Abstract
Purpose
To demonstrate a new MR imaging approach that unambiguously identifies and quantitates contrast agents based on intrinsic agent properties such as r1, r2, r2*, and magnetic susceptibility. The approach is referred to as Magnetic Barcode Imaging (MBI).
Methods
Targeted and bio-responsive contrast agents were imaged in agarose phantoms to generate T1, T2, T2*, and quantitative susceptibility maps. The parameter maps were processed by a machine learning algorithm trained to recognize the contrast agents based on these parameters. The output is a quantitative map of contrast agent concentration, identity, and functional state.
Results
MBI allowed the quantitative interpretation of intensities, removed confounding backgrounds, enabled contrast agent multiplexing, and unambiguously detected the activation and binding states of bio-responsive and targeted contrast agents.
Conclusion
MBI has the potential to overcome significant limitations in the interpretation, quantitation, and multiplexing of contrast enhancement by MR imaging probes.
Keywords: molecular imaging, magnetic resonance imaging, machine learning, contrast agent
INTRODUCTION
Magnetic resonance imaging (MRI) interrogates deep tissues at high resolution without the use of ionizing radiation. This unique combination of attributes is ideal for molecular imaging that uses specialized contrast agents to visualize cellular and biomolecular processes in whole organisms (1). Two types of contrast agents are used in molecular MRI: targeted agents with affinity for molecular markers (2-4) and bio-responsive agents that activate in response to biological cues (5-7). Current imaging methods that rely on T1 or T2 weighting limit the impact of these agents in several significant ways. First, agent contrast enhancements often appear ambiguous against complex backgrounds; second, quantitative information is lacking due to nonlinear signals; third, enhancements from unbound targeted agents lead to false detections; fourth, bio-responsive agent activation is easily confused with concentration variations because both appear as an increase in signal (8); and lastly, gray scale outputs make simultaneous probing with multiple agents extremely challenging (9,10).
A major research goal is to obtain molecular MRI that is unambiguous, multiplexed, and quantitative. These properties have been attained in the past by exploiting unique resonances provided by contrast agent platforms such as micromachined magnets (10), chemical exchange saturation transfer agents (11), and hyperpolarized xenon gas vesicles (12). However, gadolinium complexes (13) and iron oxide particles (14) continue to serve as the mainstay platforms for molecular MRI due to their ease of chemical modification, sensitivity, and long imaging time window.
A new imaging technique that overcomes the current shortcomings of gadolinium and iron oxide imaging would quickly impact a range of applications by taking advantage of the facile functionalization, high stability, and biological tolerance of established contrast agents. Progress has been made in the quantitative and specific imaging of these agents using ratiometric methods that measure two relaxation times (15-17). Beyond contrast agent imaging, a more generalized approach measuring more parameters has been long applied to the machine learning detection of tissue pathologies (18-22). Despite this history, multiparameter imaging has not penetrated in a significant way into the contrast agent community to address the shortcomings of molecular MR imaging.(23) Contrast agents present unique challenges distinct from those applicable to tissues because no dataset is readily available for machine learning, the MR parameters change as a function of agent concentration and tissue background, and the design of pulse sequence combinations cannot be based on experiences from tissues.
Here, we have developed and demonstrated a framework that combines multiparameter MR acquisitions with supervised machine learning as applied to molecular MR imaging to distinguish contrast agents by their susceptibility and relaxometric signatures (Figure 1). The approach, called Magnetic Barcode Imaging (MBI), specifically highlights theoretically any contrast agent in an image. By assigning different “colors” to individual signatures associated with contrast agents and their functional states, MBI can i. display contrast enhancement with zero background, ii. distinguish between bound and unbound states of targeted agents, iii. unambiguously detect the activation of bio-responsive agents, and iv. support the simultaneous imaging of multiple types of contrast agents in a single acquisition. Importantly, by performing regression learning, quantitation can be realized by MBI.
FIG 1. Magnetic barcode imaging.
Conventional molecular MRI utilizes (a) T1-weighted imaging for gadolinium and (b) T2-weighted imaging for iron oxide. Magnetic barcoding is a framework that identifies contrast agents by their intrinsically multiparametric barcodes. An example of a barcode comprises of T1, T2, T2* relaxivities (r1, r2, r2*) and molar susceptibility (χM). After assigning colors to barcodes, the output displays each contrast agent in the assigned color on a concentration scale with the irrelevant background removed. The framework enables unambiguous, multiplexed, and quantitative molecular MRI.
The MBI framework can be implemented in many ways depending on the pulse sequences and the machine learning strategy used. Optimized algorithms of acquisition and processing in different in vivo settings, as well as the accumulation of large training datasets that capture contrast agents in complex in vivo environments, would require a research effort across many research groups. Such an effort requires proof of concept. To this end, we prove that detection of MR contrast agent concentration, type, chemical state, and localization is possible by MBI in vitro using an implementation that barcodes by T1, T2, T2* and susceptibility maps. The capabilities of MBI were demonstrated on both gadolinium and iron oxide contrast agents across a range of applications.
METHODS
Contrast Agents
Gd(III)-daa was synthesized using a previously described protocol (24). The version of Gd(III)-daa used has a 5-carbon linker.
Synthesis of Gd(III)-biotin employed the synthetic strategy described in (25) to achieve 3-(2,4,6)-Tris(1-2(hydroxyl-3-(1H-1,2,3-triazol-1-yl(propyl)-3,7,10-tris(carboxymethyl)-1,4,7,10-tetraazacyclododecylgadolinium(III)phenoxy)-propan-1-amine (C3-amine). C3-amine (1.6 mg, 0.0008 mmol) was suspended in 200 μL, pH 9.4 bicarbonate buffer under inert atmosphere. To the stirring solution was added biotin N-hydroxysuccinimide ester (26) (1.2 mg, 0.0035 mmol) dissolved in 100 μL of DMSO. After 24 hours, the solvent was removed by lyophilization and the product purified by reverse phase HPLC using a C18 column, held at 5% for 5 minutes and eluting with a gradient of 5% to 20% acetonitrile in pH 10.38 buffered water over 21 minutes, tr= 11.8 minutes. This gave 1.1 mg of pure product as a white solid (55% yield). MS (MALDI-TOF): m/z observed = 2250.2 [M+], 2273.2 [M+ + Na+], 2289.2 [M+ + K+], 2312.2 [M+ + Na+ + K+], m/z calculated = 2250.5 [M+], 2273.5[M+ + Na+], 2289.5 [M+ + K+], 2312.5 [M+ + Na+ + K+].
