Abstract
Background and Aims Plant plastid genomes are highly conserved in size, gene content and structure; however, parasitic plants are a noticeable exception to this evolutionary stability. Although the evolution of parasites could help to better understand plastome evolution in general, complete plastomes of parasites have been sequenced only for some lineages so far. Here we contribute to filling this gap by providing and analysing the complete plastome sequence of Cytinus hypocistis, the first parasite sequenced for Malvales and a species suspected to have an extremely small genome.
Methods We sequenced and assembled de novo the plastid genome of Cytinus hypocistis using a shotgun approach on genomic DNA. Phylogenomic analyses based on coding regions were performed on Malvidae. For each coding region present in Cytinus, we tested for relaxation or intensification of selective pressures in the Cytinus lineage compared with autotrophic Malvales.
Key Results Cytinus hypocistis has an extremely divergent genome that is among the smallest sequenced to date (19·4 kb), with only 23 genes and no inverted repeat regions. Phylogenomic analysis confirmed the position of Cytinus within Malvales. All coding regions of Cytinus plastome presented very high substitution rates compared with non-parasitic Malvales.
Conclusions Some regions were inferred to be under relaxed negative selection in Cytinus, suggesting that further plastome reduction is occurring due to relaxed purifying selection associated with the loss of photosynthetic activity. On the other hand, increased selection intensity and strong positive selection were detected for rpl22 in the Cytinus lineage, which might indicate an evolutionary role in the host–parasite arms race, a point that needs further research.
Keywords: Chloroplast genome, Cytinaceae, Cytinus hypocistis, Malvales, parasite, mycoheterotroph, plastome evolution, selective pressure
INTRODUCTION
The chloroplast genome (i.e. plastome) of seed plants is well known for being remarkably conserved in size, structure and gene content across the tree of life (Jansen and Ruhlman, 2012). The most striking exceptions to the evolutionary stability of the plastome have been described in parasite species (Westwood et al., 2010), which show a clear tendency towards plastome reduction (∼11–100 kb) in comparison with their autotrophic relatives (usually 120–170 kb; Ruhlman and Jansen, 2014). This reduction is more pronounced in holoparasites, which are incapable of photosynthesis and thus completely dependent on their hosts for their nutrition. For instance, the plastome of the endoparasite Pilostyles aethiopica (Apodanthaceae) is the smallest reported to date for land plants, with 11 kb and five possibly functional genes (Bellot and Renner, 2016). In Orobanchaceae, the most studied plant parasite lineage to date, some mechanisms of DNA retention have been suggested (Wicke et al., 2013), but our understanding of plastome evolution in parasites remains limited because complete plastome sequences have not been reported for some parasite lineages, and for others only one or few species have been studied. This gap is unfortunate, as it would permit a more general understanding of the evolutionary forces that shape plastomes.
Evolutionary forces driving plastome reduction
Since the main function of chloroplasts is photosynthesis, a major question is why a plastid genome is maintained in non-photosynthetic plants. Barbrook and colleagues (2006) proposed the ‘essential tRNAs’ hypothesis, which is based on the observation that the plastid trnE is involved in the synthesis of tetrapyrroles (i.e. molecules required for vital processes such as cellular respiration) and cannot be functionally replaced by any cytosolic tRNA for this function (Kumar et al., 1996). Therefore, even non-photosynthetic plants would need to retain trnE (along with the machinery for its transcription), in order to be able to synthesize essential tetrapyrroles such as the haem component of mitochondrial cytochromes and other essential oxidative enzymes (Barbrook et al., 2006). However, two recent studies based on high-throughput DNA sequencing affirmed that plastid genomes had vanished at least in two non-photosynthetic organisms – the parasitic plant Rafflesia lagascae (Molina et al., 2014) and the colourless alga Polytomella (Smith and Lee, 2014) – and a third study stated that no functional tRNAs were found in Pilostyles (Bellot and Renner, 2016). These plastome losses would mean that the essential tRNAs hypothesis does not apply for these species, and that an alternative pathway exists for haem biosynthesis (Smith and Asmail, 2014).
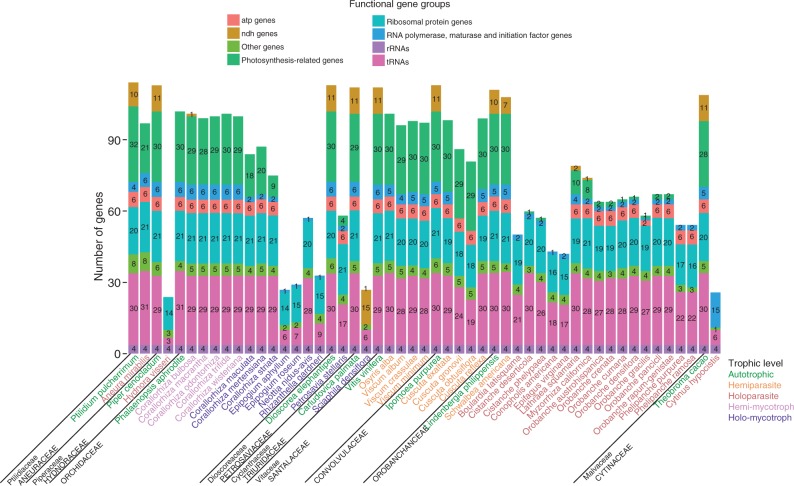
Another question related to plastome maintenance in parasites is why different subsets of genes are retained in the plastome of different parasitic species (Fig. 1), even within the same genus (Supplementary Data Table S1). Within Cuscuta (Convolvulaceae), plastome differences of the four species studied to date seem to be due to differences in their photosynthetic activity (Funk et al., 2007; McNeal et al., 2007). For Orobanchaceae, the whole plastome has been reported for 16 holoparasites and one hemiparasite (Wolfe et al., 1992; Li et al., 2013; Wicke et al., 2013; Cusimano and Wicke, 2015), most of them showing notable differences in plastid gene content and length and structural rearrangements. A recent study (Wicke et al., 2013) indicated that the retention of plastid DNA fragments in this family could be affected by both the proximity to genes under selection and their co-occurrence with those genes in operons. Regarding Santalales (a large order of plants consisting almost entirely of hemiparasites), only four species have had their plastome sequenced; the most prominent plastome modifications were found in the species with the highest nutritional dependence on the host (Petersen et al., 2015). Concerning mycoheterotrophs (i.e. species that parasitize other plants using fungi as vectors), several monocot lineages have been studied, showing very different degrees of plastome reduction, especially in orchids (Delannoy et al., 2011; Logacheva et al., 2011, 2014; Barrett and Davis, 2012; Braukmann and Stefanović, 2012; Barrett et al., 2014; Lam et al., 2015; Schelkunov et al., 2015). In summary, all these studies converge in showing that the plastome structure of parasitic plants is often radically altered in comparison with photosynthetic plants, with extensive gene losses and rearrangements in gene order. However, parasitism has independently appeared at least 12 times in seed plants (Barkman et al., 2007; Su et al., 2015), and the fact that the entire plastome is still not known for several parasite lineages limits our understanding of the macroevolutionary context of the emergence of parasitic lifestyles.
Fig. 1.
Number of unique plastid genes in each functional group of parasite and mycoheterotroph species and their closest autotrophic relative for which the whole plastome has been sequenced (Ptilidium: Forrest et al., 2011; Piper: Cai et al., 2006; Phalaenopsis: Chang et al., 2006; Dioscorea: Hansen et al., 2007; Carludovica: Lam et al., 2015; Vitis: Jansen et al., 2006; Ipomoea: McNeal et al., 2007; Lindenbergia: Wicke et al., 2013; Theobroma: Kane et al., 2012; all references concerning parasitic species are cited in the main text). The species are grouped by the family to which they belong (indicated in grey capital letters). The family of the autotrophic relative is indicated in lower-case letters when it belongs to a different family.
