Abstract
Hydrocarbon-stapled peptides are a class of bioactive alpha-helical ligands developed to dissect and target protein interactions. While there is consensus that stapled peptides can be effective chemical tools for investigating protein regulation, their broader utility for therapeutic modulation of intracellular interactions remains an active area of study. In particular, the design principles for generating cell-permeable stapled peptides are empiric, yet consistent intracellular access is essential to in vivo application. Here, we used an unbiased statistical approach to determine which biophysical parameters dictate the uptake of stapled peptide libraries. We found that staple placement at the amphipathic boundary combined with optimal hydrophobic and helical content are the key drivers of cellular uptake, whereas excess hydrophobicity and positive charge at isolated amino acid positions can trigger membrane lysis at elevated peptide dosing. Our results provide a design roadmap for maximizing the potential to generate cell-permeable stapled peptides with on-mechanism cellular activity.
INTRODUCTION
The therapeutic impact of targeting yet undruggable intracellular protein interactions could be transformative and continues to inspire new chemical approaches to developing potent and specific compounds that feature (1) the large and complex surface binding capacity of antibodies with (2) the intracellular access of small molecules. Hydrocarbon-stapled peptides have emerged as one possible solution in that they recapitulate the structure and specificity of bioactive α-helices, resist proteolytic degradation in vivo and, when appropriately designed, gain entrance to the cell by a macropinocytotic mechanism1. The original proof-of-concept for synthesizing structurally-reinforced and protease-resistant “stapled” α-helices derived from the insertion of α,α-disubstituted non-natural amino acids bearing olefin tethers into an RNAse A peptide template at i, i+4 or i, i+7 positions, followed by ruthenium-catalyzed olefin metathesis2. The first cellular application of hydrocarbon-stapled α-helices, which were modeled after the BCL-2 homology 3 (BH3) domain of pro-apoptotic BID, revealed their capacity for cellular uptake by an energy-dependent macropinocytotic mechanism, resulting in activation of the apoptotic signaling cascade3. The subsequent development of a library of stapled α-helical peptides modeled after the p53 transactivation domain highlighted that not all stapled peptides are cell permeable; in fact, none of the stapled peptides in the original p53 panel were cell penetrant4. Based on our prior experience in enhancing the cellular uptake of stapled BID peptides by optimizing α-helicity and adjusting the overall peptide charge from −2 to 0 or +1, we generated a revised p53 panel bearing E to Q and D to N mutations, yielding cell penetrant analogs capable of reactivating the p53 pathway through targeted inhibition of HDM24 and HDMX5. Iteration of this compound to mitigate serum binding and further improve potency6 resulted in the development of the first stapled peptide drug to be evaluated in ongoing clinical trials for targeting an intracellular protein interaction (ClinicalTrials.gov identifier: NCT02264613).
Despite the remarkable promise of stapled peptides as a novel class of compounds for dissecting and targeting protein interactions5,7–12, the criteria for generating cell penetrant analogs is unknown, with design strategies driven by trial-and-error or cumulative empiric observations. Whereas such factors as α-helicity, positive charge, peptide sequence, staple composition and placement, and membrane interaction have all been invoked as contributing factors for cell uptake propensity1,13–15, an unbiased biostatistical approach has not been applied to formally interrogate the determinants of stapled peptide uptake. This lack of clarity presents a roadblock to the broader utility of stapled peptides for cellular and in vivo analyses. Indeed, the use of cell impermeable stapled peptides in cellular studies has led to faulty conclusions about stapled peptide uptake and activity16–18. Conversely, the application of supraphysiologic doses of aggregation-prone constructs can lead to cytotoxicity misinterpreted as mechanistically on-target rather than nonspecific membrane lysis1,19.
Having recently confirmed by electron microscopy that cell penetrant stapled peptides can reach their intracellular site of biological activity without plasma membrane disruption20, here we applied high content microscopy and biophysical characterization of staple scanning and point mutation libraries of stapled BIM BH3 peptides to identify those parameters that dictate cellular penetrance using unbiased statistical methods. We found that placement of the all-hydrocarbon staple at the amphipathic boundary, which extends the hydrophobic surface, in addition to optimal peptide hydrophobicity and alpha-helicity, are the key drivers of cellular uptake. In addition, the combination of excess overall hydrophobicity and positive charge that derives from specific amino acid positions can be a risk factor for cell membrane lysis at elevated peptide dosing. We envision that implementing these design principles will expedite the advancement of cell-penetrating stapled peptides with on-mechanism biological and therapeutic activities.
RESULTS
High-stringency assessment of stapled peptide uptake
Our first goal was to harness high-throughput epifluorescence microscopy and the ImageXpress Micro (IXM) Widefield High Content Analysis System to develop a rigorous quantitation platform for measuring cellular uptake of stapled peptides derivatized at the N-terminus with a FITC fluorophore. We chose FITC-BIM SAHBA1 to benchmark the microscopic analyses, since its cellular uptake has been documented across a variety of diverse experimental methods, including fluorescence scan of electophoresed lysates from treated cells, confocal microscopy, and electron microscopy10,20. We utilized Custom Module Editor to create a custom module (CM) for analysis based on the following principles: (1) define cellular uptake in mouse embryonic fibroblasts (MEFs) based on visualizing cellular and nuclear boundaries using CellMask Deep Red (CMDR) and Hoechst 33342, respectively, thereby excluding extracellular aggregates and autofluorescent debris (Fig. 1a), (2) contract the resultant cellular mask by a defined pixel boundary to avoid quantitation of extracellular, membrane-adherent peptide, (3) exclude FITC signal outliers that reflect out-of-focus fluorescence, (4) acquire images for analysis at 20× and 40× magnification to ensure precise resolution of fluorescence on a per cell basis, (5) visualize distinct central locations of the cellular field to achieve unbiased sampling and avoid edge artifact (Supplementary Results, Supplementary Fig. 1), and (6) maximize signal-to-noise ratios to ensure optimal sensitivity and specificity of internal FITC-peptide signal (Supplementary Fig. 2). Using our CM, we confirmed that fixation with 4% paraformaldehyde had no effect on our total internalized FITC intensity (TIFI) measurements when compared to live cell imaging, enabling us to eliminate any variability that could arise from subjecting live cells to acquisition times lasting up to 2 hours (Supplementary Fig. 3).
