Abstract
Head and neck squamous cell carcinoma (HNSCC) is a major public health concern. Recent data indicate the presence of cancer stem cells (CSC) in many solid tumors, including HNSCC. Here, we assessed the stem cell (SC) characteristics, including cell surface markers, radioresistance, chromosomal instability, and in vivo tumorigenic capacity of CSC isolated from HNSCC patient specimens. We show that spheroid enrichment of CSC from early and short-term HNSCC cell cultures was associated with increased expression of CD44, CD133, SOX2 and BMI1 compared with normal oral epithelial cells. On immunophenotyping, five of 12 SC/CSC markers were homogenously expressed in all tumor cultures, while one of 12 was negative, four of 12 showed variable expression, and two of the 12 were expressed heterogeneously. We showed that irradiated CSCs survived and retained their self-renewal capacity across different ionizing radiation (IR) regimens. Fluorescence in situ hybridization (FISH) analyses of parental and clonally-derived tumor cells revealed different chromosome copy numbers from cell to cell, suggesting the presence of chromosomal instability in HNSCC CSC. Further, our in vitro and in vivo mouse engraftment studies suggest that CD44+/CD66− is a promising, consistent biomarker combination for HNSCC CSC. Overall, our findings add further evidence to the proposed role of HNSCC CSCs in therapeutic resistance.
Keywords: Cancer stem cells, cell surface markers, chromosomal instability, head and neck squamous cell carcinoma, radioresistance
Introduction
Head and neck squamous cell carcinoma (HNSCC) is an epithelial tumor caused by multiple genetic alterations. Risk factors for HNSCC include tobacco smoking, alcohol consumption, and/or human papillomavirus infection [1, 2]. The number of new cases of HNSCC in the USA in 2016 is estimated to be more than 61,000 and HNSCC is estimated to result in 3,000 deaths [3]. Prognosis remains poor, with approximately a 50% 5-year overall survival (OS) rate [4, 5]. This low survival rate is due to a number of factors, including local recurrence, distant metastasis, and therapeutic resistance [2, 6].
The cancer stem cell theory suggests that a subpopulation of cells in the tumor possesses stem cell properties with the potential to self-renew and generate the entire heterogeneous tumor bulk in a unique ‘hierarchic’ pattern [7, 8]. The ‘hierarchic’ model of CSC suggests that CSCs are a distinct subpopulation of cells that drive the tumor. These cells reside on the top of the tumor cell hierarchy and divide symmetrically and asymmetrically in a similar pattern to normal stem cells (SC) [8, 9]. However, another model, the stochastic model, has been proposed to explain how cancer drives tumor development. This model proposes that a tumor forms as a result of random oncogenic mutations [8, 10]. We and others believe that the hierarchic and stochastic models of cancer are not mutually exclusive, adding another level of complexity to our understanding of the biology of cancer [11]. CSCs were shown first in hematopoietic cancers [12] and later in solid tumors, such as glioma [13], lung cancer [14], breast cancer [15], colon cancer [11, 16] and HNSCC [6, 17, 18]. Various terms have been used to describe cancer stem cells, including cancer initiating cells, functional tumor stem cells, tumor initiating cells, and cancer stem-like cells.
The literature reveals that several different markers have been used to identify CSC in HNSCC. No single cell surface marker or approach has been shown to be optimal for identifying CSC in HNSCC. However, a number of markers have been applied by multiple research groups, including CD133 [19-23], CD44 [6, 17, 24-27] and ALDH1 enzymatic activity [18, 20, 28, 29]. CD44 has been the most common marker used to identify HNSCC CSC [17, 27]. CD133 has been shown by some researchers to be a consistent marker [19, 20]. ALDH1 is a promising marker verified by a number of groups [18, 20, 28-30]. Overall, the variability of cell surface markers used in CSC research has led many researchers to focus on the identification of other antigens and antibodies that might be more reliable and consistent biomarkers of CSC.
Radiotherapy is currently an important treatment modality for HNSCC, used either alone or in combination with chemotherapy for primary and recurrent cancers [31]. Radiotherapy will remain a crucial component of therapy for HNSCC because these tumors tend to locally infiltrate important surrounding structures. Surgery is often difficult due to the anatomical location. We and others have shown that the markers, CD133, CD44, and SOX2 are biomarkers of radioresistance in tumors [32-34]. Breast CSC have been shown to survive various radiation therapy protocols, and subsequent to some protocols, show higher self-renewal [35]. The side population (SP) cells from breast cancer cell lines were found to be more resistant to radiation than the non-SP cells [36]. SP isolation depends on the characteristic property of CSC of Hoechst dye efflux by multidrug resistance transporters [36, 37]. Bao et al. showed that CD133-positive glioma CSCs repaired radiation-induced DNA damage more efficiently than CD133-negative cells, and might be the source of tumor recurrence after radiation therapy [13]. CD133-positive non-small cell lung carcinoma (NSCLC) CSCs are also radioresistant, due to DNA repair defects manifested as increased basal γH2AX staining and decreased phosphorylation of various protein kinases, including ATM and KAP1/TRIM28 [38]. In prostate cancer, CD44-positive CSCs are radioresistant compared to parental cancer cells [39]. CD44 knockdown enhances radiosensitivity [34]. Furthermore, Ghisolfi et al. showed that irradiation enriched CSCs in hepatocellular carcinoma cell lines [40]. A few groups have begun to examine radioresistance in HNSCC CSC, showing radioresistance using a number of different markers. Chen et al. found evidence that an ALDH1-positive subpopulation exhibited increased radioresistance compared to ALDH1-negative cells [30]. On the other hand, Wilson et al. showed no significant differences in radiosensitivity between CSC isolated from HNSCC cell lines using several different CSC isolation techniques [17]. We have recently demonstrated that CSCs (SOX2+) showed fewer and different types of chromosomal segregation defects after ionizing radiation treatment than non-CSCs (SOX2−), suggesting that CSCs may undergo altered behavior, including therapeutic resistance as a result of chromosomal instability [32]. Overall, radioresistance in HNSCC CSCs is a promising field of research that we chose to explore.
To characterize CSC from primary tumors, we enriched CSC sequentially on stromal feeder layers and later on Matrigel™-coated culture dishes or ultra-low attachment plates. The stromal feeder layer provides cellular interaction for CSC and promotes epithelial cell growth. This system has been previously used by our group to isolate and expand CSC from human metastatic colon cancer for at least five passages [11]. To understand and characterize CSC populations within HNSCC, we examined cells derived from five HNSCC. We examined the expression of 21 different cell surface markers, assessed self-renewal, chromosomal instability (CIN), radioresistance of CSC in vitro, and confirmed the tumorigenicity of HNSCC CSC in vivo.
Materials and Methods
Tumor cell acquisition, preparation and establishment of short-term cell strains
This study was approved by the University of Pittsburgh Institutional Review Board and the University of Pittsburgh Animal Care and Use Committee. Following informed consent, HNSCC were collected after surgical removal from five different patients. Tumor specimens were minced into multiple small fragments with scalpels and digested using a two-step collagenase dissociation method. Tissues were further dissociated with a 20 min trypsin incubation, followed by tumor cell collection. Four specimens were analyzed by flow cytometry. Later, tumor cells were cultured on stromal monolayers of previously irradiated (80 Gy) rodent epithelial feeder layer cells in SC medium composed of serum-free Dulbecco's minimum essential (DMEM/F12) medium supplemented with N2, B27, L-glutamine, Gentamicin (20 mg/ml) (all from Thermo Fisher Scientific, Pittsburgh, PA), human recombinant epidermal growth factor (EGF; 20 ng/ml, Sigma-Aldrich), and human basic fibroblast growth factor (bFGF; 20 ng/ml, Sigma-Aldrich). TERT-transfected human oral keratinocytes (OKF6/TERT-1), were kindly provided by Dr. James Rheinwald, Brigham and Women's Hospital, Harvard Institutes of Medicine [41] and were cultured in Keratinocyte-SFM supplemented with 25 μg/ml bovine pituitary extract, 0.2 ng/ml EGF, 0.3 mM CaCl2, and penicillin-streptomycin (Thermo Fisher Scientific).
Spheroid self-renewal assessment
Primary/secondary spheroid enrichment analysis was done to compare the frequencies of primary spheroids formed from short-term cell tumor cultures compared to secondary spheroids formed from dissociated primary spheroids in order to assess the self-renewal capacity of the enriched CSC. Equal numbers of cells derived from short-term HNSCC CSC cultures (primary) and cells derived from primary enriched spheroids (secondary) were suspended in SC medium on ultra-low attachment six-well plates (Corning, New York). To form secondary spheroids, primary non-adherent spheroids were selected using a 37 μm Reversible Strainer (STEMCELL Technologies Inc., Vancouver, British Columbia, Canada), centrifuged at 800×g for 4 min, and then mechanically dissociated and re-plated. Large spheroids (more than 100 cells) from primary and secondary cultures were counted and photographed using a phase contrast photomicroscope (Leica Microsystems, Wetzlar, Germany).