A new insightful case study in Cytinaceae
Our study focuses on Cytinus hypocistis, which belongs to Cytinaceae. Cytinus was long considered to belong to Rafflesiaceae; Bouman and Meijer (1994) highlighted morphological differences in the reproductive traits of Cytinus compared with other members of this family and described a new tribe (Cytineae), but maintained the familial placement. Later, molecular analyses supported the placement of Cytinus in a separate family (Cytinaceae) within Malvales (Nickrent et al., 2004; Barkman et al., 2007; Nickrent, 2007). However, these phylogenetic studies were only based on two mitochondrial regions (matR, atp1) and the nuclear 18S rDNA sequence for Cytinus. This plant family with only two genera (Cytinus and Bdallophytum) shows one of the most extreme manifestations of parasitism: holoparasitic species whose vegetative body is reduced to an endophytic system living within its host root, which only emerges during the blooming period when flowers arise from host tissues. The genus Cytinus contains about six species, distributed in the Mediterranean Basin, South Africa and Madagascar. In the Mediterranean Basin, two species are commonly distinguished, C. hypocistis and C. ruber; however, a recent study showed that C. ruber is no more differentiated genetically than three subspecies of C. hypocistis (de Vega et al., 2008). Cytinus hypocistis parasitizes exclusively roots of Cistaceae (Maire, 1961; de Vega et al., 2010) with a striking specialization at the host level, with distinct genetic races associated with different host plant species (De Vega et al., 2008). This species is monoecious and presents short spikes of basal female flowers and distal male flowers that are mostly ant-pollinated (De Vega et al., 2009).
Here, we provide the first complete plastid genome sequence for a member of Cytinaceae (Cytinus), built from Illumina short-read sequencing data using a shotgun approach on genomic DNA. Our first objective was to confirm the phylogenetic placement of Cytinaceae. Second, we aimed to identify which genes have been lost or show evidence of pseudogene formation compared with related photosynthetic species. Given that Cytinus is a holoparasite, we expected all genes related to photosynthesis to be missing, or at least pseudogenized. Third, we investigated the evolutionary trajectories taken by conserved coding genes in Cytinus. To do so, we analysed whether different past selective pressures can be detected in Cytinus compared with its autotrophic relatives. More specifically, we expected some coding genes to show evidence of relaxed selective constraints compared with their autotrophic relatives. We did so with a formal test that detects relaxation or intensification of selective pressures in a comparative phylogenetic framework. Finally, we compared our study case with other studies conducted to date with the aim of providing a better understanding of plastome evolution in parasites and highlighting the main research perspectives.
MATERIALS AND METHODS
DNA extraction and sequencing
One sample of Cytinus hypocistis was collected in south-eastern France (44.326894° N, 4.819041° E) by Jérémie Van Es (voucher deposited at the herbarium of the Station Alpine Joseph Fourier [GR]). Total genomic DNA was extracted from silica-dried material of one individual using the DNeasy Plant Kit (Qiagen) and subsequently quantified using the NanoDrop spectrophotometer. Genomic DNA (250 ng) was used for Illumina library preparation based on a semi-automatized protocol using the SPRIWorks Library Preparation System and a SPRI TE instrument (Beckmann Coulter), according to the manufacturer’s instructions. DNA fragments were amplified by 12 PCR cycles using a Platinum Pfx Taq Polymerase Kit (Life Technologies) and Illumina adapter-specific primers, and size-selected on 2 % agarose gel (∼600 bp). After library profile analysis conducted using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and qPCR quantification (MxPro, Agilent Technologies, USA), the library was sequenced using 101 base-length read chemistry in a paired-end flow cell on the Illumina HiSeq2000 sequencer (Illumina, USA). The raw sequences have been deposited in the Sequence Read Archive (accession number PRJEB9903).
De novo assembly and annotation of the complete plastome of C. hypocistis
The sequence was assembled de novo into a single circular contig using ORG.Asm, an assembler specially developed for the assembly of organelle genomes (available at http://metabarcoding.org/org-asm), based on a De Bruijn graph, and already used for the assembling of several plastomes (e.g. Besnard et al., 2014; Malé et al., 2014; Colli et al., 2015; Kocher et al., 2015). To assess the quality of the reconstructed sequence, raw reads were mapped against the reconstructed sequence with the software BWA (Li and Durbin, 2009). The mean coverage of the reconstructed sequence was 138×, ranging from 78× to 350×. Pair-end information was consistent with the assembled sequence, and the insert size was in accordance with library specifications.
Protein-encoding genes were identified based on the detection of open reading frames. For genes that included an intron or, like rps12, were trans-spliced, boundaries of exons were defined based on similarities with already annotated genes available in GenBank. Functional annotations of the encoded proteins were assigned by similarities and manually curated. The 23S, 16S and 5S rRNA genes were identified using rRNAmer (Lagesen et al., 2007), the 4.5S rRNA gene was identified using BLAST by searching against a local rRNA database constituted by a set of 4.5 rRNA genes extracted from complete plastomes downloaded from the NCBI ftp site (http://ftp.ncbi.nlm.nih.gov/refseq/release/plastid/). Transfer RNA genes were annotated with tRNAscan-SE (Lowe and Eddy, 1997). We looked for the inverted repeat regions using RepSeek (Achaz et al., 2007). The plastid genome map was drawn with OGDraw (Lohse et al., 2013). The assembled sequence has been deposited in GenBank under the accession number KT335971.
Phylogenomic analyses
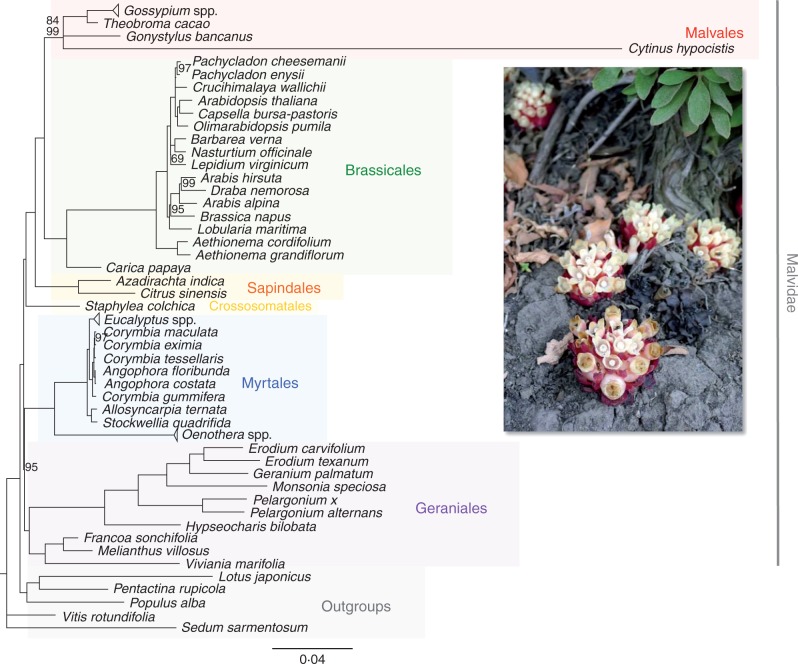
In order to confirm the phylogenetic position of Cytinus within Malvales (as proposed by Nickrent et al., 2004) and obtain a phylogenetic tree for molecular evolution analyses (see below), we recovered all chloroplast genome sequences available in GenBank for all Malvidae s.l. (i.e. including Geraniales, Myrtales, Crossosomatales, Sapindales, Brassicales and Malvales) plus several levels of outgroup taxa: three rosid species belonging to Malpighiales, Fabales and Rosales (Populus alba, Lotus japonicus and Pentactina rupicola, respectively), one species of Saxifragales (Sedum sarmentosum) and one species from Vitales (Vitis rotundifolia). Sequences were downloaded on 17 June 2014. When more than one genome sequence was available for a species, we kept only one (species list and accession numbers are provided in Supplementary Data Table S2). We then extracted coding regions with OBITools (Boyer et al., 2015). We retrieved all coding regions and kept only those that were present at least in 50 % of all study species, resulting in 78 regions. When a coding region was not present, it was coded as missing data.
