Abstract
Background
Transmitted by the whitefly Bemisia tabaci, tomato yellow leaf curly virus (TYLCV) has posed serious threats to plant growth and development. Plant innate immune systems against various threats involve WRKY Group III transcription factors (TFs). This group participates as a major component of biological processes in plants.
Results
In this study, 6 WRKY Group III TFs (SolyWRKY41, SolyWRKY42, SolyWRKY53, SolyWRKY54, SolyWRKY80, and SolyWRKY81) were identified, and these TFs responded to TYLCV infection. Subcellular localization analysis indicated that SolyWRKY41 and SolyWRKY54 were nuclear proteins in vivo. Many elements, including W-box, were found in the promoter region of Group III TFs. Interaction network analysis revealed that Group III TFs could interact with other proteins, such as mitogen-activated protein kinase 5 (MAPK) and isochorismate synthase (ICS), to respond to biotic and abiotic stresses. Positive and negative expression patterns showed that WRKY Group III genes could also respond to TYLCV infection in tomato. The DNA content of TYLCV resistant lines after SolyWRKY41 and SolyWRKY54 were subjected to virus-induced gene silencing (VIGS) was lower than that of the control lines.
Conclusions
In the present study, 6 WRKY Group III TFs in tomato were identified to respond to TYLCV infection. Quantitative real-time–polymerase chain reaction (RT-qPCR) and VIGS analyses demonstrated that Group III genes served as positive and negative regulators in tomato–TYLCV interaction. WRKY Group III TFs could interact with other proteins by binding to cis elements existing in the promoter regions of other genes to regulate pathogen-related gene expression.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-3123-2) contains supplementary material, which is available to authorized users.
Keywords: TYLCV, WRKY Group III TFs, VIGS, Regulation mechanism, Tomato
Background
Microbial pathogens, including fungi, bacteria, and viruses, have disrupted plant growth, development, and yields. For instance, tomato yellow leaf curly virus (TYLCV) adversely affects the growth and development of tomato. This virus contains a circular single-stranded DNA (ssDNA) with 2.7–2.8 kb molecules. The TYLCV genome is composed of six open reading frames (ORFs), virion sense strands V1 and V2, and complementary sense strands C1–C4 [1]. First identified in Israel in 1930, TYLCV has invaded and damaged new areas, including the Mediterranean, Asian countries, and the US [2].
TYLCV is transmitted by the whitefly Bemisia tabaci in a persistent and circulative manner [3]. Intact virions pass through the food canal in the stylet of B. tabaci and reach the esophagus; they then move through the plant phloem as B. tabaci inserts its proboscis and feeds on infected tomato leaves [4, 5]. Upon entering plant host cells, the DNA of TYLCV replicates via a rolling circular mechanism [6]. Host enzymes can convert the incoming geminivirus ssDNA to double-stranded DNA (dsDNA) that serves as a transcription template. A complex composed of ssDNA and coat protein (CP) forms and enters the host nucleus. Host nucleus provides the factors for viral genome replication and transcription. The ssDNA:CP complex can then be transferred to an adjacent cell via two proposed routes: one route involves PreCP, which can bind to the ssDNA:CP complex and target the cytoplasm via the plasmodesmata and the other route includes CP, which participates in the nuclear export of the ssDNA:CP complex [7]. After TYLCV is released into the phloem of a young leaf by the feeding B. tabaci, this virus can systemically invade most plant organs above and below ground within 1–2 weeks [8]. TYLCV has threatened up to 20 different plant species. TYLCV infection-induced damages have also been exacerbated because of the uncontrollable spread of B. tabaci.
WRKY transcription factors (TFs) family, named from a highly conserved WRKY domain, is a large TF family distributed widely among plants [9]. The WRKY domain contains approximately 60 amino acids, comprising a highly conserved short peptide WRKYGQK at the N-terminus, and a C2H2 or C2HC zinc-binding motif at the C-terminus [10]. WRKY TFs can be divided into three groups (Groups I, II, and III) on the basis of the type of zinc-finger structures and the number of WRKY domains. SPF1, which encoded a WRKY factor, was cloned in Ipomoea batatas in 1994 [11]. Since then, numerous WRKY genes have been identified in different species, including terrestrial plants and green algae [12]. For instance, 72 WRKY members have been found in Arabidopsis [13], 95 members have been detected in Daucus carota [10], 50 members have been identified in Camellia sinensis [14], 100 members have been documented in Oryza sativa [15] and 145 members have been observed in Brassica rapa ssp. pekinensis [16].
Compared with Group I and II WRKY TFs, Group III TFs alter C2H2 to C2HC zinc-finger motif C-X7-C-X23-H-X1-C. Among the three groups, Group III has been considered the most adaptable and most advanced in monocot evolution [17, 18]. WRKY Group III TFs have also been identified in many plants: 13 members in Arabidopsis [19], 25 members in Chinese cabbage [16], 28 members in rice, 6 members in grape and 10 members in Populus [20]. WRKY Group III TFs participate in defense against pathogens. For example, resistance to Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae in rice is strong because of WRKY45 overexpression [21]. ClWRKY70 and FcWRKY70, respectively identified in Citrullus lanatus and Fortunella crassifolia, are also implicated in plant disease resistance against pathogenic infections [22, 23]. Group III members in Arabidopsis are also involved in different plant defense signaling pathways; this phenomenon indicates that Group III TFs have evolved as a result of increasing biological requirements [19]. Furthermore, WRKY Group III TFs initiate transcriptional activation during pathogen invasion and function as essential components of plant innate immune systems, including basal defense and systemic acquired resistance [19, 24, 25]. WRKY TFs can bind to W/W-like box type cis elements or other cis elements in their promoters or in other gene promoters [26]. WRKY TFs also play a role in the interaction of salicylic acid (SA) and jasmonic acid (JA)-mediated signaling, which is involved in pathogen-associated molecular pattern (PAMP) triggered immunity and effector triggered immunity [27, 28]. WRKY TFs can also serve as major targets of perturbation-activated mitogen-activated protein kinase (MAPK) cascades [29, 30]. Therefore, certain WRKY Group III TFs significantly affect plant immunity through autoregulation, cross regulation, and transcriptional activation.
Tomato (Solanum lycopersicum) is an important vegetable that provides nutrients, including vitamins, dietary fiber, and sugars. As a model system, tomato has been used to investigate fruit development and ripening, domestication, and defense regulation [31–33]. Tomato was mostly produced in Asia from 1993 to 2014, followed by America and Europe (http://faostat.fao.org). Tomato production in Asia accounted for 51.2 % yield. However, pathogens have severely threatened tomato development and production. For instance, TYLCV infection has adversely affected various plant aspects, such as yield. In tomato, TYLCV infection has caused evident symptoms of mottling and yellowing of young leaves, upward curling of leaf margins, stunted growth, and flower abscission [34].
Tomato is a valuable vegetable worldwide but is prone to pathogenic infections, including TYLCV. Huang et al. conducted a genome-wide analysis of tomato WRKY genes, identified 81 WRKY TFs in tomato, and classified these TFs into three main groups, namely, Groups I, II, and III [35]. However, the roles of WRKY TFs, especially those in Group III, in tomato remain unclear. Our study aimed to obtain novel insights into the interaction of WRKY Group III TFs with TYLCV infection in tomato. Symptom characteristics, interaction network, expression profiles, and virus-induced gene silencing (VIGS) were also discussed to elucidate the response to TYLCV infection of WRKY Group III TFs. Our study further aimed to determine the functional mechanism of WRKY Group III TFs involved in defense against TYLCV and to improve the resistance to TYLCV infection of tomato.
Methods
Plant material and TYLCV infection
Derived from T5678161-1-1-2-2 and T07-018, ‘Zheza-301’ was bred to resist to TYLCV infection with the Ty-2 locus. ‘Jinpeng-1’, a hybrid of Holland tomato cultivar 99-13A and 9708B from America, was susceptible to TYLCV infection [6]. Seeds of ‘Jinpeng-1’ and ‘Zheza-301’ were obtained from Xi’an Jinpeng Seed Co., Ltd. and the Institute of Vegetables, Zhejiang Academy of Agricultural Sciences, respectively [6]. The seeds of TYLCV resistant tomato cultivar ‘Zheza-301’ and TYLCV susceptible cultivar ‘Jinpeng-1’ were sown in an artificial chamber (Nanjing Agricultural University (32°02’N, 118°50’E)) at 25 °C/18 °C for 12 h/12 h conditions with 300 μmol · m−2 s−1 light intensity.
Tomatoes were grown in hole trays (6 × 12 holes) filled with a mixture of vermiculite, organic solid, and perlite (1:2:1, v/v/v). Viruliferous whiteflies Bemisia tabaci (Hemiptera: Aleyrodidae) were allowed to feed on tomato plants in an insect-proof greenhouse in the Provincial Key Laboratory of Agrobiology, Jiangsu Academy of Agricultural Sciences (Nanjing, China). Upon reaching the two-leaf stage, tomato seedlings were exposed to viruliferous whiteflies, which fed on tomato plants to infect TYLCV. After 0, 2, 4, 6, 8, and 10 d of TYLCV infection, the leaves were collected and frozen in liquid nitrogen for RNA extraction. Tobacco (Nicotiana tabacum) from the genus Nicotiana was widely used as a premier plant cell biology model [36]. Tobacco seeds were also grown in the same artificial chamber to prepare for subcellular localization analysis.