Extreme limiting dilution analyses of CSC
To determine the frequency of CSCs capable of colony formation in vitro, we used extreme limiting dilution analysis as described previously [11]. Tumor cells were stained with HLA-ABC and distributed by flow cytometry to 96-well plates starting at one cell per well to 1000 cells per well in 100 μl of medium and cultured on irradiated stromal cells. After three to four weeks of culture, micro-cultures were scored for the presence of colonies by direct microscopic examination. Determination of the frequency of colony-forming CSCs was obtained by the minimum Chi square method derived from the Poisson distribution relationship between the colony-positive CSC wells and the logarithm of the percentage of the colony-negative CSC wells. The L-Calc statistical software program from STEMCELL Technologies Inc. (http://www.stemcell.com/technical/28425_L-Calc.pdf) was used to calculate the frequency of colony forming tumor cells.
Immunostaining and Imaging
Suspension immunofluorescence (IF) was used to assess CSC in non-adherent spheroids. Spheroids were enriched in SC medium on ultra-low attachment six-well plates. The spheroid suspension was transferred to a microcentrifuge tube and centrifuged at 800×g for 3 min and re-suspended in 1 ml PBS. The spheroid suspension was centrifuged at 800×g for 3 min, and the supernatant was then discarded. The spheroid pellets were re-suspended in 160 μl Phosphate-buffered saline (PBS)/20 μl 37% Paraformaldehyde (PF) for 10 min. Spheroid suspensions were centrifuged, PF was then discarded, and the pellet was re-suspended in 1 ml PBS. The spheroid pellet was permeabilized with 0.3% Triton X-100/1x PBS for 10 min, centrifuged at 800×g for 3 min, and the supernatant was then discarded. The spheroid pellet was treated with blocking buffer (1% Bovine serum albumin (BSA) or 3% rabbit serum, 0.2 M glycine or Image iT FX Signal Enhancer (Life Technologies) for 30 - 60 min at RT. The spheroid suspension was centrifuged at 800×g, the supernatant was discarded, and the pellet was mixed with 100 μl primary antibody: CD44 (1:100, 550392, BD), CD133 (1:200, 19898, Abcam), BMI1 (1:400, 6362A, Imgenex) or SOX2 (1:400, 6507A, Imgenex), thermo-mixed for 30 sec, and then incubated at room temperature (RT) for 1 hr. The spheroid suspension was centrifuged at 800×g, the supernatant was discarded, and the pellet was washed three times with PBS. 100 μl secondary antibody, goat anti-mouse Alexa Fluor 488 (1:500, A-11001, Life Technologies) or goat anti-rabbit Alexa Fluor 546 (1:500, A-11010, Life Technologies) was added to the spheroid pellet, thermo-mixed for 30 sec and then incubated at RT for 1 hr. The spheroid suspension was centrifuged at 800×g, the supernatant was discarded, and the pellet was washed three times with PBS. Cells were counterstained with 4',6-diamidino-2-phenylindole (DAPI) for 5 min at RT. The spheroid suspension was centrifuged at 800×g, the supernatant was then discarded, and the pellet was mounted using ProLong Gold Antifade Reagent (P10144, Life Technologies). Slides were examined and photographed using a fluorescence photomicroscope (Leica DM5000B; Leica Microsystems, Richmond, IL, USA).
Flow cytometry for cell surface markers
Flow cytometry was used to study CSC markers in HNSCC cultures. Cells were harvested, counted and suspended in ice cold PBS, 10% fetal calf serum, 1% sodium azide at 106 cells per tube and incubated with monoclonal antibodies prepared in ice cold reagents/sodium azide at the appropriate dilution for 30 min. Unstained controls were prepared by omitting the staining step. CD antibodies, primary and secondary, were obtained from BD Biosciences, except for CD133 (Miltenyi Biotec, San Diego, CA) and HLA-ABC (Ancell, Bayport, MN). Dead cells were detected with propidium iodide (10 μg/ml). Post-acquisition analysis of the fluorescence-activated cell sorting data and desktop publishing were accomplished using the third-party flow cytometry software FlowJo (Tree star, Ashland, Oregon). The cells were kept in the dark on ice or at 4°C in a refrigerator until analysis. For extended storage, cells without PI were fixed in 4% PF to prevent deterioration.
Clonal expansion
HNSCC tumor cells from HN-SCC#13 were stained with cell surface markers and sorted for the desired phenotype utilizing flow cytometry. The sorted populations were resuspended in 2 mL of medium and deposited at different concentrations in a six-well plate. Clonogenic expansion was observed using an inverted microscope at 100× magnification. Single cells were individually aspirated using a micropipette and manually deposited individually into a 96-well plate lined previously with a stromal monolayer of irradiated (80 Gy) rodent epithelial feeder cells. Three to four weeks later, plates were screened for colonies.
Fluorescence in situ hybridization (FISH) to assess chromosomal instability in CSC
To study chromosome copy number variation, chromosome enumeration probes hybridizing to the centromeric region of six different chromosomes were prepared and fluorescence in situ hybridization carried out as described previously [42]. Plasmid DNA containing chromosome-specific DNA segments were obtained as a generous gift from Dr. Mariano Rocchi, Bari, Italy. Probes for the centromeric region of chromosomes 4 (p4n1/4), 17 (pZ17-14), and 20 (pZ20) were labeled with Spectrum Orange®, and chromosomes 6 (pEDZ6), 7 (pZ7.5), and 9 (pMR9A) were labeled with Spectrum Green® using a nick translation kit (Abbott Molecular Inc., Des Plaines, IL). Probe specificity was tested by FISH of normal lymphocyte metaphase chromosomes. FISH was done on CSC isolated from HNSCC#13 (non-clonal) and HNSCC#13 (clonal, clone E8). One Spectrum Orange® probe and one Spectrum Green® probe were co-hybridized in each area of the slide. All FISH analyses and imaging were carried out using an Olympus BX61 epifluorescence microscope (Olympus Microscopes, Melville, NY) and CytoVision Genus v3.6 software (Leica Microsystems, San Jose, CA).
Spheroid survival assay
To assess the radioresistance of HNSCC CSC, spheroid survival was examined. 5×104 cells were plated in triplicate in ultra-low attachment six-well plates in SC media. After 7–11 days in culture, colonies with >50 cells were counted. To determine the spheroid survival of CSC in response to ionizing irradiation (IR), cells were treated with 2.5 Gy IR every other day (EOD) ×3 and 2.5 Gy IR EOD ×6 of γ-irradiation from a Gammacell 1,000 Elite Irradiator (Nordion International, Ottawa, Canada) with a 137Cs source at a dose rate of 2.83 Gy/min. Results were reported as ‘Surviving Fraction,’ which is the ratio of the number of spheroids observed at a particular dose to that observed in the untreated control, represented as a percentage. It is calculated using the following formula:
Surviving Fraction, SF = (spheroids counted in treated / spheroids counted in untreated) × 100.
Cell viability assay
Cell viability assays were used to assess the survival of CSC post-irradiation at 2.5 and 5 Gy. Equal volumes of cell suspension and 0.4% trypan blue (Life Technologies) were thoroughly mixed and assessed using a hemocytometer. Cell viability was calculated as the number of viable cells divided by the total number of cells within the grids on the hemocytometer. If cells stained blue due to uptake of trypan blue dye, they were considered non-viable.
Spheroid migration zone assessment
To assess the effect of IR on spheroids, the spheroid migration zone was measured. Spheroid migration zone assessment was modified as previously described by Vinci et al. [43]. A fixed endpoint (96 hrs post-plating) was utilized. Plates were coated with Matrigel™ (BD, Franklin Lakes, NJ) for at least 2 hr before use. The spheroids were grown in SC medium on ultra-low attachment plates; centrifuged at 800×g for 30 min and allowed to attach on Matrigel™-coated plates overnight, followed by the IR treatment the following morning. Four days post-treatment, plates were fixed with 70% ethanol and stained with Giemsa (Sigma, St. Louis, MO). Plates were then photographed (Leica Microsystems, Germany) and the spheroid migration measurement were carried out using ImageJ (NIH, Bethesda, MD). The area covered by the spheroids at t = 0 hr and the area covered by the cells that have migrated from the spheroids at t = 96 hr was determined. Data were normalized to the original spheroid recorded using the following formula:
% migration = (migrated area (t = 96 hr) / original spheroid (t = 0 hr)) × 100.
Xenografting in mice
Six- to 8-wk-old Rag-2/γc−/− mice were anesthetized and injected subcutaneously into the neck region with CSC. CSC directly isolated from primary tissue or after short-term culture expansion were mixed with medium/Matrigel™ at a ratio of 1:1 and injected. The CSC tumorigenicity was determined by transplanting tumor cells (106); specific tumorigenicity of sorted CD44 (CSC marker) and CD66 (non-CSC/mature cancer cell marker) was done to assess differences in tumorigenicity. Injected mice were followed for up to several months and sacrificed by CO2 euthanasia when tumors reached a diameter of 15 mm. The tumors removed at that time were subjected to immunohistochemical analyses. Samples were embedded in Tissue-Tek optimum cutting temperature (OCT) compound (Tissue-Tek, VW International) and frozen. All experiments involving the use of animals were performed in accordance with the University of Pittsburgh Institutional animal welfare guidelines after Institutional Animal Care and Use Committee approval.
