed region (ABS, BS and BBS), with an average of 197–383 well-resolved spots, ranging in Mr from about 97 to 14·4 kDa (Supplementary Data Fig. S4).
The comparison of the 2-D electrophoresis maps through PDQuest-assisted analysis revealed a total of 66 protein spots differentially expressed (P < 0·01) among all samples, representing 33·5 % of all resolved spots. These protein spots, indicated by arrows in the master gel (Fig. 5), were further identified by MALDI-TOF-MS or nanoLC-ESI-LIT-MS/MS (Table 2). Only one spot (spot 39) was not associated with a protein component.
Fig. 5.
Master gel. Map showing the 66 proteins differentially represented in unstressed, and in convex and concave sides of three bent sectors (ABS, BS and BBS). Arrows indicate the position of each protein spot and corresponding spot ID reported in Table 2.
Table 2.
Differentially-represented proteins in control roots and in concave and convex sides of mechanically stressed poplar woody taproots
| Spot ID | Protein name | NCBI accession | Organism | Theor. pI/Mr (kDa) | Exp. Mr (kDa) | ID method | Unique peptides | Sequence coverage (%) | Mascot score | Annotated NCBI accession with the highest BLAST score (% sequence identity) | Protein name | Function |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Predicted protein | 222846818 | P. trichocarpa | 4·80/65 | 63 | TMS | 2 | 4 | 91 | 255537297 (100) | Ara4-interacting protein | Intracellular traffic |
| 2 | Predicted protein | 224121826 | P. trichocarpa | 5·16/90 | 67 | PMF | 14 | 26 | 177 | 108706222 (96) | Cell division cycle protein 48 | Cell growth/division/development |
| 3 | Putative dehydrin | 29120045 | P. canadensis | 6·12/69 | 85 | TMS | 15 | 16 | 535 | Disease/defence | ||
| 4 | Putative dehydrin | 29120045 | P. canadensis | 6·12/69 | 83 | TMS | 7 | 7 | 209 | Disease/defence | ||
| 5 | Predicted protein | 224063066 | P. trichocarpa | 4·76/56 | 59 | PMF | 21 | 48 | 261 | 255545368 (95) | Protein disulphide isomerase | Protein destination/storage |
| 6 | Predicted protein | 222838558 | P. trichocarpa | 5·20/59 | 58 | TMS | 3 | 9 | 126 | 13752562 (100) | Importin alpha2 | Transporters |
| 7 | Unknown | 118486025 | P. trichocarpa | 7·58/58 | 61 | TMS | 14 | 29 | 610 | 255537027 (98) | Heat shock protein 70-interacting protein | Disease/defence |
| 8 | Predicted protein | 222840955 | P. trichocarpa | 6·17/66 | 61 | TMS | 3 | 6 | 202 | 255537027 (100) | Heat shock protein 70-interacting protein | Disease/defence |
| 9 | DHN1 | 29120043 | P.×canadensis | 5·17/26 | 50 | PMF | 15 | 33 | 176 | Disease/defence | ||
| 10 | Mitochondrial beta subunit of F1 ATP synthase | 224111564 | P. trichocarpa | 5·96/60 | 55 | PMF | 17 | 44 | 244 | Energy | ||
| 11 | Predicted protein | 224099437 | P. trichocarpa | 5·91/60 | 56 | PMF | 13 | 32 | 192 | 255582911 (96) | ATP synthase beta subunit | Energy |
| 12 | Predicted protein | 224068957 | P. trichocarpa | 5·56/48 | 54 | PMF | 14 | 44 | 196 | 255575355 (100) | Enolase | Energy |
| 13 | Predicted protein | 224136806 | P. trichocarpa | 5·67/48 | 56 | TMS | 6 | 25 | 358 | 255539693 (100) | Enolase | Energy |
| 14 | Predicted protein | 224136806 | P. trichocarpa | 5·67/48 | 55 | PMF | 27 | 67 | 325 | 255539693 (100) | Enolase | Energy |
| 15 | Predicted protein | 224136806 | P. trichocarpa | 5·67/48 | 54 | PMF | 13 | 43 | 157 | 255539693 (100) | Enolase | Energy |
| 16 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 912530 | T. hamiltonii | 6·32/49 | 54 | PMF | 10 | 17 | 134 | Energy | ||
| 17 | Predicted protein | 222847266 | P. trichocarpa | 5·97/54 | 55 | TMS | 3 | 8 | 139 | 255554609 (100) | Methylmalonate semialdehyde dehydrogenase | Secondary metabolism |
| 18 | Mitochondrial lipoamide dehydrogenase | 134142802 | P. tremuloides | 7·25/54 | 56 | PMF | 10 | 28 | 101 | Energy | ||
| 19 | Serine hydroxymethyltransferase | 134142065 | P. tremuloides | 7·59/52 | 54 | PMF | 8 | 18 | 111 | Metabolism | ||
| 20 | Predicted protein | 222864715 | P. trichocarpa | 6·92/54 | 54 | TMS | 3 | 8 | 179 | 71842524 (100) | Alanine aminotransferase 1 (AlaT1) | Metabolism |
| 21 | Cytosolic phosphoglycerate kinase 1 | 3738259 | P. nigra | 5·70/43 | 51 | PMF | 14 | 39 | 185 | Energy | ||
| 22 | DHN1 | 29120043 | P.×canadensis | 5·17/26 | 46 | PMF | 16 | 44 | 230 | Disease/defence | ||
| 23 | Bark storage protein B | 728987 | P. deltoides | 6·90/34 | 47 | PMF | 8 | 31 | 113 | Protein destination/storage | ||
| 24 | Predicted protein | 224104631 | P. trichocarpa | 5·53/27 | 43 | TMS | 5 | 27 | 219 | 161778778 (100) | Cytosolic ascorbate peroxidase | Disease/defence |
| 25 | Predicted protein | 222841191 | P. trichocarpa | 5·76/31 | 43 | TMS | 4 | 15 | 184 | 327344121 (82) | Putative protodermal factor 1 (PDF1)-interacting protein 3 | Cell growth/division/development |
| 26 | Unknown | 118482960 | P. trichocarpa | 5·31/35 | 42 | TMS | 2 | 9 | 133 | 255548505 (99) | Protein disulphide isomerase | Protein destination/storage |
| 27 | Predicted protein | 224136806 | P. trichocarpa | 5·67/48 | 53 | PMF | 5 | 21,1 | 333 | 255575355 (100) | Enolase | Energy |
| 28 | Predicted protein | 118488824 | P. trichocarpa | 7·67/37 | 54 | TMS | 6 | 24 | 377 | 255540341 (99) | Glyceraldehyde 3-phosphate dehydrogenase | Energy |
| 29 | Unknown | 118489199 | P. trichocarpa×P. deltoides | 7·01/39 | 45 | PMF | 8 | 29 | 122 | 255575381 (100) | Fructose-bisphosphate aldolase | Energy |
| 30 | Predicted protein | 224082494 | P. trichocarpa | 5·69/39 | 45 | PMF | 9 | 24 | 128 | 14031049 (95) | Peroxidase | Disease/defence |
| 31 | Predicted protein | 224126027 | P. trichocarpa | 8·63/39 | 45 | PMF | 9 | 36 | 140 | 255575381 (100) | Fructose-bisphosphate aldolase | Energy |
| 32 | Predicted protein | 224082494 | P. trichocarpa | 5·69/39 | 41 | PMF | 16 | 32 | 172 | 14031049 (95) | Peroxidase | Disease/defence |