Sequence alignment and phylogenetic analysis of WRKY Group III TFs
The WRKY Group III TFs identified in rice, tomato, and Arabidopsis were downloaded from the Phytozome10. The WRKY Group III TFs sequences in Chinese cabbage were obtained from the Brassica Database (BRAD, http://brassicadb.org/brad/blastPage.php). PlantTFDB (Plant Transcription Factor Database) provided the WRKY TFs of other species [37]. Multiple sequence alignment was conducted using Clustal X, and MEGA 5 was used to conduct the phylogenetic tree based on the neighbor-joining method [38, 39].
Exon/intron structure and promoter region analysis of WRKY Group III TFs
Gene Structure Display Server (GSDS v2.0, http://gsds.cbi.pku.edu.cn/index.php) was used to analyze the exon and intron structures of each WRKY Group III TF in rice, tomato, and Arabidopsis [40]. Approximately 1.5 kb of DNA sequence upstream from the codons of 6 WRKY Group III TFs in response to TYLCV infection was downloaded from the genome Solanaceae Genomics Network, (SGN, Release 2.3, http://solgenomics.net/organism/solanum_lycopersicum/genome). PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to analyze the promoter region of WRKY Group III TFs [41].
Identification of orthologous and paralogous gene and interaction network analysis of WRKY Group III TFs
OrthoMCL software was used to identify the orthologous and paralogous genes of WRKY TFs in tomato, Arabidopsis, and rice [42]. The relationship between orthologous and paralogous genes was displayed by Circos software [43]. The functional interaction network of WRKY Group III TFs in tomato was conducted. STRING software was responsible for the conduction of the interaction network [44].
Subcellular localization analysis of WRKY Group III TFs in tomatoes
The full-length cDNA of SolyWRKY41 and SolyWRKY54 without stop codons were amplified into the pA7 vector by using specific primers in accordance with a previously described method to determine the subcellular localization of Group III TFs in tomato [10]. Empty vector pA7 containing 35S::GFP (green fluorescent protein) fusion protein was used as control. The constructed plasmids were then transferred into N. benthamiana leaves, which were placed on Murashige and Skooog (MS) medium via a biolistics procedure by using a helium-driven particle accelerator (PDS-1000, Bio-Rad). N. benthamiana leaves were then incubated for 24 h in the dark; the transient expression of GFP was then obtained using the LSM 780 confocal microscopy imaging system (Zeiss, Germany) [45]. The primers are shown in Table 1.
Table 1.
Primers used in the text
| Gene | Direction | Sequence(5'-3') | Function |
|---|---|---|---|
| SolyWRKY41 | Forward | ATGGAGAAAGTTAAAAGTATGGA | Full lengths clone |
| Reverse | TTAAATGAAGAATTCTTCAATGTC | ||
| SolyWRKY54 | Forward | ATGGATTGTGGATTCAATTATGAAT | Full lengths clone |
| Reverse | TTATCTGAAAAAATCAGAGAAATTTG | ||
| SolyWRKY41 | Forward | CACCATCACCATCACGCCATGATGGAGAAAGTTAAAAGTATGGA | Subcellular localization |
| Reverse | CACTAGTACGTCGACCATGGCAATGAAGAATTCTTCAATGTC | ||
| SolyWRKY54 | Forward | CACCATCACCATCACGCCATGATGGATTGTGGATTCAATTATGAAT | Subcellular localization |
| Reverse | CACTAGTACGTCGACCATGGCTCTGAAAAAATCAGAGAAATTTG | ||
| SolyWRKY41 | Forward | GTGAGTAAGGTTACCGAATTCGCAACACCAAACCATAACGCTGAA | VIGS vector conduction |
| Reverse | GGGACATGCCCGGGCCTCGAGAGGAATTTGAAATCGAAGTCGGAGT | ||
| SolyWRKY54 | Forward | GTGAGTAAGGTTACCGAATTCCCACAGAAACATAAACACAAGAAAC | VIGS vector conduction |
| Reverse | GGGACATGCCCGGGCCTCGAGAGGAGAGACAAAAGATGGTGAATAA | ||
| SolyWRKY41 | Forward | GCAACACCAAACCATAACGCTGAA | RT-qPCR |
| Reverse | AGGAATTTGAAATCGAAGTCGGAGT | ||
| SolyWRKY42 | Forward | AAGAAACTGTTCCTTTCACTCCACT | RT-qPCR |
| Reverse | GAGCTGAACACAATACGATCCGATT | ||
| SolyWRKY53 | Forward | CCACAACCAACATCGCCAGAGAA | RT-qPCR |
| Reverse | ACGGTGAATAGCCGCTACCTATCA | ||
| SolyWRKY54 | Forward | CCACAGAAACATAAACACAAGAAAC | RT-qPCR |
| Reverse | AGGAGAGACAAAAGATGGTGAATAA | ||
| SolyWRKY80 | Forward | TCACTGTCCAACTTCAAACTCTACT | RT-qPCR |
| Reverse | ACGGTCTTGCTTCTCCTTCTTCT | ||
| SolyWRKY81 | Forward | GCAATAGAAGGTTTAATTCGTGGTC | RT-qPCR |
| Reverse | GTAGCGACGACGACATCAGA | ||
| SolyWRKY41 | Forward | AGTGACATATAAAGGAAGGCACAGT | RT-qPCR for VIGS |
| Reverse | CCCTTCTCCACCAAATGAGGAATT | ||
| SolyWRKY54 | Forward | GCAAGTGCAGAGGTCTGATGATGA | RT-qPCR for VIGS |
| Reverse | TGTTGATGAAGTGTTGGCTGAGAAC | ||
| TYLCV02 | Forward | CGCCCGTCTCGAAGGTTC | RT-qPCR for VIGS |
| Reverse | GCCATATACAATAACAAGGC | ||
| TYLCV01 | Forward | ATGTCGAAGCGACCAGGCGATATAAT | Detection for TYLCV DNA |
| Reverse | TTAATTTGATATTGAATCATAGAAAT | ||
| Tubulin | Forward | TGACGAAGTCAGGACAGGAA | Reference gene |
| Reverse | CTGCATCTTCTTTGCCACTG |
Construction of VIGS vector and agroinfiltration
The fragments of SolyWRKY41 (262 bp) and SolyWRKY54 (243 bp) genes were also amplified with specific primers and constructed into pTRV2 vectors from Antoine Bouteilly (Centre National de la Recherche Scientifique). Recombinant plasmids of SolyWRKY41 and SolyWRKY54 were then transformed into Agrobacterium tumefaciens strain GV3101 by electroporation, respectively. Agrobacterium GV3101 cells were cultured in liquid YEB medium with 50 mg/L rifampin and 50 mg/L kanamycin overnight at 28 °C; cells were then collected and resuspended in infiltration media (10 mM MgCl2, 10 mM MES, and 200 mM acetosyringone) with an OD (optical density) value of 2.0. After being cultured at 28 °C for 4 h, the cotyledons of two tomato cultivar seedlings were infiltrated into the Agrobacterium [34]. Agroinfiltration of pTRV1 and pTRV2 served as negative control and pTRV2-PDS with pTRV1 were used as positive control.
For VIGS experiments, new emerging leaves from ‘Zheza-301’ were collected to extract RNA and determine the expression level of the target genes via quantitative real-time–polymerase chain reaction (RT-qPCR) after seven days of agroinfiltration. Seedlings of ‘Zheza-301’ were then transferred into the insect-proof greenhouse to feed the virusliferous whiteflies. After one week, the DNA was extracted from TYLCV-infected leaves to determine TYLCV DNA accumulation. Primers of SolyWRKY41 and SolyWRKY54 for RT-qPCR were designed to anneal outside the region targeted for silencing to ensure that only the endogenous gene would be detected [46].
Quantitative real-time PCR analysis
To detect whether the DNA of TYLCV accumulated in resistant and susceptible tomato cultivars, we conducted a PCR analysis by using primer TYLCV01-F/R (Table 1). For PCR analysis, DNA was extracted from the two tomato cultivars after they were exposed to TYLCV infection for approximately 7 days. For VIGS experiments, total RNA and DNA were extracted from non-silenced and silenced plant leaves by using RNA kit (RNA simple total RNA kit) and DNA kits (DNA secure Plant kit) (Tiangen, Beijing, China) in accordance with the manufacturer’s protocols. To detect TYLCV accumulation, specific primers (TYLCV02-F/R) were used to conduct the RT-qPCR analysis (Table 1).
For RT-qPCR, SYBR Premix Ex Taq kit (TaKaRa, Dalian, China), iQTM5 software, and iQTM5 Real-time PCR System were used to complete the following procedure: 95 °C for 30 s initially, followed by 40 cycles at 95 °C for 5 s; 60 °C for 30 s and melting curve analysis (61 cycles) at 65 °C for 10 s. Three technical repeats were performed with each RNA sample of two tomato cultivars. A-Tubulin was used to regulate the expression level. 2-ΔΔCT method was used to measure the RNA level, which were expressed relative to the Tubulin gene [47].