Super paramagnetic iron oxide SPIO was synthesized using a published method (27). The SPIO used was surface-functionalized with diethylene glycol. Feridex was purchased from Bayer HealthCare (Wayne, NJ). MPIO was purchased from Bangs Laboratories (Cat. No. ME03F, Fishers, IN). Concentrations of agents were measured using ICP-MS. Iron oxide particle concentrations reported are Fe concentrations. Gd(III)-biotin concentrations reported are molecular concentrations. MPIO concentrations reported in μg/mL include polymeric weight and were based on supplier label.
Training Agarose Phantoms
All agarose phantoms used for radial basis function neural network (RBFNN) training were prepared in a similar fashion, consisting of contrast agent-doped 1% low melting point (lowMP) agarose (Life Technologies, Carlsbad, CA) embedded within blank 1% regular agarose. 2% agarose in milliQ water and a buffer were mixed in equal parts to produce 1% agarose. The buffer used was 10 mM HEPES for Gd(III)-daa, 10 mM PBS for Gd(III)-biotin, and milliQ water for SPIO, Feridex, and MPIO in the multiplexing experiment. MPIO training for the zero-background experiment was performed using 10% serum-supplemented minimum essential media mixed with 2% agarose in equal parts (media-agarose). All samples were prepared in 15 mL conical tubes. 4 mL of 1% agarose was gelled at room temperature around a 5 mm OD NMR tube. The tube was later removed to create a cylindrical bore of approximately 1 cm in depth. The bore was filled with 220 μL of 1% lowMP agarose doped with the appropriate concentration of contrast agent. Concentrations are specified in the methods of each individual experiment. After filling, the tube was cooled to 4 °C for 5 minutes to solidify the doped lowMP agarose. To the same tube, 2 mL of 1% agarose was added and solidified for 5 minutes at 4 °C to completely enclose the doped lowMP agarose. Approximately 5 mL of the same buffer used to prepare the agarose was diluted to 10 mL with milliQ water and added on top of the agarose plug to completely fill the tube. Diffusion in agarose occurred for small molecule contrast agents. Therefore, imaging was performed immediately after preparation of the phantom.
Magnetic Barcode Acquisition
Imaging was performed at 25°C on a Bruker Pharmscan 7 T imaging spectrometer equipped with shielded gradient coils (Bruker BioSpin, Billerica, MA). A 300 MHz 1H 89mm/23mm quadrature transceiver volume coil was used. The Bruker MapShim method was used for shimming. T1 measurements were performed using a Rapid Acquisition with Refocused Echoes (RARE) pulse sequence. T2 measurements were performed using a Multi-Spin Echo (MSE) pulse sequence. T2* measurements were performed using a Multi-Gradient Echo (MGE) pulse sequence. Field map measurements used for susceptibility mapping were performed using the Bruker FieldMap macro based on a double gradient echo pulse sequence. Unless otherwise specified, the geometries for T1, T2, and T2* acquisitions were FOV = 15×15 mm2, slice thickness = 0.9375 mm, and matrix = 128×128. The parameters for field map acquisitions were TR = 29.6 ms, TE = 2.3, 27.1 ms, NEX = 1, FOV = 22.5×22.5×22.5 mm3, matrix size = 96×96×96, and scan time = 4 min 33 s. Other pulse sequence parameters are detailed in the methods for each individual experiment.
Barcode Processing and Machine Learning
T1 and T2 maps were generated using custom programs written in Matlab (version R2010b, MathWorks, Natick, MA). For T1, each voxel in a repetition time (TR) series of images was fitted to equation S = M·[1-exp(-TR/T1)]. S is signal, and M is equilibrium magnetization. S and TR were known. M and T1 were fitted. For T2, each voxel in an echo time (TE) series of images was fitted to equation S = M·exp(-TE/T2)+C. C is an arbitrary constant that is fitted. T2* was fitted in the same way as T2. Susceptibility maps were generated based on field maps using the Quantitative Susceptibility Mapping (QSM) algorithm described by de Rochefort, et al (28). Field map voxels with SNR < 6 were set to 0. Regions-of-interest (ROI) were defined by the non-zero regions in the field map. Manual corrections were performed to include air bubbles within the sample into the ROI. Top and bottom 5 rows of the field maps were removed to minimize interference from field inhomogeneity during QSM processing. Tuning of the conjugate gradient loop stop criterion and the edge regularization parameter was performed manually; values that result in stable solutions that do not vary significantly with the parameters were selected. These parameters were kept the same across training and testing for each experiment. Procedures for automatic parameter selection are available (28) but were not implemented. RBFNN training and image processing were performed in Matlab using the neural network toolbox. 200 voxels from each training agarose phantom were randomly selected and aggregated to form the training set for each contrast agent or agent functional state. For Gd(III)-daa and Gd(III)-biotin, 200 additional training data points were generated by interpolation to provide coverage for partial Gd(III)-daa activation and partial Gd(III)-biotin binding. Each training set was normalized by its own mean and standard deviation. An empirical relationship between susceptibility, as obtained by QSM, and concentration, as measured by ICP-MS, was then found by linear fitting. The empirical equation was used to calculate the answer vector containing concentrations for multivariate regression training. The number of nodes in the RBFNN was restricted to less than 500. A spread of 0.1-0.3 and a mean error goal of < 10% were used. These parameters were not optimized, and the accuracy of RBFNN regression can be further improved by techniques such as cross-validation and ROC curve analysis.