| 33 | Unknown | 118485585 | P. trichocarpa | 9·21/36 | 40 | TMS | 5 | 17 | 310 | 255554971 (99) | Pectinesterase precursor | Cell structure |
| 34 | Unknown | 118486511 | P. trichocarpa | 6·34/36 | 39 | PMF | 7 | 27 | 114 | 4775662 (94) | Major storage protein | Protein destination/storage |
| 35 | Unknown | 118485585 | P. trichocarpa | 9·21/36 | 39 | TMS | 5 | 17 | 310 | 255554971 (99) | Pectinesterase precursor | Cell structure |
| 36 | Predicted protein | 222849325 | P. trichocarpa | 4·44/24 | 34 | TMS | 3 | 13 | 133 | 1705811 (100) | Acidic endochitinase WIN6 | Disease/defence |
| 37 | Predicted protein | 222841154 | P. trichocarpa | 5·01/21 | 30 | TMS | 3 | 19 | 190 | 255583930 (73) | Endomembrane-associated protein | Cell structure |
| 38 | Predicted protein | 224099437 | P. trichocarpa | 5·91/60 | 33 | PMF | 10 | 25 | 140 | 255582911 (96) | ATP synthase beta subunit/probable proteolytic fragment | Energy |
| 39 | Non-identified | 30 | ||||||||||
| 40 | Predicted protein | 224138586 | P. trichocarpa | 5·48/28 | 30 | PMF | 7 | 45 | 133 | 161778778 (99) | Cytosolic ascorbate peroxidase | Disease/defence |
| 41 | Predicted protein | 222854239 | P. trichocarpa | 5·74/27 | 30 | TMS | 7 | 25 | 311 | 255583952 (100) | Proteasome subunit alpha type | Protein destination/storage |
| 42 | Predicted protein | 224098421 | P. trichocarpa | 6·00/27 | 30 | PMF | 8 | 37 | 144 | 255584863 (99) | Triosephosphate isomerase | Energy |
| 43 | Unknown | 118484162 | P. trichocarpa | 8·50/28 | 30 | TMS | 5 | 30 | 216 | 225438529 (100) | Probable ATP synthase 24 kDa subunit, mitochondrial | Energy |
| 44 | Unknown | 118481011 | P. trichocarpa | 6·45/27 | 30 | PMF | 14 | 61 | 213 | 255584863 (99) | Triosephosphate isomerase | Energy |
| 45 | Predicted protein | 222849184 | P. trichocarpa | 8·06/35 | 30 | TMS | 3 | 17 | 171 | 255569554 (100) | Class I chitinase | Disease/defence |
| 46 | Unknown | 118488495 | P. trichocarpa | 8·98/28 | 31 | TMS | 2 | 13 | 120 | 255558968 (88) | Tropinone reductase | Secondary metabolism |
| 47 | Unknown | 118487320 | P. trichocarpa | 8·29/28 | 30 | TMS | 2 | 8 | 119 | 255581099 (99) | Zeamatin precursor | Disease/defence |
| 48 | Predicted protein | 224123646 | P. trichocarpa | 7·78/26 | 31 | PMF | 20 | 55 | 264 | 225426050 (99) | V-type proton ATPase subunit E | Energy |
| 49 | Predicted protein | 224095604 | P. trichocarpa | 9·26/32 | 37 | PMF | 10 | 55 | 167 | 255553601 (100) | Prohibitin | Disease/defence |
| 50 | Predicted protein | 224065729 | P. trichocarpa | 5·52/25 | 28 | PMF | 8 | 34 | 133 | 283135880 (100) | Phi class glutathione transferase GSTF2 | Disease/defence |
| 51 | Predicted protein | 224098421 | P. trichocarpa | 6·00/27 | 28 | PMF | 11 | 48 | 193 | 255584863 (99) | Triosephosphate isomerase | Energy |
| 52 | Predicted protein | 224065727 | P. trichocarpa | 6·32/24 | 28 | PMF | 7 | 33 | 123 | 283135878 (100) | Phi class glutathione transferase GSTF1 | Disease/defence |
| 53 | Predicted protein | 224132348 | P. trichocarpa | 5·73/24 | 27 | PMF | 7 | 38 | 120 | 283135884 (94) | Phi class glutathione transferase GSTF5 | Disease/defence |
| 54 | Predicted protein | 224132348 | P. trichocarpa | 5·73/24 | 27 | PMF | 8 | 52 | 113 | 283135884 (94) | Phi class glutathione transferase GSTF5 | Disease/defence |
| 55 | Predicted protein | 224132348 | P. trichocarpa | 5·73/24 | 27 | TMS | 7 | 38 | 345 | 283135884 (94) | Phi class glutathione transferase GSTF5 | Disease/defence |
| 56 | Predicted protein | 222863969 | P. trichocarpa | 6·30/19 | 25 | TMS | 7 | 54 | 345 | 255556338 (100) | Protein translocase | Transporters |
| 57 | Glutathione peroxidase | 224071850 | P. trichocarpa | 9·42/28 | 24 | TMS | 5 | 17 | 203 | Disease/defence | ||
| 58 | Peroxiredoxin | 19548660 | P. tremula xP. tremuloides | 5·56/18 | 20 | TMS | 2 | 18 | 116 | Disease/defence | ||
| 59 | Predicted protein | 222867006 | P. trichocarpa | 5·97/20 | 18 | TMS | 6 | 28 | 232 | 68299221 (100) | Putative ethylene-responsive protein | Disease/defence |
| 60 | Predicted protein | 224127037 | P. trichocarpa | 8·49/23 | 21 | PMF | 13 | 54 | 138 | 47027028 (96) | CBS 1 protein | Metabolism |
| 61 | Unknown | 118486489 | P. trichocarpa | 6·90/36 | 19 | PMF | 11 | 25 | 110 | 4775662 (94) | Major storage protein/probable proteolytic fragment | Protein destination/storage |
| 62 | Predicted protein | 224060951 | P. trichocarpa | 8·57/18 | 20 | TMS | 2 | 20 | 132 | 163914225 (100) | Putative pathogenesis-related protein 1 | Disease/defence |
| 63 | Predicted protein | 224140323 | P. trichocarpa | 5·20/18 | 16 | PMF | 12 | 67 | 132 | 297820136 (98) | Universal stress protein family protein | Disease/defence |
| 64 | Predicted protein | 224136442 | P. trichocarpa | 6·09/16 | 16 | PMF | 7 | 47 | 115 | 255571035 (100) | Nucleoside diphosphate kinase | Signal transduction |
| 65 | Unknown | 118484128 | P. trichocarpa | 6·31/16 | 16 | PMF | 10 | 64 | 192 | 255571035 (100) | Nucleoside diphosphate kinase | Signal transduction |
| 66 | Predicted protein | 224054740 | P. trichocarpa | 8·99/15 | 14 | TMS | 4 | 26 | 225 | 1370287 (99) | Core protein | Transporters |
The list includes: Spot ID, assigned spot number on the master gel (see Fig. 5); protein name; accession number in the NCBI database; corresponding organism; theoretical pI and Mr (kDa) values; experimental Mr (kDa) values; identification method: tandem mass spectrometry (TMS), peptide mass fingerprint (PMF); number of unique peptides identified; percentage sequence coverage; Mascot score; annotated NCBI accession with the highest BLAST score (sequence identity %), the corresponding protein name and function of each identified protein provided according to Bevan et al. (1998).