Immunohistochemistry and confocal microscopy of xenografts
Primary antibodies, CD66 (BD Biosciences), CK5/14 (Santa Cruz Biotechnology), Involucrin (Santa Cruz Biotechnology), transglutaminase (Santa Cruz Biotechnology) and filagrin (Santa Cruz Biotechnology) were incubated for an hour followed by chicken anti-mouse Alexa Fluor (488 or 594, Invitrogen). HLA-biotin (Ancell) was followed by streptavidin-FITC (PharMingen). Specimens were examined and photographed using a fluorescence photomicroscope (Leica DM5000B; Leica Microsystems, Richmond, IL, USA).
Statistical methods
All studies were done in triplicate and the results compared by the most appropriate statistical methods. Most data are presented as the mean ± SEM, unless otherwise specified. Any p value < 0.05 was considered statistically significant.
Results
HNSCC CSC self-renewal and clonogenic potential
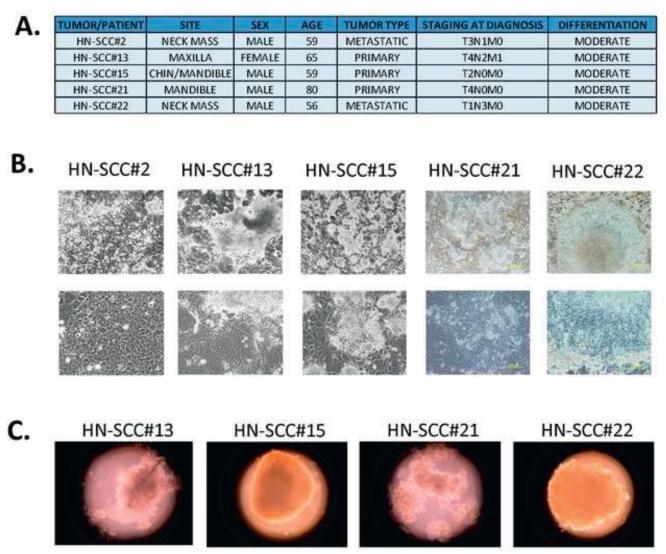
Most human epithelial CSCs can form spheroids in vitro in enriched SC culture medium. All five specimens were successfully cultured on mouse feeder layers utilizing enriched SC culture medium (Figures 1A, B) Four CSC cultures were further examined for spheroid forming capacity in ultra-low attachment plates. The CSC formed sphere-like or grape-like spheroids in enriched SC media within 7–14 days (Figure 1C). The self-renewal capacity of two of the HNSCC CSC cultures was assessed further by primary/secondary spheroid forming capacity. The two primary CSC cultures showed a statistically significant increase in secondary spheroid formation when compared to primary spheroid formation, confirming that the CSC enhanced self-renewal capacity (Figures 1C, D).
FIGURE 1. CSC were enriched and cultured in SC enrichment medium on mouse stromal layers or ultra-low attachment plates.
1A. Epidemiological, clinical and histopathological characteristics of HNSCC specimens.
1B. CSC from the same patient were grown and expanded on mouse feeder layers for more than 5 months without any alterations in morphology (scale, 200 μm).
1C. Representative images of spheroids derived from HNSCC after culture on ultra-low attachment plates in CSC enrichment medium.
1D. Primary and secondary spheroid-forming capacity of HN-SCC#13 and HN-SCC#21. The two primary CSC cultures both showed statistically significant increases in secondary spheroid formation compared to primary spheroid formation, confirming that the conditions favored CSC selection.
1E. CSC frequency as assessed in vitro by extreme limiting dilution analysis in vitro (+/−1SE). The values represent the estimation of CSC frequency with upper limit (UL) and lower limit (LL) values setting the range.
To investigate the tumorigenic potential of single CSCs from HNSCCs, we performed extreme limiting dilution assessment (ELDA) (Figure 1E), an in vitro SC method described by our group and others to assess presence of SCs in cultures or sorted cell populations [11]. The four primary CSC cultures showed colony formation at extreme dilutions ≈2-3 weeks post-plating. The estimation of colony forming efficiency (CFE) through extreme limiting dilution ranged from 1/5 for HN-SCC#2 to 1/52 for HN-SCC#13 (Figure 1E). Overall, our results suggest that we cultured cells with self-renewal and colony forming capacity at extremely low dilutions, both unique characteristics of SC. Furthermore, our unique cell culture enrichment approach successfully maintained this CSC population for as many as 8 passages in culture.
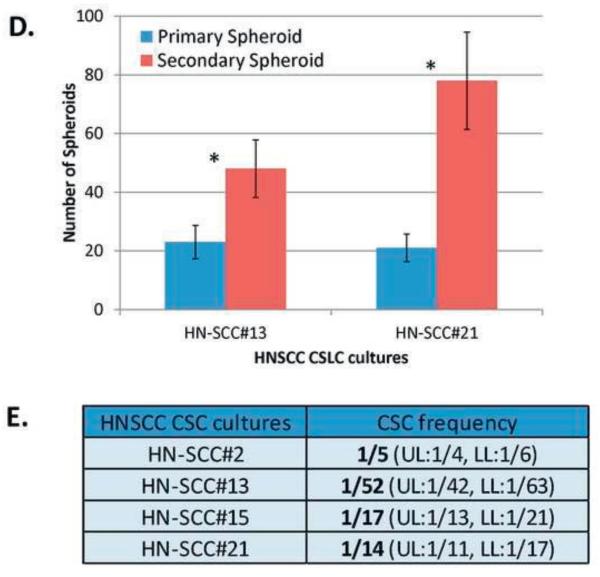
Staining of the spheroids using stem cell markers (CD44, CD133, SOX2 and BMI1) showed heterogeneous expression. However, dispersed cell sheets were largely negative (Figure 2A). These results further confirm the observations that our cell culture system enriches for CSC. Assessing CSC markers in a normal oral epithelial cell line (OKF6/TERT-1) [41] showed that CD133, SOX2 and BMI1 were not expressed; but CD44 was expressed rarely (Figure 2B). These results suggest that these markers may be rare or absent in normal epithelial cell cultures and relatively expanded in cancerous tissues. Further studies are warranted to confirm our observations.
FIGURE 2. Immunofluorescence (IF) staining for selected SC/CSC markers in enriched CSCs and the control cell line, OKF6/TERT-1.
2A. HN-SCC#13 showed a heterogeneous pattern for SC/CSC markers CD44, CD133, BMI1 and SOX2.
2B. OKF6/TERT-1 expressed the SC/CSC marker, CD44 in a rare population of cells, and did not stain with the other three SC/CSC markers, CD133, SOX2, and BMI1.
CSC expanded in vitro are enriched for cells expressing SC markers
Although several markers have been used to identify CSC in HNSCC tumors, no single cell surface has been shown to be optimal. We analyzed 21 cell surface markers selected to identify stem cell/cancer stem cell (SC/CSC) or to distinguish epithelial or mesenchymal lineages in primary/expanded CSC cultures. Flow cytometric analyses of primary and expanded HNSCC cells at first expansion (passage 0) showed that the feeder cell culturing approach enriches for cells with an epithelial phenotype; non-epithelial lineage markers were largely absent (Table 1). Furthermore, analyses of short-term cultures (passages 2-7) showed stable expression of these different cell surface markers among the cultured CSC populations. The cell surface markers, CD26, CD29, CD44, CD54, CD49b, CD49f CD133, CD90 and CD326 (EpCam) have been reported to be present on human SC and CSC in various solid tumors [44-51]. CD24 and CD66 have been shown to identify mature/differentiated cells in normal and cancer tissues, including breast cancer [52, 53]. CD49c, CD81, CD104 and EGFR have been considered to be cancer or epithelial cancer cell markers [54-57]. CD34, CD49d and CD56 have been shown to identify non-epithelial lineage cells [58-60].
Table 1.