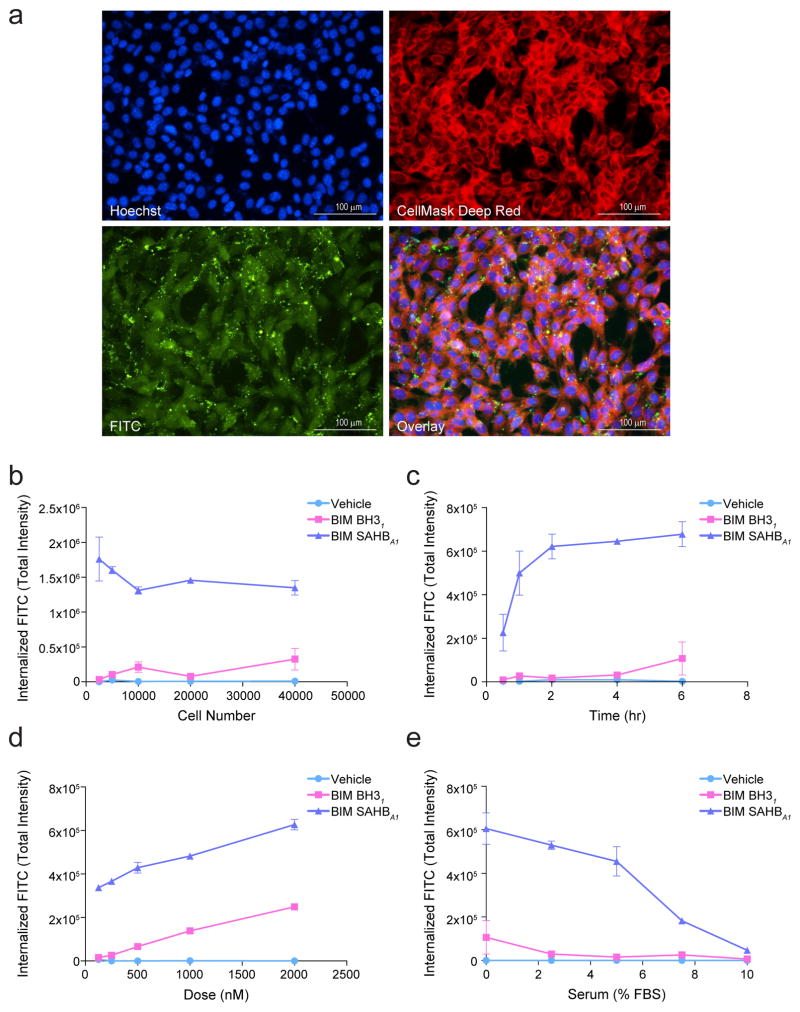
Figure 1. Microscopy-based quantitation of stapled peptide uptake.
(a) Exemplary images of mouse embryonic fibroblasts (MEFs) treated with FITC-BIM SAHBA1 (green) and stained with Hoechst (blue) and CellMask Deep Red (red) to monitor the localization of FITC-peptide with respect to cellular architecture (overlay). Images were acquired by IXM at 20× magnification. Bar, 100 μm (b–e) Total internalized fluorescence intensity (TIFI) on a per cell basis was monitored upon varying (b) cell density (500 nM peptide, 4 hr, 0% FBS), (c) acquisition time (500 nM peptide, 2×104 cells, 0% FBS), (d) FITC-peptide dose (2×104 cells, 4 hr, 0% FBS), and (e) percent added FBS (500 nM peptide, 2×104 cells, 4 hr). Data are mean ± s.e.m. for experiments performed in duplicate wells with 4 image acquisitions per well. Three biological replicates were performed for each experiment with similar results.
We then sought to determine what cellular density, acquisition time point, and peptide dose was optimal for measuring internalized FITC-peptide. Comparing vehicle, the unmodified template peptide FITC-BIM BH31 (aa 146–166) and the corresponding stapled analog BIM SAHBA1 at 500 nM dosing for 4 hours, we detected robust TIFI only for BIM SAHBA1, with the signal essentially constant on a per cell basis across the 2.5×103 to 4.0×104 range of cellular densities tested (Fig. 1b). We next followed TIFI over time upon treating MEFs at a density of 2×104 cells with 500 nM peptides, and observed time-responsive uptake for BIM SAHBA1 that peaked by 2 hours and remained stable thereafter (Fig. 1c). Using a plating density of 2×104 and time point of 2 hours, we varied the dose of applied peptide and observed a linear, dose-responsive increase in TIFI for FITC-BIM BH31 and FITC-BIM SAHBA1, with the stapled analog consistently showing markedly enhanced uptake over the entire dose range (Fig. 1d). To avoid any differences in peptide internalization based on variability in serum binding, the above analyses were conducted in serum-free medium. However, given the importance of understanding the influence of serum exposure on the cellular activity of stapled peptides, we tested the effect of fetal bovine serum (FBS) on peptide internalization over a 0% to 10% dose range. Consistent with our prior observations that FBS can suppress the cellular uptake and biological activity of BIM SAHBA18,10, here we found that the serum-suppressive effect was dose-responsive, with near complete elimination of cellular uptake at 10% FBS but relative preservation of cellular uptake up to 5% of added FBS (Fig. 1e).
A recent report suggested that stapled peptides may gain cellular entry by inducing plasma membrane lysis and that FBS suppressed this uptake by preserving membrane integrity19. To explore this hypothesis, we subjected MEFs at a plating density of 2×104 to a serial dilution of FITC-BIM SAHBA1 starting at 20 μM in the absence of serum and monitored for cellular lysis by LDH release over time. Importantly, we observed no LDH release across the entire dose range at either 30 min or 180 min post-treatment (Supplementary Fig. 4). Consistent with our recent electron microscopy study20, these data rule out the possibility that FITC-BIM SAHBA1 achieves cellular entry via membrane lysis or that FBS is blocking stapled peptide import by preventing membrane lysis. Thus, based on the above results, we selected (1) 2×104 cell density, (2) 4 hour acquisition time, (3) 500 nM dosing, and (4) 0% FBS as treatment parameters for our study in order to maximize sensitivity and specificity of our detection method while avoiding the variability introduced by live cell imaging and differential effects of serum on the diverse compositions within our stapled peptide libraries.
Cell import determinants for a staple scanning library
When designing stapled peptides for biochemical and biological studies, one of the first questions to address is where to install the staple. Here, we generated a library of stapled BIM BH3 peptides by performing a “staple scan” that sequentially places the staple along the length of the template peptide, yielding 17 i, i+4 stapled peptides. We monitored the cellular uptake of the FITC-derivatized stapled peptides by IXM using our CM. Importantly, of the cellular fluorescent dyes measured in our microscopy assay (i.e. Hoechst, CMDR, FITC), only the FITC intensity varied with stapled peptide composition (Supplementary Fig. 5). Our prototype BIM SAHBA1 peptide that bears a staple flanking IGD emerged as the clear winner (TIFI >3.0×106), with four additional constructs containing staples flanking QEL, ELR, AYY, and LRR also demonstrating notable uptake at TIFIs of 2.16×106, 1.21×106, 1.07×106, and 0.88×106, respectively (Fig. 2a). We verified the reproducibility of both our microscopy method and TIFI values for comparative uptake analyses by performing biological replicates, which showed strong mutual association (Spearman’s correlation p <0.0001) (Supplementary Fig. 6a). To examine whether the TIFI hierarchy observed in MEFs was recapitulated in a different cell line, we repeated the analyses in HeLa cells and again observed strong association (Spearman’s correlation p = 0.0002) (Supplementary Fig. 6b), reinforcing the technical and biological reproducibility of our quantitative approach.