Relaxivity and Susceptibility Measurement
The r1, r2, and r2* of contrast agents were obtained from their respective training set. From the T1, T2, and T2* maps of each agent at each concentration, the relaxation time values were recorded from voxels near the center of the agarose phantom to minimize errors introduced by agent diffusion. r1, r2, and r2* were found as the slopes of linear fits of 1/T1, 1/T2, and 1/T2*, respectively, against concentration in units of mM−1s−1. The χM of each agent was obtained in a fashion similar to how relaxivities were measured, except that χM was found as the slope of χ linearly fitted against concentration in units of ppm/M. χM were calibrated against the theoretical Gd(III) value of 326 ppm/M (29) using a calibration factor obtained by comparing the Gd(III)-daa data to the theoretical value.
Zero-Background Imaging
The test phantom was prepared in a 4 mL 15 × 45 mm glass vial. 2 mL of 1% media-agarose warmed at 70 °C was added to the vial. As the agarose gels, 10 μL of each of 8 μg/mL MPIO and 2.3 mM FeCl3 in 1% media-lowMP agarose warmed at 60 °C was carefully infused in two separate spots. 10 μL of dodecanol was infused in a third location. The 1% media-lowMP agarose was colored by 2.5% v/v trypan blue for visibility. The sample was solidified for 5 minutes at 4 °C before 2 mL more of 1% media-agarose was added and cooled at 4 °C for 5 minutes. Media diluted 2X by milliQ water was added at the end to completely fill the vial. RBFNN training was performed using 0, 2, 4, 6, 8, 10 μg/mL MPIO. Detailed imaging parameters are as follows: for T1 measurements, TR = 770, 1300, 1900, 2700, 3700, 5000, 7400, 17500 ms, effective TE = 36 ms, RARE factor = 8, NEX = 1, and scan time = 10 min 44 s. For T2 measurements, TR = 3500 ms, TE = 11, 22, 33...358 ms in 11 ms increments, NEX = 1, and scan time = 7 min 28 s. For T2* measurements, TR = 3000 ms, TE = 5, 13, 21...317 ms in 8 ms increments, matrix = 256 × 256, NEX = 1, and scan time = 12 min 48 s. For T2-weighted imaging, TR = 3500 ms, TE = 88 ms, and NEX = 1.
Bio-responsive and Targeted Agent Imaging
Test phantoms were prepared in a similar method as described for the training phantoms. An additional step was included for mixtures of Gd(III)-daa with ZnCl2, and Gd(III)-biotin with avidin (Molecular Probes Cat. No. A887, Eugene, OR) or human serum albumin (Sigma-Aldrich Cat. No. A9511, St. Louis, MO). Specifically, these solutions were left undisturbed for 30 min at RT to ensure complete activation or binding before doping into lowMP agarose. Bio-responsive activation of Gd(III)-daa occurs when the diacetate domain binds Zn2+, opening a coordination site on the Gd(III) and concomitantly increasing r1 and r2. Gd(III)-biotin is targeted to avidin. When incubated with the protein, the tumbling rate of the agent decreases and its relaxivities change accordingly. For Gd(III)-daa, training was performed using 0, 0.25, 0.75, 1.25, 2 mM Gd(III)-daa with and without 2 equivalents of ZnCl2. For Gd(III)-biotin, training was performed using 0, 50, and 100 μM molecule without avidin, and 33.3, 66.7, 83.3 μM molecule with 1/3 equivalents of avidin. Detailed imaging parameters for Gd(III)-daa are as follows: for T1 measurements, TR = 49, 57, 66, 75, 84, 94, 105, 117, 130, 145, 160, 175, 190, 210, 230, 260, 300, 350, 450, 750 ms, effective TE = 9 ms, RARE factor = 2, NEX = 1, and scan time = 4 min 15 s For T1-weighted imaging, TR = 100 ms, TE = 9 ms, RARE factor = 1, and NEX = 1. For T2 measurements, TR = 800 ms, TE = 11, 22, 33...358 ms in 11 ms increments, NEX = 1, and scan time = 1 min 42 s. Parameters for Gd(III)-biotin are as follows: for T1 measurements, TR = 62, 72, 84, 104, 124, 144, 164, 190, 220, 250, 280, 320, 360, 400, 460, 520, 600, 700, 900, 1500 ms, effective TE = 9 ms, RARE factor = 2, NEX = 1, and scan time = 7 min 57 s. For T1-weighted imaging, TR = 400 ms, TE = 9 ms, RARE factor = 1, and NEX = 1. For T2 measurements, TR = 800 ms, TE = 11, 22, 33...358 ms in 11 ms increments, NEX = 1, and scan time = 1 min 42 s. Display intensities of the T1-weighted images were normalized by equalizing the blank agarose intensity across images.
Multiplexed Imaging
The test phantom was prepared in a 15 mL conical tube. 12 mL of 1% agarose was gelled at RT around five 16 gauge needles that were later removed to create cylindrical bores in the agarose. 1% lowMP agarose warmed at 60 °C and doped with various concentrations of SPIO, Feridex, and MPIO were used to fill the cylindrical bores. Contrast agent concentrations are indicated in the relevant figure. The volume used for each bore was 25 μL. The sample was cooled at 4 °C for 5 min to solidify the lowMP agarose. 2 mL more of 1% agarose was added and solidified at 4 °C for 5 min to completely encapsulate the embedded contrast agent. The remainder of the tube was filled with milliQ water. RBFNN training was performed using 240 and 320 μM Fe SPIO, 60 and 80 μM Fe Feridex, 60 and 80 μM Fe MPIO, 120/30 and 160/40 μM Fe SPIO/Feridex mixture, and 120/30 and 160/40 μM Fe SPIO/MPIO mixture. Additionally, blank 1% agarose was included in the training set to define the zero point for regression training. Detailed imaging parameters are as follows: For T1 measurements, TR = 315, 400, 480, 570, 650, 750, 850, 1000, 1100, 1200, 1400, 1600, 1800, 2000, 2300, 2600, 3000, 3500, 4400, and 7500 ms, effective TE = 36 ms, RARE factor = 8, NEX = 1, and scan time = 9 min 58 s. For T2 measurements, TR = 3500 ms, TE = 11, 22, 33...358 ms in 11 ms increments, NEX = 1, and scan time = 7 min 28 s. For T2* measurements, TR = 3500 ms, TE = 5, 13, 21...317 ms in 8 ms increments, NEX = 1, and scan time = 7 min 28 s. For T2-weighted imaging, TR = 3500 ms, TE = 176 ms, and NEX = 1.