Results
Detection of TYLCV DNA accumulation in resistant and susceptible tomato cultivars
Two tomato cultivars (‘Zheza-301’ and ‘Jinpeng-1’) showed different symptoms after TYLCV infection. ‘Jinpeng-1’ was susceptible to TYLCV infection and the symptom in ‘Jinpeng-1’ was obvious than in ‘Zheza-301’ during the process of TYLCV infection. After TYLCV infection for 40 d, ‘Jinpeng-1’ showed more obvious symptom than ‘Zheza-301,’ which showed yellow and curly leaves, dwarfed plants, withered flowers, and small and shriveled newly formed leaves (Fig. 1). PCR analysis detected TYLCV DNA accumulation in both tomato cultivars. As shown in Fig. 2, in the control plants, there was no TYLCV accumulation in either ‘Zheza-301’ or ‘Jinpeng-1’. After TYLCV infection for 7 d, expression of TYLCV DNA was detected both in ‘Jinpeng-1’ and ‘Zheza-301’. There was higher expression of TYLCV DNA in ‘Jinpeng-1’than in ‘Zheza-301’.
Fig. 1.

Symptom characteristics of ‘Zheza-301’ and ‘Jinpeng-1’ after TYLCV infection about 40 d
Fig. 2.
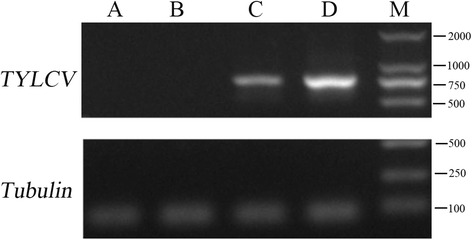
Detection of TYLCV DNA accumulation in control and TYLCV infection tomato cultivars by PCR. a Control plants: Zheza-301, b Control plants: Jinpeng-1, c Treatment plant: Zheza-301 after TYLCV infection at 7 d, d Treatment plant: Jinpeng-1 after TYLCV infection at 7 d, m marker
Members of tomato WRKY Group III TFs are involved in response to TYLCV infection
Comparative transcriptome profiling of resistant and susceptible tomato cultivars in response to TYLCV infection has been conducted by Chen et al. [48]. According to the transcriptome database (SRP028618), 6 WRKY TFs (SolyWRKY41, SolyWRKY42, SolyWRKY53, SolyWRKY54, SolyWRKY80, and SolyWRKY81) were identified to respond the TYLCV infection. The expression levels of these 6 WRKY genes in susceptible tomato cultivar were down regulated; by contrast, their expression did not significantly differ in the resistant tomato cultivar. These 6 WRKY TFs were classified into Group III according to the phylogenetic tree analysis, suggesting the importance of Group III TFs in the plant-pathogen interaction.
Phylogenetic analysis of WRKY Group III TFs
To investigate the phylogenetic relationship of WRKY Group III TFs in tomato, comparative analysis from three other sequenced plant genomes including the Arabidopsis, rice, and Chinese cabbage, was conducted using MEGA 5 (Additional file 1: Table S1) [16]. Clade 1 had the most Group III TFs members (28), followed by clade 4 (24), and clade 3 (17); clade 2 contained the least WRKY Group III TFs members (8) (Fig. 3). Clades 1, 3, and 4 contained WRKY Group III TFs in all 4 species. WRKY Group III TFs in tomato and rice were not classified into clade 2 which consisting of Arabidopsis and Chinese cabbage; this distribution may be related to some special circumstances (the split of different families in dicotyledon). Among those WRKY Group III TFs that responded to TYLCV infection in tomato, 2 members (SolyWRKY80, SolyWRKY81) and 4 members (SolyWRKY53, SolyWRKY54, SolyWRKY41, and SolyWRKY42) were classified into clades 1 and 3, respectively.
Fig. 3.
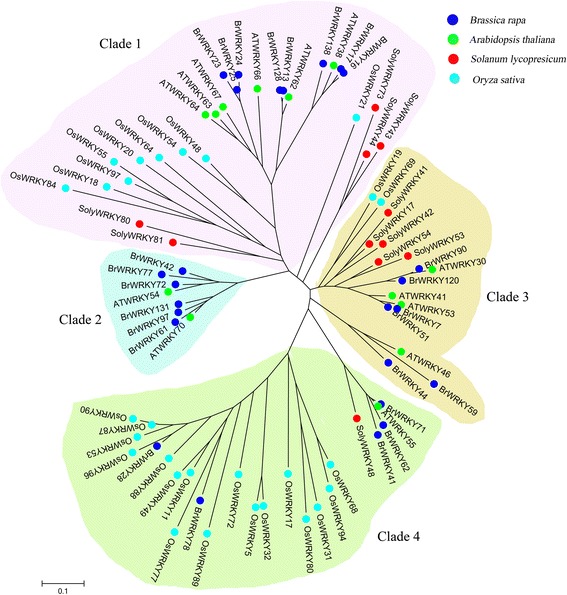
Phylogenetic tree of WRKY Group III TFs of tomato, Arabidopsis, Chinese cabbage, and rice
Exon/intron structure analysis of each WRKY Group III TF
Gene structure including exon/intron structural diversification plays important roles in the evolution of multigene families [20]. To understand the structural diversity of WRKY Group III TFs, exon/intron structure analysis was performed on 81 WRKY TFs in tomato that were identified by Huang et al. [35]. As shown in Additional file 2: Figure S1, the exon/intron structures in the same group were similar. Group III TFs contained 11 members; 9 genes had two introns. For Group II, most genes had 3 exons in Group IIc and Group IId. By comparison, most Group IIa and IIb genes had at least 3. Of the 17 genes in Group IIe, 8 (SolyWRKY18, SolyWRKY25, SolyWRKY39, SolyWRKY40, SolyWRKY36, SolyWRKY37, SolyWRKY38, and SolyWRKY40) showed the same intron/exon structures.
Exon/intron organization maps from each Group III TF among Arabidopsis, rice, and tomato were generated (Fig. 4). Classified into 4 clades, 52 Group III TFs contained different exon numbers, ranging from 2 to 6. A total of 32 genes had 3 exons, followed by 2 exons (10), 4 exons (4) and 6 exons (4). OsWRKY53 and OsWRKY90 had 5 exons. Exon numbers of most genes in clade 4 were from 4 to 6. Among 11 WRKY Group III TFs in tomato, 9 genes contained 3 exons and 2 introns. SolyWRKY73 of clade 1 only had 2 exons and 1 intron. AtWRKY55/SolyWRKY48, an orthologous pair according to the cluster at the terminal branch of the phylogenetic tree, showed the change of exon/intron numbers. Compared with AtWRKY55, SolyWRKY48 gained 2 exons. The difference could also be found among those orthologous pairs AtWRKY41/53, OsWRKY69/19, and OsWRKY53/87. During WRKY Group III TFs evolution, exon/intro losses and gains have always occurred, which may be associated with the functional diversity among closely related Group III TFs.
Fig. 4.
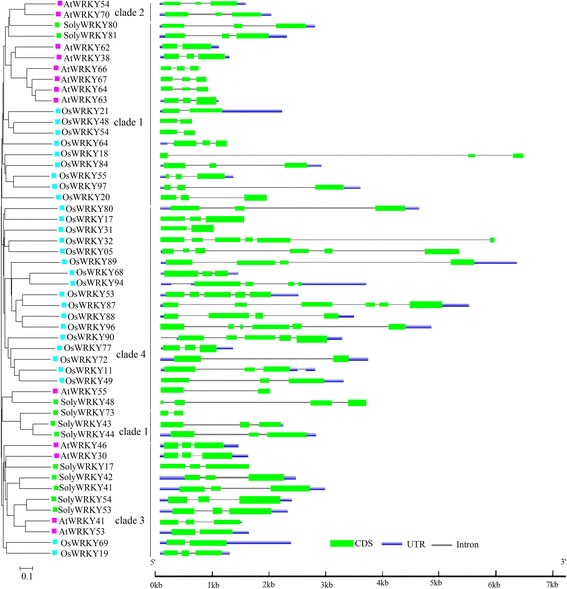
Exon/intron structure and phylogenetic relationship of WRKY Group III TFs from tomato, Arabidopsis, and rice. Green rectangles and thin lines represent the exons and introns, respectively. UTRs (untranslated regions) are showed by thick blue lines
Promoter region analysis of SolyWRKY Group III TFs in response to TYLCV infection
In WRKY TF promoters, a series of W or W-like boxes suggested complex interaction of WRKY TFs with each other by binding with W-box elements [49, 50]. Some WRKY proteins could regulate their own gene expression by binding with cis elements in their own promoters [51, 52]. To determine if auto-regulation and cross-regulation existed in WRKY Group III TFs, promoter region, a sequence that could regulate and initiate gene transcription, was identified using approximately 1.5 kb DNA sequences upstream from the codons of 6 SolyWRKY Group III TFs (Fig. 5). Approximately 12 cis elements were found in most SolyWRKY Group III TFs, including W-box, TATA-box, TC-rich, and HSE-element. Some basic elements, including Box 4 (part of a conserved DNA module involved in light response), Box I (light-responsive element), Skin 1 motif (cis-acting regulatory element required for endosperm expression), and TATA elements (core promoter element around −30 of transcription start), existed in the promoter regions of all 6 SolyWRKY Group III TFs.
Fig. 5.