Flow cytometric results of HNSCC CSC isolated from primary cultures grown on irradiated mouse feeder layers.
| HN-SCC#2 | HN-SCC#13 | HN-SCC#15 | HN-SCC#21 | |
|---|---|---|---|---|
| STEM CELL/CANCER STEM CELL MARKER | ||||
| CD9 (p24) | POSITIVE | POSITIVE | POSITIVE | POSITIVE |
| CD26 (DPPIV) | NEGATIVE | HETERO | NO DATA | HETERO |
| CD29 (β1 integrin) | POSITIVE | POSITIVE | POSITIVE | POSITIVE |
| CD44 (H-CAM) | HETERO | HETERO | HETERO | HETERO |
| CD49b (α2 integrin) |
POSITIVE | POSITIVE | POSITIVE | POSITIVE |
| CD49f (α6 integrin) | POSITIVE | POSITIVE | POSITIVE | POSITIVE |
| CD54 (ICAM) | POSITIVE | HETERO | POSITIVE | POSITIVE |
| CD90 (Thy1) | NEGATIVE | HETERO | NEGATIVE | NEGATIVE |
| CD133/1 (prominin1) |
NEGATIVE | NEGATIVE | NEGATIVE | NEGATIVE |
| CD133/2 (prominin1) |
NEGATIVE | HETERO (few) | NEGATIVE | NEGATIVE |
| CD227 (Muc1) | HETERO | HETERO (few) | HETERO | HETERO |
| CD326 (EpCAM) | POSITIVE | POSITIVE | POSITIVE | POSITIVE |
| DIFFERENTIATION MARKER | ||||
| CD24 (HSA) | POSITIVE | HETERO | POSITIVE | HETERO |
| CD66 (CEACAM) | HETERO | HETERO | HETERO | HETERO |
| CANCER/EPITHELIAL MARKER | ||||
| CD49c (α3 integrin) | POSITIVE | POSITIVE | POSITIVE | POSITIVE |
| CD81 (TAPA1) | POSITIVE | POSITIVE | NO DATA | POSITIVE |
| CD104 (β4 integrin) |
POSITIVE | POSITIVE | POSITIVE | POSITIVE |
| EGFR | POSITIVE | POSITIVE | NO DATA | NO DATA |
| NON-EPITHELIAL LINEAGE CELLS | ||||
| CD34 (Anti-HPCA) | NEGATIVE | NEGATIVE | NEGATIVE | NEGATIVE |
| CD49d (α4 integrin) |
NEGATIVE | NEGATIVE | NEGATIVE | NEGATIVE |
| CD56 (NCAM) | NEGATIVE | NEGATIVE | HETERO | NEGATIVE |
Samples were cultured on stroma and were stained with markers specific for SC/CSCs. Samples show substantial similarity. The Table summarizes flow cytometric results for a panel of 21 cell surface markers. The cells expressed CSC markers including CD44, CD133.2, CD90, CD29 and CD56 in variable forms.
Five of twelve SC/CSC markers were expressed homogenously in all tumor specimens, while one of twelve was largely negative, four of twelve showed variability and two of the twelve were heterogeneous (Table 1). The expression of CD44 and CD227 were consistently heterogeneously expressed in the primary cultured HNSCC epithelial cells. We found CD66 to be a consistent differentiation marker when compared to CD24. Most cancer/epithelial markers were positive and most lineage/non-epithelial cell markers were negative (Table 1). Overall, based on our flow cytometric results, many cell surface markers identified on SC/CSC were expressed on the CSCs that we propagated in vitro. Based on our results and a review of the literature, we chose to focus on CD44, as this marker is the most consistent marker used for HNSCC CSC identification to date.
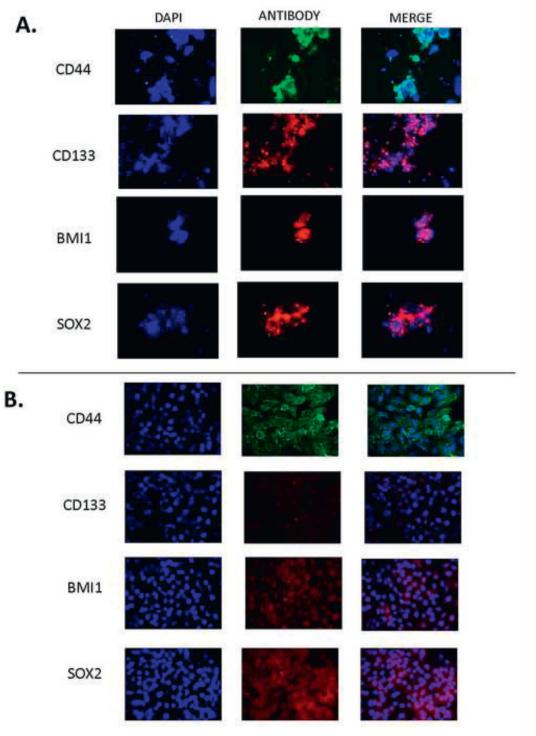
Chromosomal instability is present in HNSCC CSC
To assess chromosomal instability in CSC, centromeric FISH analysis was performed to enumerate copy number alterations in the short-term HN-SCC#13 and a clonally-derived CSC culture HN-SCC#13(clone E8). Our group has previously shown that chromosomal instability is present in metastatic colorectal carcinoma cells [11]. We observed variability in the number of chromosomes in CSC from the parental HNSCC#13 and cloned HN-SCC#13(clone E8) (Figure 3), suggesting the presence of chromosomal instability in CSC from which tumors develop in vitro. As a control, we cultured normal human fetal liver cells under similar conditions. After several passages in culture, no chromosomal abnormalities were detected in the ‘normal’ human fetal liver cell cultures. Overall, our results confirm that the in vitro culture conditions do not cause chromosomal instability in the CSC cultures.
FIGURE 3.
Chromosomal instability in enriched CSC using multi-color FISH analysis of HN-SCC#13 and HN-SCC#13 (clone E8). FISH analysis of CSC isolated from HN-SCC#13 and HN-SCC#13 (clone E8) showed variability in the number of chromosomes in both the parental and cloned CSC cultures, suggesting the presence of chromosomal instability in CSCs.
Radioresistance of HNSCC CSC
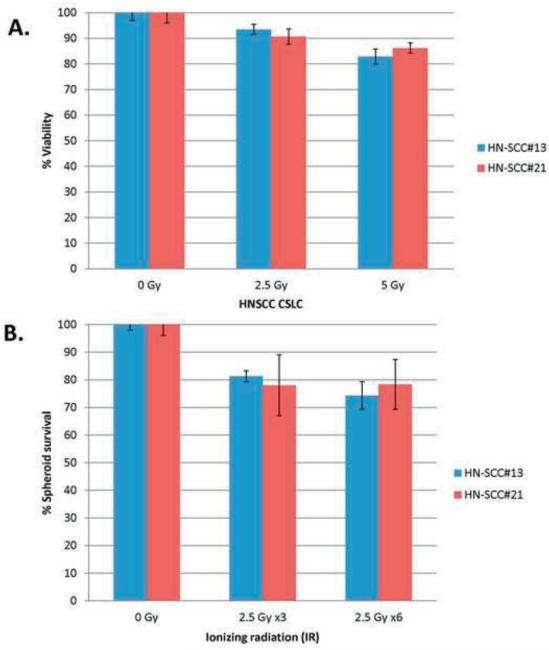
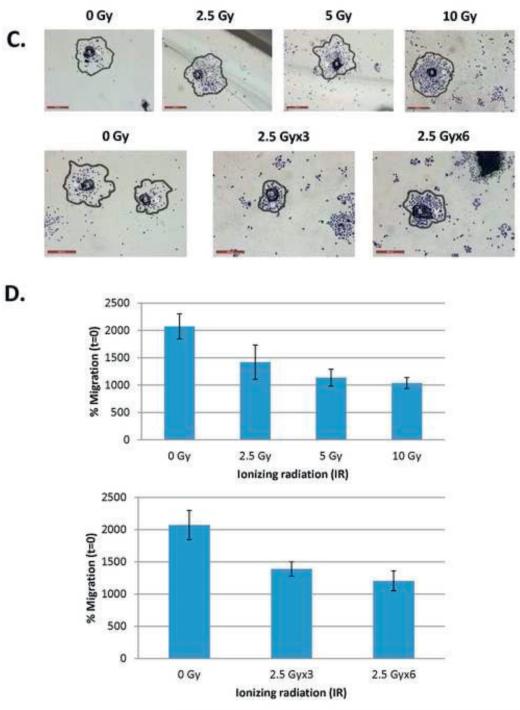
Few studies have addressed how CSC contribute to radioresistance in HNSCC. Therefore, we analyzed the response of HNSCC CSC cells to IR by viability, spheroid survival and spheroid migration (Figure 4). Cells were treated with doses of 2.5 Gy or 5 Gy IR, based on previous studies in our lab [61, 62]. For the spheroid survival assay, results were reported as ‘Surviving Fraction’ at each IR dose after normalization against the untreated cells (0 Gy). We observed high survival of the CSC by cell viability assays in response to 2.5 or 5 Gy IR (Figure 4A). In an effort to replicate standard radiation therapy in which patients receive multiple doses of IR (2.5 Gy) over a period of weeks, we irradiated CSC with 2.5 Gy EOD for 1 or 2 weeks [35]. There was no significant difference in the surviving fraction of spheroids at the different total IR doses (2.5 Gy x3 vs. 2.5 Gy x6), suggesting a possible contributory role of CSC to radioresistance (Figure 4B). Next, we assessed spheroid migration, a standardized approach that tests the therapeutic response of CSC and mimics the initiation of spread of metastases occurs in vivo [43]. The spheroids from HN-SCC#13, 21 and 22 did not migrate on Matrigel™ or poly-L-lysine-coated plates; only HN-SCC#15 formed a migration zone and therefore, was used for our experiments. The assay showed mild to moderate reduction in spheroid migration in response to 2.5 Gy, 5 Gy and 10 Gy compared to the untreated control (0 Gy). Interestingly, there were no statistically significant changes in migration zone based on the different IR doses (up to 10 Gy), confirming that CSC are extremely radioresistant (Figures 4C,D). Spheroid CSC treated with one (2.5 Gy x3/EOD) or two (2.5 Gy x6/EOD) week long IR regimens also showed similar trends, confirming our prior observations (Figures 4C,D). Overall, our results from several experimental approaches suggest that HNSCC CSC play a role in radioresistance (Figure 4).