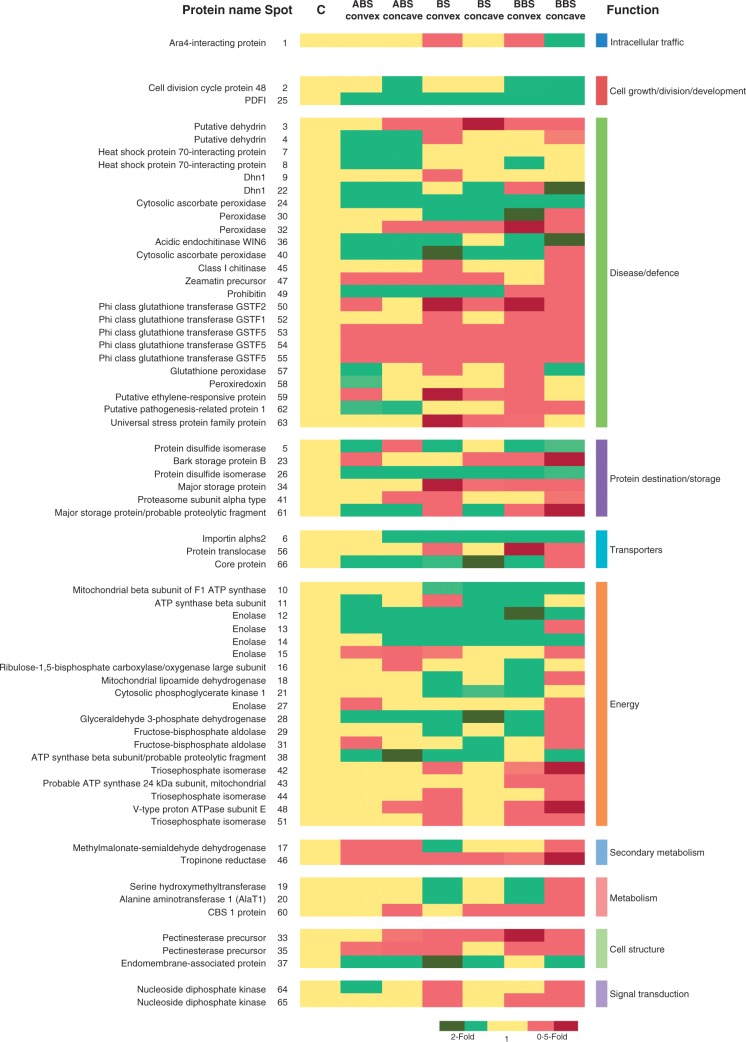
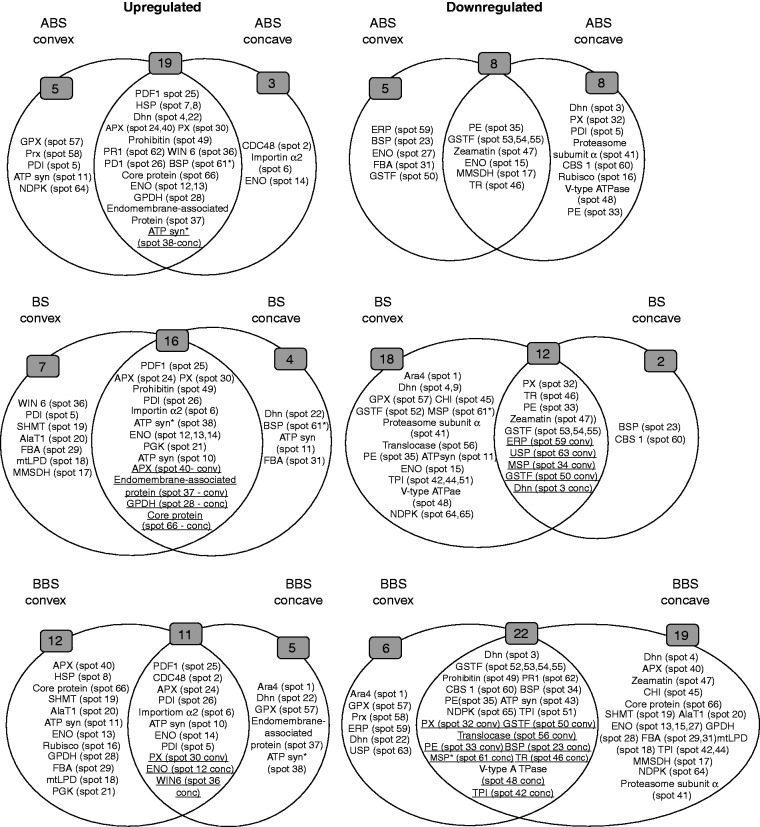
The expression profiles of all identified proteins were represented in a heat map (Fig. 6) that shows the corresponding over- or under-representation in convex and concave sides of the three bent sectors, compared with the control. This figure also catalogues proteins with respect to their predominant function according to Bevan et al. (1998). Furthermore, heat map information was summarized in two-way diagrams shown in Fig. 7. In detail, the two-way diagram groups proteins that, compared with the control, are over- or under-represented: (1) in both convex and concave sides of a bent sector (ABS, BS and BBS), reported in the overlapping regions of the diagrams; or (2) in either the convex or concave side of each bent sector. The results clearly indicate that proteins are asymmetrically expressed in the convex and concave sides of each bent root sector (Fig. 7).
Fig. 6.
Heat map. The map reports the expression level of proteins differentially represented in control, and in convex and concave sides of three bent sectors. Proteins are grouped on the basis of assigned functional classification (Table 2). Light green and red block colours indicate, respectively, over- (2-fold) or under- (0·5-fold) representation of each protein in both the convex and concave sides of the three bent sectors, compared with the control. Dark green and dark red block colours indicate, respectively, proteins over- (2-fold) or under- (0·5-fold) represented in a specific side, compared with the opposite side of the same sector. ABS, above bending sector; BS, bending sector; BBS, below bending sector.
Fig. 7.
Two-ways diagram. Proteins are grouped according to their over- or under-representation in the convex and concave sides of the three bending sectors (ABS, BS and BBS). Overlapping regions include proteins over- or under-represented in both sides of a bent region, compared with control; underlined proteins were found to be over- or under-represented in either the convex or the concave side of the same sector.
In ABS, 19 (spots 4, 7, 8, 12, 13, 22, 24, 25, 26, 28, 30, 36, 37, 38, 40, 49, 61, 62 and 66) and eight protein species (spots 15, 17, 35, 46, 47, 53, 54 and 55) were over- or under-represented, respectively, in both the concave and convex sides. Among the protein components induced at both sides of ABS, an ATP synthase β-subunit fragment (ATPsyn*, spot 38) was more abundant in the concave than in the convex side. Several proteins were specifically over- or under-represented in either the convex or concave side of ABS. In detail, five protein species were induced in the convex side of ABS, namely a protein disulphide isomerase isoform (PDI, spot 5), ATP synthase β-subunit (ATPsyn, spot 11), glutathione peroxidase (GPX, spot 57), peroxiredoxin (Prx, spot 58) and a nucleoside diphosphate kinase isoform (NDPK, spot 64), while another five were under-represented, i.e. bark storage protein B (BSP, spot 23), an enolase isoform (ENO, spot 27), a fructose-bisphosphate aldolase isoform (FBA, spot 31), phi class glutathione transferase 2 (GSTF2, spot 50) and a putative ethylene-responsive protein (ERP, spot 59). At the concave side of ABS, three protein species were over-represented, namely cell division cycle protein 48 (CDC48, spot 2), importin alpha2 (spot 6) and an ENO isoform (spot 14), whereas eight proteins showed reduced levels, i.e. a dehydrin 1 isoform (DHN1, spot 3), a PDI isoform (spot 5), ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (Rubisco large subunit, spot 16), a peroxidase isoform (PX, spot 32), a pectinesterase precursor isoform (PE, spot 33), proteasome subunit alpha type (spot 41), V-type proton ATPase subunit E (V-type ATPase, spot 48) and cystathionine-β-synthase 1 protein (CBS 1, spot 60).