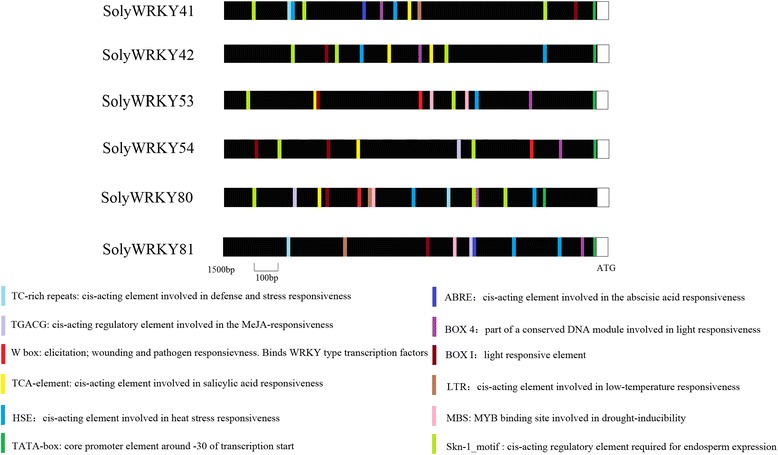
Protomer region analysis of six SolyWRKY Group III TFs involved in TYLCV defense. Different colors indicate different cis elements existing in the promoter region
In the study, elements related to hormone regulation, such as ABRE (cis-acting element involved in abscisic acid response), TCA (cis-acting element involved in salicylic acid response), and TGACG element (cis-acting element involved in the MeJA response) were identified in Group III TF promoter region. Cis elements related to abiotic stress were found in several SolyWRKY TFs promoter regions; for example, HSE/LTR-elements, which are involved in high/low temperature responsiveness, suggested that upstream genes could regulate WRKY gene expression by binding to the corresponding cis elements. Several SolyWRKY TFs (SolyWRKY53, SolyWRKY54, and SolyWRKY80) contained the W or W-box element in their promoter regions, suggesting that these SolyWRKY TFs could be regulated by interaction with each other or by auto-regulation (Fig. 5).
Microsynteny analysis of WRKY Group III TFs
Two consecutive genome triplications have happened in the Solanum lineage [53]. In the study, Arabidopsis and rice were selected to make the microsynteny with tomato using whole-genome sequence according to the homologous gene locations. First, we analyzed the relationship of WRKY TFs in the three species. As shown in Fig. 6, 47 and 12 pairs of orthologous WRKY TFs were identified between tomato and Arabidopsis and between tomato and rice, respectively (Additional file 1: Table S2). Moreover, tomato, Arabidopsis, and rice contained 28, 19, and 39 paralogous WRKY TFs pairs (Additional file 1: Table S3).
Fig. 6.
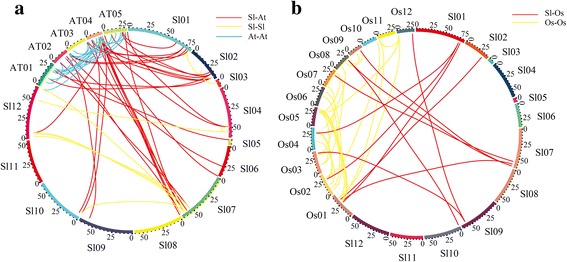
Comparative analysis of WRKY TFs in tomato, Arabidopsis, and rice. a Microsynteny of WRKY TFs across tomato and Arabidopsis. Tomato chromosomes label Sl01- Sl12. AT01-AT05 represents the Arabidopsis chromosomes. Red lines represent the orthologous WRKY genes among tomato and Arabidopsis. Yellow and blue lines represent paralogous WRKY genes in tomato and Arabidopsis, respectively. b Microsynteny of WRKY TFs across tomato and rice. Tomato chromosomes label Sl01- Sl12. OS01-Os12 represents the rice chromosomes. Red lines represent the orthologous WRKY genes among tomato and Arabidopsis. Yellow lines represent paralogous WRKY genes rice
Eleven WRKY Group III TFs from tomato were distributed across 7 chromosomes. SolyWRKY43, SolyWRKY44, and SolyWRKY73 were located in chromosome 05. Chromosomes 03 and 08 each contained 2 members of WRKY Group III TFs, followed by chromosomes 01 (1), 06 (1), 09 (1), and 10 (1) (Additional file 3: Figure S2). To analyze the orthologous and paralogous relationships of WRKY Group III TFs between eudicots and monocots, the microsynteny analyses of Arabidopsis, tomato, and rice were conducted on the basis of WRKY Group III TF locations of orthology and paralogy gene pairs. As shown in Fig. 7, 7 collinear gene pairs were identified in the tomato genome. Four WRKY Group III TFs, including SolyWRKY80, SolyWRKY81, SolyWRKY48, and SolyWRKY43, from tomato were absent in any microsynteny. This finding indicated that independent duplication events, except whole genome duplication, occurred.
Fig. 7.
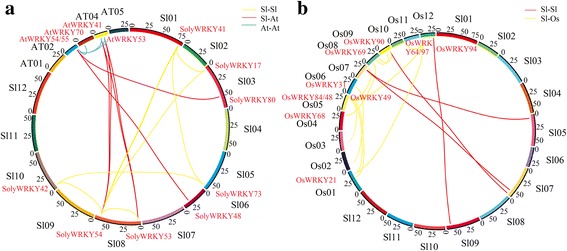
Microsynteny of WRKY Group III TFs in tomato, Arabidopsis, and rice. a Microsynteny of WRKY TFs across tomato and Arabidopsis. Tomato chromosomes label Sl01- Sl12. AT01-AT05 represents the Arabidopsis chromosomes. Red lines represent the orthologous WRKY genes among tomato and Arabidopsis. Yellow and blue lines represent paralogous WRKY genes in tomato and Arabidopsis, respectively. b Microsynteny of WRKY Group III TFs across tomato and rice. Tomato chromosomes label Sl01- Sl12. OS01-Os12 represents the rice chromosomes. Red lines represent the orthologous WRKY genes among tomato and Arabidopsis. Yellow lines represent paralogous WRKY genes rice
The corresponding interspecies relationships of microsynteny were also analyzed. Six orthologous gene pairs, namely, SolyWRKY53/AtWRKY53, SolyWRKY53/AtWRKY41, SolyWRKY54/AtWRKY53, SolyWRKY54/AtWRKY41, SolyWRKY48/AtWRKY55, and SolyWRKY80/AtWRKY54, were found between tomato and Arabidopsis. Four orthologous gene pairs existed between tomato and rice. This finding suggested that the relationship between tomato and Arabidopsis was closer than that between tomato and rice. Some collinear gene pairs were present in tomato and Arabidopsis but were absent in tomato and rice because rice diverged from the common ancestor of tomato and Arabidopsis. Among 11 SolyWRKY Group III TFs, SolyWRKY81 showed no linkage with other WRKY TFs, which may take part in the expansion of WRKY Group III TFs.
Evolution of WRKY Group III TFs among different species
WRKY TFs are common not only in land plants but in algae as well [10]. To analyze the evolution of WRKY Group III TFs, a schematic of the phylogenetic tree was conducted (Fig. 8). A total of 1,722 WRKY TFs among 28 species were identified. As shown in Fig. 8, numbers of WRKY TFs in land plants are larger than in algae, which contain only 2 WRKY TFs in Chlamydomonas reinhardtii and Ostreococcus lucimarinus. In algae, C. reinhardtii showed no Group III TFs. WRKY Group III TFs exist in most land plants, except Picea abies. The numbers of WRKY Group III TFs in monocots were more than in eudicots. Percentage of Group III TFs in monocots was almost at 20.00 %–36.94 %. Group III TFs in Triticum aestivum occupied 36.94 % in monocots; by contrast, A. lyrata yielded the highest percentage (21.12 %) of Group III TFs in eudicots. These results indicated that numerous diversifications for WRKY TFs, particularly WRKY Group III TFs, occurred after the divergence of the monocots and eudicots. Notably, Group I TFs exists in all the land plants, suggesting that Group I may have evolved earlier.
Fig. 8.

Comparison of WRKY family transcription factors among different plants
Subcellular localization analysis of SolyWRKY Group III TFs involved in TYLCV infection
In tomato, 4 out of 6 SolyWRKY Group III TFs in response to TYLCV infection were classified into clade 3. Two clade 3 TFs pairs (SolyWRKY53/54 and SolyWRKY41/42) were clustered together (Fig. 3). To determine the subcellular localization of SolyWRKY Group III TFs, constructions of two clade 3 members (SolyWRKY41-GFP and SolyWRKY54-GFP) and empty vectors containing 35S::GFP were introduced into the N. benthamiana leaves. N. benthamiana leaf cells of the control with 35S::GFP showed GFP fluorescence throughout the cell. In contrast, confocal laser scanning microscopy showed that SolyWRKY41-GFP and SolyWRKY54-GFP exhibited clear signals in the nucleus of the N. benthamiana leaf cells (Fig. 9). Results indicated that SolyWRKY41 and SolyWRKY54 were nuclear proteins in vivo.
Fig. 9.