Codon-based alignments of each coding region were generated with MACSE (Ranwez et al., 2011). MACSE is able to handle frameshifts in the reading frames, which can be artefacts (e.g. sequencing errors) or of biological origin (it has been shown that reading frame shifts can be tolerated during translation; Farabaugh, 1996). Resulting alignments were visually checked. Poorly aligned regions were removed using Gblocks (Castresana, 2000) with the ‘codon’ option activated (i.e. selected blocks contain only complete codons). To check for anomalous sequences, we built maximum likelihood (ML) trees from each of the alignments with RAxML v.8 (Stamatakis, 2014), partitioning first, second and third codon positions separately, and performed bootstrap analyses with the automatic stopping criteria implemented with the option autoMRE. We detected two sequences in obvious spurious phylogenetic locations (rps2 in Lobularia maritima and rps12 in Arabis alpina). We removed those anomalous sequences from the alignments and realigned them following the steps described above. Definitive alignments were concatenated using FASconCAT (Kück and Meusemann, 2010). We used the ‘greedy’ algorithm implemented in PartitionFinder v.1.1.1 (Lanfear et al., 2012) to choose the optimal partition scheme (according to the Bayesian information criterion) for phylogenetic inference starting from an initial partition scheme based on gene and codon position. We ran 200 ML searches from different starting trees using RAxML with the concatenated dataset, applying the GTRCAT model, the rapid hill-climbing algorithm (Stamatakis et al., 2008) and the partition scheme selected with PartitionFinder (21 partitions). Bootstrap searches (1000 replicates) were executed separately using the standard bootstrap option in RAxML. Bootstrap results were drawn in the best ML tree obtained in the previous searches (Fig. 2). The same analyses were performed with a DNA matrix including only the coding regions present in Cytinus, with the aim of checking whether the missing data for Cytinus (i.e. genes that have been lost) have an impact on phylogenetic inference.
Fig. 2.
Chloroplast gene phylogeny of Malvidae: phylogram of the maximum likelihood tree determined by RAxML with 200 independent searches. Numbers associated with branches indicate bootstrap support values obtained with 1000 replicates; unnumbered branches had 100 % support. Scale indicates substitutions per site. Collapsed monophyletic clades correspond to five Oenothera, 20 Gossypium and 31 Eucalyptus species. The inserted image shows Cytinus hypocistis parasitizing a Cistus sp. (Stromboli, Aeolian Islands; © Cristina Roquet).
Test for selective pressures and estimation of substitution rates
For each potential coding region, we tested for potential relaxed or intensified selection in Cytinus compared with related autotrophic taxa using the hypothesis-testing framework RELAX (Wertheim et al., 2015) available on the Datamonkey webserver (www.datamonkey.org/RELAX). Importantly, RELAX overcomes the statistical limitations of existing methods aiming to identify targets of natural selection. Most of these methods are based on the estimation of the ratio of non-synonymous (dN) and synonymous (dS) substitutions (ω = dN/dS) to test for deviations of the neutral expectation ω = 1. RELAX tests whether selection is relaxed or intensified in a subset of predefined test branches in a tree (compared with reference branches) with the selection intensity parameter k, which distinguishes whether selective strength on the test subset of branches is compressed towards or repelled away from neutrality compared with the reference subset. In the RELAX null model, k is constrained to 1 for all branches, whereas in the alternative one it is allowed to differ between reference and test branches. A likelihood-ratio test is implemented to accept or reject the alternative model, and values of the Akaike information criterion with correction for finite sample size (AICc) are also provided. For both models, three categories of ω are estimated (one for sites under purifying selection, ω ≪ 1; another for nearly neutral ones, ω ≃ 1; and a third for sites under diversifying selection, ω ≫ 1), together with the relative proportion of sites they represent. Additionally, RELAX fits the partitioned MG94xREV model for descriptive purposes and to provide a goodness-of-fit reference for the hypothesis-testing framework: in this model separate ω distributions are estimated for test and reference branches without accounting for the selection intensity parameter.
RELAX requires a priori specification of the test and reference branches. Because our a priori hypothesis was that some coding regions might have experienced a different selective pressure in Cytinus because of parasitism, we set the Cytinus branch as the test one. All RELAX analyses were run only with Malvales (plus an outgroup from Brassicales, Carica papaya) because exploratory analyses with the PAML (phylogenetic analysis by maximum likelihood) package (Yang, 2007; results not shown) with all Malvidae taxa included in the phylogenetic analyses yielded very high values of ω in very short branches, which is an indication that too divergent sequences are being compared. The analyses were run providing as input the best ML tree obtained with RAxML, as described in the previous section, after pruning all non-Malvales taxa except Carica.
As a complement to RELAX analyses, we also fitted site and branch-site codon models implemented in the software Codeml included in the package PAML v4.8 (Yang, 2007). These models estimate rates of synonymous and non-synonymous substitutions (dS and dN) and the ω ratio (dN/dS), taking into account potential rate differences in individual codon sites (site models) and also in specific lineages (branch-site models). Full methodological details are provided in Supplementary Data Methods.
RESULTS
Gene content and organization of C. hypocistis plastome
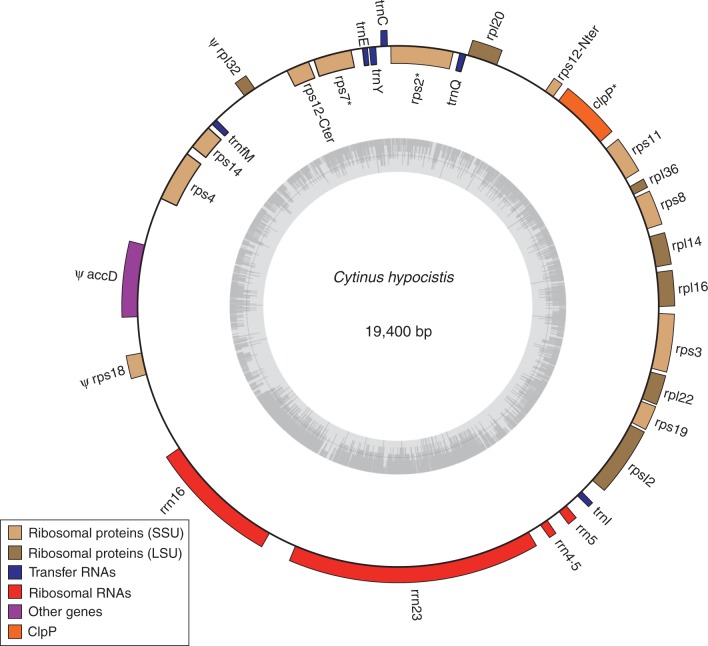
The complete plastid genome of C. hypocistis is 19 400 bp in length, and it encodes a total of 26 genes, including 13 protein-coding genes (5501 bp), six tRNAs (453 bp) and four rRNAs (4573 bp) (Table 1, Fig. 3). Nucleotides corresponding to coding regions account for 28·4 % of the genome. Three additional genes (clpP, rps2 and rps7) show strong sequence divergence compared with other Malvales and could therefore be pseudogenes; we thus did not include them in the set of coding regions for subsequent analyses. Three other regions have certainly become pseudogenes (and hence were also excluded from subsequent analyses): rpl32 encodes only the first 28 amino acids; accD lacks the sequence encoding the 200 first amino acids; and in rps18 the first 20 amino acids are replaced by 30 others and the last 20 amino acids are missing.