In BS, 16 proteins were found to be over-represented (spots 6, 10, 12, 13, 14, 21, 24, 25, 26, 28, 30, 37, 38, 40, 49 and 66) and 12 under-represented (spots 3, 32, 33, 34, 46, 47, 50, 53, 54, 55, 59 and 63) at both sides (convex and concave). Among the protein species induced in both sides of BS, glyceraldehyde 3-phosphate dehydrogenase (GPDH, spot 28) and core protein (spot 66) were more abundant in the concave than in the convex side; in contrast, endomembrane-associated protein (spot 37) and ascorbate peroxidase (APX, spot 40) were more abundant in the convex than in the concave side. In the case of under-represented proteins, the level of a DHN1 isoform (spot 3) was higher in the concave than in the convex side, in contrast to GSTF2 (spot 50), major storage protein (MSP, spot 34), ERP (spot 59) and universal stress protein (USP, spot 63) which were more abundant in the convex than in the concave side. At the convex side of BS, seven protein species were over-represented, namely a PDI isoform (spot 5), methylmalonyl semialdheyde dehydrogenase (MMSDH, spot 17), mitochondrial lipoamide dehydrogenase (mtLPD, spot 18), serine hydroxymethyltransferase (SHMT, spot 19), alanine aminotransferase 1 (AlaT1, spot 20), an FBA isoform (spot 29) and acidic endochitinase (WIN6, spot 36), while 18 components showed reduced levels, i.e. ara4-interacting protein (Ara4, spot 1), two DHN1 isoforms (spots 4 and 9), ATPsyn (spot 11), an ENO isoform (spot 15), a PE isoform (spot 35), proteasome subunit alpha type (spot 41), three triosephosphate isomerase isoforms (TPI, spots 42, 44 and 51), class I chitinase (CHI, spot 45), V-type ATPase (spot 48), phi class glutathione transferase 1 (GSTF1, spot 52), protein translocase (spot 56), GPX (spot 57), an MSP proteolytic fragment (MSP*, spot 61) and two NDPK isoforms (spots 64 and 65). At the concave side of BS, four protein species were induced, namely ATPsyn (spot 11), a DHN1 isoform (spot 22), an FBA isoform (spot 31) and MSP* (spot 61), whereas two were under-represented, i.e. BSP (spot 23) and CBS 1 (spot 60).
In BBS, 11 (spots 2, 5, 6, 10, 12, 14, 24, 25, 26, 30 and 36) and 22 protein species (spots 3, 23, 32, 33, 34, 35, 42, 43, 46, 48, 49, 50, 51, 52, 53, 54, 55, 56, 60, 61, 62 and 65) were found to be over- or under-represented, respectively, on both sides. Among the induced proteins, WIN6 (spot 36) was more abundant in the concave than in the convex side; in contrast, an ENO isoform (spot 12) and a PX isoform (spot 30) presented higher levels in the convex than in the concave side. Among the under-represented proteins, BSP (spot 23), a TPI isoform (spot 42), tropinone reductase (spot 46), V-type ATPase (spot 48) and MSP* (spot 61) were more abundant in the concave than in the convex side, whereas a PX isoform (spot 32), a PE isoform (spot 33), GSTF2 (spot 50) and traslocase (spot 56) were more abundant in the convex than in the concave side. At the convex side of BBS, 12 protein species were specifically over-represented, i.e. a heat shock protein isoform (HSP, spot 8), ATPsyn (spot 11), an ENO isoform (spot 13), Rubisco large subunit (spot 16), mtLPD (spot 18), SHMT (spot 19), AlaT1 (spot 20), phosphoglycerate kinase (PGK, spot 21), GPDH (spot 28), an FBA isoform (spot 29), APX (spot 40) and core protein (spot 66), whereas six components were under-represented, namely Ara4 (spot 1), a DHN1 isoform (spot 22), GPX (spot 57), Prx (spot 58), ERP (spot 59) and USP (spot 63). At the concave side of BBS, five protein species were induced, namely Ara4 (spot 1), a DHN1 isoform (spot 22), endomembrane-associated protein (spot 37), ATPsyn* (pot 38) and GPX (spot 57), whereas 19 components were under-represented, i.e. a DHN1 isoform (spot 4), three ENO isoforms (spots 13, 15 and 27), MMSDH (spot 17), mtLPD (spot 18), SHMT (spot 19), AlaT1 (spot 20), GPDH (spot 28), FBA (spots 29 and 31), APX (spot 40), proteasome subunit alpha type (spot 41), TPI (spots 42 and 44), CHI (spot 45), zeamatin precursor (spot 47), an NDPK isoform (spot 64) and core protein (spot 66).
DISCUSSION
Anatomical changes and lignin content
The characteristics of reaction wood induced by mechanical stress in plant roots are still largely unknown. The results obtained in the present study seem to indicate that they are significantly different from those reported for the stem. In hardwood species, reaction wood tends to form in zones of the tree held in tension (the upper side of a leaning stem), and is characterized by lower values of vessel area and number, and poor lignification (reviewed in Plomion et al., 2001). Conversely, in the case of poplar bent taproots, anatomical analyses revealed that reaction wood is produced at the compressed concave side of the three bent sectors, with the most significant difference in BS. Indeed, the highest values of cambial cell number and relative xylem thickness were measured in the concave side of BS. The high number of cambial cells appears to be directly related to cambium cell activity. Indeed, we observed that fibres and vessels near the cambial zone had a thinner wall compared with those at a centripetal position, suggesting the occurrence of different maturation stages. It has been reported that a different distribution of environmental stress in roots might lead to asymmetrical responses (Lux et al., 2011; Líŝka et al., 2016). Furthermore, effects of compression forces on cambium cells have been reported for Phaseolus vulgaris and A. thaliana, where a compressive force on the stem determined the differentiation of secondary vascular cambium and subsequent formation of secondary xylem (Biro et al., 1980; Ko et al., 2004). Similarly, our data suggest that in th4e concave side of BS compression forces enhance cambium cell activity and the differentiation of reaction wood with specific features, such as high relative xylem thickness and low specific vessel number and area. Moreover, despite the lower specific vessel number and area, relative vessel number and area were found to be comparable with those measured in the control. Based on these data, it is reasonable to suggest that besides achieving its biomechanical function, the significant increase in relative xylem thickness measured in BS concave may represent a way for the root to balance water uptake and transport (Christensen-Dalsgaard et al., 2008).
The above-mentioned anatomical features have been previously described for poplar stem TW (Jourez et al., 2001). However, here we show that in bent poplar root, differently from the stem, reaction wood is produced in compressed sectors (BS and BBS concave sides) and characterized by high lignin content, similarly to gymnosperm CW.
In the BS convex side subjected to high tension forces, OW was formed with a smaller mean vessel area and a higher specific vessel number. Despite the higher specific vessel number, values of relative xylem thickness, relative vessel area and number in the BS convex side remained significantly lower than in the BS concave side and in the control. These anatomical features suggest that, in contrast to reaction wood in the BS concave side, OW may not counteract the effects of tension forces on water transport efficiency. Thus, the increase of lateral root formation reported on the BS convex side (Trupiano et al., 2012b) may be a strategy to safeguard water uptake on this side.
Hormones
In poplar bent root, the most significant differences in IAA and ABA distribution were observed between BS and BBS concave and convex sides. Indeed, IAA and ABA levels in BS and BBS concave sides were significantly higher than in the corresponding convex sides and in the control. The functional role of auxin in plant response to mechanical stress has been an active area of research on arabidopsis roots and poplar stems. In fact, Ditengou et al. (2008) and Richter et al. (2009) demonstrated that bending of arabidopsis roots causes the initiation of lateral root primordia toward the convex side. In gymnosperm and angiosperm trees, IAA controls the extent of cambial growth (Sundberg et al., 2000; Schrader et al., 2003), reaction wood formation (Funada et al., 1990; Sundberg et al., 1994; Du et al., 2004) and vessel density (Aloni et al., 2006). However, despite these findings, the relationship between endogenous auxin levels in the cambial region and the formation of TW and CW remains to be elucidated (Du and Yamamoto, 2007). Hellgren et al. (2004) found that the formation of TW and CW in poplar and pine bent stems is not mediated by changes in the IAA level in the cambial tissues, whereas Funada et al. (1990) and Du et al. (2004) detected higher amount of endogenous IAA at the side of the cambial region forming CW. The auxin accumulation in the BS concave side reported here provides additional evidence for the hypothesis that poplar stem and root respond differently to bending stress. Indeed, similarly to the bent stem of gymnosperms, but differently from that of hardwood species, reaction wood formation in bent polar root is induced by compression forces and mediated by high levels of IAA which may enhance cambium activity and cell wall stiffening by means of lignin deposition.