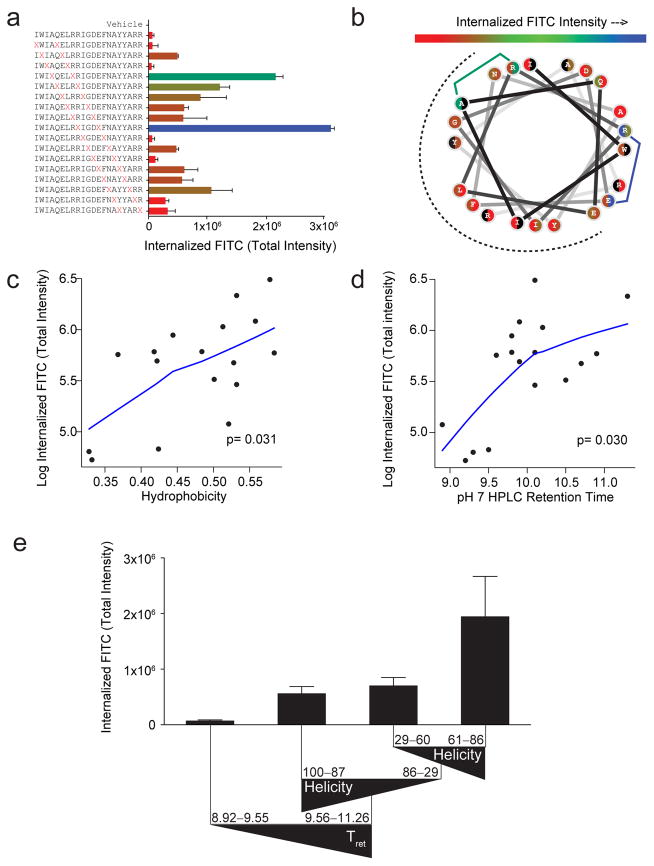
Figure 2. Determinants of cellular uptake for differentially stapled BIM BH3 peptides.
(a) Range of TIFI for BIM BH3 peptides bearing sequentially placed i, i+4 staples (red X pairs) for MEFs (2×104/well) treated with 500 nM peptides and measured by IXM (20x) at 4 hours. Data are mean ± s.d. for experiments performed in duplicate wells with 4 image acquisitions per well. Three biological replicates were performed with similar results (see Supplementary Fig. S6a). X, S-pentenyl alanine (b) Wheel depiction of the BIM BH3 α-helix, with the hydrophobic interaction face indicated by the dotted surface and stapling amino acid pairs color-coded based on level of measured TIFI. Residues that participate in two distinct staples are shown as split circles, with the left and right colors corresponding to their roles in N- vs. C-terminal staples, respectively. (c–d) Single variable plots for TIFI vs. (c) calculated hydrophobicity and (d) HPLC retention time (pH 7), as assessed by Spearman’s rank correlation. p-values were calculated using the permutation test included in the R package pvrank, and line fitting was performed using a loess smoother. (e) The tree resulting from recursive partitioning depicts the influence of principal components (hydrophobicity/HPLC retention time, α-helicity, and pI) on TIFI outcome. Triangles reflect the directionality of parameter values. Retention time and α-helicity are indicated in minutes and percent, respectively. Data are mean ± s.d. for each peptide subpopulation.
In examining the staple distribution of the most penetrant constructs along the amphipathic helix using a colorimetric scale, we found that staple positions located at the hydrophobic/hydrophilic boundary were the most favorable for cellular uptake (Fig. 2b). These data suggest that extending the hydrophobic surface of stapled peptides, such as beyond 180 degrees in the case of BIM BH3, may be a critical design feature of cell penetrant stapled peptides. Indeed, all six BIM SAHB constructs bearing a staple restricted to the hydrophobic interaction surface of the helix were among the least cell permeable peptides.
To expand this type of know-how beyond empiric observation, we next compiled a table of calculated and experimental parameters that define key biophysical properties of the stapled peptide library, including calculated hydrophobicity, HPLC column retention time (pH 7), percent α-helicity (circular dichroism), and calculated net charge and pI (Supplementary Table 1). In examining individual parameter associations with TIFI, we found that only the calculated hydrophobicity and experimentally-determined HPLC retention time (pH 7) demonstrated a statistically-significant relationship with cellular uptake (Spearman’s correlation p = 0.031 and 0.030, respectively), with the association following a piecewise linear trajectory (Fig. 2c–d, Supplementary Fig. 7). Of note, we determined HPLC retention times at pH 7 to gauge peptide behavior at physiologic pH, but also found that the retention times at pH 4 – the more typical pH for HPLC purifying peptides – were strongly associated with the pH 7 values (Spearman’s correlation p <0.001), thereby allowing retention time analyses for these purposes to be conducted under routine conditions (Supplementary Fig. 8). Because calculated hydrophobicity and HPLC retention time also correlated extremely well with one another (Spearman’s correlation p = 0.009) (Supplementary Fig. 9), our data suggest that staple placement at the amphipathic boundary of the binding interface combined with prioritization of constructs based on relatively high hydrophobicity as determined from calculations alone, reflect a critical first step in designing a cell penetrant stapled peptide. Indeed, we verified these design conclusions using a different sequence template bearing less amphipathic character and an alternative i, i+7 staple composition, namely stabilized alpha-helices of SOS1 (SAH-SOS1). Again, we found that cellular uptake, as reflected by TIFI measurement, was highly associated with hydrophobicity (Spearman’s correlation p = 0.035) (Supplementary Fig. 10a) and all three of the staples that extended the hydrophobic surface beyond the target binding interface were among the constructs yielding the highest TIFI values (Supplementary Fig. 10b).
The overall variability of the data was initially investigated using a principal components analysis (PCA)21–23 that included 9 potential explanatory variables and TIFI. The measures of hydrophobicity (calculated hydrophobicity, HPLC retention time), charge state (pI, net charge), and α-helicity were identified as the primary features of the overall variability. Therefore, we performed a subsequent PCA using representative covariates for these quantities. Because overall charge and pI are both calculated and strongly associated values (Spearman’s correlation p <0.001) (Supplementary Fig. 11), we selected pI to represent peptide charge state for our analysis. Both HPLC retention time and calculated hydrophobicity contributed strongly to principal component 1, whereas percent α-helicity and pI defined principal components 2 and 3, respectively (Supplementary Table 2). Stapled peptide hydrophobicity and retention time represent the largest portion of the observed data variability (44%) by PCA and are the only individual parameters to demonstrate a statistically-significant relationship with TIFI (Fig. 2c–d,), yet the PCA also suggests that α-helical content and pI serve as key secondary contributors to the data variability (28% and 21%, respectively).
To delve further into how hydrophobicity, α-helical content and pI together influenced the cellular uptake of our staple scanning library, we next constructed a regression tree using a recursive partitioning algorithm24, which serves as an exploratory statistical tool for identifying key subpopulations within the peptide dataset through a series of binary queries. As expected, the first breakpoint for distinguishing between cell permeable and impermeable peptides was based on HPLC retention time, a measure of hydrophobicity (Fig. 2e). All peptides with a retention time of less than 9.56 minutes uniformly demonstrated little to no cellular uptake. However, of the stapled peptides that exhibited 9.56 minutes or greater retention time (i.e. the more hydrophobic subgroup), percent α-helicity then became a major driver. Interestingly, those stapled peptides with α-helicities of >87% or <60% demonstrated only moderate cellular uptake and notably less than constructs with α-helicities in the 61–86% range (Fig. 2e). Thus, the sweet spot for cellular uptake of stapled BIM BH3 peptides appears to be dictated by relatively high hydrophobicity combined with elevated, but not excessive, α-helical content, an important subtlety masked by other modes of analysis.