Table 1.
Gene content and characteristics of the chloroplast genome of Cytinus hypocistis
| Gene class | Gene | Size (bp) | GC content (%) |
|---|---|---|---|
| Protein genes | accDΨ | 867 | 28·6 |
| clpP* | 58 | 35·2 | |
| rpl2 | 804 | 34·7 | |
| rpl14 | 369 | 28·7 | |
| rpl16 | 405 | 33·8 | |
| rpl20 | 360 | 23·3 | |
| rpl22 | 348 | 26·4 | |
| rpl32Ψ | 147 | 23·8 | |
| rpl36 | 111 | 25·2 | |
| rps2* | 756 | 26·1 | |
| rps3 | 660 | 23·6 | |
| rps4 | 621 | 27·9 | |
| rps7* | 465 | 29·2 | |
| rps8 | 408 | 26·7 | |
| rps11 | 423 | 31·9 | |
| rps12 | 404 | 72·3 | |
| rps14 | 303 | 32 | |
| rps18Ψ | 270 | 22·6 | |
| rps19 | 285 | 24·2 | |
| rRNA | rrn4.5 | 98 | 35·7 |
| rrn5 | 118 | 39 | |
| rrn16 | 1485 | 50 | |
| rrn23 | 2872 | 46·9 | |
| tRNA | trnC-GCA | 72 | 51·4 |
| trnE-UUC | 73 | 52·1 | |
| trnI-CAU | 74 | 46 | |
| trnfM-CAU | 74 | 48·6 | |
| trnQ-UUG | 72 | 59·7 | |
| trnY-GUA | 88 | 51·1 |
ΨPseudogenes.
*Genes that need empirical confirmation of their functionality.
Fig. 3.
Chloroplast genome map of Cytinus hypocistis showing annotated genes. The grey circle indicates the GC content and the line marks the 50 % threshold. Ψ indicates pseudogenes and * indicates genes that might be pseudogenes.
The reconstructed genome lacks inverted repeat (IR) copies. The overall GC content of the whole genome is 29·9 %. This value is slightly lower in protein-encoding genes (29·3 %) and much lower in intergenic spacer regions (16·9 %), while tRNA and rRNA show higher GC values, with 51·4 % and 47·5 % respectively. The six tRNAs found in the plastid genome match six different codons corresponding to six amino acids: cysteine, glutamic acid, glutamine, isoleucine, N-formylmethionine and tyrosine.
Phylogenomic analysis
The final concatenated alignment used for analyses was 58 863 bp in length and corresponded to 78 coding regions (of which only 13 were present as likely coding genes for Cytinus). The proportion of gaps and undetermined characters was 6·24 % (5·39 % excluding Cytinus). The likelihoods of the 200 ML trees obtained varied only slightly (−424 574·81 to −424 575·18 for the best ML tree). The bootstrap analysis showed that most of the nodes were highly supported (Fig. 2): 85 nodes obtained very high support (86–100 % bootstrap support [BS]), five nodes received strong support (71–85 % BS), five received moderate support (50–70 % BS) and only four did not receive any support (<50 % BS). The nodes with moderate or no support corresponded to intrageneric relationships for Gossypium and Eucalyptus taxa, except for the node relating the genus Lepidium to Nasturtium and Barbarea taxa (Brassicaceae) (69 % BS). Phylogenetic analyses provided strong support (84 % BS) for a sister relationship between Thymelaeaceae and Malvaceae, and for a Malvales clade also containing Cytinaceae (99 % BS, Fig. 2). The Malvales clade was sister to the clade corresponding to Brassicales (100 % BS). Phylogenetic analyses based on the reduced matrix (only including the coding genes present in Cytinus) yielded congruent results and similar branch lengths compared with the complete DNA matrix (Supplementary Data Fig. S1).
Signatures of selection in plastid genes and substitution rates
For each putative coding region found in the Cytinus plastome, we tested whether different selective pressures can be detected on Cytinus compared with autotrophic Malvales. We did so by comparing two models implemented in RELAX, which makes it possible to investigate whether the selection intensity (k) is different between the test (here, Cytinus) and reference branches. Model parameter estimates are summarized in Table 2. For the majority of the regions, we did not find significant differences in selection patterns between Cytinus and other Malvales, i.e. the null model (which constrains the selection intensity to 1 for all branches) could not be rejected, and a low ω ratio was estimated for the majority of the sites (Table 2). The null model was rejected in favour of the alternative one only for five regions: rpl16, rps3, rps11, rps19 and rpl22. The region rpl22 showed a significant high increase in selection intensity (k ≫ 1, P = 0·04) for the Cytinus lineage, with a small proportion of its sites (6 %) under positive selection in Cytinus while evolving neutrally in other Malvales. In contrast, the other four regions showed a significant relaxation of selection (k < 1). Specifically, we found weaker purifying selection for a notable proportion of sites of these genes, with ω values that are higher (and nearer to the neutral expectation of ω = 1) in Cytinus than in Malvales.
Table 2.