Unlike auxin, the role of ABA in the response of woody species to bending has rarely been investigated. ABA is known to be present in cambial tissues (Funada et al., 2005) and has been reported to act as an antagonist to IAA in xylem differentiation (Sundberg et al., 2000; Mellerowicz et al., 2001; Muday and DeLong, 2001). However, ABA is a well-known mediator of the plant response to environmental stresses such as drought (Zhu, 2002). The significant increase of ABA observed in poplar bent root may be related to a water stress response induced by compression forces, particularly intense in BS and BBS concave sides (Trupiano et al., 2012b). CW was found to be particularly vulnerable to drought (Mayr and Cochard, 2003) due to the deformation of conduits and changes in hydraulic conductivity (Tyree and Zimmermann, 2002; Christensen-Dalsgaard et al., 2007, 2008).
Other hormones, including GAs, cytokinins and ethylene, have also been implicated in stem reaction wood formation and are potentially involved in bending stress response, but a comprehensive understanding of their role remains incomplete. GAs regulate early stages of xylem differentiation and cell elongation (Israelsson et al., 2005), while in vitro studies suggested an important function of cytokinins in regulating tracheid differentiation and lignin biosynthesis (Little and Savidge, 1987; Savidge, 1988). Ethylene is involved in both TW and CW formation (Telewski and Jaffe, 1986; Yamamoto and Kozlowski, 1987; Rinne, 1990; Little and Eklund, 1999; Eklund and Klintborg, 2000; Andersson-Gunnerås et al., 2003; Du and Yamamoto, 2003) and also seems to affect cell wall composition, by altering deposition of polysaccharides and lignification (Roberts and Miller, 1983; Miller et al., 1985; Eklund, 1991; Ingemarson et al., 1991; Abeles et al., 1992). However, while some data clearly indicate that ethylene reduces gravitropic responses by altering flavonoid synthesis and IAA polar transport (Buer et al., 2006), others showed an increase of gravitropic curvature by cell elongation inhibition (Chang et al., 2004).
The lack of specific information on the role of these hormones in the root response to bending stress together with the contradictory results often reported in the literature make it difficult to interpret their role in modulating the asymmetrical response at the two sides of the three bent sectors, creating a demand for further investigations.
Proteomic analysis
Proteomic analysis confirmed our previous results which demonstrated that the general response to bending, independently of the intensity of mechanical forces, involves the differential expression of proteins involved in carbohydrate and energy metabolism, defence machinery and cell wall/membrane stability (Di Michele et al., 2006; Trupiano et al., 2012a, b, 2013b, 2014; Rossi et al., 2015).
Here we report that tension and compression forces determine an asymmetrical expression at the convex and concave side of each bent sector of several proteins involved in xylem differentiation, lignin production and lateral root formation. Ara4-interacting protein (Ara4, spot 1) was found to be present at a higher level at concave sides compared with convex sides of BS and BBS. Ara4 has been suggested to be a part of the gene network regulating secondary xylem development and differentiation and the formation of secondary cell walls and lignification (Brembu et al., 2005). Similarly, cystathionine-β-synthase 1 (CBS1) (spot 60) was found to be less abundant in ABS and BS concave compared with the opposite sides and in both sides of BBS. Evidence has been presented for the involvement of this enzyme in ethylene synthesis (Wang et al., 1982) and stress tolerance (Luhua et al., 2008), and in the negative regulation of lignin accumulation (Yoo et al., 2011). However, although several cystathionine-β-synthases have been identified in plants, their functions remain largely unknown.
Mitochondrial lipoamide dehydrogenase (mtLPD, spot 18), alanine aminotransferase (AlaT1, spot 20) and serine hydroxymethyltransferase (SHMT, spot 19) were more abundantly represented in BS and BBS convex sides, compared with concave sides. mtLPD is part of four multienzyme complexes which produce NADH as the main source of reductive potential energy further harvested by oxidative phosphorylation to generate ATP (Chen et al., 2014). SHMT is an important enzyme that produces N5,N10-methylene tetrahydrofolate and glycine from serine and tetrahydrofolate; it generates carbon units for cellular use (Wang et al., 2015). AlaT1 converts pyruvate and glutamate to alanine and 2-oxoglutarate, thereby ensuring the efficient use of nitrogen and facilitating the maintenance of carbon–nitrogen homeostasis (Suarez et al., 2002; Miyashita et al., 2007).
An efficient use of carbon and nitrogen in BS and BBS convex sides regulated by the above-mentioned enzymes may be a mechanism to regulate osmotic potential and turgor in corresponding concave sides subjected to high compression forces and xylem cavitation. Indeed, it has been hypothesized that solutes might move radially along the ray cell walls, enter the embolized xylem conduits and increase the solute concentration of the residual water within them, thus promoting xylem refilling by altering osmoticum, as has been shown to occur during drought (Salleo et al., 2009; Secchi et al., 2011).
Factors involved in signal transduction and secondary metabolism were also asymmetrically expressed. Two nucleotide disphosphate kinase isoforms (NDPK, spots 64, 65) accumulated differently in convex and concave sides of the three bent sectors. NDPK belongs to a multifunctional gene family with phosphodiesterase, peroxidase, ROS signalling, F-actin binding and Ca2+ channel activities, and involved in a wide range of functions, such as root growth, lateral root development and the response to adverse conditions, including mechanical stress (Bassani et al., 2004; Clark et al., 2005a, b; Mortimer et al., 2008). Methylmalonate semialdehyde dehydrogenase (MMSDH, spot 17), an enzyme of valine catabolism that catalyses the conversion of methylmalonate semialdehyde into propionyl-CoA and of malonate semialdehyde into acetyl-CoA, was found to be most abundant in BS convex and almost completely absent in BBS concave. Oguchi et al. (2004) and Tanaka et al. (2005) indicated a role for MMSDH in auxin-mediated lateral root formation, and evidenced that high MMSDH expression levels induced by auxin were maintained over time. In BS convex, where a low IAA content was found, we hypothesize that MMSDH overexpression could have been initially induced by IAA and maintained over time (in an IAA-independent manner) to guarantee lateral root formation. Indeed, Richter et al. (2009) demonstrated that in arabidopsis bent roots, auxin dynamics preceded and were correlated with curve-dependent lateral root initiation. However, with time progressing, mechanical force alone was responsible for lateral root formation at the convex side of the curve.
Two factors controlling cambial cell division, ethylene-responsive protein (ERP, spot 59) and cell division control protein 48 (CDC48, spot 2), were also found asymmetrically distributed between the concave and convex sides of bent sectors. Indeed, ERP was found to be less abundant in the convex side of all three bent sectors, compared with the concave side. ERP has already been implicated in many plant functions, such as cellular proliferation and lateral root formation (Trupiano et al., 2013a), hormonal signal transduction (Ohme-Takagi and Shinshi, 1995), response to biotic or abiotic stresses (Gu et al., 2000; Dubouzet et al., 2003) and regulation of metabolism (van der Fits and Memelink, 2000; Zhang et al., 2005). Recent studies showed that ERPs are required for the increase of vascular cambial cell division in poplar (Vahala et al., 2013), suggesting that its low expression in the bent root convex sides may be associated with a decrease in cambial cell division. CDC48 was more abundant in ABS concave than in the opposite convex side, while it was found to be highly accumulated in both sides of BBS. We previously suggested a role for this protein in the control of cambial growth resumption (Trupiano et al., 2012a). However, besides its control of cell cycle/proliferation, CDC48 protein has been found to regulate cell expansion and differentiation (Rancour et al., 2002, 2004; Park et al., 2008) and to preserve cell wall and plasma membrane integrity (Shi et al., 1995; Rancour et al., 2002). A similar function may be associated with the high abundance of CDC48 in ABS concave and both sides of BBS. This might be particularly true for BBS, in which a gravitropic response possibly requires the control of cell elongation and cell wall elasticity for organ curvature.