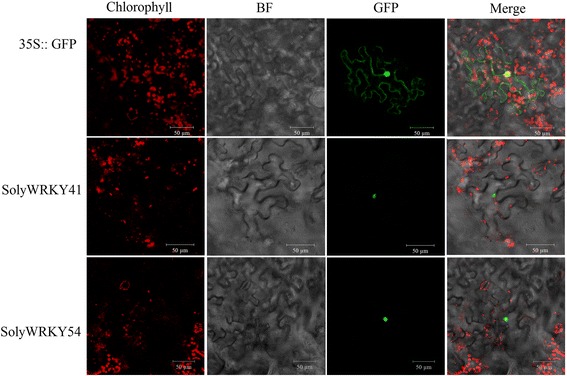
Subcellular localization of SolyWRKY Group III genes in response to TYLCV infection. Confocal images showing the localization of SolyWRKY Group III genes and terminal proteins transiently expressed in N. benthamiana leaves. Bar 50 μm. Chlorophyll: Image after chlorophyll autofluorescence; BF: Image after bright field. GFP: Image after green fluorescence. Merge represents the Image after merger of chlorophyll, bright field, and green fluorescence
Interaction network of WRKY Group III proteins in tomatoes
Interaction network of WRKY Group III TFs was analyzed to further understand about the regulation mechanism. As shown in Fig. 10, 5 Group III proteins (SolyWRKY17, SolyWRKY41, SolyWRKY53, SolyWRKY54, and SolyWRKY80) showed interactions with other proteins in tomato genome. SolyWRKY80 showed co-expression relationships with 2 WRKY Group I proteins (SolyWRKY68 and SolyWRKY49), and 2 WRKY Group IIa proteins (SolyWRKY50 and SolyWRKY62). SolyWRKY80 could also interact with MAPK5. SolyWRKY41was identified to interact with other WRKY proteins (SolyWRKY24, SolyWRKY62, and SolyWRKY50), suggesting SolyWRKY80 and SolyWRKY42 had the similar functions in the regulation network. Isochorismate synthase (ICS, LOC778225), could interact with SolyWRKY41 protein. The WRKY Group III proteins, SolyWRKY80 and SolyWRKY53 showed significant correlations with SolyWRKY17.
Fig. 10.
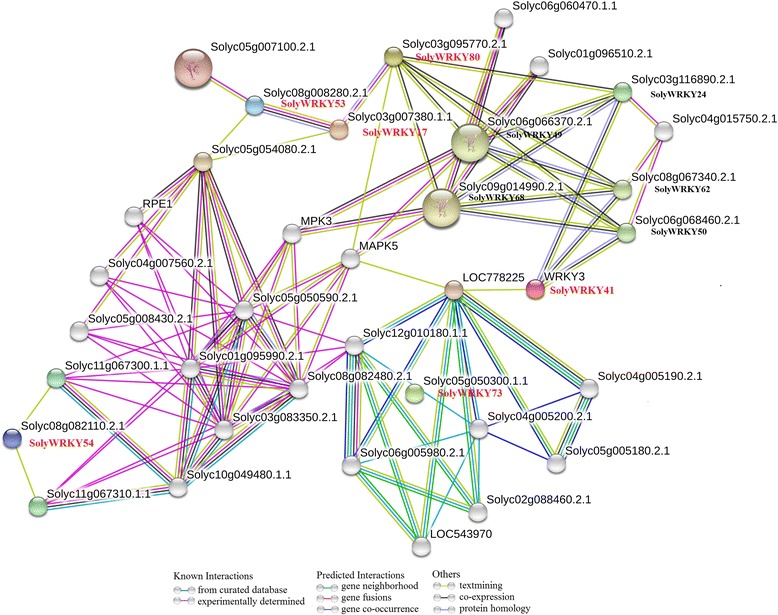
Interaction network of six Soly WRKY Group III TFs in response to TYLCV infection
Expression profiles of SolyWRKY Group III genes in response to TYLCV infection
To elucidate the expression patterns of SolyWRKY Group III genes involved in TYLCV response in resistant and susceptible tomato cultivars, SolyWRKY41, SolyWRKY42, SolyWRKY54, SolyWRKY53, SolyWRKY80, and SolyWRKY81 were selected for RT-qPCR analysis after TYLCV infection at different times in resistant and susceptible tomato cultivars (Fig. 11). The expression levels of 6 SolyWRKY Group III genes increased in the susceptible tomato cultivar ‘Jinpeng-1’ after TYLCV infection; and in the resistant tomato cultivar ‘Zheza-301’, 4 Group III genes showed down-regulation, except for SolyWRKY42 and SolyWRKY80. In ‘Jinpeng-1,’ after 6 d of TYLCV infection, the expression level of SolyWRKY53 increased by approximately 8.0-fold. The highest expression peaks of other five genes were observed after TYLCV infection for 4 d, with approximately 13-fold in SolyWRKY80, 5-fold in SolyWRKY81, and 3-fold in SolyWRKY42 (Fig. 11). In ‘Zheza-301,’ SolyWRKY80 showed rapid increase after 6 d and the expression peaked at 23-fold after TYLCV infection for 10 d. Expression of SolyWRKY42 showed the highest expression level at 4 d with 5-fold.
Fig. 11.
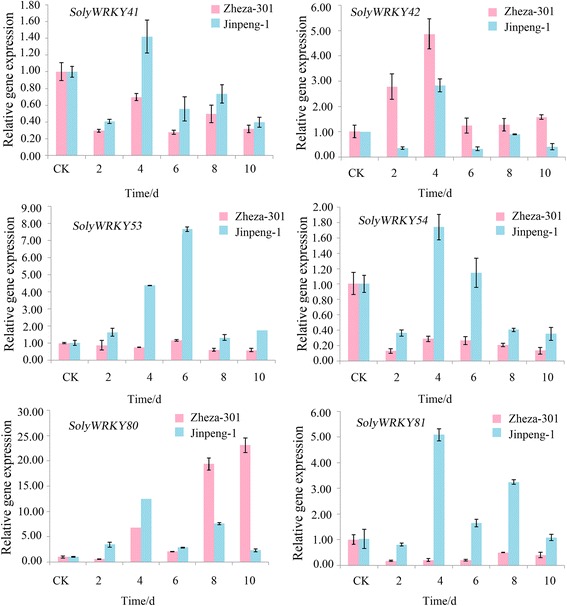
Expression levels of six Soly WRKY Group III genes after TYLCV infection in tomatoes
VIGS validation of WRKY Group III genes in response to TYLCV infection in tomatoes
To further analyze the function of SolyWRKY Group III genes in response to TYLCV infection, VIGS validation was conducted in resistant tomato cultivar ‘Zheza-301’. Two SolyWRKY Group III genes (SolyWRKY41 and SolyWRKY54), classified in clade 3, were knocked down by the tobacco rattle virus (TRV) mediated VIGS system (Fig. 3) [54].
As shown in Fig. 12a, after agroinfiltration for 3 weeks, positive control plant with pTRV1 and pTRV2-PDS showed leached areas in leaves. Expression levels of SolyWRKY41 and SolyWRKY54 after silencing were decreased by approximately 20 % and 50 %, respectively, compared with those in the negative control (Fig. 12b). Meanwhile, TYLCV DNA accumulation was detected after TYLCV infection for one week. As shown in Fig. 12c, TYLCV DNA accumulation decreased in silencing SolyWRKY41 and SolyWRKY54, compared with those in the control plants. Results indicated that SolyWRKY41 and SolyWRKY54 negatively regulated response to TYLCV infection.
Fig. 12.
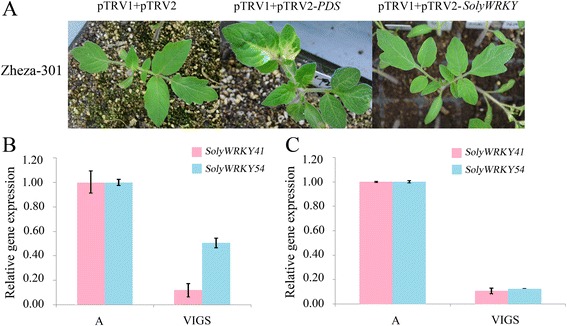
Validation of putative Soly WRKY Group III genes by virus induced gene silencing (VIGS). a Cotyledon agroinfiltration of pTRV vectors was conducted in the resistant cultivar ‘Zheza-301’ at the cotyledon stage. After three weeks, leaves with pTRV1 and pTRV2-SolyWRKY showed normal phenotype. Leaves of ‘Zheza-301’ with pTRV1 and pTRV2-PDS showed bleached areas. b The expression level of two SolyWRKY Group III genes in the VIGS treated plants one week after agroinfiltration. A-Tubulin (Solyc04g077020.2) was used as an internal reference. ‘A’ represented the ‘Zheza-301’ treated with pTRV1 and pTRV2 (negative control), ‘VIGS’ represented the ‘Zheza-301’ with pTRV1 and pTRV2-SolyWRKY. c The accumulation of TYLCV DNA in the SolyWRKY Group III genes silenced ‘Zheza-301’ plants. ‘A’ represented the ‘Zheza-301’ treated with pTRV1 and pTRV2 (negative control), ‘VIGS’ represented the ‘Zheza-301’ with pTRV1 and pTRV2-SolyWRKY
Discussion
Plants are constantly exposed to various adverse stress conditions, such as pathogen infection and abiotic stress, because of their sessile growth habit. During pathogen infection, genes could activate inducible defense responses in host plants by transcriptional regulation [55]. A large number of WRKY TFs that contain WRKY domains participate in pathogen infection and other treatments, such as SA, wounding, or senescence [56–58]. As an important group of WRKY TFs, Group III TFs are involved in plant innate immune systems; however, limited information is available regarding the biological functions of WRKY Group III TFs in response to TYLCV infection of tomato. In our study, expression analysis and VIGS-based knockdown demonstrated that WRKY Group III TFs functioned as positive and negative regulators in defense response against TYLCV infections in tomatoes.