Test for selective pressures on plastid genes of Cytinus compared with its Malvales autotrophic relatives (‘reference group’). The parameter k is the estimated selection intensity; values under and above 1 indicate relaxed and intensified selection in Cytinus, respectively. The P value is indicated in bold for cases in which the null model was rejected (P < 0·05). Columns and indicate ω values calculated for Cytinus and reference branches under the MG94xREV model, respectively. Columns ω1, ω2 and ω3 indicate three categories of ω values (ω1 for sites under purifying selection; ω2 for nearly neutral ones; ω3 for sites under diversifying selection) estimated with the alternative or the null model depending on which is most suitable according to P value; values in parentheses correspond to the relative proportion of sites they represent
| Gene | k | P | AICcNull | AICcAlt. | ωaCytinus | ωaref. | ω1Cytinus | ω1 ref. | ω2Cytinus | ω2 ref. | ω3Cytinus | ω3ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rpl2 | 0·96 | 0·90 | 3234·1 | 3236·2 | 0·25 | 0·27 | 0·17 (84 %) | 0·17 (84 %) | 0·7 (16 %) | 0·7 (16 %) | – | – |
| rpl14 | 0·85 | 0·44 | 1718 | 1719·6 | 0·15 | 0·09 | – | – | 0·10 (100 %) | 0·10 (100 %) | – | – |
| rpl16 | 0·47 | 0·04 | 1925·2 | 1923·4 | 0·20 | 0·08 | 0(57 %) | 0(57 %) | 0·44 (43 %) | 0·18 (43 %) | ||
| rpl20 | 0·06 | 0·15 | 1953·9 | 1954 | 0·22 | 0·24 | <0·01 (64 %) | <0·01 (64 %) | 0·73 (36 %) | 0·73 (36 %) | – | – |
| rpl22 | 49·5 | 0·04 | 1089·3 | 1087·6 | 0·47 | 0·33 | 0(50 %) | 0(50 %) | 1(44 %) | 1(44 %) | 337(6 %) | 1·12 (6 %) |
| rpl36 | 1·06 | 0·94 | 571·4 | 574 | 0·12 | 0·12 | <0·01 (49 %) | <0·01 (49 %) | 0·21 (51 %) | 0·21 (51 %) | – | – |
| rps3 | 0·48 | 0·02 | 3555·8 | 3552·4 | 0·27 | 0·14 | <0·01 (39 %) | <0·01 (39 %) | 0·47 (61 %) | 0·21 (61 %) | – | – |
| rps4 | 23·4 | 0·08 | 1599·7 | 1598·9 | 0·29 | 0·20 | 0(76 %) | 0(76 %) | – | – | 1·4 (24 %) | 1·4 (24 %) |
| rps8 | 0·2 | 0·08 | 2305 | 2304 | 0·29 | 0·3 | 0(34 %) | 0(34 %) | 0·33 (64 %) | 0·33 (64 %) | 9·57 (2 %) | 9·57 (2 %) |
| rps11 | 0·2 | <0·01 | 1968·6 | 1958·5 | 0·29 | 0·05 | 0·08 (67 %) | <0·01 (67 %) | 0·68 (33 %) | 0·14 (33 %) | – | – |
| rps12 | 1·36 | 0·49 | 1378·2 | 1379·9 | 0·18 | 0·29 | – | – | 0·17 (100 %) | 0·17 (100 %) | – | – |
| rps14 | 0·65 | 0·12 | 1521·1 | 1520·9 | 0·24 | 0·11 | – | – | 0·17 (100 %) | 0·17 (100 %) | – | – |
| rps19 | 0·25 | 0·02 | 1298·4 | 1294·9 | 0·19 | 0·06 | 0·01 (73 %) | <0·01 (73 %) | 0·65 (27 %) | 0·19 (27 %) | – | – |
The fitted site and branch-site models implemented in PAML provided congruent results with those obtained with RELAX concerning selective constraints on each putative coding plastid region found in Cytinus. A summary of the compared models and the parameter estimates of the best model according to a likelihood ratio test are provided in Supplementary Data Table S3. For rpl22, the branch-site model MA fitted better than the null MAn (P = 0·0172), which indicates that some sites (specifically, 9 %) evolve under positive selection only on the Cytinus branch. For four regions (rps11, rps3, rps19 and rpl16) a branch-site model also fitted better than site models according to the AIC (Supplementary Data Table S4), with a considerable proportion of sites inferred to be under neutral evolution in the Cytinus branch (17–39 %) while evolving under strong negative selection in the other lineages. For all other regions, the null models provided a better fit to the data, and the low ω ratio estimated for the majority of the sites indicated that these genes are evolving under purifying selection. Synonymous (dS) and non-synonymous (dN) substitution rate values estimated for Cytinus and other Malvales are provided in Supplementary Data Table S5.
DISCUSSION
General features of the C. hypocistis plastome
The Cytinus plastome reported here represents the first complete sequence for one of the 12 clades where parasitism evolved independently in angiosperms. This genome shows a spectacular reduction to only 19·4 kb, confirming previous indications from Southern blots of digested genomic DNA, in which plastid gene probes hybridized to a fragment of ∼20 kb in length (Nickrent et al., 1997b). This plastome is extremely reduced both in size and gene content, with only 16 protein-coding genes (all corresponding to 50S or 30S ribosomal proteins, except clpP, which encodes ATP-dependent protease), three pseudogenes, four rRNAs and six tRNAs (Table 1). Three of the potentially coding genes are highly divergent and thus need further confirmation from empirical research. To date, the smallest known plastomes correspond to the endoparasitic Apodanthaceae Pilostyles aethiopica and Pilostyles hamiltonii (five genes and 11–15 kb, respectively; Bellot and Renner, 2016), followed by the fully mycoheterotrophic orchids Epipogium roseum (19 kb and 29 genes) and Epipogium aphyllum (31 kb and 27 genes; Schelkunov et al., 2015), the strange holoparasite Hydnora visseri (27 kb and 24 genes; Naumann et al., 2016), the parasitic green alga Helicosporidium sp. (37·5 kb and 54 genes; De Koning and Keeling, 2006) and the Orobanchaceae Conopholis americana (45·5 kb and 43 genes; Wicke et al., 2013).
Compared with the closest photosynthetic relatives of Cytinus, ∼80 % of plastid genes have been lost, including all genes required for photosynthesis, those coding for the plastid RNA polymerase (PEP) and the maturase-like protein matK, seven genes encoding ribosomal proteins and all but six tRNAs. However, we cannot exclude the possibility that some missing genes have been transferred to the nuclear genome, as the low coverage of this genome in our data makes it impossible to test this scenario. It is particularly intriguing why only six tRNAs were retained in Cytinus while the rest were lost. The tRNAs present in Cytinus are found in all parasites sequenced to date, with the following exceptions: trnC-GCA is missing in Conopholis, Hydnora and Phelipanche purpurea; trnQ-UUG is not present in Epipogium aphyllum and Hydnora; and trnY-GUA is not found in Sciaphila and Hydnora (Supplementary Data Table S1). In other words, half of the tRNAs conserved in Cytinus (trnE-UUC, trnfM-CAU, trnI-CAU) are present in almost all sequenced parasitic and fully mycoheterotrophic plants (no functional tRNAs have been found in Pilostyles), and it is noteworthy that they correspond to the three unique tRNAs found in Hydnora (Naumann et al., 2016). Among them, two of the three tRNAs carrying the CAU anticodon are kept. They load the isoleucine and formyl-methionine needed to ensure correct translation initiation without conflict with the isoleucine codon translation in chloroplasts and bacteria (Kashdan and Dudock, 1982). The third tRNA (CAU) dedicated to the classical methionine amino acid is absent. It has been proposed that nuclear-encoded tRNAs might be imported from the cytosol into chloroplast to compensate for missing genes (Morden et al., 1991), and indeed this has been shown experimentally to occur in plant mitochondria (e.g. Dietrich et al., 1992), but not in plastids so far. If this was the case, the question then arises as to whether the remaining tRNAs are maintained in the plastome because they have not yet been subjected to deletion, or whether a few specific tRNAs have an essential function and cannot be imported from the cytoplasm for some particular reasons, as has been proposed in the essential tRNAs hypothesis (Barbrook et al., 2006). Our results (together with data on the other known parasitic plastomes to date) point to the latter possibility, despite the notable exceptions of Rafflesia lagascae and the alga Polytomella, in which the plastome is apparently entirely absent (Molina et al., 2014; Smith and Lee, 2014), and two endoparasitic species of Pilostyles (Apodanthaceae) for which no trace of functional tRNAs has been found in their extremely reduced plastome (Bellot and Renner, 2016).