CONCLUSION
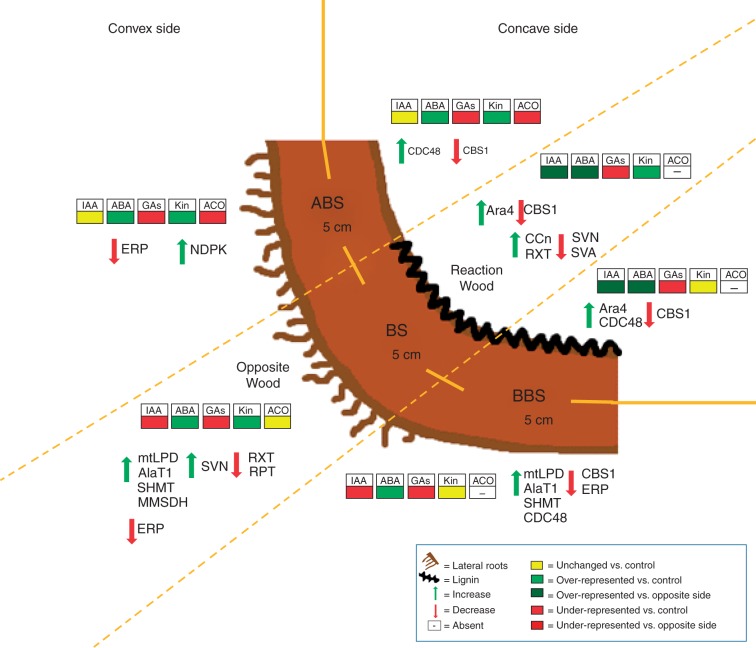
This study reports that differences in intensity of tension and compression mechanical forces induce specific responses in the convex and concave sides of bent poplar root. The results obtained demonstrate, for the first time to our knowledge, that in poplar bent taproot, in contrast to what happens in the stem, reaction wood is produced at the compressed side, showing features similar to that observed in gymnosperm CW in the stem. The key results of this paper have been summarized in the model presented in Fig. 8 which illustrates that the induction of root reaction wood by compression forces is characterized by an increase in xylem thickness, a low vessel density, a high lignin content and an induction of cambium cell activity. All these responses may be triggered by auxin and involve protein factors that control cell wall deformation, lignification and xylem differentiation. Furthermore, besides ensuring biomechanical functions, the bent root may use two different strategies to maintain water uptake and transport in a deforming condition induced by tension and compression forces: increasing xylem thickness at the compressed side and enhancing lateral root formation at the tension site.
Fig. 8.
Model summarizing the main anatomical, phytohormonal and proteomical changes observed in the convex and concave sides of three bent taproot sectors (ABS, BS and BBS). Phytohormone (IAA, ABA, GAs, Kin and ethylene) changes are represented by diverse coloured blocks. Proteomic and anatomical changes are indicated by arrows. Zones with the highest lateral root number and lignin content are also reported. ABS, above bending sector; AlaT1, alanine aminotransferase 1 (spot 20); Ara4, ara4-interacting protein (spot 1); BS, bending sector; BBS, below bending sector; CBS1, cystathionine-β-synthase 1 (spot 60); CCN, cambial cell number; CDC48, cell division cycle protein 48 (spot 2); ERP, ethylene-responsive protein; MMSDH, methylmalonate semialdehyde dehydrogenase (spot 17); NDPK, nucleoside diphosphate kinase (spot 64); RPT, relative phloem thickness; RXT, relative xylem thickness; SHMT, serine hydroxymethyltransferase (spot 19); SVA, specific vessel area; SVN; specific vessel number.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: simulation of mechanical bending stress in Populus nigra root. Figure S2: anatomical measurements of Populus nigra bent root. Figure S3: ACO sequence. Figure S4: two-dimensional proteomic maps of Populus nigra woody taproots in control and mechanical stress conditions.
ACKNOWLEDGEMENTS
This work was supported in part by grants from the MIUR (PRIN 2008 n. 223), the University of Insubria (FAR) and the EC FP7 Project ZEPHYR-308313.
LITERATURE CITED
- Abedini R, Clair B, Pourtahmasi K, Laurans F, Arnould O. 2015. Cell wall thickening in developing tension wood of artificially bent poplar trees. IAWA Journal 36: 44–57. [Google Scholar]
- Abeles FB, Morgan PW, Saltveit ME., Jr. 1992. Ethylene in plant biology, 2nd edn. San Diego: Academic Press. [Google Scholar]
- Aloni R, Aloni E, Langhans M, Ullrich CI. 2006. Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Annals of Botany 97: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson-Gunnerås S, Hellgren JM, Björklund S, Regan S, Moritz T, Sundberg B. 2003. Asymmetric expression of a poplar ACC oxidase controls ethylene production during gravitational induction of tension wood. The Plant Journal 34: 339–349. [DOI] [PubMed] [Google Scholar]
- Andersson-Gunnerås S, Mellerowicz EJ, Love J, et al. 2006. Biosynthesis of cellulose-enriched tension wood in Populus: Global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. The Plant Journal 45: 144–165. [DOI] [PubMed] [Google Scholar]
- Bassani M, Neumann PM, Gepstein S. 2004. Differential expression profiles of growth-related genes in the elongation zone of maize primary roots. Plant Molecular Biology 56: 367–380. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, et al. 1998. Analysis of 1·9 Mb contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391: 485–488. [DOI] [PubMed] [Google Scholar]
- Biro RL, Hunt ER, Erner Y, Jaffe MJ. 1980. Thigmomorphogenesis: changes in cell division and elongation in the internodes of mechanically-perturbed or ethrel-treated bean plants. Annals of Botany 45: 655–664. [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive for the quantization of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brembu T, Winge P, Bones AM. 2005. The small GTPase AtRAC2/ROP7 is specifically expressed during late stages of xylem differentiation in Arabidopsis. Journal of Experimental Botany 56: 2465–2476. [DOI] [PubMed] [Google Scholar]
- Buer CS, Sukumar P, Muday GK. 2006. Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiology 140: 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Kim YS, Lee JY, et al. 2004. Brassinolide interacts with auxin and ethylene in the root gravitropic response of maize (Zea mays). Physiologia Plantarum 121: 666–673. [Google Scholar]
- Chen W, Taylor NL, Chi Y, Harvey Millar A, Lambers H, Finnegan PM. 2014. The metabolic acclimation of Arabidopsis thaliana to arsenate is sensitized by the loss of mitochondrial LIPOAMIDE DEHYDROGENASE2, a key enzyme in oxidative metabolism. Plant, Cell & Environment 37: 684–695. [DOI] [PubMed] [Google Scholar]
- Chiatante D, Scippa GS, Di Iorio A, De Micco V, Sarnataro M. 2007. Lateral root emission in woody taproots of Fraxinus ornus L. Plant Biosystems 141: 204–213. [Google Scholar]
- Christensen-Dalsgaard KK, Fournier M, Ennos AR. 2007. Changes in vessel anatomy in response to mechanical loading in six species of tropical trees. New Phytologist 176: 610–622. [DOI] [PubMed] [Google Scholar]
- Christensen-Dalsgaard KK, Ennos AR, Fournier M. 2008. Interrelations between hydraulic and mechanical stress adaptations in woody plants. Plant Signaling and Behavior 7: 463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GB, Cantero-Garcia A, Butterfield T, Dauwalder M, Roux SJ. 2005a. Secretion as a key component of gravitropic growth: implications for annexin involvement in differential growth. Gravitational and Space Biology Bulletin 18: 113–114. [PubMed] [Google Scholar]
- Clark GB, Lee DW, Dauwalder M, Roux SJ. 2005b. Immunolocalization and histochemical evidence for the association of two different Arabidopsis annexins with secretion during early seedling growth and development. Planta 220: 621–631. [DOI] [PubMed] [Google Scholar]
- Dejardin A, Leple JC, Lesage-Descauses MC, Costa G, Pilate G. 2004. Expressed sequence tags from poplar wood tissues – a comparative analysis from multiple libraries. Plant Biology 6: 55–64. [DOI] [PubMed] [Google Scholar]
- Di Iorio A, Lasserre B, Petrozzi L, Scippa GS, Chiatante D. 2008. Adaptive longitudinal growth of first-order lateral roots of a woody species (Spartium junceum) to slope and different soil conditions – upward growth of surface roots. Environmental and Experimental Botany 63: 207–215. [Google Scholar]
- Di Michele M, Chiatante D, Plomion C, Scippa GS. 2006. A proteomic analysis of Spanish broom (Spartium junceum L.) root growing on a slope condition. Plant Science 170: 926–935. [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, et al. 2008. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proceedings of the National Academy Sciences, USA 105: 18818–18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doster MA, Bostock RM. 1988. Quantification of lignin formation in almond bark in response to wounding and infection by Phytophthora species. Phytopathology 78: 473–477. [Google Scholar]