Influence of point mutagenesis on cellular uptake
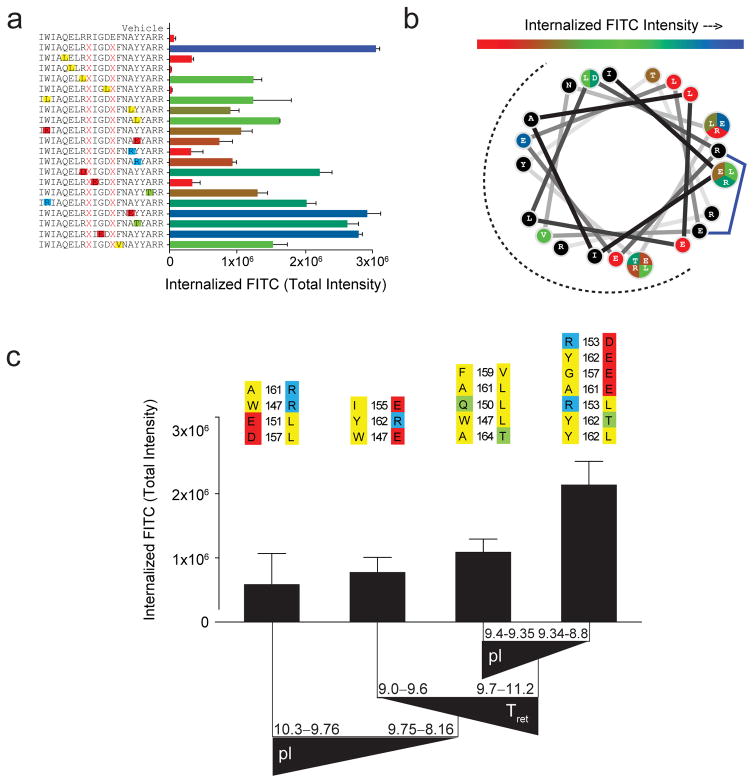
Once a lead stapled peptide emerges based on staple placement and the above-described hydrophobicity and α-helicity considerations, it may be desirable to generate an expanded cluster of leads by fine tuning amino acid composition, including evaluating the impact of adjusting overall charge, as has been pursued for a variety of stapled peptide applications3,4,9,11,12. To better understand the influence of charge in the context of an already well-optimized stapled peptide lead, we generated a BIM SAHBA1 library composed of a series of point mutations that altered charge and hydrophobicity at discrete positions (Fig. 3a, Supplementary Table 3). Cellular uptake analysis of this library revealed a remarkable influence of single point mutations on TIFI (Fig. 3a–b). Consistent with a multifactorial role of biophysical influences on stapled peptide uptake, no single parameter independently showed an association with TIFI for this point mutant library (Supplementary Fig. 12). Like the previous case, PCA analysis revealed that hydrophobicity and retention time explain the largest amount of variability (component 1, 47%), but the contribution of pI (component 2, 32%) surpassed that of α-helicity (component 3, 17%) in this analysis (Supplementary Table 4). Interestingly, recursive partitioning demonstrated that mutations that either increased the pI (>9.75) or reduced the retention time (<9.7) relative to BIM SAHBA1, impaired cellular uptake, whereas constructs that maintained or increased retention time (9.7–11.2) and even lowered the overall pI (8.8–9.34) preserved high level cellular uptake (Fig. 3c). These data are inconsistent with the notion that positive charge is an independent, mechanism-based requirement for stapled peptide import14, as evidenced by retention of high level TIFI upon R153D, G156E, and A161E mutagenesis of BIM SAHBA1. Taken together, we find that the cellular uptake of stapled BIM BH3 peptides is predominantly driven by hydrophobicity and α-helicity, yet because of multifactorial influences that include pI, expanding and fine tuning a pool of lead compounds can be achieved by judicious sequence modification.
Figure 3. Impact of point mutagenesis on the cellular uptake of BIM SAHBA1.
(a) Range of TIFI values for a point mutant library of BIM SAHBA1 (BIM SAHB9) peptides, as measured by IXM (20x) in MEFs (2×104 cells/well) treated with 500 nM peptides for 4 hours. Mutations are colored in accordance with their properties (yellow, hydrophobic; red, negatively charged; blue, positively charged; green, hydrophilic). Data are mean ± s.d. for experiments performed in duplicate wells with 4 image acquisitions per well. Three biological replicates were performed with similar results. X, S-pentenyl alanine (b) Wheel depiction of the BIM SAHBA1 α-helix, with the hydrophobic interaction face indicated by the dotted surface and mutant sites color-coded based on level of measured TIFI. Residue positions subjected to differential mutation are partitioned and colored according to their level of TIFI. (c) The tree resulting from recursive partitioning depicts the influence of principal components (hydrophobicity/HPLC retention time, pI, and α-helicity) on TIFI outcome. Triangles reflect the directionality of parameter values. The point mutants that comprise each data bar are indicated above and color-coded as in (a). Retention time is indicated in minutes. Data are mean ± s.d. for each peptide subpopulation.
Identifying stapled peptides with biological function
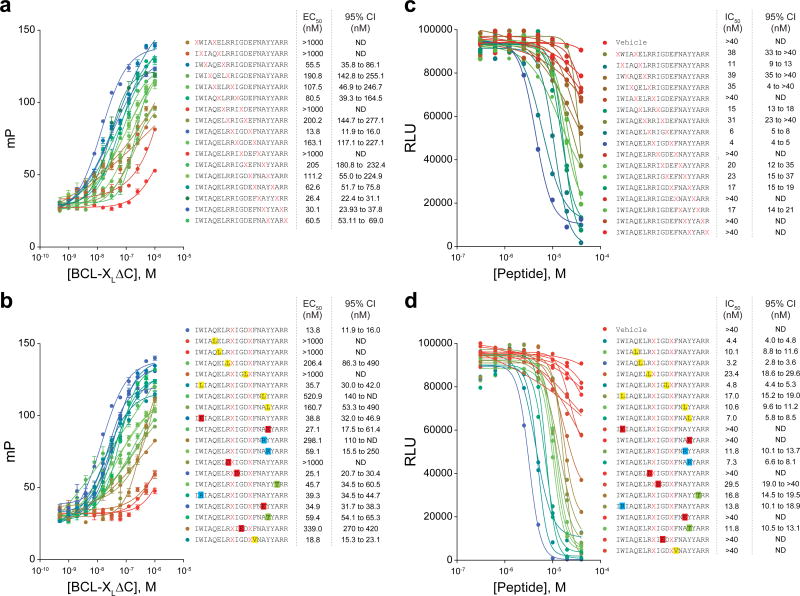
Because staple insertion and point mutagenesis, by definition, alter the native sequence of the template peptide, biochemical and biological testing is essential to identifying cell-penetrant stapled peptides that are also bioactive. For example, when we subjected our staple scanning and point mutation libraries of FITC-BIM BH3 to fluorescence polarization binding analysis against an established BCL-2 family target, such as BCL-XL, we identified a spectrum of binding affinities ranging from 14 nM (BIM SAHBA1) to over 1 μM. The observed losses in binding activity are consistent with disrupting the critical hydrophobic interaction surface and/or mutating specific residues known to be essential for BCL-XL interaction (e.g. L152, G156) (Fig. 4a–b). We then screened our libraries for cytotoxic activity in a leukemia cell line engineered to be dependent on BCL-XL for survival (BCL-XL-reconstituted p185+Arf−/−/Mcl-1-deleted B-cell acute lymphoblastic leukemia)25,26, and again obtained a spectrum of cytotoxic activity, with IC50s ranging from 3 to >40 μM (Fig. 4c–d). In order to identify compounds whose cytotoxicity correlates with the capacity for cellular uptake and intracellular target engagement, it is important to first rule out nonspecific cytotoxicity due to potential plasma membrane lysis. We screened for this undesirable activity by exposing cells to high micromolar dosing of stapled peptides, followed by measurement of LDH release after 30 min of treatment in the absence of serum. Twenty-eight of 36 constructs (~80%) showed no lytic activity when applied to the B-ALL cells at 10 μM dosing for 30 min, with 3 constructs demonstrating low grade lytic activity and 5 constructs identified as clearly lytic (Fig. 5a–b). Of the five notably lytic stapled peptides, three had in common the conversion of E151 to a hydrophobic residue (i.e., stapling amino acid substitution or leucine mutation) and the remaining two featured the mutations W147R or A164T. Thus, lytic properties can be attributed to very specific, focal changes in sequence composition, rather than reflect a general liability of installing hydrocarbon staples.