RESULTS
MBI uniquely identifies contrast agents and their functional states by signatures that are encoded in the magnetic properties and the molecular dynamics of each agent. To capture these signatures, a MBI implementation is comprised of a selection of acquisition pulse sequences, a pre-processing protocol, and a machine learning strategy (Figure 2). Together, these components determine the barcode space dimensionality, the accuracy and precision of the measured parameters, and the classification and quantitation accuracy. The implementation reported here uses multi-gradient echo and Carr-Purcell-Meiboom-Gill multi-spin echo sequences to measure T1, T2, T2*, and phase-based susceptibility (χ) in a voxel-by-voxel manner (30). This combination of pulse sequences captures the corresponding agent relaxivities and molar susceptibility (r1, r2, r2*, χM). For pre-processing, relaxation times were estimated by least-squares fitting to known signal equations, and susceptibility was obtained using a morphology-enabled dipole inversion method devised by de Rochefort, et al. (28) Machine learning was performed by a radial basis function neural network (31) (RBFNN) trained on contrast agent phantoms prepared at a range of concentrations (Figure 2a). RBFNN was chosen because it is amenable to training without negative examples, outputs posterior Bayesian probabilities, and handles regression problems well. Depending on the training set used, the RBFNN can learn to highlight a single contrast agent from the background, or use different colors to represent different agents or different functional states of a single agent. The different RBFNN configurations used to achieve the different outputs are described in the results for each experiment.
FIG. 2. Prototype implementation and zero-background imaging.
MBI specifically identifies contrast agents by combining multiparametric acquisition with machine learning. The prototype consists of T1, T2, T2*, and χ map acquisitions in combination with a RBFNN. The approach is demonstrated using the example of MPIO in the context of zero-background imaging. (a) The RBFNN is trained to recognize MPIO using a 4-dimensional training set generated by imaging agarose phantoms doped with 0-10 μg/mL MPIO. Each voxel at each concentration represents a data point for regression training. (b) Zero-background imaging is tested by embedding 8 μg/mL MPIO, fat-mimicking dodecanol, and FeCl3 simulating a hemorrhage into agarose. T1, T2, T2*, and χ maps are used as the input to generate the barcode image. (c) By T2-weighted MRI, the three hypo-intensities are indistinguishable. By MBI, background and confounding features are removed, leaving only the MPIO contrast displayed on a quantitative scale.
Zero-background Imaging
Micron-sized Superparamagnetic Iron Oxide Particles (MPIOs) are customarily used in T2-weighted MRI. These agents generate negative contrast that are often confounded by hypo-intense features such as air, hemorrhage, iron-rich tissues, or calcification (32). In theory, a RBFNN trained to recognize MPIOs would differentiate the contrast agent from the confounding features. To test this hypothesis, MBI was performed on an agarose phantom embedding 8 μg/mL MPIO, fat mimicking dodecanol, and 2.3 mM FeCl3 simulating a hemorrhage. Training of the RBFNN was performed using 0-10 μg/mL MPIO.
MBI successfully distinguished MPIO from dodecanol and Fe3+ whereas T2-weighted imaging showed all three as undifferentiated hypo-intensities (Figure 2c). On MBI, the MPIO contrast is displayed as a quantitative intensity without background. MBI recapitulates the major advantages of quantitation and zero-background in nuclear imaging – considered the gold standard in clinical molecular imaging – without the radiation risk and at a higher resolution.
Detection of Bio-responsive Agent Activation
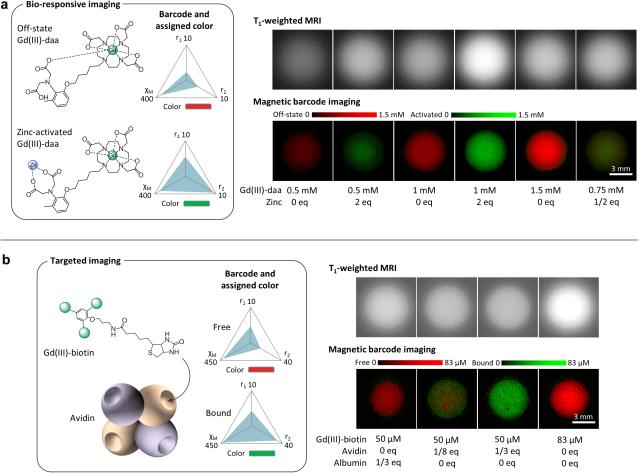
To test MBI on a bio-responsive agent, Gd(III)-diaminoacetate (Gd(III)-daa) was used as a model system. Gd(III)-daa is a Zn2+ sensor that allows water coordination to Gd(III) upon Zn2+ binding (24,33). The binding event leads to an increase in the r1 and r2 to the agent's barcode (Figure 3a). r2* is degenerate with r2 in Gd(III)-daa and not used. The RBFNN for this system was constructed with two output nodes to represent the concentrations of Gd(III)-daa in its off and activated state, respectively. The training performed on the agent was with and without Zn2+ at a range of concentrations. Once the training was complete, testing was performed using concentrations not seen during training to demonstrate external validity.
FIG 3. Color switching by functional state.