- Du S, Yamamoto F. 2003. Ethylene evolution changes in the stems of Metasequoia glyptostroboides and Aesculus turbinate seedlings in relation to gravity-induced reaction wood formation. Trees 17: 522–528. [Google Scholar]
- Du S, Yamamoto F. 2007. An overview of the biology of reaction wood formation. Journal of Integrative Plant Biology 49: 131−143. [Google Scholar]
- Du S, Uno H, Yamamoto F. 2004. Roles of auxin and gibberellin in gravity-induced tension wood formation in Aesculus turbinate seedlings. International Association of Wood Anatomists Journal 25: 337–347. [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, et al. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal 33: 751–763. [DOI] [PubMed] [Google Scholar]
- Eklund L. 1991. Relations between indoleacetic-acid, calcium-ions and ethylene in the regulation of growth and cell-wall composition in Picea abies. Journal of Experimental Botany 42: 785–789. [Google Scholar]
- Eklund L, Klintborg A. 2000. Ethylene, oxygen and carbon dioxide in woody stems during growth and dormancy In: Savidge R, Barnett J., Napier R. eds. Cell and molecular biology of wood formation. Oxford: BIOS Scientific Publishers, 43–56. [Google Scholar]
- van der Fits L, Memelink J. 2000. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297. [DOI] [PubMed] [Google Scholar]
- Foti M, Dhoa S, Fusconia A. 2014. Root plasticity of Nicotiana tabacum in response to phosphorus starvation. Plant Biosystems 150: 429–435. [Google Scholar]
- Funada R, Mizukami E, Kubo T, Fushitani M, Sugiyama T. 1990. Distribution of indole-3-acetic acid and compression wood formation in the stems of inclined Cryptomeria japonica. Holzforschung 44: 331–334. [Google Scholar]
- Funada R, Kubo T, Tabuchi M, Sugiyama T, Fushitani M. 2005. Seasonal variations in endogenous indole-3-acetic acid and abscisic acid in the cambial region of Pinus densiflora Sieb. et Zucc. stems in relation to earlywood–latewood transition and cessation of tracheid production. Holzforschung 55: 128–134. [Google Scholar]
- Gerttula S, Zinkgraf M, Muday G, et al. 2015. Transcriptional and hormonal regulation of gravitropism of woody stems in Populus. The Plant Cell 27: 2800–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara VK, Zhou J, Martin GB. 2000. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. The Plant Cell 12: 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellgren JM, Olofsson K, Sundberg B. 2004. Patterns of auxin distribution during gravitational induction of reaction wood in poplar and pine. Plant Physiology 135: 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge A, Berta G, Doussan C, Merchan F, Crespi M. 2009. Plant root growth, architecture and function. Plant and Soil 321: 153–187. [Google Scholar]
- Ingemarson BSM, Eklund L, Eliasson L. 1991. Ethylene effects on cambial activity and cell-wall formation in hypocotyls of Picea abies seedlings. Physiologia Plantarum 82: 219–224. [Google Scholar]
- Israelsson M, Sundberg B, Moritz T. 2005. Tissue-specific localization of gibberellins and expression of gibberellin-biosynthetic and signaling genes in wood-forming tissues in aspen. The Plant Journal 44: 494–504. [DOI] [PubMed] [Google Scholar]
- Jourez B, Riboux A, Leclercq A. 2001. Anatomical characteristics of tension wood and opposite wood in young inclined stems of poplar (Populus euramericana cv ‘Ghoy’). IAWA Journal 22: 133–157. [Google Scholar]
- Ko JH, Han KH, Park S, Yang J. 2004. Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiology 135: 1069–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarguette F, Leple JC, Dejardin A, et al. 2004. Poplar genes encoding fasciclin-like arabinogalactan proteins are highly expressed in tension wood. New Phytologist 164:107–121. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, et al. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biology 6: e307. doi: 10.1371/journal.pbio.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Líŝka D, Martinka M, Kohanov J, Lux A. 2016. Asymmetrical development of root endodermis and exodermis in reaction to abiotic stresses. Annals of Botany 118:667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little CHA, Eklund L. 1999. Ethylene in relation to compression wood formation in Abies balsamea shoots. Trees 13: 173–177. [Google Scholar]
- Little CHA, Savidge RA. 1987. The role of plant growth regulators in forest tree cambial growth. Plant Growth Regulation 6: 137–169. [Google Scholar]
- Luhua S, Ciftci-Yilmaz S, Harper J, Cushman J, Mittler R. 2008. Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiology 148: 280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux A, Martinka M, Vaculík M, White PJ. 2011. Root responses to cadmium in the rhizosphere: a review. Journal of Experimental Botany 62: 21–37. [DOI] [PubMed] [Google Scholar]
- Mauriat M, Leplé J-C, Claverol S, et al. 2015. Quantitative proteomic and phosphoproteomic approaches for deciphering the signaling pathway for tension wood formation in poplar. Journal of Proteome Research 8(14). doi:10.1021/acs.jproteome.5b00140. [DOI] [PubMed] [Google Scholar]
- Mayr S, Cochard H. 2003. A new method for vulnerability analysis of small xylem areas reveals that compression wood of Norway spruce has lower hydraulic safety than opposite wood. Plant, Cell & Environment 26: 1365–1371. [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W. 2001. Unravelling cell wall formation in the woody dicot stem. Plant Molecular Biology 47: 239–274. [PubMed] [Google Scholar]
- Mihr C, Braun HP. 2003. Proteomics in plant biology In: PM Conn, ed. Handbook of proteomics. Totowa, NJ: Humana Press, 409–416. [Google Scholar]
- Miller AR, Crawford DL, Roberts LW. 1985. Lignification and xylogenesis in Lactuca pith explants cultured in vitro in the presence of auxin and cytokinin: a role for endogenous ethylene. Journal of Experimental Botany 36: 110–118. [Google Scholar]
- Miyashita Y, Dolferus R, Ismond KP, Good AG. 2007. Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. The Plant Journal 49: 1108–1121. [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Weisenseel MH, Gilroy S. 2009. Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. The Plant Cell 21: 2341–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Terzaghi M, Di Iorio A, Scippa GS, Chiatante D. 2012a. Fine-root morphological and growth traits in a Turkey-oak stand in relation to seasonal changes in soil moisture in the Southern Apennines, Italy. Ecological Research 27: 1015–1025. [Google Scholar]
- Montagnoli A, Terzaghi M, Di Iorio A, Scippa GS, Chiatante D. 2012b. Fine-root seasonal pattern, production and turnover rate of European beech (Fagus sylvatica L.) stands in Italy Prealps: possible implications of coppice conversion to high forest. Plant Biosystems 146: 1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Di Iorio A, Terzaghi M, Trupiano D, Scippa GS, Chiatante D. 2014. Influence of soil temperature and water content on fine-root seasonal growth of European beech natural forest in Southern Alps, Italy. European Journal of Forest Research 133: 957–968. [Google Scholar]
- Morel H, Mangenet T, Beauche J, Ruelle J, Nicolini E, Heuret P, Thibaut B. 2015. Seasonal variations in phenological traits: leaf shedding and cambial activity in Parkia nitida Miq. and Parkia velutina Benoist (Fabaceae) in tropical rainforest. Trees 29: 973–984. [Google Scholar]
- Mortimer JC, Laohavisit A, Macpherson N, et al. 2008. Annexins: multi-functional components of growth and adaptation. Journal of Experimental Botany 59: 533–544. [DOI] [PubMed] [Google Scholar]
- Muday GK, DeLong A. 2001. Polar auxin transport: controlling where and how much. Trends in Plant Science 6: 535–542. [DOI] [PubMed] [Google Scholar]