Figure 4. Target binding affinity and cytotoxicity of stapled BIM BH3 libraries.
(a–b) BCL-XLΔC binding affinities as determined by fluorescence polarization assay of (a) staple scan and (b) point mutant FITC-BIM BH3 peptide libraries (peptide, 25 nM; protein 0.5 nM–1 μM). Binding isotherms are color coded based on relative binding potency (blue-green-red scale: blue, best binders; red, worst binders). Data are mean ± s.e.m. for experiments performed in technical quadruplicate and repeated twice. (c–d) The effect of stapled peptide treatment (0.3 μM–40 μM) on cell viability of BCL-XL-reconstituted p185+Arf−/−Mcl-1del B-ALL cells, as measured by CellTiter Glo assay at 24 hours. Data represent the mean of technical duplicates. The experiments were repeated three times with similar results.
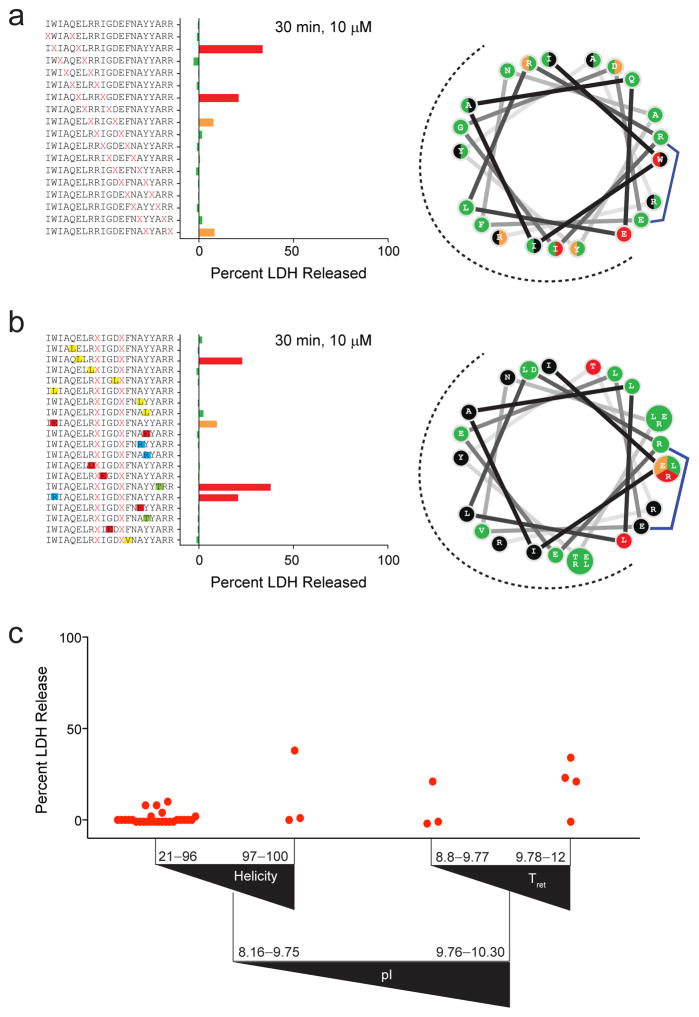
Figure 5. Determinants of cellular lysis for stapled BIM BH3 peptides.
(a) Staple scanning and (b) point mutant BIM BH3 peptide libraries were screened for membrane lytic properties by LDH release assay, performed on BCL-XL-reconstituted p185+Arf−/−Mcl-1del B-ALL cells (2×104 cells/well) treated with 10 μM peptide for 30 minutes in the absence of added serum. Data are normalized based on the response to treatment with 1% Triton X-100 (100% release) and media alone (0% LDH release). Data represent the mean of technical duplicates. The experiments were repeated twice with similar results. The wheel depictions to the right demonstrate the discrete staple insertion (a) and point mutation (b) sites that give rise to lysis (green, no lysis; orange, low grade lysis; red, higher grade lysis). (c) Recursive partitioning analysis depicting the influence of pI, α-helicity, and HPLC retention time on LDH release outcome. Triangles reflect the directionality of parameter values and red dots are the mean percent LDH release values for each peptide.
To better predict what biophysical parameters give rise to stapled peptides that are lytic, we applied the recursive partitioning algorithm using LDH release as the undesirable outcome (Fig. 5c). Interestingly, pI served as the first breakpoint for classifying stapled peptides as at risk for causing lysis or not. Among the 28 of 36 constructs that displayed no lytic activity, 23 have a pI <9.76 and α-helical content spanning a broad range, from 21% to 96%. Only 1 of 29 constructs with pI <9.76 caused prominent membrane lysis, and it belonged to the subgroup of three peptides with α-helicities of >97%, suggesting that near-perfect α-helical stabilization could be detrimental. Of those stapled peptides with pIs in the 9.76–10.30 range, 1 of 3 peptides with retention times of 8.8–9.77 min and 3 of 4 peptides with retention times of 9.78–12 min were lytic. Thus, the combination of relatively high positive charge and hydrophobicity represent the greatest risk factors for generating stapled peptides with the propensity to lyse cellular membranes at high doses. To test this conclusion, we performed LDH release assays using our distinct i, i+7-stapled SOS1 library and found that none of the SAH-SOS1 peptides induced plasma membrane lysis, consistent with all constructs having a pI of <9.76 (Supplementary Fig. 13).