Magnetic barcodes can be defined by the functional state of an agent. (a) Gd(III)-daa is a Zn2+ sensor whose barcode increases in r1 and r2 upon Zn2+ binding. The agent brightens on T1-weighted imaging upon activation, but the increased signal is difficult to distinguish from higher concentrations of “off-state” Gd(III)-daa (no Zn2+ present). MBI removes this ambiguity by displaying a switch from red to green based on the agent's barcode upon Zn2+ binding. Partial activation by a half equivalent of Zn2+ results in a partial conversion to green. The intensities for both the off and activated states are quantitative. (b) Gd(III)-biotin is a targeted agent. Its barcode changes upon binding to avidin due to slower molecular tumbling. T1-weighted imaging provides no information on the binding status of Gd(III)-biotin. MBI specifically detects binding by distinguishing between free Gd(III)-biotin, Gd(III)-biotin in mixture with human serum albumin, and Gd(III)-biotin fully or partially bound to avidin. Avidin equivalents of less than 1/4 results in partial binding because avidin has 4 binding sites. r1 and r2 are in mM−1s−1. χM is in ppm/M.
The standard method for imaging bio-responsive Gd(III) agents is T1-weighted imaging. However, a major limitations of this method is evidenced by its inability to distinguish an activation event from an increase in agent concentration, as Gd(III)-daa produces residual contrast in its “off” (absence of Zn2+) state (Figure 3a). MBI, on the other hand, unambigusouly differentiates between the functional states of Gd(III)-daa regardless of concentration. Concentration is quantitatively displayed, allowing for the detection of partial activation when less than one equivalent Zn2+ is present. Therefore, MBI removes the obstacle of ambiguity in the imaging of Gd(III) bio-responsive agents.
Detection of Targeted Agent Binding
MBI can detect the binding of a targeted agent to the selected molecular target, analogous to the way differentiation is achieved with bio-responsive agent activation. To test this capability, an avidin-targeted agent (Gd(III)-biotin) was used. Using the same barcode design as Gd(III)-daa, Gd(III)-biotin phantoms with and without avidin across a range of concentrations were imaged for training. Upon Gd(III)-biotin binding to avidin, the molecular rotation slowed, decreasing r1 and increasing r2 (Figure 3b). As before, testing was performed using concentrations that differ from those in the training set.
The goal of a targeted agent is to delineate the presence of a molecular marker from the background tissues. However, non-specific retention can compromise interpretation of the results. Under standard T1-weighted imaging, the binding status of the agent is unidentifiable (Figure 3b). In comparison, MBI quantitatively displays the binding status of the contrast agent as a change in color by differentiating between the barcodes of the bound and unbound states. Further, MBI distinguishes between partial binding and saturation depending on whether Gd(III)-biotin or avidin is in excess. When human serum albumin instead of avidin was mixed with Gd(III)-biotin, MBI accurately identified the agent as unbound. The ability to switch color upon protein binding of a small molecule targeted contrast agent is a unique capability of MBI that has not been demonstrated with any other imaging modality.
Multiplexing of Iron Oxide Particles
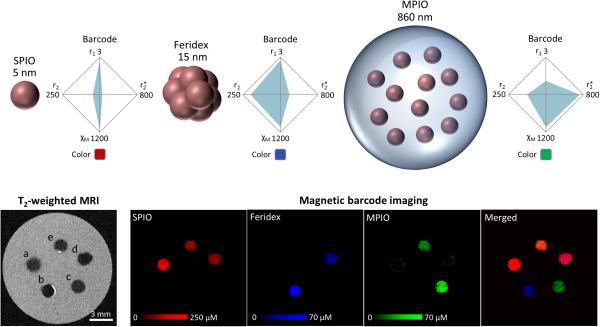
Multiplexing in molecular imaging refers to the simultaneous visualization of multiple imaging agents (34). This capability is important because there is increasing realization that no single biomarker is likely to provide sufficient characterization of a disease process (35). Magnetic particles have the potential to serve as a platform for MR multiplexing because their magnetic and dynamic properties change with size, composition, and geometry (36). However, this potential has not been fully realized.
Using MBI, multiplexing was tested with superparamagnetic iron oxide nanoparticles (SPIOs), Feridex, and MPIOs. RBFNN training was performed on the individual particles as well as mixtures of particles. All three particles used were Fe3O4 in material, but of different sizes and configurations, producing distinct barcodes (Figure 4). By T2-weighted MRI, the three particles embedded in agarose produce hypo-intensities that are indistinguishable from each other. When the same specimen is imaged by MBI, each particle is displayed as a different color on a quantitative scale, even in mixtures. This result supports the notion that magnetic particles can be multiplexed via the tuning of their size and shape.
FIG. 4. Multiplexing.
Three iron oxide particles are imaged, namely SPIOs (5 nm), Feridex (15 nm clusters of 5 nm particles), and MPIOs (heterogeneous particles in polymeric microspheres). The different structures result in different barcodes. T2-weighted imaging shows contrast from all three particles as indistinguishable hypo-intensities. (a) 280 μM SPIO (b) 70 μM Feridex (c) 70 μM MPIO (d) 140 μM SPIO and 35 μM Feridex (e) 140 μM SPIO and 35 μM MPIO. MBI enables multiplexing, displaying each particle in a different color based on its barcode. Particles in mixtures are accurately identified. Merged image uses a common scale of 150 μM. r1, r2, r2* are in mM−1s−1. χM is in ppm/M.
DISCUSSION
We have demonstrated a proof-of-concept of the magnetic barcoding framework across a range of targeted, bio-responsive, gadolinium, and iron oxide contrast agents. By abandoning the treatment of T1 and T2 as a dichotomy, barcoding opens up several opportunities for MR contrast agent design. For example, an emerging strategy to enable quantitation and specific detection in molecular MRI is to synthesize multimodal probes (8,37). MBI helps to achieve the same goal while allowing the contrast agent design to remain simple. Another burdensome task in traditional contrast agent development is its optimization for different field strengths. MBI alleviates this need as evidenced by its successful detection of Gd(III)-biotin binding at 7 T even though the agent was optimized for 1.5 T and lost its property of binding-induced r1 enhancement at the higher field strength. Further opportunities lie in the parameter-tuning of contrast agents. MBI turns the old design rule of maximizing or minimizing r1/r2 ratio upside down as sensitivity and specificity are maximized with a balanced ratio in a multi-parameter framework. Similarly, engineering the traditionally ignored r2* becomes valuable in MBI because it adds a dimension to barcode space, as shown by MPIO. These properties imply that the performance of MBI is further optimized when matched to specifically designed contrast agents.