TYLCV infection in tomato cultivars
In this study, viruliferous whiteflies spread the virus by feeding on tomato leaves. Studies showed that whiteflies could carry and transmit TYLCV and other viruses, including cucurbit leaf crumple virus and tobacco curly shoot virus [59, 60]. PCR analysis was used to detect TYLCV DNA accumulation in tomato leaves. As shown in Fig. 2, compared with control plants, TYLCV DNA showed high expression levels in both two tomato leaves after TYLCV infection for 7 d. Numerous studies have conducted the analyses between plants and TYLCV infection using the whiteflies as the medium, suggesting their feasibility in conducting the analysis of plant-TYLCV interaction [3, 61–63].
Gene structure and evolution of WRKY Group III TFs
Multigene family evolution can be driven by gene structural diversity [20]. Exon/intron organization mapping of WRKY Group III TFs from tomato, Arabidopsis, and rice was conducted to better understand the structural diversity. A total of 52 WRKY Group III TFs contained different numbers of exon/intron. Gene structural diversity existed in these three species. AtWRKY53 and OsWRKY69 in clade 3 contained 2 exons; by comparison, 3 exons existed in other WRKY Group III TFs in this clade. Similar exon/intron structures could be found in most recent paralogs. AtWRKY55/SolyWRKY48, an ortholog pair, showed change of exon/intron numbers, indicating the exon/intron loss or gain during the long evolutionary period [64]. Huang et al. have analyzed the conserved motifs of WRKY Group III TFs, which were also important to diversified functions of Group III proteins [35]. Different exon/intron structures existed among 11 SolyWRKY Group III TFs, such as SolyWRKY73 and SolyWRKY48, although similar motif structures were identified [35]. Differences in exon/intron structures and motif characteristics indicated functional diversity of WRKY Group III TFs.
To explore the evolution of WRKY Group III TFs, we first analyzed the relationship between tomato with Arabidopsis and rice by microsynteny. Difference in the orthologous gene pairs between tomato/Arabidopsis (6) and tomato/rice (4) indicated the closer relationship of tomato and Arabidopsis. Independent duplication also existed in the process of WRKY Group III TFs. During species evolution, gene duplication and loss have always taken place. A model diagram among plantae plants was then built to further analyze the origin and evolution of WRKY Group III TFs. As shown in Fig. 8, WRKY Group I TFs exist in almost all species, even in C. reinhardtii. However, Groups II and III TFs are identified in most land plants. These results showed that Group I had early origin, but Groups II and III TFs expanded with the evolution of higher plants [17, 65]. Compared with algae, land plants have a relatively large number of Group III TFs. Significant difference in the number of Group III TFs between monocotyledonous and dicotyledonous plants suggested higher function and more active duplication in monocots than in eudicots.
WRKY Group III TFs involved in TYLCV infection in tomatoes
Six SolyWRKY TFs belonging to Group III are involved in response to TYLCV infection according to a previously described database [48]. The cis-regulatory elements were identified in the promoter regions of 6 SolyWRKY Group III proteins involved in different functions, such as abiotic stress (LTR, HSE elements), hormone regulation (ABER,TCA, and MeJA elements), and disease resistance (W-box elements). WRKY TFs could be regulated by other WRKY TFs and proteins with special binding to different cis elements. WRKY proteins could bind with W or W-box (C/T)TGAC(T/C) elements to regulate the pathogen defense-related genes [10]. In rice, 2 cis elements, PRE2 and PRE4 (harboring the W box), were identified from the promoter region of OsWRKY13, which could positively regulate OsWRKY13 gene expression after pathogen-induced protein binding [66]. WRKY13 could bind to W-box in WRKY42 and regulate its expression [26]. There were W boxes existed in the promoter regions of SolyWRKY53, SolyWRKY54, and SolyWRKY80, suggesting these WRKY Group III TFs can participate in TYLCV defense process by autoregulation or cross-regulation with each other [49, 51].
In plant immunity, WRKY TFs could form a complex interconnected regulatory network at several different levels [49, 67]. In the study, an interaction network of SolyWRKY Group III proteins was conducted, which showed that Group III proteins could interact with a series of defense proteins. For example, MAPK5 could phosphorylate some TFs including WRKY to activate the transcription of other genes and interact with SolyWRKY80 [68, 69]. ICS is required to synthesize SA for plant defense [70]. SolyWRKY41 showed significant interaction with ICS by binding the W-box elements existed in the promoter region of ICS. Interaction relationships also existed among WRKY proteins, even in the same group. Two WRKY Group III proteins (SolyWRKY80 and SolyWRKY53) both showed co-expression with SolyWRKY17 (Fig. 10).
Expression profiles of SolyWRKY Group III genes in response to TYLCV infection
In the study, 6 SolyWRKY Group III genes (SolyWRKY41, SolyWRKY42, SolyWRKY53, SolyWRKY54, SolyWRKY80, and SolyWRKY81) were selected to conduct RT-qPCR and VIGS validation analyses (Figs. 11 and 12). WRKY Group III genes in tomato could be implicated in TYLCV defense by inducing positive and negative expression patterns in resistant and susceptible tomato cultivars. Two clade 3 WRKY TFs (SolyWRKY41 and SolyWRY54) were chosen for subcelluar localization. Subcellular localization analysis by using tomato is not yet mature. Numerous studies on tomatoes have successfully used tobacco to conduct subcellular localization [71, 72]. SolyWRKY41 and SolyWRKY54 were located in the nucleus of the N. benthamiana leaf cells. Expression levels of SolyWRKY41 and SolyWRKY54 were decreased compared with those of the control after VIGS. The TYLCV DNA expression also decreased in the silencing SolyWRKY41 and SolyWRKY54 plants, suggesting SolyWRKY41and SolyWRKY54 were down-regulated upon TYLCV infection in resistant tomato cultivar ‘Zheza-301.’
During the process of induced disease resistance, transcriptional regulation of defense gene expression appeared to play a central role [73]. Overexpression of WRKY TFs genes has been shown to confer disease resistance and induce a number of defense-related genes in systemic acquired resistance, including pathogenesis-related (PR) genes (PR1, PR2, and PR5) [74]. Overexpression of AtWRKY70 increased the resistance to virulent pathogens and induced expression of SA-induced pathogenesis-related genes [73]. AtWRKY53 was implicated as positive regulators of senescence [75]. Overexpression of WRKY12 resulted in reduced soft symptoms by Pectobacterium carotovorum in Arabidopsis and Chinese cabbage [76]. In tomato, silencing of SlDRW1, the defense-related WRKY1 gene, resulted in increased severity caused by Botrytis cinerea, suggesting the positive regulation of SlDRW1. Overexpression of WRKY TFs genes could induce a series of related protein expression to regulate resistance to abiotic and biotic stresses, as well as plant growth. In the study, SolyWRKY42 and SolyWRKY80 served as positive regulators in the defense from TYLCV infection process (Fig. 11). These genes increased the resistance in tomato by inducing expression of resistant-related genes or involving SA or JA signaling.
In A. thaliana, 13 Group III members out of 74 WRKY TFs could respond to different plant defense signaling pathways. Orthologous genes of SolyWRKY80, SolyWRKY81, SolyWRKY53, and SolyWRKY54, AtWRKY53 (At4g23810) and AtWRKY70 (At3g56400) were important in plant-pathogen interaction [19]. In addition to WRKY TFs, AP2/ERF TFs could respond to TYLCV infection, and 5 different SolyERF genes (Soly19, Soly36, Soly106, Soly67, and Soly66) are involved in TYLCV defense with a complex network through positive and negative expression [77]. Negative and positive regulations were both important for gene expression [78]. Wang showed that SlybHLH131 was up-regulated in resistant tomato line and down-regulated in susceptible tomato line after TYLCV infection, which was similar to the expression patterns of SolyWRKY53 and SolyWRKY80 [79]. In TYLCV defense, SolyWRKY Group III TFs could serve as positive and negative regulators in the plant defense network and help orchestrate a broad and spatially controlled response.
A probable function mechanism of WRKY Group III involved in TYLCV defense
Transcriptional regulation of defense gene expression is pivotal in inducing disease resistance [80]. A probable regulation network on WRKY-TYLCV infection was conducted [26, 67, 68, 73, 81]. During pathogen infection, WRKY Group III TFs may respond to adverse stress by participating in various signaling pathways, such as hormone signaling, pathogen-induced defense program (Fig. 13). There is complex connection between MAPKs and TFs including WRKY [10, 76]. A MAPK cascade may activate the expression of some TFs (AP2/ERF, WRKY) [68]. SolyWRKY80 showed significant interaction with MAPK5, which was identified to involve in the defense against fungal and viral attacks [82, 83]. AP2/ERF TFs participate in many processes in plant development and induce PR protein expression [81]. Autoregulation and cross-regulation also occur in WRKY TFs by binding to specific cis elements, such as W-box (Fig. 5). WRKY45-2, WRKY13, and WRKY42 could form a transcriptional regulatory cascade in rice resistance to fungal pathogen [26]. WRKY13 could suppress WRKY42 expression by binding the W-box with the promoter region [26].
Fig. 13.