FIGURE 4. Radioresistance of CSCs in HNSCC.
4A. Cell viability post-IR in HN-SCC#13 and HN-SCC#21, showing relatively high survival in response to 2.5 Gy and 5 Gy IR compared to untreated cells (0 Gy).
4B. Spheroid survival assay post-IR in HN-SCC#13 and HN-SCC#21 showing relatively high survival in response to 2.5 Gy x3 and 2.5 Gy x6 compared to untreated cells (0 Gy).
4C. Images showing migration of HN-SCC#15 spheroids in response to radiation treatment: 0 Gy (untreated), 2.5 Gy, 5 Gy, 10 Gy , 2.5 Gy x3 and 2.5 Gy x6 (stained with Giemsa stain). Images showing spheroid-based cell migration marked as S (Spheroid Zone) and M (Migration Zone) (scale, 500 μm).
4D. Line graphs showing quantification of HN-SCC#15 CSC migration on Matrigel™-coated plates 96 hr after IR treatment. Data are mean ± SEM of replicate spheroids.
The tumorigenic potential of HNSCC CSC after in vivo engraftment
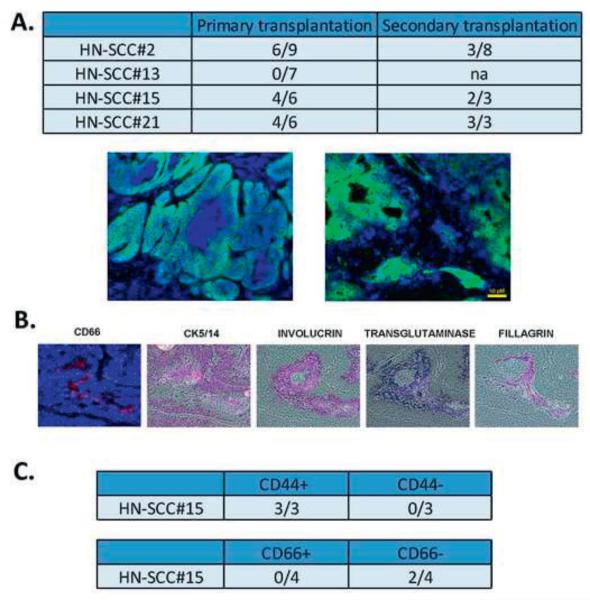
One of the characteristics of CSC is the ability to initiate tumors in mouse xenografts. We studied the potential of expanded CSCs derived from HNSCCs to initiate tumors after primary and serial transplantation. CSC isolated from fresh HNSCC specimens were transplanted under the skin of the neck region of immunodeficient Rag-2/γc−/− mice (Figures 1, 5). The mice were sacrificed and found to carry human tumors except for HN-SCC#13 that did not grow (Figure 5A). All other xenografts were morphologically similar to the patients’ original tumors. Histopathologic analysis of the xenografts demonstrated moderately differentiated squamous cell carcinoma tissue morphology without evidence of local infiltration (Figure 5A). The isolated CSC from primary xenografts were serially transplanted and shown to be tumorigenic upon secondary transplantation (Figure 5A). Xenografts derived from CSC were analyzed further for lineage differentiation markers including, basal epithelial layer (CK 5/14), epithelial differentiation (involucrin), keratin fibers (filaggrin), epithelial tumor progression (CD66), and foreign tissue reaction (transglutaminase) [63-66]. The xenografts showed markers of squamous cell tissue differentiation CK 5/14, involucrin, and filaggrin as shown in Figure 5B, suggesting that they resemble the original human tumors [6]. Further, the xenografts expressed CD66, a marker of epithelial tumor progression, as well as transglutaminase, a marker of foreign tissue growth of human tissue in the mouse (Figure 5B). Overall, our results show clearly that CSC express in vivo tumorigenic potential, a unique characteristic of CSC.
FIGURE 5. Tumorigenicity of CSC in HNSCC.
5A. 106 tumor cells were injected subcutaneously into the neck region of immunodeficient mice. Tumors were harvested and stained by immunohistochemistry. HLA-ABC (green) staining specifically identified the human tumor cells in the mouse. The Figure represents two distinct tumors, each showing different architecture, similar to the original tumors.
5B. IHC showing reconstitution of the original heterogeneity in HN-SCC#2. The xenograft was stained with antibodies against CD66, CK5/14, involucrin, transglutaminase and filagrin. Histopathologic analysis of the xenografts demonstrated moderately differentiated squamous cell carcinoma-like tissue morphology without evidence of invasion.
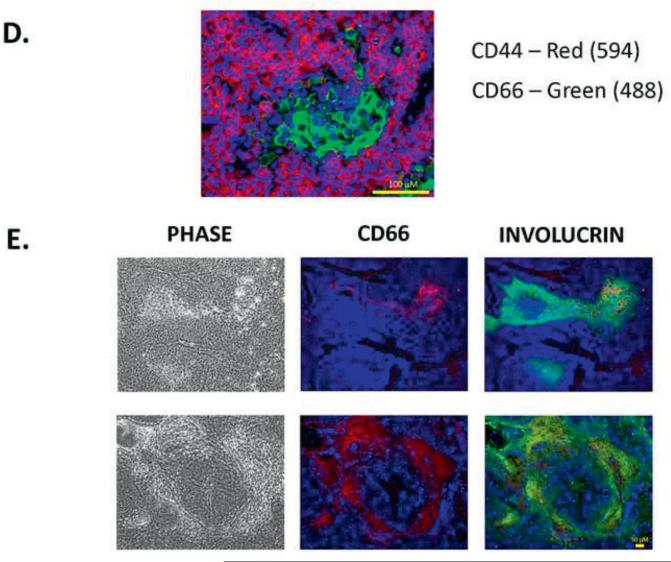
5C. CD44 and CD66 subpopulations were sorted and injected into immunodeficient mice. CD44 sorted cells show tumorigenicity when compared to CD66 cells.
5D. Double immunostaining of xenografts derived from HN-SCC#2 (CD44, CD66). The staining shows distinct areas of CD44+ and CD66+, suggesting that CSC and differentiating cells are mutually exclusive.
5E. Double immunostaining of xenografts, derived from HN-SCC#2 (CD66, involucrin). The staining shows co-localization of CD66+ and involucrin+ in the center of the squamous tumor islands, confirming that CD66 is a differentiation marker in HNSCC.
5F. Flow cytometric analyses of CD44 and CD66 staining in four early passage HNSCC cultures. The results suggest that our culture approach selects for a CD44+/CD66− subpopulation of cells.
5G. CD44 subpopulations or CD66 subpopulations were sorted, plated on 96-well plates, and assessed for CSC frequency by limiting dilution analysis. The results show higher colony forming units (CFU) for both the CD44+ and CD66− sorted populations compared to the CD44− and CD66+ sorted populations.
CD44+/CD66− CSCs possess tumorigenic potential upon in vivo and in vitro engraftment
The hierarchic CSC theory states that only a subset of tumor cells has the ability to initiate tumors [6, 15]. To determine if our in vitro culture expansion system could generate similar results to those reported previously for HNSCC CSC, we flow sorted CSC based on CD44 (a CSC marker) and CD66 (a differentiation marker). Further, based on our extensive flow cytometric results and the literature, we decided to investigate whether CD44+/CD66− is a useful antibody combination to identify CSC. To this end, we sorted CSC for CD44 and CD66 and transplanted these cells into immunodeficient Rag-2/γc−/− mice. The CD44+, CD44−, CD66+ or CD66− CSC were transplanted to the neck region of immunodeficient mice. Six to twelve weeks after transplantation, only the site injected with CD44+ cells developed tumors, confirming previous reports that CD44+ HNSCC CSC can grow tumors after xenograft transplantation (Figure 5C). On the other hand, the CD66+ cells did not initiate tumors, while the CD66− cells developed tumors (Figure 5C). Furthermore, dual staining of CD44/CD66 showed that these markers do not co-localize in xenografts (Figure 5D). Instead, CD66/involucrin dual staining showed co-localization in the center of the squamous tumor islands, confirming that CD66 is associated with cellular differentiation in HNSCC (Figure 5E). Our flow cytometric results showed that a CD44+/CD66− subpopulation exists in all of our HNSCC cultures (Figure 5F). Furthermore, our in vitro ELDA showed that CD44+ and CD66− populations show higher tumorigenic capacity when compared to CD44− and CD66+, thereby confirming our in vivo results (Figure 5G). Overall, our results suggest that CD44+/CD66− is a promising marker combination for CSC in HNSCC.
Discussion
CSCs contribute to therapeutic resistance, recurrence and metastasis. If the CSCs in tumors can be targeted, then the mortality rates of patients may decrease. For a cell subpopulation to be defined as CSCs, it should show certain characteristics. First, it should possess specific cell surface markers. Second, the CSC population should be able to give rise to phenotypically diverse tumor cells. Third, CSC should be tumorigenic, that is, able to generate tumors in immunodeficient mice [67, 68]. Last, serial transplantation through multiple generations should be proven, thereby showing that these cells possess the ability to self-renew [11]. This study reports the identification, isolation and expansion of tumor cells from human HNSCC with properties of CSCs including in vivo tumor initiation. We have also demonstrated that HNSCC CSC exhibit all of the criteria of CSC [7], including the expression of the SC/CSC markers, CD44, SOX2, BMI1 and others (Figures 1,2); self-renewal as assessed by primary/secondary spheroid formation (Figure 1) and in vivo tumor formation in mice after primary or serial transplantation (Figure 5). We also showed that CSC can be isolated by flow sorting utilizing the dual markers CD44+/CD66− (Figure 5); show high radioresistance (Figure 4); and chromosomal instability after cloning (Figure 3).