To translate the proof-of-concept results described here in vivo, further iterative development of the technique will need to focus on two areas. First, acceleration is needed because motion and time-varying contrast agent concentrations in vivo necessitate scan times on the order of a minute. In comparison, an acquisition by the current implementation takes approximately 30 minutes. Incorporation of compressed sensing (38), steady-state free precession (39), or parallel imaging (40) into future iterations can shorten scan times by a factor of between 10-100 fold.
Second, the additional complexities associated with tissue background in vivo need to be addressed. Fundamentally, T1, T2, T2*, and χ are determined by both the contrast agent and the tissue background. To account for the greatly increased number of combinations in vivo compared to in phantoms, the training set used for machine learning would need to be expanded. One way to perform the expansion is to computationally add the relaxation rates and susceptibilities of blank tissues to those of the contrast agents; alternatively, training can be performed by imaging a collection of contrast-infused tissues ex vivo. The latter strategy would be preferred if the agent barcode changes substantially in the tissue microenvironment. The likelihood of such a change depends on the barcode design, the contrast agent used, the compartmentalization of the contrast agent, and tissue properties such as diffusion. Therefore, the extensiveness and the method of MBI training will vary with the imaging problem being addressed. Continual advancement of MBI in the two areas of speed and training set development is expected to enable its use in vivo.
Other important areas of future MBI development will include strategies for handling highly heterogeneous backgrounds, new barcode designs, and applications of various machine learning algorithms. While multi-parametric MR approaches have been explored extensively for tissue imaging, the unique challenges and opportunities of the approach in molecular MRI warrant a separate effort. For example, variations due to agent concentration and the numerous possible agent-tissue combinations are not a concern in tissue imaging. This property adds to the complexity of MBI machine learning when applied to a highly heterogeneous background. In addition to obtaining large training sets as already discussed, a strategy for addressing this complexity would be to exploit pulse sequences that are more sensitive to magnetic materials than tissues, such as QSM or off-resonance imaging (41,42). This strategy circumvents the more cumbersome method of acquiring pre-scans for background removal, and illustrates the unique opportunities to innovate in multi-parameter contrast agent imaging that is distinct from the challenges of tissue imaging.
In relation to other approaches that similarly aim to address the shortcomings of T1- and T2-weighted contrast agent imaging, we view MBI as a complementary rather than a competing approach. At its core, MBI is a framework for integrating multi-dimensional image data into an output with emergent attributes that are highly desirable in a molecular imaging study. The use of machine learning circumvents the challenge of having to reinvent image analysis every time a new parameter is added to the acquisition. Pulse sequences such as T1ρ mapping (43), intermolecular multiple quantum coherences (44), ultrashort echo time relaxometry (45), and dispersion imaging (46), can be employed under the MBI framework to further improve multiplexing degree, sensitivity, and accuracy. Similarly, signal intensities rather than fitted parameters can be explored as barcode components; this type of implementation has the advantage of improved accuracy when there are heterogeneous tissue types within a voxel. Furthermore, a wide array of machine learning (47-49) and susceptibility mapping algorithms (50-52) exist that can be applied within the MBI framework to reduce processing artifacts and improve quantitation and classification accuracy. As an example, in the current implementation, the quantitative maps produced are multiplicative products of concentration and Bayesian probability. That is, voxels with low classification confidence display underestimated concentrations in exchange for fewer false positives. In our dataset, the largest underestimation seen is with the SPIOs at 15%, while the false positive rate as determined by the percentage of voxels with grossly over-estimated concentrations is < 10%. This type of trade-off can be modified by the processing algorithm design.
CONCLUSIONS
MBI uniquely enables the multiplexing, quantitation, and specific detection of MRI contrast agents, conferring on them the advantages that were once exclusive to optical and nuclear imaging. The framework, when implemented fully, will remove a significant barrier towards the routine use of molecular MRI and opens new possibilities for contrast agent design, biomarker quantitation, and multi-marker detection of pathologies.
ACKNOWLDEGMENTS
We thank Professor Warren Warren, Ryan Davis, Dr. Thomas Theis, Yi Han, Dr. Alex Waters, Dr. Chad Haney, Dr. Keith MacRenaris, Luke Vistain, and Adam Preslar for helpful discussions. We acknowledge support by NIH Grants R01EB005866, R01EB014806, U54CA151880, and T32GM008152. We are grateful to the Integrated Molecular Structure Education and Research Center (IMSERC), the Quantitative Bioelemental Imaging Center (QBIC), and the Center for Advanced Molecular Imaging (CAMI) at Northwestern University. CAMI was supported by NCI CCSG P30 CA060553 and NCRR 1S10RR025624-01.