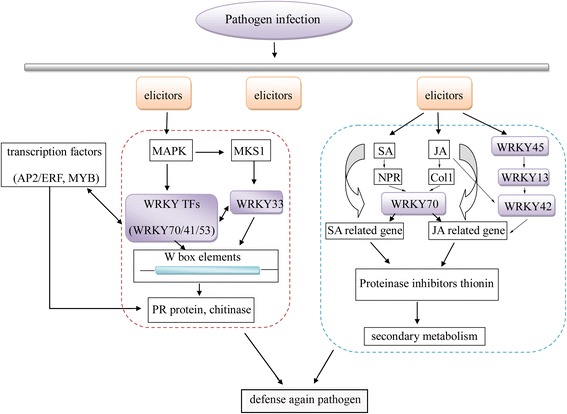
A possible function network of SolyWRKY Group III TFs in response to TYLCV infection
SolyWRKY Group III TFs not only responded to TYLCV infection by negative and positive regulation patterns, but also induced a series of defense protein expression (PR protein, chitinase) and hormone signaling (SA, abscisic acid, gibberellin acid) by binding cis elements [84]. SolyWRKY41 protein could interact with ICS protein by recognizing and binding W-box to regulate pathogen and stress response [70]. WRKY70 was demonstrated to be a common component in SA- and JA-mediated signal pathways [73]. Hormone signaling pathway would induce relative protein expressions, including proteinase inhibitors and thionin, to regulate secondary metabolism. Cross talk existed among signal transduction pathways, which could be important in plant immunity [78]. The extensive cross-regulation mechanism of WRKY Group III TFs is important in tomato–TYLCV defense.
Conclusion
In this study, six WRKY Group III TFs in tomato were identified, and these TFs responded to TYLCV infection. Microsynteny revealed the independent duplication events, except whole-genome duplication, of WRKY Group III TFs. The phylogenetic tree of 28 species also showed that WRKY Group III TFs diversified after monocots and eudicots diverged. WRKY Group III TFs served as positive and negative regulators in tomato–TYLCV interaction. The expression levels of SolyWRKY42 and SolyWRKY80 were upregulated in ‘Zheza-301.’ In ‘Jinpeng-1,’ the expression patterns of the 6 WRKY Group III genes were positive. RT-qPCR and VIGS analyses indicated that SolyWRKY41 and SolyWRKY54 served as negative regulators in the resistance to TYLCV infection. WRKY Group III TFs could also interact with other proteins by binding to cis elements in the promoter region of other genes to regulate pathogen-related gene expression. Our study provided novel insights into the interaction of WRKY Group III TFs members with TYLCV infection in tomato.
Acknowledgements
Not applicable.
Funding
The research was supported by Jiangsu Natural Science Foundation (BK20130027), New Century Excellent Talents in University (NCET-11-0670), Priority Academic Program Development of Jiangsu Higher Education Institutions Project.
Availability of data and materials
The data sets supporting the conclusions of this article are included within the article and its additional files.
Authors’ contributions
Conceived and designed the experiments: ASX, YH. Performed the experiments: YH, PW, ZSX, LMY, FQ, FW. Analyzed the data: YH, ASX. Contributed reagents/materials/analysis tools: ASX. Wrote the paper: YH. Revised the paper: ASX, YH, MYL. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- ABRE
Cis-acting element involved in abscisic acid response
- CP
Coat protein
- dsDNA
Double-stranded DNA
- GFP
Green fluorescent protein
- HSE
Cis-acting element involved in heat stress response
- ICS
Isochorismate synthase
- JA
Jasmonic acid
- MAPK
Mitogen-activated protein kinase
- MeJA
Methyl jasmonate
- MS
Murashige and Skooog
- OD
Optical density
- ORF
Open reading frame
- PAMPs
Pathogen associated molecular patterns
- PCR
Polymerase chain reaction
- PR
Pathogenesis-related
- RT-qPCR
Quantitative real-time–polymerase chain reaction
- SA
Salicylic acid
- Soly
Solanum lycopersicum
- ssDNA
Single-stranded DNA
- TF
Transcription factor
- TRV
Tobacco rattle virus
- TYLCV
Tomato yellow leaf curly virus
- VIGS
Virus-induced gene silencing.
Additional files
Table S1. WRKY transcription factors name in rice, Arabidopsis, and tomato. Red color in tomato represented the SolyWRKY Group III TFs. Table S2. The orthologous WRKY genes in tomato, Arabidopsis, and rice. Red represented the orthologous genes of Group III genes responded to TYLCV infection in tomato. Table S3. The paralogous WRKY genes in tomato, Arabidopsis, and rice. Red represented the paralogous genes of Group III genes responded to TYLCV infection in tomato. (XLS 48 kb)
Figure S1. Structural analysis of WRKY transcription factors in tomato. A: Phylogenetic tree of SolyWRKY transcription factors. B: Exon/intrion structure analysis of WRKY transcription factors in tomato. (TIF 1982 kb)
Figure S2. Chromosome location of SolyWRKY TFs in tomato. Box represented the SolyWRKY Group III TFs in response to TYLCV infection. (TIF 5463 kb)
References
- 1.Dry IB, Rigden JE, Krake LR, Mullineaux PM, Rezaian MA. Nucleotide sequence and genome organization of tomato leaf curl geminivirus. J Gen Viro. 1993;74(1):147–151. doi: 10.1099/0022-1317-74-1-147. [DOI] [PubMed] [Google Scholar]
- 2.Akad F, Eybishtz A, Edelbaum D, Gorovits R, Dar-Issa O, Iraki N, Czosnek H. Making a friend from a foe: expressing a GroEL gene from the whitefly bemisia tabaci in the phloem of tomato plants confers resistance to tomato yellow leaf curl virus. Arch Virol. 2007;152(7):1323–1339. doi: 10.1007/s00705-007-0942-0. [DOI] [PubMed] [Google Scholar]
- 3.Pakkianathan BC, Kontsedalov S, Lebedev G, Mahadav A, Zeidan M, Czosnek H, et al. Replication of tomato yellow leaf curl virus in its whitefly vector, Bemisia tabaci. J Virol. 2015;89(19):9791–9803. doi: 10.1128/JVI.00779-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghanim M, Morin S, Czosnek H (2001) Rate of tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the Whitefly Bemisia tabaci. Phytopathology 91(2):188–196 [DOI] [PubMed]
- 5.Ghanim M. A review of the mechanisms and components that determine the transmission efficiency of tomato yellow leaf curl virus (Geminiviridae; Begomovirus) by its whitefly vector. Virus Res. 2014;186:47–54. doi: 10.1016/j.virusres.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Ma HY, Huang W, Wang F, Xiong AS. Comparative proteomic analysis provides novel insight into the interaction between resistant vs susceptible tomato cultivars and TYLCV infection. BMC Plant Biol. 2016;16(1):162. doi: 10.1186/s12870-016-0819-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gafni Y, Epel BL. The role of host and viral proteins in intra- and inter-cellular trafficking of geminiviruses. Physiol Mol Plant P. 2002;60(5):231–241. doi: 10.1006/pmpp.2002.0402. [DOI] [Google Scholar]
- 8.Wege C (2007). Movement and localization of tomato yellow leaf curl viruses in the infected plant. In: Czosnek H editor. Tomato Yellow Leaf Curl Virus Disease. Springer Netherlands. p. 185–206.