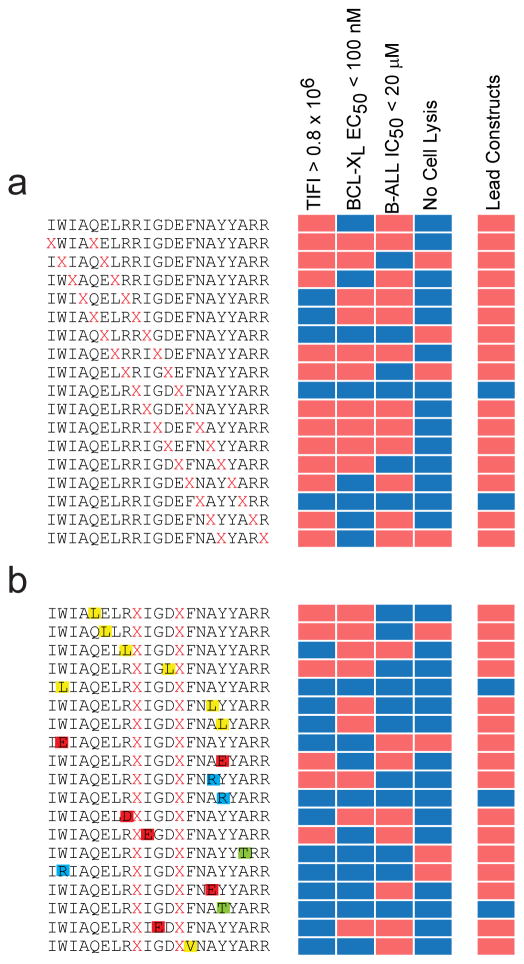
With cellular uptake, biochemical, and cellular data in hand, desired thresholds can then be applied to prioritize lead compounds for in vitro and in vivo application. For example, using selection criteria of TIFI >0.8×106, BCL-XL binding activity of <100 nM, and cellular IC50 <20 μM, BIM BH3 peptides bearing XIGDX- and XAYYX-positioned staples emerged as the most promising constructs for BCL-XL targeting (Fig. 6). Indeed, the XIGDX staple (our reported “A” position) has been the most successful to date when applied in the context of BID3, BIM8,10, BAD27,28, and PUMA29 BH3 peptides, whereas installing a staple at the corresponding XAYYX position in MCL-130 BH3 yielded an optimal construct for structural and cellular work. Importantly, these analyses can also inform the selection of negative control point mutants that maintain efficient cellular uptake but lose binding and cellular activity as a result of disrupting important amino acid contacts for protein target interaction, as exemplified by the R153D10 and G156E mutations in BIM BH3. Constructs that manifest cellular uptake, target binding affinity, and cytotoxicity but also induce membrane lysis can be identified and promptly disqualified from further development (e.g. XLRRX staple in BIM BH3). Alternatively, compounds that are taken up by cells and bind the target but have weak cellular activity (e.g. XELRX position) or that exhibit relatively weak binding affinity but manifest cytotoxicity in the absence of significant membrane lysis, could warrant further affinity optimization or investigation of other cellular target(s), respectively. Taken together, these data provide a practical and unbiased approach for identifying a lead peptide such as BIM SAHBA1 (TIFI>3×106, target binding EC50 of 14 nM, cellular IC50 of 4 μM, and no membrane lysis), for advancement as both a chemical tool and prototype therapeutic.
Figure 6. Identification of lead stapled peptide constructs for cellular and in vivo application.
(a) Lead constructs are identified by establishing exemplary thresholds for cellular uptake, binding activity, cytotoxicity, and absence of lytic properties. BIM BH3 peptides that contained staples flanking IGD and AYY emerged as the most favorable constructs. (b) Evaluation of point mutants revealed ideal negative control compounds that retain cellular penetrance without membrane lysis, yet lose target binding activity and cytotoxicity by disrupting the interaction surface (e.g. R153D, G156E). Mutations are colored in accordance with their properties (yellow, hydrophobic; red, negatively charged; blue, positively charged; green, hydrophilic).
DISCUSSION
Structural stabilization of bioactive α-helices by insertion of all-hydrocarbon staples has yielded a large and growing diversity of compounds for scientific investigation and potential clinical development1. Among the most exciting features of stapled peptides are their capacity to recapitulate the structure and function of evolutionarily-honed motifs for protein modulation, and substantially resist proteolysis in vitro and in vivo3,31, overcoming a key liability of therapeutic peptides. Whereas stapled peptides have been developed to address a series of extracellular targets3,31,32, including potential use as structured antigens for vaccination33, the capacity of appropriately-designed constructs to access the intracellular environment offers a new opportunity to potentially target a host of yet undruggable protein interactions. Indeed, when compared head-to-head with unmodified peptide sequences bearing cell-penetrating leader sequences such as TAT, structured and proteolytically-resistant stapled peptides can achieve robust and more sustained intracellular levels9,12. Therefore, understanding just how to design cell penetrant stapled peptides for clinical development remains a high priority goal.
In our earliest work on examining the effects of BID BH3 stapled peptides on treated leukemia cells, we observed energy-dependent macropinosomal import followed by time-dependent pinosomal release and concentration at the mitochondrial outer membrane, a physiologic site of ligand-target engagement3. Our recent electron microscopy studies of BIM BH3 stapled peptides confirmed vesicular import in the absence of plasma membrane disruption, with compound accumulation at the mitochondria20. Fluorescence correlation microscopy has recently been applied to track the import and cytosolic release of stapled p53 peptides, with the extent of intracellular access shown to correlate with compound efficacy in cell-based assays4,34. These high resolution imaging studies clearly document that appropriately-designed stapled peptides can penetrate intact cells and access the cytoplasm. Indeed, drawing conclusions about stapled peptide permeability without actually measuring cellular uptake can lead to faulty assertions18,19. Nevertheless, what specific properties confer cellular penetrance has remained enigmatic. A recent survey of a large and eclectic library of stapled peptides suggested that the staple itself along with overall positive peptide charge were the two key properties that dictated cellular penetrance14. Yet, stapled peptides with significantly net positive (+3 to +5) and negative (−4) charge have been reported to exhibit intracellular activity, in addition to the cell penetrant stapled peptides with near-neutral net charge (−1 to +2) that predominate in the literature16.
To address this critical gap in understanding, we developed a rigorous quantitative approach for measuring intracellular stapled peptides, and generated a comprehensive staple scanning library to prospectively determine what biophysical factors, singly or in combination, correlate with cellular uptake using a series of statistical methods. We find that the extent of peptide hydrophobicity, as determined by calculation or by measuring HPLC retention time, is the major driver of cellular penetrance, which is also influenced by α-helicity and pI. For the BIM BH3 template, retention times of 9.67–11.26 minutes, coupled with α-helicities of 61–86%, represented the sweet spot for cellular uptake. Importantly, we found that stapled peptides with a pI in the 9.76–10.3 range and retention times of 9.78–12.0 minutes, were at greatest risk of inducing nonspecific cellular lysis. Thus, to achieve cellular uptake without membrane disruption, the combination of excessive positive charge and hydrophobicity should be avoided. In generating and analyzing a point mutant library of our lead BIM BH3 stapled peptide, BIM SAHBA1, we observed that cellular uptake can be effectively preserved by lowering the pI to the 8.8–9.34 range while maintaining elevated hydrophobic content (retention times of 9.7–11.2). Thus, in contrast to other classes of cell-penetrating peptides, such as poly-Arg and TAT constructs, positive charge is not a design requirement for generating cell penetrant stapled peptides. Indeed, the first stapled peptide to emerge as a clinical candidate for intracellular targeting has a net charge of −16.
Consistent with the importance of hydrophobicity in driving the cellular uptake feature of stapled peptides, those BIM BH3 constructs containing staples at the amphipathic boundary were the most penetrant compounds. This suggests that creating an expanded hydrophobic surface on the peptide α-helix is a key design strategy for generating cell permeable stapled peptides, as also evidenced by our cellular uptake analysis of SAH-SOS1 stapled peptides that are composed of a different and less amphipathic amino acid sequence, and bear an alternate staple type. Indeed, increasing the hydrophobic contact surface may facilitate plasma membrane tropism for cellular import15. A fortuitous benefit of installing hydrocarbon staples at the boundary of the binding interface is the opportunity for the staple itself to make additional hydrophobic contacts with the protein target. This phenomenon has been observed for a series of cell penetrant, bioactive stapled peptides as demonstrated by the crystal structures of the MCL-1 SAHBD/MCL-130, SAH-p53–8/HDM235, and ATSP-7041/HDMX6 complexes; in each case, staple engagement increased binding affinity without compromising selectivity. Taken together, we find that a stapled peptide design approach that incorporates the described principles of staple placement and tuning hydrophobicity, α-helicity, and pI provides the best chance for rapidly identifying lead compounds for cellular and clinical application.