REFERENCES
- 1.James ML, Gambhir SS. A molecular imaging primer: modalities, imaging agents, and applications. Physiol Rev. 2012;92(2):897–965. doi: 10.1152/physrev.00049.2010. [DOI] [PubMed] [Google Scholar]
- 2.Overoye-Chan K, Koerner S, Looby RJ, Kolodziej AF, Zech SG, Deng Q, Chasse JM, McMurry TJ, Caravan P. EP-2104R: a fibrin-specific gadolinium-based MRI contrast agent for detection of thrombus. J Am Chem Soc. 2008;130(18):6025–6039. doi: 10.1021/ja800834y. [DOI] [PubMed] [Google Scholar]
- 3.Serres S, Soto MS, Hamilton A, McAteer MA, Carbonell WS, Robson MD, Ansorge O, Khrapitchev A, Bristow C, Balathasan L. Molecular MRI enables early and sensitive detection of brain metastases. Proc Natl Acad Sci U S A. 2012;109(17):6674–6679. doi: 10.1073/pnas.1117412109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strauch RC, Mastarone DJ, Sukerkar PA, Song Y, Ipsaro JJ, Meade TJ. Reporter protein-targeted probes for magnetic resonance imaging. J Am Chem Soc. 2011;133(41):16346–16349. doi: 10.1021/ja206134b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louie AY, Hüber MM, Ahrens ET, Rothbächer U, Moats R, Jacobs RE, Fraser SE, Meade TJ. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18(3):321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 6.Li W-H, Fraser SE, Meade TJ. A calcium-sensitive magnetic resonance imaging contrast agent. J Am Chem Soc. 1999;121(6):1413–1414. [Google Scholar]
- 7.Moats RA, Fraser SE, Meade TJ. A “smart” magnetic resonance imaging agent that reports on specific enzymatic activity. Angew Chem Int Ed. 1997;36(7):726–728. [Google Scholar]
- 8.Frullano L, Catana C, Benner T, Sherry AD, Caravan P. Bimodal MR–PET agent for quantitative pH imaging. Angew Chem Int Ed. 2010;49(13):2382–2384. doi: 10.1002/anie.201000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sosnovik DE, Weissleder R. Emerging concepts in molecular MRI. Current opinion in biotechnology. 2007;18(1):4–10. doi: 10.1016/j.copbio.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Zabow G, Dodd S, Moreland J, Koretsky A. Micro-engineered local field control for high-sensitivity multispectral MRI. Nature. 2008;453(7198):1058–1063. doi: 10.1038/nature07048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahon MT, Gilad AA, DeLiso MA, Cromer Berman SM, Bulte JW, van Zijl P. New “multicolor” polypeptide diamagnetic chemical exchange saturation transfer (DIACEST) contrast agents for MRI. Magn Reson Med. 2008;60(4):803–812. doi: 10.1002/mrm.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro MG, Ramirez RM, Sperling LJ, Sun G, Sun J, Pines A, Schaffer DV, Bajaj VS. Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat Chem. 2014;6:629–634. doi: 10.1038/nchem.1934. [DOI] [PubMed] [Google Scholar]
- 13.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Gadolinium (III) chelates as MRI contrast agents: structure, dynamics, and applications. Chem Rev. 1999;99(9):2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 14.Gupta AK, Gupta M. Synthesis and surface engineering of iron oxide nanoparticles for biomedical applications. Biomaterials. 2005;26(18):3995–4021. doi: 10.1016/j.biomaterials.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Alberti D, van't Erve M, Stefania R, Ruggiero MR, Tapparo M, Geninatti Crich S, Aime S. A Quantitative relaxometric version of the ELISA test for the measurement of cell surface biomarkers. Angewandte Chemie International Edition. 2014;53(13):3488–3491. doi: 10.1002/anie.201310959. [DOI] [PubMed] [Google Scholar]
- 16.Catanzaro V, Gringeri CV, Menchise V, Padovan S, Boffa C, Dastrù W, Chaabane L, Digilio G, Aime S. A R2p/R1p Ratiometric Procedure to Assess Matrix Metalloproteinase-2 Activity by Magnetic Resonance Imaging. Angewandte Chemie International Edition. 2013;52(14):3926–3930. doi: 10.1002/anie.201209286. [DOI] [PubMed] [Google Scholar]
- 17.Hu F, Jia Q, Li Y, Gao M. Facile synthesis of ultrasmall PEGylated iron oxide nanoparticles for dual-contrast T1-and T2-weighted magnetic resonance imaging. Nanotechnology. 2011;22(24):245604. doi: 10.1088/0957-4484/22/24/245604. [DOI] [PubMed] [Google Scholar]
- 18.Clarke SE, Hammond RR, Mitchell JR, Rutt BK. Quantitative assessment of carotid plaque composition using multicontrast MRI and registered histology. Magn Reson Med. 2003;50(6):1199–1208. doi: 10.1002/mrm.10618. [DOI] [PubMed] [Google Scholar]
- 19.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng Y, Jiang Y, Antic T, Giger ML, Eggener SE, Oto A. Validation of quantitative analysis of multiparametric prostate MR images for prostate cancer detection and aggressiveness assessment: a cross-imager study. Radiology. 2014;271(2):461–471. doi: 10.1148/radiol.14131320. [DOI] [PubMed] [Google Scholar]
- 21.Ecker C, Marquand A, Mourão-Miranda J, Johnston P, Daly EM, Brammer MJ, Maltezos S, Murphy CM, Robertson D, Williams SC. Describing the brain in autism in five dimensions—magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. The Journal of Neuroscience. 2010;30(32):10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs MA, Knight RA, Soltanian-Zadeh H, Zheng ZG, Goussev AV, Peck DJ, Windham JP, Chopp M. Unsupervised segmentation of multiparameter MRI in experimental cerebral ischemia with comparison to T2, diffusion, and ADC MRI parameters and histopathological validation. Journal of Magnetic Resonance Imaging. 2000;11(4):425–437. doi: 10.1002/(sici)1522-2586(200004)11:4<425::aid-jmri11>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 23.Villringer A, Rosen BR, Belliveau JW, Ackerman JL, Lauffer RB, Buxton RB, Chao YS, Wedeenand VJ, Brady TJ. Dynamic imaging with lanthanide chelates in normal brain: contrast due to magnetic susceptibility effects. Magnetic resonance in medicine. 1988;6(2):164–174. doi: 10.1002/mrm.1910060205. [DOI] [PubMed] [Google Scholar]