Regarding its genomic structure, the plastome of C. hypocistis does not present the typical IR regions, resulting in the inability to define large single copy (LSC) and small single copy (SSC) regions. Absence of IRs has been reported for the parasites Conopholis americana and Phelipanche ramosa (Orobanchaceae), and also for certain non-parasites, including a big monophyletic clade of Fabaceae (the ‘inverted repeat-lacking’ clade; Wojciechowski et al., 2000), certain conifers (Strauss et al., 1988) and a few genera of Ericaceae (Fajardo et al., 2013; Martinez-Alberola et al., 2013), Poaceae (Guisinguer et al., 2010) and Geraniaceae (Guisinger et al., 2011). Inverted repeats are thought to help stabilize the plastid genome structure (Palmer and Thompson, 1982), and their disappearance might accelerate gene loss for regions that are not functionally important any more due to parasitism. Further work on the comparative structure of plastomes in relation to species divergence time from autotrophic relatives should allow the identification of the parts of the plastome that are lost first following the loss of autotrophy, and in particular whether the loss of IRs triggers plastome reduction. The Cytinus plastome also presents repeated DNA motifs. In particular, we found a motif of ∼100 bp repeated six times with a similarity of 70–90 %, and some of these repetitions are contiguous to sequences that could correspond to truncated coding regions. These areas could result from plastome rearrangements related to the deletion of some parts of the genome. The C. hypocistis plastome possesses a relatively low GC content of 29·9 % (non-parasitic species usually have 35–40 % GC content). It has been shown that the reduction of the GC content in the plastome coincides with the transition to heterotrophy and increases gradually in holoparasitic Orobanchaceae (Wicke et al., 2013); low GC content was also found in plastid-derived 16S rRNA for seven holoparasites belonging to different lineages (Nickrent et al., 1997a). These results suggest a relationship between GC downward bias and relaxation of selective pressures linked to the loss of photosynthesis.
Selective pressures on plastome coding regions of C. hypocistis
The establishment of obligate parasitism could trigger the relaxation of selective constraints on chloroplast coding regions that are not functionally important any more. We conducted a formal test that estimates whether selection intensity on plastid coding regions differs in Cytinus compared with its autotrophic relatives. According to our results, all but five protein coding regions present in Cytinus are likely evolving under purifying selection, with selective intensity similar to that in its autotrophic relatives (Table 2). These regions may thus remain functionally important in all the lineages studied here. Similarly, the remaining genes of the highly divergent Epipogium orchids do not show any signs of relaxed selection (Schelkunov et al., 2015). On the other hand, relaxed negative selection was inferred in the Cytinus branch for rpl16, rps3, rps11 and rps19, which implies that the importance of these regions has been reduced without fitness consequences in the holoparasite.
For the region rpl22, high intensification of selective pressure was inferred in the Cytinus branch, with 6 % of sites evolving under strong positive selection for Cytinus, and the remaining sites evolving neutrally (44 %) or under strong negative selection (50 %). In the other branches, half of the sites were inferred to be completely conserved and the other half to be evolving neutrally. Given the small number of positions inferred to be under positive selection and the small size of the gene, we cannot fully discard the idea that this region is diverging in Cytinus just as a result of plastome degeneration, since the region rpl22 is missing or pseudogenized in some parasites (Table S1). It is, however, known that plant lineages can differ in plastid gene needs, and indeed, rpl22 is essential for tobacco (Fleischmann et al., 2011) whereas it is not present in the legume plastome (Doyle et al., 1995). On the other hand, a plausible explanation for positive selection in this lineage would be that this gene is linked somehow to the host–parasite evolutionary arms race (Haraguchi and Sasaki, 1996). Plastid housekeeping genes are not known to be specifically connected to host–parasite interaction, but chloroplast genomes contain genes that are related to key metabolic roles (Barbrook et al., 2006; Krause, 2008), and many aspects of parasite physiology and development could be key to attack hosts. Positive selection has also been suggested in strictly heterotrophic Corallorhiza orchids (Barrett et al., 2014) for atp genes, which are known to be involved in photosynthesis but have been suggested to play additional (and yet unknown) roles as they are highly preserved in some holoparasites (Wicke et al., 2013).
PAML site models consistently show that C. hypocistis has much faster rates of molecular evolution than other Malvales, with dS and dN values at least one order of magnitude larger (Table S5). This result is strongly congruent with previous studies that showed that parasitic plants have raised rates of molecular evolution compared with their close non-parasitic relatives (Nickrent and Starr, 1994; Bromham et al., 2013). Higher mutation rates can be derived from small effective population size (Ebert, 1998; Cutter and Payseur, 2013). This is a plausible explanation for this species, which always occurs in very sparse and disconnected populations: De Vega et al. (2008) reported low levels of historical gene flow in C. hypocistis populations of the western Mediterranean Basin, with distinct genetic races associated with different hosts. However, we cannot discard the possibility that raised rates are only due to changing selective pressures in the plastid genome; to verify this, nuclear and/or mitochondrial data would be required, as small population size should affect all genomes equally.
Plastome evolution in parasitic plants
Highly diverged plastomes are found in all non-photosynthetic species sequenced to date, with the exception of holomycotroph Corallorhiza orchids (Barrett et al., 2014) and the holomycotroph liverwort Aneura mirabilis (Wickett et al., 2008), which show only a slight reduction compared with autotrophic relatives and few pseudogenes. The most common trend among parasites is that plastome reduction tends to increase with host dependence: within the obligate hemiparasite species of Cuscuta, the species with lower photosynthetic activity have smaller plastomes (with fewer genes and smaller intergenic regions) than those with higher photosynthesis (Funk et al., 2007; McNeal et al., 2007); in Santalales, obligate parasites of Viscum have lost more functional genes than the facultative parasite Osyris (Petersen et al., 2015); and in Orobanchaceae, a strong correlation between gene loss or pseudogenization and degree of host dependence has been reported (Wicke et al., 2013). Additionally, most photosynthetic genes are either absent or have become pseudogenes in non-green parasites.
Only a subset of housekeeping genes are commonly found in parasites and fully mycoheterotrophic plants sequenced to date, which includes six genes encoding ribosomal protein subunits (rpl2, rpl16, rpl36, rps8, rps11, rps14), three tRNAs (trnI-CAU, trnE-UUC, trnfM-CAU) and the usual four rRNAs (Table S1). The extremely reduced Pilostyles plastomes differ from this subset, as their genomes only include the genes accD, rps3 and rps4, plus two rRNAs (Bellot and Renner, 2016). The number and type of genes lost seem to be idiosyncratic, but a general path of plastome evolution for parasites has been proposed (Barrett and Davis, 2012; Barrett et al., 2014). According to this model, plastid genome degradation in heterotrophic plants would follow four main stages: (1) degradation of the ndh complex; (2) degradation in ndh and other photosynthesis-related genes; (3) degeneration of all plastid gene systems, including housekeeping genes; and (4) complete or nearly complete loss of the plastid genome (as has been suggested for Rafflesia lagascae; Molina et al., 2014). Following this model, Cytinus is somewhere between stages 3 and 4 (Fig. 1). As stated by Barrett et al. (2014), some clear deviations from this model are found in Cuscuta. Additionally, this model does not explain why positive selection has been detected for some genes in certain parasites. Anyway, the fact that the majority of studies have included one or few parasite species per lineage (and that not all lineages have been studied) still hampers a broader understanding of the mechanisms and pace of plastome reduction.
Concerning the fate of vanished plastid genes, a recent study on seven non-green Orobanchaceae (Cusimano and Wicke, 2015) identified nuclear and mitochondrial insertions for most of the genes lost in the plastome (some corresponding to full copies, others just to fragments), likely as a result of multiple intracellular transfers accumulated over time. However, the majority of these insertions showed high sequence drift and many indels, indicating that they might not be functional.
Phylogenetic implications
Our phylogenomic analyses based on coding regions confirm the placement of Cytinaceae within Malvales with strong support, with Malvaceae and Thymelaeaceae as the closest relatives to Cytinaceae. We included here only two Malvales families apart from Cytinaceae because they were the only ones to have complete plastomes (or with a high number of coding regions) available in GenBank. Our results suggest that Malvaceae and Thymeleaceae are more closely related to each other than to Cytinaceae, a new result not reported before (some interfamilial relationships of Malvales are still unclear today); however, a broader taxon sampling at the family level is needed to confirm this point. Regarding higher taxonomic scale relationships, we here confirm interordinal relationships reported for Malvidae by Ruhfel et al. (2014).