- 9.Agarwal P, Reddy M P, Chikara J (2011) WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol Biol Rep 38(6):3883-3896. [DOI] [PubMed]
- 10.Li MY, Xu ZS, Tian C, Huang Y, Wang F, Xiong AS. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci Rep. 2016;6:23101. doi: 10.1038/srep23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiguro S, Nakamura K. Characterization of a cdna-encoding a novel DNA-binding protein, Spf1, that recognizes Sp8 sequences in the 5' upstream regions of genes-coding for sporamin and beta-amylase from sweet-potato. Mol Gen Genet. 1994;244(6):563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 12.Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7(5):491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Wang QS, Wang MH, Zhang XZ, Hao BJ, Kaushik SK, Pan YC. WRKY gene family evolution in Arabidopsis thaliana. Genetica. 2011;139(8):973–983. doi: 10.1007/s10709-011-9599-4. [DOI] [PubMed] [Google Scholar]
- 14.Wu ZJ, Li XH, Liu ZW, Li H, Wang YX, Zhuang J. Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Mol Genet Genomics. 2016;291(1):255–269. doi: 10.1007/s00438-015-1107-6. [DOI] [PubMed] [Google Scholar]
- 15.Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12(1):9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- 16.Tang J, Wang F, Hou XL, Wang Z, Huang ZN. Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassica rapa ssp pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol Biol Rep. 2014;32(4):781–795. doi: 10.1007/s11105-013-0672-2. [DOI] [Google Scholar]
- 17.Zhang YJ, Wang LJ. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 2005;5:1–12. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 19.Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant Microbe In. 2003;16(4):295–305. doi: 10.1094/MPMI.2003.16.4.295. [DOI] [PubMed] [Google Scholar]
- 20.Wang YY, Feng L, Zhu YX, Li Y, Yan HW, Xiang Y. Comparative genomic analysis of the WRKY III gene family in populus, grape, Arabidopsis and rice. Biol Direct. 2015;10:48. doi: 10.1186/s13062-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama A, Fukushima S, Goto S, Matsushita A, Shimono M, Sugano S, et al. Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol. 2013;13:1–11. doi: 10.1186/1471-2229-13-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho SM, Kang EY, Min KH, Lee YK, Kim YC, Yang KY, et al. A positive regulatory role of the watermelon ClWRKY70 gene for disease resistance in transgenic Arabidopsis thaliana. Biol Plantarum. 2012;56(3):560–565. doi: 10.1007/s10535-012-0070-x. [DOI] [Google Scholar]
- 23.Gong XQ, Zhang JY, Hu JB, Wang W, Wu H, Zhang QH, et al. FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ. 2015;38(11):2248–2262. doi: 10.1111/pce.12539. [DOI] [PubMed] [Google Scholar]
- 24.Jiang YZ, Duan YJ, Yin J, Ye SL, Zhu JR, Zhang FQ, et al. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J Exp Bot. 2014;65(22):6629–6644. doi: 10.1093/jxb/eru381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rushton PJ, Somssich IE, Ringler P, Shen QXJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Cheng HT, Liu HB, Deng Y, Xiao JH, Li XH, Wang SP. The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 2015;167(3):1087–1099. doi: 10.1104/pp.114.256016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shim JS, Jung C, Lee S, Min K, Lee YW, Choi Y, et al. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013;73(3):483–495. doi: 10.1111/tpj.12051. [DOI] [PubMed] [Google Scholar]
- 28.Atamian HS, Eulgem T, Kaloshian I. SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta. 2012;235(2):299–309. doi: 10.1007/s00425-011-1509-6. [DOI] [PubMed] [Google Scholar]
- 29.Kumar K, Srivastava V, Purayannur S, Kaladhar VC, Cheruvu PJ, Verma PK. WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum): comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene(s) DNA Res. 2016;23(3):225–239. doi: 10.1093/dnares/dsw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishihama N, Yoshioka H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol. 2012;15(4):431–437. doi: 10.1016/j.pbi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, et al. Mode of inheritance of primary metabolic traits in tomato. Plant Cell. 2008;20(3):509–523. doi: 10.1105/tpc.107.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy GG. Tomato, pests, parasitoids, and predators: Tritrophic interactions involving the genus Lycopersicon. Annu Rev Entomol. 2003;48:51–72. doi: 10.1146/annurev.ento.48.091801.112733. [DOI] [PubMed] [Google Scholar]
- 34.Kil EJ, Kim S, Lee YJ, Byun HS, Park J, Seo H et al (2016) Tomato yellow leaf curl virus (TYLCV-IL): a seed-transmissible geminivirus in tomatoes. Sci Rep 6:19013. [DOI] [PMC free article] [PubMed]
- 35.Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol Genet Genomics. 2012;287(6):495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- 36.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol Plant Microbe In. 2008;21(8):1015–1026. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- 37.Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42:1182–1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Peterson D, Peterson N, Stercher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;2028(2010):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rombauts S, Dehais P, Van Montagu M, Rouze P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27(1):295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shu Y, Tao Y, Wang S, Huang L, Yu X, Wang Z, et al. GmSBH1, a homeobox transcription factor gene, relates to growth and development and involves in response to high temperature and humidity stress in soybean. Plant Cell Rep. 2015;34(11):1927–1937. doi: 10.1007/s00299-015-1840-7. [DOI] [PubMed] [Google Scholar]
- 46.Yan HX, Fu DQ, Zhu BZ, Liu HP, Shen XY, Luo YB. Sprout vacuum-infiltration: a simple and efficient agroinoculation method for virus-induced gene silencing in diverse solanaceous species. Plant Cell Rep. 2012;31(9):1713–1722. doi: 10.1007/s00299-012-1285-1. [DOI] [PubMed] [Google Scholar]
- 47.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen TZ, Lv YD, Zhao TM, Li N, Yang YW, Yu WG, et al. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS One. 2013;8(11):e80816. doi: 10.1371/journal.pone.0080816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10(4):366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 50.Tao Z, Kou Y, Liu H, Li X, Xiao J, Wang S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot. 2011;62(14):4863–4874. doi: 10.1093/jxb/err144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao J, Cheng HT, Li XH, Xiao JH, Xu CG, Wang SP. Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 2013;163(4):1868–1882. doi: 10.1104/pp.113.226019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Gene Dev. 2002;16(9):1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu YL, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31(6):777–786. doi: 10.1046/j.1365-313X.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 55.Liu B, Hong YB, Zhang YF, Li XH, Huang L, Zhang HJ, et al. Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 2014;227:145–156. doi: 10.1016/j.plantsci.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Hinderhofer K, Zentgraf U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta. 2001;213(3):469–473. doi: 10.1007/s004250000512. [DOI] [PubMed] [Google Scholar]
- 57.Chen CH, Chen ZX. Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol Biol. 2000;42(2):387–396. doi: 10.1023/A:1006399311615. [DOI] [PubMed] [Google Scholar]
- 58.Xu XP, Chen CH, Fan BF, Chen ZX. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18(5):1310–1326. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nyoike TW, Liburd OE, Webb SE. Suppression of whiteflies, Bemisia tabaci (Hemiptera : Aleyrodidae) and incidence of cucurbit leaf crumple virus, a whitefly-transmitted virus of zucchini squash new to Florida, with mulches and imidacloprid. Fla Entomol. 2008;91(3):460–465. doi: 10.1653/0015-4040(2008)91[460:SOWBTH]2.0.CO;2. [DOI] [Google Scholar]
- 60.Webb SE, Akad F, NyoikeT W, Liburd E, Polston JE. Whitefly-transmitted cucurbit leaf crumple virus in Florida. Entomol Nematol. 2008;91(3):460–465. [Google Scholar]
- 61.Wang JY, Yu WG, Yang YW, Li X, Chen TZ, Liu TL, et al. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci Rep. 2015;5:16946. doi: 10.1038/srep16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorovits R, Czosnek H. Expression of stress gene networks in tomato lines susceptible and resistant to tomato yellow leaf curl virus in response to abiotic stresses. Plant Physiol Bioch. 2008;46(4):482–492. doi: 10.1016/j.plaphy.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Sade D, Sade N, Shriki O, Lerner S, Gebremedhin A, Karavani A, et al. Water balance, hormone homeostasis, and sugar signaling are all involved in tomato resistance to tomato yellow leaf curl virus. Plant Physiol. 2014;165(4):1684–1697. doi: 10.1104/pp.114.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou M, Yan J, Ma ZW, Zhou Y, Abbood NN, Liu JF, et al. Comparative and evolutionary analysis of the HES/HEY gene family reveal exon/intron loss and teleost specific duplication events. PLoS One. 2012;7(7):e40649. doi: 10.1371/journal.pone.0040649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brand LH, Fischer NM, Harter K, Kohlbacher O, Wanke D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013;41(21):9764–9778. doi: 10.1093/nar/gkt732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai M, Qiu DY, Yuan T, Ding XH, Li HJ, Duan L, et al. Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 2008;31(1):86–96. doi: 10.1111/j.1365-3040.2007.01739.x. [DOI] [PubMed] [Google Scholar]
- 67.Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150(4):1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang SQ, Klessig DF. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001;6(11):520–527. doi: 10.1016/S1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- 69.Cvetkovska M, Rampitsch C, Bykova N, Xing T. Genomic analysis of MAP kinase cascades in Arabidopsis defense responses. Plant Mol Biol Rep. 2005;23(4):331–343. doi: 10.1007/BF02788882. [DOI] [Google Scholar]
- 70.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414(6863):562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 71.Li S, Li K, Ju Z, Cao DY, Fu DQ, Zhu HL, et al. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genomics. 2016;17:1–16. doi: 10.1186/s12864-015-2294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J, Kristiansen KA, Hansen BG, Halkier BA. Cellular and subcellular localization of flavin-monooxygenases involved in glucosinolate biosynthesis. J Exp Bot. 2011;62(3):1337–1346. doi: 10.1093/jxb/erq369. [DOI] [PubMed] [Google Scholar]
- 73.Li J, Brader G, Palva ET. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16(2):319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen CH, Chen ZX. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002;129(2):706–716. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot. 2012;63(7):2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim HS, Park Y H, Nam H, Lee YM, Song K, Choi C et al (2014). Overexpression of the Brassica rapa, transcription factor WRKY12 results in reduced soft rot symptoms caused by Pectobacterium carotovorum, in Arabidopsis, and Chinese cabbage. Plant Biol 16(5):973-981 [DOI] [PubMed]
- 77.Huang Y, Zhang B, Sun S, Xing G, Wang F, Li M, Tian Y, Xiong A. AP2/ERF transcription factors involved in response to tomato yellow leaf curly virus in tomato. Plant Genome. 2016;9:1–15. doi: 10.3835/plantgenome2015.09.0082. [DOI] [PubMed] [Google Scholar]
- 78.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 79.Wang JY, Hu ZZ, Zhao TM, Yang YW, Chen TZ, Yang ML, et al. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum) BMC Genomics. 2015;16:1–14. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nimchuk Z, Eulgem T, Holt BE, Dangl JL. Recognition and response in the plant immune system. Annu Rev Genet. 2003;37:579–609. doi: 10.1146/annurev.genet.37.110801.142628. [DOI] [PubMed] [Google Scholar]
- 81.Zhang GY, Chen M, Li LC, Xu ZS, Chen XP, Guo JM, Ma YZ. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60(13):3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Desikan R, Hancock JT, Ichimura K, Shinozaki K, Neill SJ. Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol. 2001;126(4):1579–1587. doi: 10.1104/pp.126.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nuhse TS, Peck SC, Hirt H, Boller T. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem. 2000;275(11):7521–7526. doi: 10.1074/jbc.275.11.7521. [DOI] [PubMed] [Google Scholar]
- 84.Park CJ, Shin YC, Lee BJ, Kim KJ, Kim JK, Paek KH. A hot pepper gene encoding WRKY transcription factor is induced during hypersensitive response to tobacco mosaic virus and Xanthomonas campestris. Planta. 2006;223(2):168–179. doi: 10.1007/s00425-005-0067-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the conclusions of this article are included within the article and its additional files.