- 24.Matosziuk LM, Leibowitz JH, Heffern MC, MacRenaris KW, Ratner MA, Meade TJ. Structural Optimization of Zn(II)-Activated Magnetic Resonance Imaging Probes. Inorg Chem. 2013;52(21):12250–12261. doi: 10.1021/ic400681j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison VS, Carney CE, Macrenaris KW, Meade TJ. A multimeric MR-optical contrast agent for multimodal imaging. Chemical Communications. 2014;50(78):11469–11471. doi: 10.1039/c4cc05651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura Y, Ito S, Shimizu Y, Kanai M. Catalytic anomeric aminoalkynylation of unprotected aldoses. Organic letters. 2013;15(16):4130–4133. doi: 10.1021/ol401810b. [DOI] [PubMed] [Google Scholar]
- 27.Hu F, MacRenaris KW, Waters EA, Liang T, Schultz-Sikma EA, Eckermann AL, Meade TJ. Ultrasmall, water-soluble magnetite nanoparticles with high relaxivity for magnetic resonance imaging. J Phys Chem C. 2009;113(49):20855–20860. doi: 10.1021/jp907216g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Rochefort L, Liu T, Kressler B, Liu J, Spincemaille P, Lebon V, Wu J, Wang Y. Quantitative susceptibility map reconstruction from MR phase data using bayesian regularization: validation and application to brain imaging. Magn Reson Med. 2010;63(1):194–206. doi: 10.1002/mrm.22187. [DOI] [PubMed] [Google Scholar]
- 29.de Rochefort L, Brown R, Prince MR, Wang Y. Quantitative MR susceptibility mapping using piece-wise constant regularized inversion of the magnetic field. Magn Reson Med. 2008;60(4):1003–1009. doi: 10.1002/mrm.21710. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein MA, King KF, Zhou XJ. Handbook of MRI pulse sequences. Elsevier; 2004. [Google Scholar]
- 31.Park J, Sandberg IW. Universal approximation using radial-basis-function networks. Neural Comput. 1991;3(2):246–257. doi: 10.1162/neco.1991.3.2.246. [DOI] [PubMed] [Google Scholar]
- 32.Shin T-H, Choi J-s, Yun S, Kim I-S, Song H-T, Kim Y, Park KI, Cheon J. T1 and T2 dual-mode MRI contrast agent for enhancing accuracy by engineered nanomaterials. ACS nano. 2014;8(4):3393–3401. doi: 10.1021/nn405977t. [DOI] [PubMed] [Google Scholar]
- 33.Major JL, Parigi G, Luchinat C, Meade TJ. The synthesis and in vitro testing of a zinc-activated MRI contrast agent. Proc Natl Acad Sci U S A. 2007;104(35):13881–13886. doi: 10.1073/pnas.0706247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22(8):969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 35.Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, Natan MJ, Gambhir SS. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proc Natl Acad Sci U S A. 2009;106(32):13511–13516. doi: 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J-H, Huh Y-M, Jun Y-w, Seo J-w, Jang J-t, Song H-T, Kim S, Cho E-J, Yoon H-G, Suh J-S. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat Med. 2006;13(1):95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 37.Frullano L, Meade TJ. Multimodal MRI contrast agents. JBIC Journal of Biological Inorganic Chemistry. 2007;12(7):939–949. doi: 10.1007/s00775-007-0265-3. [DOI] [PubMed] [Google Scholar]
- 38.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 39.Deoni SC, Peters TM, Rutt BK. High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn Reson Med. 2005;53(1):237–241. doi: 10.1002/mrm.20314. [DOI] [PubMed] [Google Scholar]
- 40.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952–962. [PubMed] [Google Scholar]
- 41.Sherry AD, Woods M. Chemical exchange saturation transfer contrast agents for magnetic resonance imaging. Annu Rev Biomed Eng. 2008;10:391. doi: 10.1146/annurev.bioeng.9.060906.151929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khemtong C, Kessinger CW, Ren J, Bey EA, Yang S-G, Guthi JS, Boothman DA, Sherry AD, Gao J. In vivo off-resonance saturation magnetic resonance imaging of αvβ3-targeted superparamagnetic nanoparticles. Cancer Res. 2009;69(4):1651–1658. doi: 10.1158/0008-5472.CAN-08-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wheaton AJ, Borthakur A, Kneeland JB, Regatte RR, Akella SV, Reddy R. In vivo quantification of T1ρ using a multislice spin-lock pulse sequence. Magn Reson Med. 2004;52(6):1453–1458. doi: 10.1002/mrm.20268. [DOI] [PubMed] [Google Scholar]
- 44.Warren WS, Ahn S, Mescher M, Garwood M, Ugurbil K, Richter W, Rizi RR, Hopkins J, Leigh JS. MR imaging contrast enhancement based on intermolecular zero quantum coherences. Science. 1998;281(5374):247–251. doi: 10.1126/science.281.5374.247. [DOI] [PubMed] [Google Scholar]
- 45.Liu W, Dahnke H, Rahmer J, Jordan EK, Frank JA. Ultrashort T2* relaxometry for quantitation of highly concentrated superparamagnetic iron oxide (SPIO) nanoparticle labeled cells. Magn Reson Med. 2009;61(4):761–766. doi: 10.1002/mrm.21923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alford JK, Rutt BK, Scholl TJ, Handler WB, Chronik BA. Delta relaxation enhanced MR: Improving activation-specificity of molecular probes through R1 dispersion imaging. Magn Reson Med. 2009;61(4):796–802. doi: 10.1002/mrm.21933. [DOI] [PubMed] [Google Scholar]
- 47.Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
- 48.Suykens JA, Vandewalle J. Least squares support vector machine classifiers. Neural Process Lett. 1999;9(3):293–300. [Google Scholar]
- 49.Wu X, Kumar V, Quinlan JR, Ghosh J, Yang Q, Motoda H, McLachlan GJ, Ng A, Liu B, Philip SY. Top 10 algorithms in data mining. Knowl Inf Syst. 2008;14(1):1–37. [Google Scholar]
- 50.Liu J, Liu T, de Rochefort L, Ledoux J, Khalidov I, Chen W, Tsiouris AJ, Wisnieff C, Spincemaille P, Prince MR. Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. Neuroimage. 2012;59(3):2560–2568. doi: 10.1016/j.neuroimage.2011.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu T, Wisnieff C, Lou M, Chen W, Spincemaille P, Wang Y. Nonlinear formulation of the magnetic field to source relationship for robust quantitative susceptibility mapping. Magn Reson Med. 2013;69(2):467–476. doi: 10.1002/mrm.24272. [DOI] [PubMed] [Google Scholar]
- 52.Liu T, Spincemaille P, de Rochefort L, Wong R, Prince M, Wang Y. Unambiguous identification of superparamagnetic iron oxide particles through quantitative susceptibility mapping of the nonlinear response to magnetic fields. Magn Reson Imaging. 2010;28(9):1383–1389. doi: 10.1016/j.mri.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]