Future research perspectives in parasitic plastome evolution
Recent years have seen an upsurge of sequenced plastomes of plant parasites and mycoheterotrophs. However, to really understand the genomic consequences of niche shifts from autotrophy to heterotrophy in plants, a much broader taxon sampling is needed, including all trophic levels of major plant parasite lineages. The information derived from an extensive sampling would likely illuminate how genetic changes in parasites are correlated with the degree of host dependence, time since trophic shift, and gene function. Wider knowledge of the selective pressures on plastome genes of parasites compared with their autotrophic relatives would also help to unravel whether evolution to parasitism happened stepwise or in a more continuous way; for instance, rapid deletion of large gene blocks has been suggested in Cuscuta, whereas in Orobanchaceae slow plastome degradation appears to be more likely (Cusimano and Wicke, 2015).
Another interesting research area that remains to be explored more deeply is horizontal gene transfer (HGT) between parasitic plants and their hosts, which is expected to occur thanks to direct physical contact. Indeed, HGT has already been identified in some cases for nuclear and mitochondrial genomes between parasites and host in both directions (e.g. Mower et al., 2004; Davis et al., 2005; Xi et al., 2013), but no plastid-to-plastid transfer has yet been reported to our knowledge. Broader research with genomic and transcriptomic approaches in this area is necessary to unravel the evolutionary consequences derived from HGT and also to identify the mechanisms that facilitate it. Genomic and transcriptomic research could also help to resolve other open questions, such as why plastome genomes have been retained in plant parasites and whether (and which) missing plastid genes have been integrated in the nuclear or mitochondrial genomes.
Concluding remarks
We provide here the complete plastid genome sequence of C. hypocistis, the first parasite genome sequenced for Malvales. This genome is extremely diverged and harbours a spectacular reduction in size and gene number: it mostly encodes ribosomal proteins, tRNAs and rRNAs, and presents no IR regions. We detected very high substitution rates for all coding regions of Cytinus compared with its non-parasitic relatives, which might be associated with small effective population size and/or changing selective pressures. As expected, some regions were inferred to be under relaxed negative selection. On the other hand, strong positive selection was inferred for some sites of the region rpl22 in the Cytinus lineage, which could be explained by a role in the host–parasite evolutionary arms race. However, given the small size of this region, we cannot fully discard the possibility that the divergence found is only the result of plastome degeneration. Phylogenomic analysis of all coding regions confirmed the position of Cytinus within the order Malvales. Future research should incorporate a broad taxon sampling of parasite lineages, including species with different trophic levels, in order to enhance our understanding of mechanisms driving plastome evolution.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Supplementary Methods: Evolutionary codon models. Figure S1: chloroplast gene phylogeny of Malvidae based on a reduced DNA matrix (only including coding genes present in Cytinus); numbers associated with branches indicate bootstrap support values obtained with 1000 replicates; collapsed monophyletic clades correspond to five Oenothera, 20 Gossypium and 31 Eucalyptus species. Table S1: comparison of the gene content of Cytinus hypocistis with parasitic plants and fully mycoheterotrophic plants sequenced to date. Table S2: list of species included in the phylogenomic analysis and the corresponding GenBank accession numbers. Table S3: summary of the fitted null and alternative codon-substitution models implemented in PAML. Table S4: Akaike information criterion (AIC) scores obtained for site and branch-site models for each putative coding region. The model with the lowest AIC is indicated in bold. Table S5: synonymous (dS) and non-synonymous (dN) substitution rate values of Cytinus versus other Malvales.
ACKNOWLEDGEMENTS
This work received support from the CNRS-INEE (research call in Environmental Genomics ‘APEGE-INEE’, 2012) for the sequencing aspects. The research leading to this paper had received funding from the European Research Council under the European Community’s Seven Framework Programme FP7/2007–2013 Grant Agreement no. 281422 (TEEMBIO).
LITERATURE CITED
- Achaz G, Boyer F, Rocha EPC, Viari A, Coissac E. 2007. Repseek, a tool to retrieve approximate repeats from large DNA sequences. Bioinformatics 23: 119–121. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. 2006. Why are plastid genomes retained in non-photosynthetic organisms? Trends in Plant Science 11: 101–108. [DOI] [PubMed] [Google Scholar]
- Barkman TJ, McNeal JR, Lim S-H, et al. 2007. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evolutionary Biology 7: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, Davis JI. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. American Journal of Botany 99: 1513–1523. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Freudenstein JV, Li J, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Molecular Biology and Evolution 31: 3095–3112. [DOI] [PubMed] [Google Scholar]
- Bellot S, Renner SS. 2016. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biology and Evolution 8: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Christin PA, Malé P-JG, et al. 2014. From museums to genomics: old herbarium specimens shed light on a C3 to C4 transition. Journal of Experimental Botany 65: 6711–6721. [DOI] [PubMed] [Google Scholar]
- Bouman F, Meijer W. 1994. Comparative structure of ovules and seeds in Rafflesiaceae. Plant Systematics and Evolution 193: 187–212. [Google Scholar]
- Boyer F, Mercier C, Bonin A, Taberlet P, Coissac E. 2015. OBITools: a Unix-inspired software package for DNA metabarcoding. Molecular Ecology Resources 16: 176–182. [DOI] [PubMed] [Google Scholar]
- Braukmann T, Stefanović S. 2012. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Molecular Biology 79: 5–20. [DOI] [PubMed] [Google Scholar]
- Bromham L, Cowman PF, Lanfear R. 2013. Parasitic plants have increased rates of molecular evolution across all three genomes. BMC Evolutionary Biology 13: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Penaflor C, Kuehl JV, et al. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evolutionary Biology 6: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin HC, Lin IP, et al. 2006. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Molecular Biology and Evolution 23: 279–291. [DOI] [PubMed] [Google Scholar]
- Colli L, Lancioni H, Cardinali I, et al. 2015. Whole mitochondrial genomes unveil the impact of domestication on goat matrilineal variability. BMC Genomics 16: 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusimano N, Wicke S. 2015. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytologist 210: 680–693. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nature Reviews Genetics 14: 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CC, Anderson WR, Wurdack KJ. 2005. Gene transfer from a parasitic flowering plant to a fern. Proceedings of the Royal Society B: Biological Sciences 272: 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas Des Francs-Small C, Brundrett M, Small I. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Molecular Biology and Evolution 28: 2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich A, Weil JH, Maréchal-Drouard L. 1992. Nuclear-encoded transfer RNAs in plant mitochondria. Annual Review of Cell Biology 8: 115–131. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD. 1995. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Systematic Botany 20: 272–294. [Google Scholar]
- Ebert D. 1998. Experimental evolution of parasites. Science 282: 1432–1436. [DOI] [PubMed] [Google Scholar]
- Fajardo D, Senalik D, Ames M, et al. 2013. Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genetics & Genomes 9: 489–498. [Google Scholar]
- Farabaugh PJ. 1996. Programmed translational frameshifting. Annual Review of Genetics 30: 507–528. [DOI] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schöttler MA, Bock R. 2011. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. The Plant Cell 23: 3137–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest LL, Wickett NJ, Cox CJ, Goffinet B. 2011. Deep sequencing of Ptilidium (Ptilidiaceae) suggests evolutionary stasis in liverwort plastid genome structure. Plant Ecology and Evolution 144: 29–43. [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K. 2007. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biology 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. Journal of Molecular Evolution 70: 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Molecular Biology and Evolution 28: 583–600. [DOI] [PubMed] [Google Scholar]