ONLINE METHODS
Stapled Peptide Synthesis and Characterization
All-hydrocarbon stapled peptides were synthesized, derivatized at the N-terminus with FITC-βAla or acetyl, and purified to >95% homogeneity by LC/MS as previously described36. Acetylated peptides were dissolved in 10% (vol/vol) acetonitrile in water for circular dichroism analyses, performed on an Aviv Biomedical spectrophotometer, as reported36.
ImageXpress Microscopy Analysis
For high-content fluorescence microscopy analysis, the indicated cell lines were plated in black, clear bottom 96-well plates overnight at a density of 2×104 cells per well in DMEM supplemented with 10% (vol/vol) FBS, 1% penicillin/streptomycin, and 1% glutamine. The following day, cells were treated with 0.5 μM FITC-labeled peptides or the equivalent amount of vehicle (0.1% DMSO) for 4 h in serum-free DMEM, and then stained with Hoechst 33342 and CellMask Deep Red (CMDR, Invitrogen) for 10 min. The media was aspirated, and cells were fixed with 4% (wt/vol) paraformaldehyde for 10 min, washed three times with PBS and imaged by ImageXpress Microscopy (high-throughput epifluorescence microscope; Molecular Devices). Data were collected for four sites per well at 20× magnification (nine sites per well at 40× magnification can also be used), with each treatment performed in duplicate, and then analyzed and quantified using MetaXpress software. The CMDR stain was used to visualize the boundaries of the cell and to create a mask for measuring FITC-peptide inside the cell, thereby excluding fluorescent debris from the analysis. A custom module in MetaXpress was applied to incrementally recede the CMDR image mask from the cellular border, further restricting the analyzed FITC signal to internalized peptide. The analysis module was calibrated by defining uniformly negative vs. uniformly positive total well fluorescence based on a vehicle-treated field and a validated positive control field, respectively (e.g. BIM SAHBA110,20); the orthogonal measure of TIFI then determined the level of absolute fluorescence detected per cell, per peptide construct, for the intra-panel comparisons. Maximum and minimum thresholding was utilized to exclude FITC and Cy5 outliers that were much larger and brighter than average and total intensity, and average intensity per cell thresholds were set such that nearly all vehicle-treated cells scored negative by the analysis. For each comparative analysis, all stapled peptides in the panel were measured on the same day using the same plating of cells and peptide dilutions. Then, the entire experiment was repeated twice more (three biological replicates overall) on different days, with freshly plated cells and peptide dilutions.
Cell Culture
B-ALL (BCL-XL-reconstituted p185+Arf−/−Mcl-1del) 25,26 (kindly provided by Dr. Joseph Opferman, St. Jude Children’s Research Hospital) and Jurkat T-cells (ATCC, TIB-152) were maintained in RPMI 1640 (ATCC) supplemented with 10% (v/v) FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, 0.1 mM MEM nonessential amino acids, and 50 mM β-mercaptoethanol. Mouse embryonic fibroblasts (MEFs) and HeLa cells (ATCC, CRM-CCL-2) were maintained in DMEM high glucose (Invitrogen) supplemented with 10% (v/v) FBS, 100 U/mLl penicillin, 100 mg/mL streptomycin, 2 mM L-glutamine, 50 mM HEPES, 0.1 mM MEM nonessential amino acids, and 50 mM β-mercaptoethanol. Cells were verified to be mycoplasma-free using the MycoAlertTM mycoplasma detection kit (Lonza Biologics Inc).
Lactate Dehydrogenase Release Assay
MEFs were plated in 96-well format (2.0×104 cells per well), and after overnight incubation, full media was replaced with serum-free DMEM. B-ALL and Jurkat T-cells were plated in 96-well format (2×104 cells per well) in serum-free RPMI. Serial dilutions of BIM-SAHBA1 from a 10 mM DMSO stock, or vehicle, were added to the MEFs in a final volume of 100 μL and incubated at 37 °C for 0.5 or 3 h, or the peptides were added to the B-ALL or Jurkat T-cells at 10 μM and incubated for 0.5 h. The plate was spun down at 1500 rpm for 5 min at 4°C, and 80 μL of cell culture media was transferred to a clear plate (Corning), incubated with 80 μL of LDH reagent (Roche) for 15 min while shaking, and absorbance measured at 490 nm on a microplate reader (SpectraMax M5 Microplate Reader, Molecular Devices).
Recombinant Protein Production
BCL-XLΔC was expressed as a glutathione-S-transferase (GST) fusion protein in Escherichia coli BL21 (DE3) from the pGEX2T vector (Pharmacia Biotech) and purified by affinity chromatography using glutathione sepharose beads (GE Healthcare), followed by thrombin cleavage of the GST tag and gel filtration FPLC, performed as described25.
Fluorescence Polarization Binding Assay
For direct FP binding assays, FlTC-derivatized peptides (25 nM) were added to serial dilutions of recombinant protein in binding buffer (100 mM NaCl, 50 mM Tris, pH 8.0) in 96-well black opaque plates. The plates were incubated in the dark at RT and then fluorescence polarization measured at 20 min on a microplate reader (SpectraMx M5 Microplate Reader, Molecular Devices). EC50s were calculated by nonlinear regression analysis of dose−response curves using Prism software 5.0 (GraphPad).
Cell Viability
B-ALL cells (4×104/well) were seeded in 96-well opaque plates in a volume of 40 μL in serum-free RPMI using a multiwell dispenser (Apricot) to ensure consistency and reproducibility among the 5–7 plates required per panel. A 2× concentrated plate of the indicated serial dilutions of peptide (10 mM DMSO stock) or DMSO (0.4%) in serum-free RPMI in a volume of 40 μL was then transferred to the cells and the plate incubated at 37°C for 4 hours, at which time, 8.9 μL per well of 100% FBS was added back and cell viability assayed 20 h later by addition of CellTiter-Glo reagent, according to the manufacturer’s protocol (Promega). IC50s were calculated by nonlinear regression analysis of dose-response curves using Prism software 5.0 (GraphPad).
Statistical Methods
The Spearman’s correlation coefficient was used to assess the degree of the univariate relationships between the TIFI and the biophysical variables in the dataset. This quantity is based on the ranks of the data rather than on the observed data values, and consequently is less sensitive to outliers or extreme values than Pearson. The significance of the Spearman’s correlation coefficient is evaluated by means of a permutation test, with the calculations performed using the R statistical software37 package pvrank (https://cran.r-project.org/web/packages/pvrank/pvrank.pdf). Data are represented graphically by scatterplots, and lines were fit using a loess smoother.
Principal components analysis (PCA)21–23 provides a useful method to observe the data through a coordinate system that highlights variability. Retaining an interpretable number of principal components, each of which is a weighted combination of a subset of the available variables, reduces the dimensionality of the data by simplifying the interpretation of the covariance structure. The first principal component explains the largest proportion of the variance, and so on. The original covariates for the exploratory PCA included TIFI, net charge, pI, hydrophilicity, percent hydrophobic residues, hydrophobicity, hydrophobic moment (vector components of magnitude and direction), pH 7 retention time, and percent α-helicity. The calculations were performed using Stata v.13.138.