To study the properties of CSC, we isolated and enriched these cells by culturing them on feeder layers and later in ultra-low attachment condition or Matrigel™-coated plates [11, 43, 69]. We used low passage CSC cultures isolated from primary tumors to recapitulate the original tumor characteristics. Spheroid enrichment for CSC is an efficient, economic, and reliable approach for studying CSC, including the examination of therapeutic response. Spheroid enrichment conditions have been standardized, facilitating reproducibility [43, 70-72]. The three-dimensional (3D) culture of spheroids mimics the behavior of growing and metastatic cancer colonies when treated with various chemotherapeutic agents, adding another advantage to this approach. The 3D spheroid culture model can also be used to investigate CSC invasion and migration, properties associated with tumor initiation and subsequent relapse or recurrence [43]. Based on the current literature and the lack of a clear cut CSC marker for sorting these cells, spheroid enrichment emerges as a more suitable approach to study CSC [73, 74]. Culture of CSC on a mouse stromal monolayer, in suspension, or adherently on Matrigel™-coated plates were utilized in our study. This provided us the versatility of studying and assessing the properties of CSC under various conditions. Morphologically, the spheroids were either spherical or grape-like in shape. Both morphological types have been described previously in the literature [69, 75]. In gliomas, spherical vs grape-like morphologies were associated with different genetic signatures and functional capacities; the typical spherical spheroid morphology being associated with embryonic SC origin and the grape-like morphology being associated predominantly with mesenchymal SC [76]. Whether this finding is also a feature of HNSCC CSC merits further investigation.
Many markers have been used to identify CSC in epithelial tumors. No single cell surface marker has been shown to be optimal in identifying CSC in HNSCC. However, a number of markers have been replicated, including CD133, CD44 and ALDH1 enzymatic activity [6, 77, 78]. The most well-studied cell surface marker used for isolation and characterization of CSC from HNSCC is CD44 [6, 17, 24-27]. Wilson et al. compared the most common isolation techniques, such as SP, ALDH1, and CD44 expression for HNSCC, and concluded that CD44 was the most promising marker for flow-sorting CSC from HNSCC [17]. Despite the marked heterogeneity of HNSCC in tissue morphology and somatic mutations [2], evidence for the cellular origin of HNSCC is consistent with a common origin as shown here and in the literature [6]. Prince et al. were the first to isolate a CSC subpopulation from HNSCC specimens using fluorescence-activated cell sorting (FACS) analysis using an antibody against CD44; further analysis showed that the CD44-positive cells highly co-expressed BMI1, another important marker of CSC [6]. Identifying CSC through cell surface markers will aid in quantifying these cells in tumors. Our data from the four HNSCC samples analyzed showed a consistent staining pattern for multiple SC/CSC markers. Our results suggest that CD9, CD29, CD44, CD49b, CD49f, CD227 and CD326 are consistent CSC markers in HNSCCs that warrant further assessment by functional assays. Our results also suggest that CD44+/CD66− is a promising consistent marker of HNSCC CSC, based on our flow sorting, in vitro and in vivo mouse engraftment studies (Figure 5C-G). The pioneering work by Al-Hajj et al. in breast tumors clearly demonstrated that CD44 alone may not be a suitable marker of CSC, and that the addition of a differentiation marker, such as CD24 is important to identify cells with robust tumorigenic potential [15]. Furthermore, IF staining of the control cell line, OKF6/TERT-1, showed CD44 in ≈5% of cells, confirming that CD44 is not cancer-specific (Figure 2). In HNSCC, we observed CD24 to be an inconsistent marker; which may explain why the marker has not been used widely for dual staining of CSC in HNSCC, resulting in reliance on CD44 as a single CSC identification marker. However, despite the observation by Prince et al. that CD44 is heterogeneous and consistent with the basal origin layer of tumors, isolated CSC may not always reflect all biological features of primary CSC, mainly as a result of loss the niche interaction that plays a role in regulating properties of CSC, such as self-renewal and/or differentiation regulation [14]. In addition, culture adaptation and genetic alterations that might occur after long-term culture under hypoxic conditions may be lead to predominance of CSC markers in even low passage cell lines [14]. Therefore, adding a second differentiation marker seems essential. Our current work demonstrates that CD66 is expressed consistently and heterogeneously in tumors and might be a suitable partner for CD44 in identifying cells with CSC properties. Overall, further research on CD44+/CD66− and other SC identification markers is warranted in HNSCC.
The copy number variation observed within the CSC clones suggests that CSC may show different therapeutic resistance properties, a ‘Qualitative trait’ and that the frequency of stem cells, a ‘Quantitative trait’ might not be the only driving force behind CSC therapeutic resistance. Our current results showing copy number alterations in CSC may provide an explanation as to why CSC markers are not consistent prognostic markers across HNSCC because copy number alterations can change the behavior of CSC. Recently, chromosomal instability has been shown to be an important factor driving radioresistance in cancer cells [79]. Further assessment of the possible role of chromosomal instability in CSC seems crucial. Our results confirm the proposal that CSC show dynamic rather than fixed therapeutic resistance [80]. Enhanced therapeutic resistance occurs as a result of complex interaction of stemness factors, driver mutations and copy number alterations, and may contribute to the unique evolutionary survival capacity of these cells [80]. Understanding the mechanisms of chromosomal instability in HNSCC CSC will aid in devising approaches to determine which patients are more prone to therapeutic resistance and to target this resistance [79]. Overall, the genetic heterogeneity in CSC necessitates using multiple targeted therapeutic approaches as well as reassessing the genetic alterations in metastatic/recurrent tumors [80].
To assess the radiosensitivity of HNSCC CSC, we tested the effect of various IR doses on colony forming capacity, cell viability, and cell migration (Figure 4). The CSC therapeutic radioresistance that we observed seem to coincide with the patterns observed by Chen et al. [30], who showed that different IR dose regimens (up to 10 Gy) did not lead to a statistically significant reduction in OSCC CSC survival. In contrast, they showed that parental cancer cells showed an IR dose-dependent reduction in survival [30]. The results of Lagadec et al. and Ghisolfi et al. [35, 40] demonstrate that IR regimens enrich the cell cultures for CSC. Our results suggest that IR may not be a suitable CSC enrichment tool in all tumor types. This could possibly be due to the heterogeneity of solid tumors and different genetic signatures in different tumor types.
One of the limitations of the spheroid survival assay is that spheroids are motile and they might merge, leading to inaccuracies in clonal estimation [81]. To overcome this limitation, we used appropriately low clonal density for these experiments [73]. In addition, other approaches, such as cell viability and migration assays were utilized to substantiate our findings. Unlike the early days of CSC culture [81], many labs are currently using standardized spheroid culturing approaches, decreasing experimental variability. The radioresistance of CSC might be explained by different gene expression patterns in CSC compared to parental cell lines, as shown previously by Wilson et al. [17]. The high radioresistance of CSC that we and others have observed necessitates the exploration of novel strategies to inhibit CSCs [13].
Recent CSC studies have added further evidence to support the role CSCs in therapeutic resistance. The characterization of CSC in HNSCC greatly advances our understanding of these tumors. In the current study, we demonstrate that CSCs isolated from primary tumors possess the characteristics of stemness and display enhanced radioresistance, clonogenicity and tumorigenicity. Our results suggest that double marker identification, such as CD44+/CD66− might be a useful combination for identifying CSC in HNSCC. Overall, our findings add more evidence to the proposed role of HNSCC CSCs in therapeutic resistance; further characterization of CSC will provide better understanding of these cells to aid in more effective therapeutic targeting.
Highlights.
Spheroid enrichment selects cancer stem cells (CSC) from head & neck tumors (HNSCC).
Compared to normal epithelial cells, isolated CSC express increased SC/CSC markers.
Isolated CSC display enhanced radioresistance, clonogenicity and tumorigenicity.
HNSCC CSC express chromosomal instability.
CD44+/CD66− is a promising, consistent biomarker for HNSCC CSC.
Acknowledgements
This research was supported in part by a Fulbright Scholar Award to HOK, the Joan G. Gaines Cancer Research Fund at the University of Pittsburgh, and a pilot grant to EL from the University of Pittsburgh Head & Neck Specialized Program of Research Excellence (SPORE) grant (NIH P50CA097190, JR Grandis, PI).