- Hansen DR, Dastidar SG, Cai Z, et al. 2007. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Molecular Phylogenetics and Evolution 45: 547–563. [DOI] [PubMed] [Google Scholar]
- Haraguchi Y, Sasaki A. 1996. Host-parasite arms race in mutation modifications: indefinite escalation despite a heavy load? Journal of Theoretical Biology 183: 121–137. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Ruhlman TA. 2012. Plastid genomes of seed plants In: Bock R, Knoop V, eds. Genomics of chloroplast and mitochondria. New York: Springer, 103–126. [Google Scholar]
- Jansen RK, Kaittanis C, Saski C, et al. 2006. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evolutionary Biology 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane N, Sveinsson S, Dempewolf H, et al. 2012. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. American Journal of Botany 99: 320–329. [DOI] [PubMed] [Google Scholar]
- Kashdan MA, Dudock BS. 1982. The gene for a spinach chloroplast isoleucine tRNA has a methionine anticodon. Journal of Biological Chemistry 257: 11191–11194. [PubMed] [Google Scholar]
- Kocher A, Gantier J-C, Holota H, et al. 2015. Complete mitochondrial genome of Lutzomyia (Nyssomyia) umbratilis (Diptera: Psychodidae), the main vector of Leishmania guyanensis. Mitochondrial DNA 22: 1–3. [DOI] [PubMed] [Google Scholar]
- De Koning AP, Keeling PJ. 2006. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biology 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K. 2008. From chloroplasts to ‘cryptic’ plastids: evolution of plastid genomes in parasitic plants. Current Genetics 54: 111–121. [DOI] [PubMed] [Google Scholar]
- Kück P, Meusemann K. 2010. FASconCAT: convenient handling of data matrices. Molecular Phylogenetics and Evolution 56: 1115–1118. [DOI] [PubMed] [Google Scholar]
- Kumar AM, Schaub U, Söll D, Ujwal ML. 1996. Glutamyl-transfer RNA: at the crossroad between chlorophyll and protein biosynthesis. Trends in Plant Science 1: 371–376. [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam VKY, Soto Gomez M, Graham SW. 2015. The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biology and Evolution 7: 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang TC, Qiao Q, et al. 2013. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS One 8: e58747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Penin AA. 2011. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biology and Evolution 3: 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA. 2014. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biology and Evolution 6: 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research 41: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malé P-JG, Bardon L, Besnard G, et al. 2014. Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Molecular Ecology Resources 14: 966–975. [DOI] [PubMed] [Google Scholar]
- Maire R. 1961. Flore de l’Afrique du nord. Paris: Éditions Paul Lechevalier. [Google Scholar]
- Martinez-Alberola F,, del Campo EM,, Lázaro-Gimeno D, et al. 2013. Balanced gene losses, duplications and intensive rearrangements led to an unusual regularly sized genome in Arbutus unedo chloroplasts. PLoS One 8: e79685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl J V, Boore JL, de Pamphilis CW. 2007. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biology 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J, Hazzouri KM, Nickrent D, et al. 2014. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Molecular Biology and Evolution 31: 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morden CW, Wolfe KH, dePamphilis CW, Palmer JD. 1991. Plastid translation and transcription genes in a non-photosynthetic plant: intact, missing and pseudo genes. EMBO Journal 10: 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Stefanovic S, Young GJ, Palmer JD. 2004. Plant genetics: gene transfer from parasitic to host plants. Nature 432: 165–166. [DOI] [PubMed] [Google Scholar]
- Naumann J, Der JP, Wafula EK, et al. 2016. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biology and Evolution 8: 345–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL. 2007. Cytinaceae are sister to Muntingiaceae (Malvales). Taxon 56: 1129–1135. [Google Scholar]
- Nickrent DL, Starr EM. 1994. High rates of nucleotide substitution in nuclear small-subunit (18S) rDNA from holoparasitic flowering plants. Journal of Molecular Evolution 39: 62–70. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Duff RJ, Konings DA. 1997a. Structural analyses of plastid-derived 16S rRNAs in holoparasitic angiosperms. Plant Molecular Biology 34: 731–743. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Ouyang Y, Duff RJ, DePamphilis CW. 1997b. Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Molecular Biology 34: 717–729. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Blarer A, Qiu Y-L, Vidal-Russell R, Anderson FE. 2004. Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evolutionary Biology 4: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29: 537–550. [DOI] [PubMed] [Google Scholar]
- Petersen G, Cuenca A, Seberg O. 2015. Plastome evolution in hemiparasitic mistletoes. Genome Biology and Evolution 7: 2520–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP. 2011. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS One 6: e22594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Jansen RK. 2014. The plastid genomes of flowering plants In: Maliga P, ed. Chloroplast biotechnology: methods and protocols. New York City, NY: Humana Press, 3–38. [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. 2014. From algae to angiosperms - inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evolutionary Biology 14: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelkunov MI, Shtratnikova VY, Nuraliev MS, Selosse M-A, Penin AA, Logacheva MD. 2015. Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biology and Evolution 7: 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Asmail SR. 2014. Next-generation sequencing data suggest that certain nonphotosynthetic green plants have lost their plastid genomes. New Phytologist 204: 7–11. [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW. 2014. A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiology 164: 1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Strauss SH, Palmer JD, Howe GT, Doerksen AH. 1988. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proceedings of the National Academy of Sciences of the USA 85: 3898–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H-J, Hu J-M, Anderson F-E, Der JP, Nickrent DL. 2015. Phylogenetic relationships of Santalales with insights into the origins of holoparasitic Balanophoraceae. Taxon 64: 491–506. [Google Scholar]
- De Vega C, Berjano R, Arista M, Ortiz PL, Talavera S, Stuessy TF. 2008. Genetic races associated with the genera and sections of host species in the holoparasitic plant Cytinus (Cytinaceae) in the Western Mediterranean basin. New Phytologist 178: 875–887. [DOI] [PubMed] [Google Scholar]
- De Vega C, Arista M, Ortiz PL, Herrera CM, Talavera S. 2009. The ant-pollination system of Cytinus hypocistis (Cytinaceae), a Mediterranean root holoparasite. Annals of Botany 103: 1065–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vega C, Arista M, Ortiz PL, Talavera S. 2010. Anatomical relations among endophytic holoparasitic angiosperms, autotrophic host plants and mycorrhizal fungi: a novel tripartite interaction. American Journal of Botany 97: 730–737. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Molecular Biology and Evolution 32: 820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. 2010. The evolution of parasitism in plants. Trends in Plant Science 15: 227–235. [DOI] [PubMed] [Google Scholar]
- Wicke S, Müller KF, de Pamphilis CW, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. The Plant Cell 25: 3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Zhang Y, Hansen SK, et al. 2008. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Molecular Biology and Evolution 25: 393–401. [DOI] [PubMed] [Google Scholar]
- Wojciechowski MF, Sanderson MJ, Steele KP, Liston A. 2000. Molecular phylogeny of the ‘temperate herbaceous tribes’ of papilionoid legumes: a supertree approach In: Herendeen PS, Bruneau A, eds Advances in legume systematics. Kew: Royal Botanic Gardens, 277–298. [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proceedings of the National Academy of Sciences of the USA 89: 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Wang Y, Bradley RK, et al. 2013. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLOS Genetics 9: e1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]