Further analysis of the data to relate the three principal components to TIFI was accomplished using the recursive partitioning algorithm24. The result is a tree-like representation, calculated using the R package rpart (https://cran.r-project.org/web/packages/rpart/rpart.pdf), and includes relevant binary splits of the dataset. The optimal cut points for each split are evaluated by assessing sums of squares as in analysis of variance.
Supplementary Material
Acknowledgments
We thank E. Smith for graphics support, S. Rudnicki of the Institute of Chemistry and Cell Biology–Longwood for assistance with IXM screening and analysis, and J. Opferman of St. Jude Children’s Research Hospital for the BCL-XL reconstituted p185+Arf−/−Mcl-1-deleted B-ALL cells. This research was supported by NIH grant 1R35CA197583 and 1R21CA209358, a Leukemia and Lymphoma Society (LLS) Marshall A. Lichtman Specialized Center of Research project grant, the William Lawrence and Blanche Hughes Foundation, the Todd J. Schwartz Memorial Fund, the Wolpoff Family Foundation, and an LLS Scholar Award to L.D.W. E.M. and D.S.N. are supported in part by the Dana-Farber/Harvard Cancer Center Support Grant 5 P30 CA006516.
Footnotes
AUTHOR CONTRIBUTIONS
G.H.B, E.M., D.S.N., and L.D.W. designed the study; G.H.B., K. O.-N., M.G., M.A.L. and L.D.W. generated stapled peptides, performed the cellular uptake experiments, and conducted binding and cell viability analyses; E.M. and D.S.N. performed the statistical analyses; G.H.B, E.M., D.S.N., and L.D.W. analyzed the data and wrote the manuscript, which was reviewed by all co-authors.
COMPETING FINANCIAL INTERESTS
L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics.
References
- 1.Walensky LD, Bird GH. Hydrocarbon-stapled peptides: principles, practice, and progress. J Med Chem. 2014;57:6275–88. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schafmeister C, Po J, Verdine G. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- 3.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–70. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–7. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernal F, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–22. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang YS, et al. Stapled alpha-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci U S A. 2013;110:E3445–54. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barclay LA, et al. Inhibition of Pro-apoptotic BAX by a noncanonical interaction mechanism. Mol Cell. 2015;57:873–86. doi: 10.1016/j.molcel.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–81. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim W, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–50. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaBelle JL, et al. A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J Clin Invest. 2012;122:2018–31. doi: 10.1172/JCI46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leshchiner ES, et al. Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc Natl Acad Sci U S A. 2015;112:1761–6. doi: 10.1073/pnas.1413185112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takada K, et al. Targeted disruption of the BCL9/beta-catenin complex inhibits oncogenic Wnt signaling. Sci Transl Med. 2012;4:148ra117. doi: 10.1126/scitranslmed.3003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird GH, Crannell WC, Walensky LD. Chemical synthesis of hydrocarbon-stapled peptides for protein interaction research and therapeutic targeting. Curr Protoc Chem Biol. 2011;3:99–117. doi: 10.1002/9780470559277.ch110042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu Q, et al. Towards understanding cell penetration by stapled peptides. Med Chem Commun. 2015;6:111–119. [Google Scholar]
- 15.Sun TL, Sun Y, Lee CC, Huang HW. Membrane permeability of hydrocarbon-cross-linked peptides. Biophys J. 2013;104:1923–32. doi: 10.1016/j.bpj.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird GH, Gavathiotis E, LaBelle JL, Katz SG, Walensky LD. Distinct BimBH3 (BimSAHB) stapled peptides for structural and cellular studies. ACS Chem Biol. 2014;9:831–7. doi: 10.1021/cb4003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto T, et al. Further insights into the effects of pre-organizing the BimBH3 helix. ACS Chem Biol. 2014;9:838–9. doi: 10.1021/cb400638p. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto T, et al. Stabilizing the pro-apoptotic BimBH3 helix (BimSAHB) does not necessarily enhance affinity or biological activity. ACS Chem Biol. 2013;8:297–302. doi: 10.1021/cb3005403. [DOI] [PubMed] [Google Scholar]
- 19.Li YC, et al. A versatile platform to analyze low-affinity and transient protein-protein interactions in living cells in real time. Cell Rep. 2014;9:1946–58. doi: 10.1016/j.celrep.2014.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards AL, et al. Cellular Uptake and Ultrastructural Localization Underlie the Pro-apoptotic Activity of a Hydrocarbon-stapled BIM BH3 Peptide. ACS Chem Biol. 2015;10:2149–57. doi: 10.1021/acschembio.5b00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hotelling H. Analysis of a complex of statistical variables into principal components. J Edu Psych. 1933;24:417–441. 498–520. [Google Scholar]
- 22.Hotelling H. Relations between two sets of variates. Biometrika. 1936;27:321–377. [Google Scholar]
- 23.Pearson K. On lines adn planes of closest fit to systems of points in space. Philosphical Magazine. 1901;2:559–572. [Google Scholar]
- 24.Breiman L, Friedman JH, Olshen R, Stone CJ. Classification and regression trees. Wadsworth International Group; Belmont CA: 1984. [Google Scholar]
- 25.Cohen NA, et al. A competitive stapled peptide screen identifies a selective small molecule that overcomes MCL-1-dependent leukemia cell survival. Chem Biol. 2012;19:1175–86. doi: 10.1016/j.chembiol.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koss B, et al. Requirement for antiapoptotic MCL-1 in the survival of BCR-ABL B-lineage acute lymphoblastic leukemia. Blood. 2013;122:1587–98. doi: 10.1182/blood-2012-06-440230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danial NN, et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14:144–53. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walensky LD, et al. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Edwards AL, et al. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chem Biol. 2013;20:888–902. doi: 10.1016/j.chembiol.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun CR, et al. Photoreactive stapled BH3 peptides to dissect the BCL-2 family interactome. Chem Biol. 2010;17:1325–33. doi: 10.1016/j.chembiol.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird GH, et al. Mucosal delivery of a double-stapled RSV peptide prevents nasopulmonary infection. J Clin Invest. 2014;124:2113–24. doi: 10.1172/JCI71856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bird GH, et al. Stapled HIV-1 peptides recapitulate antigenic structures and engage broadly neutralizing antibodies. Nat Struct Mol Biol. 2014;21:1058–67. doi: 10.1038/nsmb.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaRochelle JR, Cobb GB, Steinauer A, Rhoades E, Schepartz A. Fluorescence correlation spectroscopy reveals highly efficient cytosolic delivery of certain penta-arg proteins and stapled peptides. J Am Chem Soc. 2015;137:2536–41. doi: 10.1021/ja510391n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baek S, et al. Structure of the stapled p53 peptide bound to Mdm2. J Am Chem Soc. 2012;134:103–6. doi: 10.1021/ja2090367. [DOI] [PubMed] [Google Scholar]
- 36.Bird GH, Bernal F, Pitter K, Walensky LD. Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:369–86. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ihaka R, Gentleman R. A language for data analysis and graphics. J Comp Graph Stat. 1996 [Google Scholar]
- 38.StataCorp. Stata Statistical Software: Release 13. College Station: T.S.L; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.