Abbreviations
- CFE
colony forming efficiency
- CIN
chromosomal instability
- CSC
cancer stem cell
- ELDA
extreme limiting dilution assessment
- FISH
fluorescence in situ hybridization
- HNSCC
head and neck squamous cell carcinoma
- IR
ionizing radiation
- NSCLC
non-small cell lung carcinoma
- OS
overall survival
- SC
stem cell
- SP
side population
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 2.Gollin SM. Cytogenetic alterations and their molecular genetic correlates in head and neck squamous cell carcinoma: A next generation window to the biology of disease. Genes Chromosomes Cancer. 2014;53:972–990. doi: 10.1002/gcc.22214. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 6.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 8.Dick JE. Looking ahead in cancer stem cell research. Nat Biotechnol. 2009;27:44–46. doi: 10.1038/nbt0109-44. [DOI] [PubMed] [Google Scholar]
- 9.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 10.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odoux C, Fohrer H, Hoppo T, Guzik L, Stolz DB, Lewis DW, Gollin SM, Gamblin TC, Geller DA, Lagasse E. A stochastic model for cancer stem cell origin in metastatic colon cancer. Cancer Res. 2008;68:6932–6941. doi: 10.1158/0008-5472.CAN-07-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 13.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 14.Wang P, Gao Q, Suo Z, Munthe E, Solberg S, Ma L, Wang M, Westerdaal NA, Kvalheim G, Gaudernack G. Identification and characterization of cells with cancer stem cell properties in human primary lung cancer cell lines. PLoS One. 2013;8:e57020. doi: 10.1371/journal.pone.0057020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 17.Wilson GD, Marples B, Galoforo S, Geddes TJ, Thibodeau BJ, Grenman R, Akervall J. Isolation and genomic characterization of stem cells in head and neck cancer. Head Neck. 2013;35:1573–1582. doi: 10.1002/hed.23184. [DOI] [PubMed] [Google Scholar]
- 18.Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H, Costea DE, Mackenzie IC. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71:5317–5326. doi: 10.1158/0008-5472.CAN-11-1059. [DOI] [PubMed] [Google Scholar]
- 19.Chen YS, Wu MJ, Huang CY, Lin SC, Chuang TH, Yu CC, Lo JF. CD133/Src axis mediates tumor initiating property and epithelial-mesenchymal transition of head and neck cancer. PLoS One. 2011;6:e28053. doi: 10.1371/journal.pone.0028053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Wu L, Shen XM, Shi LJ, Zhang CP, Xu LQ, Zhou ZT. Expression patterns of cancer stem cell markers ALDH1 and CD133 correlate with a high risk of malignant transformation of oral leukoplakia. Int J Cancer. 2013;132:868–874. doi: 10.1002/ijc.27720. [DOI] [PubMed] [Google Scholar]
- 21.Wei X, Wang J, He J, Ma B, Chen J. Biological characteristics of CD133(+) cancer stem cells derived from human laryngeal carcinoma cell line. Int J Clin Exp Med. 2014;7:2453–2462. [PMC free article] [PubMed] [Google Scholar]
- 22.Yu CC, Hu FW, Ph DC, Chou MY. Targeting CD133 in the enhancement of chemosensitivity in oral squamous cell carcinomas-derived side population cancer stem cells. Head Neck. 2014 doi: 10.1002/hed.23975. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Shi S, Yen Y, Brown J, Ta JQ, Le AD. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett. 2010;289:151–160. doi: 10.1016/j.canlet.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Harper LJ, Piper K, Common J, Fortune F, Mackenzie IC. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J Oral Pathol Med. 2007;36:594–603. doi: 10.1111/j.1600-0714.2007.00617.x. [DOI] [PubMed] [Google Scholar]
- 25.Joshua B, Kaplan MJ, Doweck I, Pai R, Weissman IL, Prince ME, Ailles LE. Frequency of cells expressing CD44, a head and neck cancer stem cell marker: correlation with tumor aggressiveness. Head Neck. 2012;34:42–49. doi: 10.1002/hed.21699. [DOI] [PubMed] [Google Scholar]
- 26.Locke M, Heywood M, Fawell S, Mackenzie IC. Retention of intrinsic stem cell hierarchies in carcinoma-derived cell lines. Cancer Res. 2005;65:8944–8950. doi: 10.1158/0008-5472.CAN-05-0931. [DOI] [PubMed] [Google Scholar]
- 27.Mack B, Gires O. CD44s and CD44v6 expression in head and neck epithelia. PLoS One. 2008;3:e3360. doi: 10.1371/journal.pone.0003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian X, Wagner S, Ma C, Klussmann JP, Hummel M, Kaufmann AM, Albers AE. ALDH1-positive cancer stem-like cells are enriched in nodal metastases of oropharyngeal squamous cell carcinoma independent of HPV status. Oncol Rep. 2013;29:1777–1784. doi: 10.3892/or.2013.2340. [DOI] [PubMed] [Google Scholar]
- 29.Visus C, Wang Y, Lozano-Leon A, Ferris RL, Silver S, Szczepanski MJ, Brand RE, Ferrone CR, Whiteside TL, Ferrone S, et al. Targeting ALDH(bright) human carcinoma-initiating cells with ALDH1A1-specific CD8(+) T cells. Clin Cancer Res. 2011;17:6174–6184. doi: 10.1158/1078-0432.CCR-11-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YC, Chen YW, Hsu HS, Tseng LM, Huang PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385:307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 31.Strojan P, Corry J, Eisbruch A, Vermorken JB, Mendenhall WM, Lee AW, Haigentz M, Jr., Beitler JJ, de Bree R, Takes RP, et al. Recurrent and second primary squamous cell carcinoma of the head and neck: When and how to reirradiate. Head Neck. 2015;37:134–150. doi: 10.1002/hed.23542. [DOI] [PubMed] [Google Scholar]
- 32.Kaseb HO, Lewis DW, Saunders WS, Gollin SM. Cell division patterns and chromosomal segregation defects in oral cancer stem cells. Genes Chromosomes Cancer. 2016;55:694–709. doi: 10.1002/gcc.22371. [DOI] [PubMed] [Google Scholar]
- 33.Piao LS, Hur W, Kim TK, Hong SW, Kim SW, Choi JE, Sung PS, Song MJ, Lee BC, Hwang D, Yoon SK. CD133+ liver cancer stem cells modulate radioresistance in human hepatocellular carcinoma. Cancer Lett. 2012;315:129–137. doi: 10.1016/j.canlet.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Xiao W, Graham PH, Power CA, Hao J, Kearsley JH, Li Y. CD44 is a biomarker associated with human prostate cancer radiation sensitivity. Clin Exp Metastasis. 2012;29:1–9. doi: 10.1007/s10585-011-9423-7. [DOI] [PubMed] [Google Scholar]
- 35.Lagadec C, Vlashi E, Della Donna L, Meng Y, Dekmezian C, Kim K, Pajonk F. Survival and self-renewing capacity of breast cancer initiating cells during fractionated radiation treatment. Breast Cancer Res. 2010;12:R13. doi: 10.1186/bcr2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han JS, Crowe DL. Tumor initiating cancer stem cells from human breast cancer cell lines. Int J Oncol. 2009;34:1449–1453. [PubMed] [Google Scholar]
- 37.Wan G, Zhou L, Xie M, Chen H, Tian J. Characterization of side population cells from laryngeal cancer cell lines. Head Neck. 2010;32:1302–1309. doi: 10.1002/hed.21325. [DOI] [PubMed] [Google Scholar]
- 38.Lundholm L, Haag P, Zong D, Juntti T, Mork B, Lewensohn R, Viktorsson K. Resistance to DNA-damaging treatment in non-small cell lung cancer tumor-initiating cells involves reduced DNA-PK/ATM activation and diminished cell cycle arrest. Cell Death Dis. 2013;4:e478. doi: 10.1038/cddis.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho YM, Kim YS, Kang MJ, Farrar WL, Hurt EM. Long-term recovery of irradiated prostate cancer increases cancer stem cells. Prostate. 2012;72:1746–1756. doi: 10.1002/pros.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghisolfi L, Keates AC, Hu X, Lee DK, Li CJ. Ionizing radiation induces stemness in cancer cells. PLoS One. 2012;7:e43628. doi: 10.1371/journal.pone.0043628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reshmi SC, Saunders WS, Kudla DM, Ragin CR, Gollin SM. Chromosomal instability and marker chromosome evolution in oral squamous cell carcinoma. Genes Chromosomes Cancer. 2004;41:38–46. doi: 10.1002/gcc.20064. [DOI] [PubMed] [Google Scholar]
- 43.Vinci M, Box C, Zimmermann M, Eccles SA. Tumor spheroid-based migration assays for evaluation of therapeutic agents. Methods Mol Biol. 2013;986:253–266. doi: 10.1007/978-1-62703-311-4_16. [DOI] [PubMed] [Google Scholar]
- 44.Dotse E, Bian Y. Isolation of colorectal cancer stem-like cells. Cytotechnology. 2016;68:609–619. doi: 10.1007/s10616-014-9806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gemei M, Di Noto R, Mirabelli P, Del Vecchio L. Cytometric profiling of CD133+ cells in human colon carcinoma cell lines identifies a common core phenotype and cell type-specific mosaics. Int J Biol Markers. 2013;28:267–273. doi: 10.5301/JBM.5000020. [DOI] [PubMed] [Google Scholar]