- Oguchi K, Tanaka N, Komatsu S, Akao S. 2004. Methylmalonate-semialdehyde dehydrogenase is induced in auxin-stimulated and zinc-stimulated root formation in rice. Plant Cell Reports 22: 848–858. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. 1995. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. The Plant Cell 7: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Keathley DE, Han KH. 2008. Transcriptional profiles of the annual growth cycle in Populus deltoides. Tree Physiology 28: 321–329. [DOI] [PubMed] [Google Scholar]
- Parker AJ, Haskins EF, Deyrup-Olsen I. 1982. Toluidine Blue: a simple, effective stain for plant tissues. The American Biology Teacher 44: 487–489. [Google Scholar]
- Plomion C, Leprovost G, Stokes A. 2001. Wood formation in trees. Plant Physiology 127: 1513–1523. [PMC free article] [PubMed] [Google Scholar]
- Rancour DM, Dickey CE, Park S, Bednarek SY. 2002. Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiology 130: 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour DM, Park S, Knight SD, Bednarek SY. 2004. Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of Arabidopsis CDC48. Journal of Biological Chemistry 279: 54264–54274. [DOI] [PubMed] [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S. 2009. Mechanical stimuli modulate lateral root organogenesis. Plant Physiology 151: 1855–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne P. 1990. Effects of various stress treatments on growth and ethylene evolution in seedlings and sprouts of Betula pendula Roth. and B. pubescens Ehrh. Scandinavian Journal of Forestry Research 5: 155–167. [Google Scholar]
- Roberts LW, Miller R. 1983. Is ethylene involved in xylem differentiation? Vistas Plant Science 6: 1–24. [Google Scholar]
- Rossi M, Trupiano D, Tamburro M, et al. 2015. MicroRNAs expression patterns in the response of poplar woody root to bending stress. Planta 242: 339–351. [DOI] [PubMed] [Google Scholar]
- Ruelle J. 2014. Morphology, anatomy, and ultrastructure of reaction wood In: B Gardiner, J Barnett, P Saranpää, J Gril, eds. The biology of reaction wood. Berlin: Springer-Verlag, 13−35. [Google Scholar]
- Salleo S, Trifilò P, Esposito S, Nardini A, Lo Gullo MA. 2009. Starch-to-sugar conversion in wood parenchyma of field-growing Laurus nobilis plants: a component of the signal pathway for embolism repair? Functional Plant Biology 36: 815–825. [DOI] [PubMed] [Google Scholar]
- Savidge RA. 1988. Auxin and ethylene regulation in diameter growth in trees. Tree Physiology 4: 401–414. [DOI] [PubMed] [Google Scholar]
- Schrader J, Baba K, May ST, et al. 2003. Polar auxin transport in the wood-forming tissues of hybrid aspen is under simultaneous control of developmental and environmental signals. Proceedings of the National Academy Sciences, USA 100: 10096–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scippa GS, Trupiano D, Rocco M, Di Iorio A, Chiatante D. 2008. Unravelling the response of poplar (Populus nigra) roots to mechanical stress imposed by bending. Plant Biosystems 142: 401–413. [Google Scholar]
- Secchi F, Gilbert ME, Zwieniecki MA. 2011. Transcriptome response to embolism formation in stems of Populus trichocarpa provides insight into signaling and the biology of refilling. Plant Physiology 157: 1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Dixon RA, Gonzales RA, Kjellbom P, Bhattacharyya MK. 1995. Identification of cDNA clones encoding valosin-containing protein and other plant plasma membrane-associated proteins by a general immunoscreening strategy. Proceedings of the National Academy Sciences, USA 92: 4457–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez MF, Avila C, Gallardo F, et al. 2002. Molecular and enzymatic analysis of ammonium assimilation in woody plants. Journal of Experimental Botany 53: 891–904. [DOI] [PubMed] [Google Scholar]
- Sundberg B, Tuominen H, Little CHA. 1994. Effects of the indole-3-acetic acid (IAA) transport inhibitors N-1-naphtylphthalamic acid and morphactin on endogenous IAA dynamics in relation to compression wood formation in 1-year old Pinus sylvestris L. shoots. Plant Physiology 106: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg B, Uggla C, Tuominen H. 2000. Cambial growth and auxin gradients In: R Savidge, J Barnett, R Napier, eds. Cell and molecular biology of wood formation. Oxford: BIOS Scientific Publishers, 169–188. [Google Scholar]
- Tanaka N, Takahashi H, Kitano H, et al. 2005. Proteome approach to characterize the methylmalonate-semialdehyde dehydrogenase that is regulated by gibberellin. Journal of Proteome Research 4: 1575–1582. [DOI] [PubMed] [Google Scholar]
- Telewski FW, Jaffe MJ. 1986. Thigmomorphogenesis: anatomical, morphological and mechanical analysis of genetically different sibs of Pinus taeda in response to mechanical perturbation. Physiologia Plantarum 66: 219–226. [DOI] [PubMed] [Google Scholar]
- Timell TE. 1986. Compression wood in gymnosperms. New York: Springer-Verlag. [Google Scholar]
- Trupiano D, Renzoni G, Rocco M, et al. 2012a. The proteome of Populus nigra woody root: response to bending. Annals of Botany 110: 415–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupiano D, Di Iorio A, Montagnoli A, et al. 2012b. Involvement of lignin and hormones in the response of woody poplar taproots to mechanical stress. Physiologia Plantarum 146: 39–52. [DOI] [PubMed] [Google Scholar]
- Trupiano D, Yordanov Y, Regan S, et al. 2013a. Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 238: 271–282. [DOI] [PubMed] [Google Scholar]
- Trupiano D, Rocco M, Renzone G, et al. 2013b. Poplar woody root proteome during the transition dormancy–active growth. Plant Biosystems 147: 1–6. [Google Scholar]
- Trupiano D, Rocco M, Renzone G, et al. 2014. Temporal analysis of poplar woody root response to bending stress. Physiologia Plantarum 150: 174–193. [DOI] [PubMed] [Google Scholar]
- Tyree MT, Zimmermann MH. 2002. Xylem structure and the ascent of sap, 2nd edn. Berlin: Springer Verlag. [Google Scholar]
- Vahala J, Felten J, Love J, et al. 2013. A genome-wide screen for ethylene-induced ethylene response factors (ERFs) in hybrid aspen stem identifies ERF genes that modify stem growth and wood properties. New Phytologist 200: 511–522. [DOI] [PubMed] [Google Scholar]
- Vascotto C, Cesaratto L, D’Ambrosio C, et al. 2006. Proteomic analysis of liver tissues subjected to early ischemia/reperfusion injury during human orthotopic liver transplantation. Proteomics 6: 3455–3465. [DOI] [PubMed] [Google Scholar]
- Wang D, Liu H, Li S, Zhai G, Shao J, Tao Y. 2015. Characterization and molecular cloning of a serine hydroxymethyltransferase 1 (OsSHM1) in rice. Journal of Integrative Plant Biology 57: 745–56. [DOI] [PubMed] [Google Scholar]
- Wang SY, Adams DO, Lieberman M. 1982. Recycling of 5-methylthioadenosine-ribose carbon atoms into methionine in tomato tissue in relation to ethylene production. Plant Physiology 70: 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto F, Kozlowski TT. 1987. Effects of flooding, tilting of stems, and ethrel application on growth, stem anatomy, and ethylene production of Acer platanoides seedlings. Scandinavian Journal of Forestry Research 2: 141–156. [Google Scholar]
- Yoo KS, Ok SH, Jeong BC, et al. 2011. Single cystathionine β-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. The Plant Cell 23: 3577–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY. 2005. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). The Plant Journal 42: 689–707. [DOI] [PubMed] [Google Scholar]
- Zhu JK. 2002. Salt and drought stress signal transduction in plants. Annual Review of Plant Physiology 53: 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel BJ, van Buijtenen JP. 1989. Wood variation: its causes and control. Berlin: Springer-Verlag. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.