- 46.Jeong Y, Rhee H, Martin S, Klass D, Lin Y, Nguyen LX, Feng W, Diehn M. Identification and genetic manipulation of human and mouse oesophageal stem cells. Gut. 2016;65:1077–1086. doi: 10.1136/gutjnl-2014-308491. [DOI] [PubMed] [Google Scholar]
- 47.Kansy BA, Dissmann PA, Hemeda H, Bruderek K, Westerkamp AM, Jagalski V, Schuler P, Kansy K, Lang S, Dumitru CA, Brandau S. The bidirectional tumor--mesenchymal stromal cell interaction promotes the progression of head and neck cancer. Stem Cell Res Ther. 2014;5:95. doi: 10.1186/scrt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim GR, Ha GH, Bae JH, Oh SO, Kim SH, Kang CD. Metastatic colon cancer cell populations contain more cancer stem-like cells with a higher susceptibility to natural killer cell-mediated lysis compared with primary colon cancer cells. Oncol Lett. 2015;9:1641–1646. doi: 10.3892/ol.2015.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raffo D, Berardi DE, Pontiggia O, Todaro L, de Kier Joffe EB, Simian M. Tamoxifen selects for breast cancer cells with mammosphere forming capacity and increased growth rate. Breast Cancer Res Treat. 2013;142:537–548. doi: 10.1007/s10549-013-2760-2. [DOI] [PubMed] [Google Scholar]
- 50.Sukowati CH, Anfuso B, Torre G, Francalanci P, Croce LS, Tiribelli C. The expression of CD90/Thy-1 in hepatocellular carcinoma: an in vivo and in vitro study. PLoS One. 2013;8:e76830. doi: 10.1371/journal.pone.0076830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zakaria N, Yusoff NM, Zakaria Z, Lim MN, Baharuddin PJ, Fakiruddin KS, Yahaya B. Human non-small cell lung cancer expresses putative cancer stem cell markers and exhibits the transcriptomic profile of multipotent cells. BMC Cancer. 2015;15:84. doi: 10.1186/s12885-015-1086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manohar R, Li Y, Fohrer H, Guzik L, Stolz DB, Chandran UR, LaFramboise WA, Lagasse E. Identification of a candidate stem cell in human gallbladder. Stem Cell Res. 2015;14:258–269. doi: 10.1016/j.scr.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 54.Banyard J, Chung I, Migliozzi M, Phan DT, Wilson AM, Zetter BR, Bielenberg DR. Identification of genes regulating migration and invasion using a new model of metastatic prostate cancer. BMC Cancer. 2014;14:387. doi: 10.1186/1471-2407-14-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen CN, Chang CC, Lai HS, Jeng YM, Chen CI, Chang KJ, Lee PH, Lee H. Connective tissue growth factor inhibits gastric cancer peritoneal metastasis by blocking integrin alpha3beta1-dependent adhesion. Gastric Cancer. 2015;18:504–515. doi: 10.1007/s10120-014-0400-0. [DOI] [PubMed] [Google Scholar]
- 56.Rikimaru K, Tadokoro K, Yamamoto T, Enomoto S, Tsuchida N. Gene amplification and overexpression of epidermal growth factor receptor in squamous cell carcinoma of the head and neck. Head Neck. 1992;14:8–13. doi: 10.1002/hed.2880140103. [DOI] [PubMed] [Google Scholar]
- 57.Yoo TH, Ryu BK, Lee MG, Chi SG. CD81 is a candidate tumor suppressor gene in human gastric cancer. Cell Oncol (Dordr) 2013;36:141–153. doi: 10.1007/s13402-012-0119-z. [DOI] [PubMed] [Google Scholar]
- 58.Baines P, Mayani H, Bains M, Fisher J, Hoy T, Jacobs A. Enrichment of CD34 (My10)-positive myeloid and erythroid progenitors from human marrow and their growth in cultures supplemented with recombinant human granulocyte-macrophage colony-stimulating factor. Exp Hematol. 1988;16:785–789. [PubMed] [Google Scholar]
- 59.Ninsontia C, Chanvorachote P. Ouabain mediates integrin switch in human lung cancer cells. Anticancer Res. 2014;34:5495–5502. [PubMed] [Google Scholar]
- 60.Yurtsever A, Haydaroglu A, Biray Avci C, Gunduz C, Oktar N, Dalbasti T, Caglar HO, Attar R, Kitapcioglu G. Assessment of genetic markers and glioblastoma stem-like cells in activation of dendritic cells. Hum Cell. 2013;26:105–113. doi: 10.1007/s13577-013-0065-8. [DOI] [PubMed] [Google Scholar]
- 61.Parikh RA, White JS, Huang X, Schoppy DW, Baysal BE, Baskaran R, Bakkenist CJ, Saunders WS, Hsu LC, Romkes M, Gollin SM. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2007;46:761–775. doi: 10.1002/gcc.20462. [DOI] [PubMed] [Google Scholar]
- 62.Sankunny M, Parikh RA, Lewis DW, Gooding WE, Saunders WS, Gollin SM. Targeted inhibition of ATR or CHEK1 reverses radioresistance in oral squamous cell carcinoma cells with distal chromosome arm 11q loss. Genes Chromosomes Cancer. 2014;53:129–143. doi: 10.1002/gcc.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Benedetto A, Qualia CM, Baroody FM, Beck LA. Filaggrin expression in oral, nasal, and esophageal mucosa. J Invest Dermatol. 2008;128:1594–1597. doi: 10.1038/sj.jid.5701208. [DOI] [PubMed] [Google Scholar]
- 64.Laakso M, Loman N, Borg A, Isola J. Cytokeratin 5/14-positive breast cancer: true basal phenotype confined to BRCA1 tumors. Mod Pathol. 2005;18:1321–1328. doi: 10.1038/modpathol.3800456. [DOI] [PubMed] [Google Scholar]
- 65.Muturi HT, Dreesen JD, Nilewski E, Jastrow H, Giebel B, Ergun S, Singer BB. Tumor and endothelial cell-derived microvesicles carry distinct CEACAMs and influence T-cell behavior. PLoS One. 2013;8:e74654. doi: 10.1371/journal.pone.0074654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watt FM. Involucrin and other markers of keratinocyte terminal differentiation. J Invest Dermatol. 1983;81:100s–103s. doi: 10.1111/1523-1747.ep12540786. [DOI] [PubMed] [Google Scholar]
- 67.Bhaijee F, Pepper DJ, Pitman KT, Bell D. Cancer stem cells in head and neck squamous cell carcinoma: a review of current knowledge and future applications. Head Neck. 2012;34:894–899. doi: 10.1002/hed.21801. [DOI] [PubMed] [Google Scholar]
- 68.Sheng X, Li Z, Wang DL, Li WB, Luo Z, Chen KH, Cao JJ, Yu C, Liu WJ. Isolation and enrichment of PC-3 prostate cancer stem-like cells using MACS and serum-free medium. Oncol Lett. 2013;5:787–792. doi: 10.3892/ol.2012.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lim YC, Oh SY, Cha YY, Kim SH, Jin X, Kim H. Cancer stem cell traits in squamospheres derived from primary head and neck squamous cell carcinomas. Oral Oncol. 2011;47:83–91. doi: 10.1016/j.oraloncology.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 70.Foty R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J Vis Exp. 2011 doi: 10.3791/2720. doi: 10.3791/2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nat Protoc. 2009;4:309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 72.Ho WY, Yeap SK, Ho CL, Rahim RA, Alitheen NB. Development of multicellular tumor spheroid (MCTS) culture from breast cancer cell and a high throughput screening method using the MTT assay. PLoS One. 2012;7:e44640. doi: 10.1371/journal.pone.0044640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shrivastava S, Steele R, Sowadski M, Crawford SE, Varvares M, Ray RB. Identification of molecular signature of head and neck cancer stem-like cells. Sci Rep. 2015;5:7819. doi: 10.1038/srep07819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hueng DY, Sytwu HK, Huang SM, Chang C, Ma HI. Isolation and characterization of tumor stem-like cells from human meningiomas. J Neurooncol. 2011;104:45–53. doi: 10.1007/s11060-010-0469-1. [DOI] [PubMed] [Google Scholar]
- 76.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, Wicha MS, Prince ME. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32:1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Damek-Poprawa M, Volgina A, Korostoff J, Sollecito TP, Brose MS, O'Malley BW, Jr., Akintoye SO, DiRienzo JM. Targeted inhibition of CD133+ cells in oral cancer cell lines. J Dent Res. 2011;90:638–645. doi: 10.1177/0022034510393511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bakhoum SF, Kabeche L, Wood MD, Laucius CD, Qu D, Laughney AM, Reynolds GE, Louie RJ, Phillips J, Chan DA, et al. Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat Commun. 2015;6:5990. doi: 10.1038/ncomms6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lagasse E. Cancer stem cells with genetic instability: the best vehicle with the best engine for cancer. Gene Ther. 2008;15:136–142. doi: 10.1038/sj.gt.3303068. [DOI] [PubMed] [Google Scholar]
- 81.Singec I, Knoth R, Meyer RP, Maciaczyk J, Volk B, Nikkhah G, Frotscher M, Snyder EY. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat Methods. 2006;3:801–806. doi: 10.1038/nmeth926. [DOI] [PubMed] [Google Scholar]